Abstract
Atrial (ANP) and B-type natriuretic peptides (BNP) modulate blood pressure and volume through the stimulation of cyclic GMP production by their guanylyl cyclase-A (GC-A) receptor. A novel isoform of GC-A has been identified that is the result of differential splicing of exon 4. The deletion of a 51-bp sequence is predicted to delete 17 amino acids (Lys314–Gln330) in the membrane-distal part of the extracellular domain. Reverse transcription-PCR analyses demonstrated low messenger RNA expression levels of spliced GC-A in all tissues. Homology modeling suggested that the alterations in the protein structure could interfere with ANP binding or signaling. Indeed, functional studies in transfected HEK 293 cells demonstrated that binding of ANP and ANP-induced cyclic GMP formation by GC-AΔLys314-Gln330 were totally abolished. Furthermore, cotransfection studies showed that this GC-A variant forms heterodimers with the wild type receptor and inhibits ligand-inducible cGMP generation. Finally, treatment of mice with angiotensin II (300 ng/kg/min during 7 days) resulted in enhanced pulmonary mRNA expression of spliced GC-A, which was concomitant to diminished GC-A/cGMP responses to ANP. We conclude that alternative splicing can regulate endogenous ANP/GC-A signaling. Angiotensin II-induced alternative splicing of GC-A may represent a novel mechanism for reducing the sensitivity to ANP.
The homodimeric transmembrane guanylyl cyclase-A (GC-A)3 receptor (also known as natriuretic peptide receptor A) produces cytoplasmic cyclic GMP from GTP on binding its extracellular ligands, atrial (ANP) and B-type natriuretic peptide (BNP) (reviewed in Ref. 1). The NP/GC-A system has a critical role in the endocrine regulation of arterial blood pressure and volume and in the local counter-regulation of cardiac hypertrophy and fibrosis. In mice, genetic ablation of ANP or GC-A both resulted in severe arterial hypertension, hypervolemia, and cardiac hypertrophy (2, 3). Gene polymorphisms of ANP or GC-A have been associated with essential hypertension (4, 5) and with increased left ventricular mass in patients with essential hypertension (6). Apart from these genetic variations, functional alterations of ANP or GC-A might also be involved in cardiovascular diseases. Hence, blunted vasodilating, diuretic/natriuretic, and cGMP responses to exogenous ANP have been reported in essential hypertension (7), in Cushing disease (8), and in patients with cardiac hypertrophy or congestive heart failure (9, 10). The latter patients exhibit a severe neurohumoral imbalance, which is characterized by elevated plasma levels of ANP and BNP, and an activated renin-angiotensinaldosterone system, among others (9). Many studies have shown that exposure of GC-A to high concentrations of ANP/BNP or to growth hormones such as angiotensin II (Ang II) or endothelin can lead to homologous versus heterologous desensitization of the receptor (Refs. 11–14 and reviewed in Ref. 15). Based on observations in vitro, in GC-A-overexpressing HEK 293 cells, this desensitization has been attributed to inactivating post-translational modifications of GC-A, in particular to dephosphorylation of the receptor within its intracellular domain (12–15). However, whether this process fully accounts for the inhibition of the ANP/GC-A system observed in vivo remains to be determined.
The basic topology of GC-A consists of an ∼450-amino acid extracellular ligand-binding domain, a 21-residue hydrophobic membrane-spanning region, and a 566-amino acid intracellular domain (reviewed in Refs. 1, 15). The latter can be further divided into a juxtamembrane region of ∼250 amino acids that is similar to known serine/threonine protein kinases, a 41-amino acid amphipathic cooled-coil hinge region, and an ∼250-amino acid C-terminal guanylyl cyclase catalytic domain. In the absence of ligand, GC-A exists as a homodimer or homotetramer, and ANP binding does not lead to further aggregation (1, 15). Multiple domains that are located both outside and inside the plasma membrane mediate the oligomerization of GC-A. The intracellular dimerization interface region has been mapped to the amphipathic sequence that bisects the kinase homology and cyclase domains (1, 15).
In this study we describe the presence of a GC-A splice variant, predicted to delete 17 amino acids (ΔLys314–Gln330) in the membrane-distal part of the extracellular domain. This splice variant has unique structural properties that impede ligand binding and ligand-induced guanylyl cyclase activity. RT-PCR analyses revealed that mRNA for this splice variant is ubiquiously expressed, although at ∼10-fold lower levels as compared with wild type GC-A. We also demonstrate that splice GC-A can form heterodimers with wild type GC-A thereby interfering with ANP signaling. Finally, our in vivo studies indicate that enhanced alternative splicing of GC-A might be one of the mechanisms leading to diminished activity of the endogenous ANP/GC-A system in response to angiotensin II.
EXPERIMENTAL PROCEDURES
Animals—Mouse F1 hybrids between 129/Sv and C57BL/6 strains were used for tissue collection to isolate genomic DNA, RNA, and protein and for angiotensin II treatment protocols (see below). The experiments were conducted in accordance with the German legislation on protection of animals and the NIH Guidelines for the Care and Use of Laboratory Animals (National Institutes of Health Publication 85-23 revised 1985). They were approved by the local governmental animal care committee.
RNA Isolation and Quantitative Reverse Transcriptase PCR—Total RNA was isolated from various tissues with TRIzol reagent as recommended by the supplier. First strand cDNA was synthesized from 1 μg of total RNA using 200 units of SuperScript II RNase H- reverse transcriptase (Invitrogen) and random primers. One microliter of cDNA pool was subjected to PCR with two specific primers flanking the alternatively spliced region: forward, 5′-AGACGATGGGCAGGATAGGAGAGC-3′; reverse, 5′ CCAGAGGGAGAAATCAGTGTC-3′ (the lengths of PCR products was 352 bp for wild type GC-A and 301 bp for GC-AΔ314–330). Forty seven cycles (including first 12 touchdown cycles of the PCR) were suitable for a comparative quantification of mRNA encoding wild type GC-A and the splice variant from each tissue. Aliquots (20 μl) of reaction mixture were electrophoresed in a 3% agarose gel and stained with ethidium bromide. Thereafter, the samples were analyzed by Southern blot with the specific probe 5′-TCAGTGTCCCGATCTCCATTT-3′, complementary to exon 5 of the murine GC-A gene.
Modeling of the Splicing Variant GC-AΔ314–330—The extracellular domain dimer of the splicing variant was modeled using the structure of the rat GC-A ectodomain (Protein Data Bank (PDB) accession number 1DP4) and the ANP-peptide receptor dimeric assembly used for generation of receptor dimers as an additional guide (PDB accession number 1YK1). The peptide segment from Lys314 to Gln330, which is missing in the splice variant, was removed manually in the structure template resulting in an almost complete loss of helix α10. The open ends were connected in the model by two different ways. In the first setup the amide of Leu313 and the carbonyl of Ala331 were connected such that helix α9 running from Pro288 to Phe305 fills in the place that is freed by the loss of α-helix 10 in the splice variant. In the second setup the α-helix 9 was kept in place and the distant ends were connected via an extended structure formed by residues Ala331 to Gln338, which are part of the α-helix 10 and are not deleted by the alternative splicing. For both alternative models, structures were refined using the software Quanta2006 and Charmm. The membrane-proximal part, which is distant from the location of the deletion, was kept fixed. For the membrane-distal part in close proximity of ΔLys314–Gln330, several rounds of energy minimization and molecular dynamic simulations were performed. Only geometrical terms were used; electrostatic terms were excluded. Identical simulations were performed for wild type GC-A and both alternative models of the splicing variant GC-AΔLys314-Gln330 to allow for comparison. First only side chain atoms were refined with the protein backbone kept fixed. Subsequently, the protein backbone of the membrane-distal part (residues Ser1 to Gly127 and Glu284 to Asp373) was also refined by stepwise removing positional harmonic constraints. The final models exhibit good backbone geometries. The putative receptor dimer assembly was obtained by fitting the models onto the human GC-A receptor dimer bound to the ANP peptide ligand (PDB entry 1YK1).
Assaying Wild Type and Spliced GC-A Activity in Transfected HEK 293 Cells—The splice mutation in GC-A (GC-AΔLys314-Gln330) was generated by site-directed mutagenesis using the wild type rat GC-A expression construct pCMV5-GC-A (kind gift from Dr. L.R. Potter, Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis). To measure ligand inducibility of the cyclase activity, HEK 293 cells were seeded to 40–45% confluency in 24-well plates in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum. The cells were transfected 2 days later with 0.2 μg/well of expression construct with FuGENE (Roche Applied Science). Transfected cells were serum-starved for 4 h prior to ANP exposure (48 h after transfection). Cells were pretreated with 0.1 mm of the phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine for 15 min and then exposed to various concentrations of ANP (rat ANP, Bachem, Heidelberg, Germany) for another 10 min. Intracellular cGMP was measured by radioimmunoassay (16). Additionally, fluorescence resonance energy transfer (FRET) was used to monitor in real time the kinetics and extent of cGMP formation in intact HEK 293 cells cotransfected with GC-A (wild type or splice form) and the cGMP indicator (pGES-DE2 (17)).
Binding Assay—HEK 293 cells expressing GC-A or GC-AΔLys314-Gln330 were washed with serum-free medium and incubated in DMEM for 20 min at 37 °C with 100 pm 125I-ANP (∼100,000 cpm/well; specific activity, 904 Ci/mmol; Bachem) and various concentrations of unlabeled ANP. The cells were washed twice with serum-free DMEM and then solubilized in 500 μl of 1 m NaOH, and radioactivity was measured in a γ-counter.
Cell Fractionation and Biotinylation—For fractionated extraction of the cytosolic, membrane, and nuclear proteins of GC-A-overexpressing HEK 293 cells, a cell fractionation kit was applied according to the manufacturer's instructions (nano-TOOLS Antikörpertechnik, Teningen, Germany). For biotin-labeling experiments, HEK 293 cells were washed three times with phosphate-buffered saline 24 h after transfection and incubated in 1 mg/ml NHS-LC-biotin (Perbio Science, Bonn, Germany) for 30 min at 4 °C (18). The cells were then washed three times in phosphate-buffered saline and lysed at 4 °C for 30 min in 200 μl of lysis buffer containing 50 mm Tris·HCl, pH 8.0, 150 mm NaCl, 2 mm EGTA, 1% Triton X-100, 5% glycerol, and supplemented with a mixture of protease inhibitors (Roche Applied Science). Biotin-conjugated cell surface proteins were purified with streptavidin-agarose (Perbio Science) (18). Affinity-purified protein complexes were denatured and analyzed by Western blotting as described below for coimmunoprecipitation protein samples.
Coexpression of Wild Type and Spliced GC-A, Coimmunoprecipitation, and Western Blot Analyses—For expression of FLAG-tagged wild type GC-A, the expression construct pCMV5-FLAG-GC-A was kindly provided by Dr. Michael Chinkers (Department of Pharmacology, University of South Alabama, Mobile). For epitope tagging of the splice variant, the HA (YPYDVP-DYA) epitope was positioned immediately after the cleavage site of the signal peptide of GC-AΔLys314-Gln330 by PCR-mediated mutagenesis. HEK 293 cells were prepared in 10-cm dishes and (co)transfected with 10 μg of plasmid as described above. After 48 h, the cells were lysed at 4 °C for 30 min in 200 μl of lysis buffer (see above). After centrifugation (3000 × g, 10 min, 4 °C), the supernatant was mixed with 20 μl of pre-equilibrated anti-FLAG M2 affinity gel beads (Sigma) and agitated at 4 °C for 2 h. The beads were washed three times, resuspended in 40 μl of electrophoresis sample buffer (50 mm Tris·HCl, pH 6.8, 2% (w/v) SDS, 10% glycerol, 100 mm dithiothreitol, 0.05% bromphenol blue), and boiled for 5 min. For Western blot analyses, protein samples were resolved by 8% SDS-PAGE. Electrophoresis and immunoblotting were performed as described previously (16). Antibodies were against GC-A (generated in our laboratory), anti-FLAG (Sigma), or anti-HA (COVANCE, Hiss Diagnostics, Freiburg, Germany).
Chronic Treatment of Mice with Exogenous Angiotensin II—Mice (10 males, 10–12 weeks old) received Ang II (Sigma) at a dose of 300 ng/kg/min during 1 week. The peptide was dissolved in 0.9% NaCl and 0.01 m acetic acid and then infused subcutaneously via osmotic minipumps (model 2002; Alzet, Colorado City, CO). For comparison, their littermates were only given vehicle. Mice were sacrificed under urethane anesthesia, and the lungs were bisected and frozen in liquid nitrogen (for RNA and protein extraction).
Expression and Activity of GC-A in Murine Tissues—Murine lungs were homogenized with a microdismembrator in homogenization buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm Na2EDTA, 10% glycerol, supplemented with a mixture of protease (Roche Applied Science) and phosphatase inhibitors (Sigma)). The membrane fraction was collected after centrifugation at 100,000 × g for 30 min at 4 °C. GC-A expression was determined by Western blot analyses. Glyceraldehyde-3-phosphate dehydrogenase was used as reference protein (antibody from Biolabs, Frankfurt, Germany) (19). ANP-dependent guanylyl cyclase activity was determined as described (20). To initiate cyclase activity, 50 μg of membrane protein was incubated in assay buffer (25 mm HEPES, 4 mm MgCl2, 1 mm isobutylmethylxanthine, 2 mm ATP, 2 mm GTP, 30 mm phosphocreatine, 400 μg/ml creatine phosphokinase (185 units/mg) and 0.5 mg/ml bovine serum albumin) at 37 °C, with or without ANP. At 10 min of incubation, the reaction was stopped by addition of ice-cold 100% (v/v) ethanol (final concentration 70%). After centrifugation (3000 × g, 5 min, 4 °C), the supernatants were dried in a speed vacuum concentrator, resuspended in sodium acetate buffer (50 mm, pH 6.0) and acetylated, and the cGMP content was determined by radioimmunoassay (20). cGMP content was normalized to protein content.
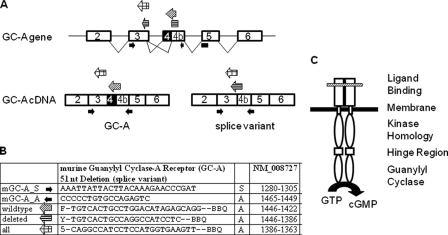
Quantitative Real Time RT-PCR—A quantitative analysis of GC-A and spliced GC-A mRNA expression was performed using the LightCycler 480 detection system (Roche Applied Science) (21). The sequences and positions of oligonucleotide primers and fluorogenic probes (TaqMan probes, all designed by TIB Molbiol, Berlin, Germany) are depicted in Fig. 1, A and B. The wild type-specific probe labeled with FAM (6-carboxyfluorescein-HX) and BlackBerry quencher binds to 16 bases of the deleted sequence, whereas the 3′-part of the deletion-specific probe with a YAK reporter dye binds to the region 5′ of the deletion (see Fig. 1B). Data were calculated using the absolute quantification/second derivative max method (Roche Applied Science LightCycler 480 software), which calculates DNA concentration by interpolation with a standard curve generated by using known amounts of the target DNA (21). Reaction was performed in 20 μl of mixture, containing Roche Applied Science probe master mix (catalog number 04887301001), 200 nm of each primer, and 100 nm FAM or YAK (Epoch Yakima Yellow)-labeled TaqMan probe. Wild type GC-A and GC-AΔLys314-Gln330 transcripts were normalized to the Mus musculus cytochrome c1 mRNA.
FIGURE 1.
Schemes illustrating relevant parts of the structure of the GC-A gene and cDNA as well as of the GC-A protein. A, structure and alternative splicing of GC-A. The exon-intron organization is shown on the top, including the splicing pattern. The positions of oligonucleotide primers and probes used for quantitative, real time RT-PCR are indicated. Their sequences and the respective nucleotide positions are shown in the table (B). C, the active GC-A receptor is formed by a homodimer and consists of an extracellular (ligand binding), a transmembrane, and three intracellular domains. The variant lacks 17 amino acids (ΔLys314–Gln330) in the membrane-distal part of the extracellular domain (marked by a hatched bar).
Data Analysis—Statistical comparisons were evaluated using Student's t test (p < 0.05). Data are given as means ± S.E.
RESULTS AND DISCUSSION
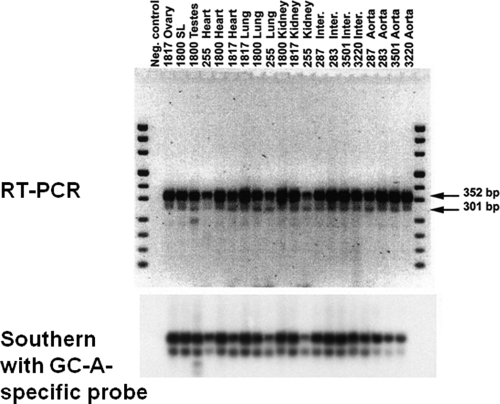
mRNA Encoding GC-AΔLys314–Gln330 Is Expressed in All Murine Organs—As an approach to study the regulation of the activity of the natriuretic peptide system, we characterized the gene sequence of the GC-A gene coding for the extracellular domain of the GC-A receptor. Nucleotide sequence analyses of PCR products amplified from mouse kidney identified a novel isoform of GC-A mRNA. This isoform results from the deletion of a 51-bp sequence in exon 4, generated by splicing a cryptic donor site to the normal acceptor site (Fig. 1) (22). This alternative splicing is predicted to delete 17 amino acids (ΔLys314-Gln330) in the membrane-distal part of the extracellular ligand-binding domain of GC-A (Fig. 1C). As shown in Fig. 2, GC-A isoform expression was detected by RT-PCR in all murine tissues examined such as ovary, spleen, brain, testes, heart, lung, kidney, aorta, as well as smaller resistance arteries such as the renal interlobar artery. The ratio of splice to wild type GC-A mRNA levels was less than 10% in all studied tissues (Fig. 2). Because total tissue RNA was used, we do not know whether this splice isoform of the GC-A gene is highly expressed in a limited number of cells or is expressed at low levels throughout these organs. This differentiation will be an important goal for our future studies.
FIGURE 2.
Relative amounts of GC-A mRNA and its splice variant quantitated by RT-PCR. First-strand cDNA was synthesized from total RNA isolated from various mouse tissues and subjected to PCR with two specific primers flanking the alternatively spliced region shown in Fig. 1. Two PCR products with the expected sizes of 352 and 301 bp were obtained which correspond to GC-A and GC-AΔLys314-Gln330. Top, ethidium bromidestained gel. Bottom, detection of the same bands by Southern blotting with a specific GC-A probe.
Isoforms of the mammalian GC-A receptor generated by alternative splicing have been described in two previous studies. mRNA encoding an isoform resulting from differential splicing of a new exon 5a (a 9-bp sequence predicted to add three amino acids to the extracellular yuxtamembrane region of GC-A) was found to be expressed mainly in renal papilla and adrenal gland (23). Another mRNA isoform lacking exon 9 (which encodes a fragment of the regulatory kinase-like domain of GC-A) was found to be ubiquitously expressed at very low levels (less than 1% of total GC-A transcripts) (24). None of these studies investigated the potential biological function of the variant. This was our question for the following investigations.
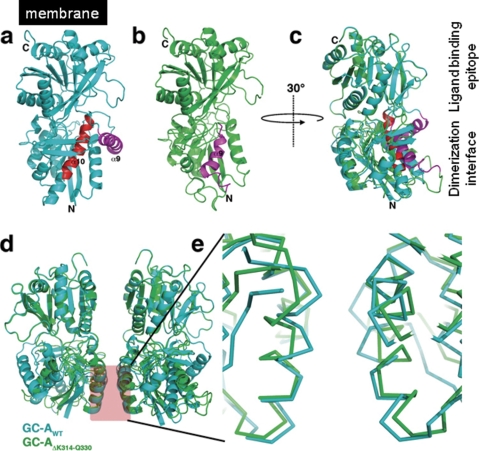
GC-AΔLys314-Gln330 Lacks ANP Binding and ANP-induced Guanylyl Cyclase Activity—Alternative splicing is common in vertebrates (5% of genes (25)) and is a useful mechanism for generating cell surface receptor isoforms with distinct functions (26). First, to obtain insights into the molecular mechanism, by which the loss of the residues Lys314 to Gln330 would influence receptor activation, we performed molecular modeling of the extracellular domain of the splicing variant. The crystal structure of rat GC-A ectodomain (PDB entry 1DP4) was used as a template, in which the residues Lys314 to Gln330 forming a major part of α-helix 10 in the membrane-distal part were removed (Fig. 3). Because the deletion is rather far off the peptide-ligand-binding site (located in the membrane-proximal part), a direct influence on ligand binding seemed unlikely. However, the membrane-distal part is involved in receptor dimer formation and possibly constrains the receptor dimer in a receptor- and signaling-competent conformation (27–30). Upon ligand binding the GC-A receptor is activated by a ligand-dependent rotation of both extracellular receptor monomer subunits (27, 30). Thus ligand binding and receptor activation are highly dependent on the proper formation of the preformed receptor dimer. The helix α10, which is almost completely removed upon alternative splicing, is located at the back of this dimerization interface. Because α-helix 10 is almost completely hidden in the hydrophobic core, loss of the helix should result in larger structural alterations (Fig. 3). Deletion of helix α10 would remove a number of hydrophobic residues, e.g. Ile316, Ile317, Phe321, Leu327, and Tyr328 that pack tightly against the central β-sheet of β-strands 1, 3, and 4. These strands are important for the structural maintenance of the dimerization interface formed by the helices 2 and 3. Thus loss of Lys314 to Gln330 might alter the dimerization properties of the GC-A ectodomain and thereby lead to inactivation of the receptor. The modeling indeed showed that the differences in the hydrophobic core propagate to the helices 2 and 3 at the dimerization epitope. In comparison with the wild type starting structure, the helix angle of helix α2 is altered with the N-terminal half of the helix moving away from the dimerization interface by almost 2 Å. This movement is coupled to a dislocation of the β-strands 1 and 3, which itself is a consequence of the changed packing in the hydrophobic core of the splicing variant. Because a defined conformation of the preformed receptor dimer is probably indispensable for ANP binding, an altered conformation of receptor extracellular domain assembly of this pre-existing receptor dimer as suggested from modeling could alter ANP binding and/or signaling.
FIGURE 3.
Modeling of the splicing variant GC-AΔLys314-Gln330. a, ribbon representation of the rat GC-A ectodomain with the deletion resulting from the alternative splicing marked in red. Helix α9, which is put into the place of α10 during modeling, is colored in magenta. b, final model of the splicing variant GC-AΔLys314-Gln330 with the new location of helix α9(magenta). c, superposition of the model structures of wild type (cyan) and splicing variant GC-A ectodomain (green). The ectodomain is rotated by 30° in the y axis compared with a and b to show the ligand binding epitope and the dimerization interface at the left-hand side. The overlay shows the structural changes induced by the deletion. d, overlay of the putative receptor dimer upon ANP ligand binding. The receptor dimer assembly was obtained by superposition of the rat GC-A wild type and splice variant onto the ligand-receptor complex of human GC-A (PDB entry 1YK1). e, zoom into the dimerization interface. The splice variant (green) shows significant changes in the α-helix 2 in the dimerization interface that might lead to alteration in the receptor dimer architecture and hence result in loss of ANP binding of the splice variant.
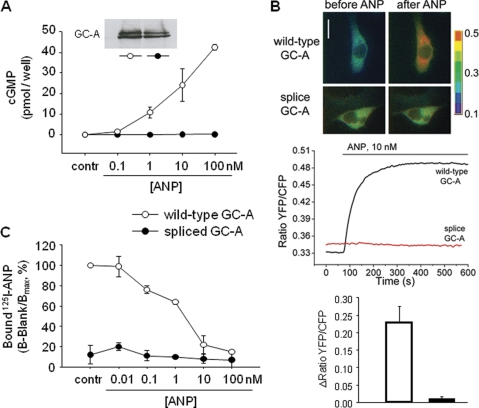
We tested the function of GC-AΔLys314-Gln330 by transfecting HEK 293 cells and measuring cGMP production under baseline conditions and after stimulation with ANP. Transfection with cDNA encoding splice GC-A produced equivalent amounts of protein by Western blot, compared with the wild type (see inset in Fig. 4A). Basal cGMP levels of cells expressing spliced GC-A were similar to those of cells expressing wild type GC-A. In the latter, intracellular cGMP levels were increased by ANP in a concentration-dependent manner, up to a 400-fold increase in cGMP in response to 100 nm ANP (Fig. 4A). ANP failed to elevate cyclic GMP concentrations in HEK 293 cells expressing splice GC-A (Fig. 4A). Additionally, FRET was used to monitor in real time the kinetics and extent of cGMP formation in single intact cells. HEK cells coexpressing the wild type GC-A receptor and a monomolecular FRET biosensor (pGES-DE2 (17)) exhibited a rapid and sustained increase in FRET upon stimulation with ANP (Fig. 4B). As described previously, pGES-DE2 responds to cGMP binding with a robust increase in FRET (17). Therefore, we can conclude that ANP elicited robust elevations in [cGMP]i via wild type GC-A receptors, whereas these responses were totally absent in cells expressing the splice variant. The same results were obtained for BNP as the second specific GC-A ligand (not shown). Finally, the transfectants were assayed for their 125I-ANP binding activity. In mock-transfected cells, negligible binding was observed. Competition experiments with unlabeled ANP or BNP showed that wild type GC-A, transiently expressed in HEK 293 cells, specifically bound 125I-ANP (Fig. 4C). In cells transfected with the splice variant, no specific binding was observed (Fig. 4C). Taken together, these data demonstrate that alternative splicing results in a GC-A variant that lacks ANP/BNP binding and NP-induced guanylyl cyclase activity. As described above, modeling suggested that mainly the membranedistal part of the ANP-receptor extracellular domain is affected by the deletion. However, the subsequent structural changes in the dimerization domain apparently provoke conformational changes in the preformed dimer which in turn prevent ANP binding.
FIGURE 4.
Guanylyl cyclase and ligand-binding properties of wild type and spliced GC-A. HEK 293 cells were transiently transfected with each GC-A isoform. A, cells were incubated with vehicle or various ANP concentrations for 10 min, and intracellular cGMP contents were then measured by radioimmunoassay (n = 4). Inset in A, Western blot analysis showing similar expression levels of wild type or spliced GC-A. B, FRET was used to monitor in real time the kinetics and extent of cGMP formation in single HEK 293 cells cotransfected with GC-A (wild type or splice form) and the cGMP indicator pGES-DE2 (17). Top, ratiometric FRET images of cells prior to and 5 min after stimulation with ANP. Bottom, representative ratiometric recordings of single cell FRET signals. C, HEK 293 cells were incubated with 125I-ANP in the presence of various concentrations of unlabeled ANP in serum-free DMEM at 37 °C for 20 min, and 125I-ANP bound was quantitated with a γ-counter. Binding to wild type GC-A in the absence of unlabeled ANP was assigned a value of 100%. Specific activity of 125I-ANP was 904 Ci/mmol (n = 4).
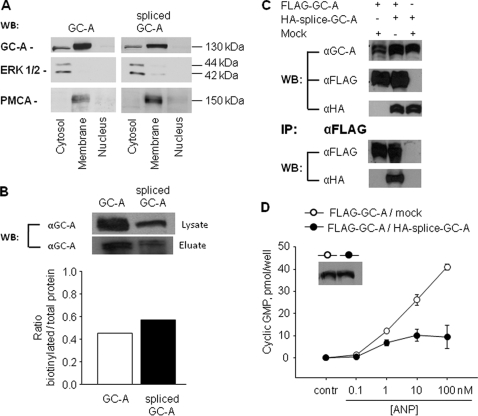
Alternative Splicing Does Not Affect Membrane Localization of GC-A—The loss of GC-A function as a result of alternative splicing could be attributed to defects in either protein expression or protein trafficking or both. To investigate these possibilities, we performed Western blot analysis after cell fractionation as well as after cell surface biotin-labeling experiments with transiently transfected HEK 293 cells. Cell fractionation showed that there were no obvious differences in the subcellular expression levels of wild type or mutant GC-A (Fig. 5A). Both the wild type and spliced form were predominantly located at the plasma membrane of HEK 293 cells. Additionally, a smaller amount of both proteins was detected in the cytosolic fraction (Fig. 5A). GC-A was absent in the nuclear fraction. In addition, the biotin-labeling experiments indicated that the levels of biotinylated GC-A protein at the plasma membrane were similar for wild type and mutant (Fig. 5B). Taken together, these results suggest that the deletion of positions ΔLys314–Gln330 did not significantly alter the protein expression or the membrane localization, and therefore it does not account for the loss of ANP binding and GC-A activity.
FIGURE 5.
Membrane localization of wild type and splice GC-A, heterodimer formation among GC-A isoforms and dominant negative effects of spliced GC-A on wild type GC-A. HEK 293 cells were transiently transfected with each or both GC-A isoforms. A, cell fractionation and Western blotting (WB). The detection of the ERK1/2 mitogen-activated protein kinase (MAPK) and of plasma membrane Ca2+-ATPase was used to characterize the cellular cytosolic and plasma membrane fractions, respectively. B, cell surface biotinylation. After cell surface biotinylation and purification of biotinylated protein with avidinagarose, Western blot analysis of whole-cell lysates (lysate) and biotin-labeled membrane proteins (eluate) from cells expressing wild type GC-A or spliced GC-A were performed using anti-GC-A antibodies. Similar results were observed in two independent experiments. C, coimmunoprecipitation experiments. Extracts from the cells transfected with FLAG-GC-A and pCMV5 (Mock) and the cells with FLAG-GC-A and HA-splice-GC-A were immunoprecipitated (IP) with the anti-FLAG antibody, and aliquots of cell lysates (before immunoprecipitation) as well as the immunoprecipitated proteins were analyzed by Western blot (WB) analyses with the anti-GC-A, anti-FLAG, and anti-HA antibodies. Similar results were observed in two independent experiments. D, guanylyl cyclase activity. Cells transfected with FLAG-GC-A and pCMV5 (Mock) and cells with FLAG-GC-A and HA-splice-GC-A were incubated with vehicle or ANP for 10 min, and intracellular cGMP contents were then measured (n = 3 experiments). Inset in D, Western blot analysis showing similar expression levels of total GC-A.
GC-AΔLys314-Gln330 Forms Heterodimers with Wild Type GC-A and Inhibits the Responsiveness of GC-A to ANP—Although the transmembrane GC-A receptor contains a single cyclase catalytic site per polypeptide chain, receptor dimerization is essential for the activation of the catalytic domain (31, 32). The introduction of mutations in individual subunits could therefore lead to the formation of nonfunctional dimers. To address this possibility, we coexpressed FLAG-tagged wild type with HA-tagged spliced GC-A in HEK 293 cells and performed coimmunoprecipitation. The same number of cells was transfected with the same amount of plasmids, and molar ratios of plasmid pairs were kept at 1:1. As shown in Fig. 5C, coexpression did not change the expression levels of either the FLAG-tagged wild type or HA-tagged mutant proteins (top). The amount of HA-tagged mutant GC-A, pulled down with the coexpressed FLAG-tagged wild type GC-A, using an anti-FLAG antibody, was similar (Fig. 5C, bottom). Taken together, these results indicate that splice GC-A is able to form heterodimers with wild type GC-A. Fig. 5D shows the functional impact of this interaction. The base-line intracellular cGMP levels of HEK 293 cells cotransfected with FLAG-GC-A and pCMV5 (mock) were similar to the levels of cells cotransfected with FLAG-GC-A and HA-splice GC-A. However, the inclusion of the splice variant resulted in much lower cGMP responses to ANP (Fig. 5D). Hence, although the variant receptor lacks ANP/BNP binding and guanylyl cyclase activity, it may be involved in the natriuretic peptide signaling system, for example as a negative regulator.
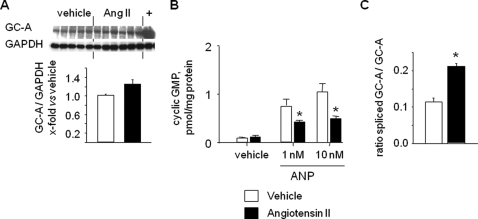
Treatment of Mice with Angiotensin II Diminishes GC-A Activity and Increases Expression of Splice GC-A—Finally, to elucidate whether increased alternative splicing of GC-A could participate in the negative regulation of the NP/GC-A system by Ang II in vivo, we examined the expression and activity of GC-A in lungs from control mice, and from mice infused with a subpressor dose of Ang II (300 ng/kg/min, n = 10) during 7 days. The expression of total GC-A protein was not altered by Ang II (Fig. 6A). Unfortunately, these immunoblot analyses did not discriminate between the protein expression levels of wild type GC-A (∼130 kDa) and the splice variant (∼128 kDa). To characterize the responsiveness of GC-A, membrane-bound ANP-dependent cGMP production was assayed. As shown in Fig. 6B, ANP-stimulated cGMP synthesis in lung membranes obtained from Ang II-treated mice was significantly blunted. Finally, we used real time RT-PCR to quantify the mRNA ratio for GC-AΔLys314-Gln330 versus wild type GC-A using probes unique for each (as depicted in Fig. 1, A and B). Notably, the ratio of splice versus wild type GC-A mRNA expression was significantly increased in the lungs of Ang II-treated mice as compared with untreated mice (Fig. 6C). Given that the splice variant serves as a dominant negative form, Ang II-induced alternative splicing of GC-A may represent a novel mechanism for reducing the sensitivity of the wild type receptor to ANP and BNP.
FIGURE 6.
Effects of chronic treatment with angiotensin II on the ANP/GC-A system in mice. A, Western blot analyses: expression of total GC-A was not different in the lungs from vehicle-treated versus Ang II-treated mice. B, determination of membrane GC-A activity. Basal (vehicle) and ANP-stimulated (1 and 10 nm) cGMP formation by lung plasma membranes were isolated from untreated and Ang II-treated mice. C, quantitative, real time RT-PCR: increased ratio of spliced versus wild type GC-A mRNA expression in the lungs of Ang II-treated as compared with untreated mice (n = 10 per group; *, p < 0.05 versus vehicle). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Our future studies will be directed to further characterize the specific regulation of alternative GC-A splicing and the possible (patho)physiological implications for the development of hypertension and cardiac hypertrophy. This type of regulation has been shown previously for soluble GC (receptor for nitric oxide) and for guanylyl cyclase B (the receptor for C-type natriuretic peptide) (33–37). For the latter receptor, splice site mutations have been shown to have a tremendous functional impact, leading to impaired skeletal growth and acromesomelic dysplasia, type Maroteaux (37). Thus, the guanylyl cyclase family, in general, may utilize such dominant negative isoforms as regulators.
This work was supported by Deutsche Forschungsgemeinschaft Sonderforschungsbereich 487 (to T. M., M. B., and M. K.) and Sonderforschungsbereich 688 (to M. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GC-A, guanylyl cyclase-A; ANP, atrial natriuretic peptide; BNP, B-type natriuretic peptide; RT, reverse transcription; Ang II, angiotensin II; DMEM, Dulbecco's modified Eagle's medium; FRET, fluorescence resonance energy transfer; PDB, Protein Data Bank; HA, hemagglutinin.
References
- 1.Kuhn, M. (2003) Circ. Res. 93 700-709 [DOI] [PubMed] [Google Scholar]
- 2.John, S. W., Krege, J. H., Oliver, P. M., Hagaman, J. R., Hodgin, J. B., Pang, S. C., Flynn, T. G., and Smithies, O. (1995) Science 267 679-681 [DOI] [PubMed] [Google Scholar]
- 3.Lopez, M. J., Wong, S. K., Kishimoto, I., Dubois, S., Mach, V., Friesen, J., Garbers, D. L., and Beuve, A. (1995) Nature 378 65-68 [DOI] [PubMed] [Google Scholar]
- 4.Rutledge, D. R., Sun, Y., and Ross, E. A. (1995) J. Hypertens. 13 953-955 [DOI] [PubMed] [Google Scholar]
- 5.Beige, J., Ringel, J., Hohenbleicher, H., Rubattu, S., Kreutz, R., and Sharma, A. M. (1997) Am. J. Hypertens. 10 1316-1318 [DOI] [PubMed] [Google Scholar]
- 6.Rubattu, S., Bigatti, G., Evangelista, A., Lanzani, C., Stanzione, R., Zagato, L., Manunta, P., Marchitti, S., Venturelli, V., Bianchi, G., Volpe, M., and Stella, P. (2006) J. Am. Coll. Cardiol. 48 499-505 [DOI] [PubMed] [Google Scholar]
- 7.Bulut, D., Potthast, R., Hanefeld, C., Schulz, T., Kuhn, M., and Mügge, A. (2003) Eur. J. Clin. Investig. 33 567-573 [DOI] [PubMed] [Google Scholar]
- 8.Sala, C., Ambrosi, B., and Morganti, A. (2001) J. Clin. Endocrinol. Metab. 86 1957-1961 [DOI] [PubMed] [Google Scholar]
- 9.Burnett, J. C., Jr., Kao, P. C., Hu, D. C., Heser, D. W., Heublein, D., Granger, J. P., Opgenorth, T. J., and Reeder, G. S. (1986) Science 231 1145-1147 [DOI] [PubMed] [Google Scholar]
- 10.Hirooka, Y., Takeshita, A., Imaizumi, T., Suzuki, S., Yoshida, M., Ando, S., and Nakamura, M. (1990) Circulation 82 147-153 [DOI] [PubMed] [Google Scholar]
- 11.Arise, K. K., and Pandey, K. N. (2006) Biochem. Biophys. Res. Commun. 349 131-135 [DOI] [PubMed] [Google Scholar]
- 12.Bryan, P. M., and Potter, L. R. (2002) J. Biol. Chem. 277 16041-16047 [DOI] [PubMed] [Google Scholar]
- 13.Fan, D., Bryan, P. M., Antos, L. K., Potthast, R. J., and Potter, L. R. (2005) Mol. Pharmacol. 67 174-183 [DOI] [PubMed] [Google Scholar]
- 14.Bryan, P. M., Smirnov, D., Smolenski, A., Feil, S., Feil, R., Hofmann, F., Lohmann, S., and Potter, L. R. (2006) Biochemistry 45 1295-1303 [DOI] [PubMed] [Google Scholar]
- 15.Potter, L. R., and Hunter, T. (2001) J. Biol. Chem. 276 6057-6060 [DOI] [PubMed] [Google Scholar]
- 16.Schreier, B., Börner, S., Völker, K., Gambaryan, S., Schäfer, S. C., Kuhlencordt, P., Gassner, B., and Kuhn, M. (2008) Endocrinology 149 4193-4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolaev, V. O., Gambaryan, S., and Lohse, M. J. (2006) Nat. Meth. 3 23-25 [DOI] [PubMed] [Google Scholar]
- 18.Mei, Z. Z., Mao, H. J., and Jiang, L. H. (2006) Am. J. Physiol. 291 C1022-C1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilic, A., Velic, A., De Windt, L. J., Fabritz, L., Voss, M., Mitko, D., Zwiener, M., Baba, H. A., van Eickels, M., Schlatter, E., and Kuhn, M. (2005) Circulation 112 2307-2317 [DOI] [PubMed] [Google Scholar]
- 20.Holtwick, R., van Eickels, M., Skryabin, B. V., Baba, H. A., Bubikat, A., Begrow, F., Schneider, M. D., Garbers, D. L., and Kuhn, M. (2003) J. Clin. Investig. 111 1399-1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potthast, R., Ehler, E., Scheving, L. A., Sindic, A., Schlatter, E., and Kuhn, M. (2001) Endocrinology 142 3087-3097 [DOI] [PubMed] [Google Scholar]
- 22.Breathnach, R., and Chambon, P. (1981) Annu. Rev. Biochem. 50 349-383 [DOI] [PubMed] [Google Scholar]
- 23.Tallerico-Melnyk, T., Watt, V. M., and Yip, C. C. (1995) Biochem. Biophys. Res. Commun. 209 930-935 [DOI] [PubMed] [Google Scholar]
- 24.Francoeur, F., Gossard, F., Hamet, P., and Tremblay, J. (1995) Clin. Exp. Pharmacol. Physiol. 22 S172-S174 [DOI] [PubMed] [Google Scholar]
- 25.Sharp, P. A. (1994) Cell 77 805-815 [DOI] [PubMed] [Google Scholar]
- 26.Breitbart, R. E., Andreadis, A., and Nadal-Ginard, B. (1987) Annu. Rev. Biochem. 56 467-495 [DOI] [PubMed] [Google Scholar]
- 27.van den Akker, F., Zhang, X., Miyagi, M., Huo, X., Misono, K. S., and Yee, V. C. (2000) Nature 406 101-104 [DOI] [PubMed] [Google Scholar]
- 28.van den Akker, F. (2001) J. Mol. Biol. 311 923-937 [DOI] [PubMed] [Google Scholar]
- 29.McNicoll, N., Gagnon, J., Rondeau, J. J., Ong, H., and DeLean, A. (1996) Biochemistry 35 12950-12956 [DOI] [PubMed] [Google Scholar]
- 30.Ogawa, H., Qiu, Y., Ogata, C. M., and Misono, K. S. (2004) J. Biol. Chem. 279 28625-28631 [DOI] [PubMed] [Google Scholar]
- 31.Wilson, E. M., and Chinkers, M. (1995) Biochemistry 34 4696-4701 [DOI] [PubMed] [Google Scholar]
- 32.Yang, R. B., and Garbers, D. L. (1997) J. Biol. Chem. 272 13738-13742 [DOI] [PubMed] [Google Scholar]
- 33.Behrends, S., Harteneck, C., Schultz, G., and Koesling, D. (1995) J. Biol. Chem. 270 21109-21113 [DOI] [PubMed] [Google Scholar]
- 34.Gupta, G., Azam, N., Yang, L., and Danziger, R. S. (1997) J. Clin. Investig. 100 1488-1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharina, I. G., Jelen, F., Bogatenkova, E. P., Thomas, A., Martin, E., and Murad, F. (2008) J. Biol. Chem. 283 15104-15113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura, N., and Garbers, D. L. (2003) J. Biol. Chem. 278 48880-48889 [DOI] [PubMed] [Google Scholar]
- 37.Bartels, C. F., Bükülmez, H., Padayatti, P., Rhee, D. K., van Ravenswaaij-Arts, C., Pauli, R. M., Mundlos, S., Chitayat, D., Shih, L. Y., Al-Gazali, L. I., Kant, S., Cole, T., Morton, J., Cormier-Daire, V., Faivre, L., Lees, M., Kirk, J., Mortier, G. R., Leroy, J., Zabel, B., Kim, C. A., Crow, Y., Braverman, N. E., van den Akker, F., and Warman, M. L. (2004) Am. J. Hum. Genet. 75 27-34 [DOI] [PMC free article] [PubMed] [Google Scholar]