Abstract
The NfuA protein has been postulated to act as a scaffolding protein in the biogenesis of photosystem (PS) I and other iron-sulfur (Fe/S) proteins in cyanobacteria and chloroplasts. To determine the properties of NfuA, recombinant NfuA from Synechococcus sp. PCC 7002 was overproduced and purified. In vitro reconstituted NfuA contained oxygen- and EDTA-labile Fe/S cluster(s), which had EPR properties consistent with [4Fe-4S] clusters. After reconstitution with 57Fe2+, Mössbauer studies of NfuA showed a broad quadrupole doublet that confirmed the presence of [4Fe-4S]2+ clusters. Native gel electrophoresis under anoxic conditions and chemical cross-linking showed that holo-NfuA forms dimers and tetramers harboring Fe/S cluster(s). Combined with iron and sulfide analyses, the results indicated that one [4Fe-4S] cluster was bound per NfuA dimer. Fe/S cluster transfer from holo-NfuA to apo-PsaC of PS I was studied by reconstitution of PS I complexes using P700-FX core complexes, PsaD, apo-PsaC, and holo-NfuA. Electron transfer measurements by time-resolved optical spectroscopy showed that holo-NfuA rapidly and efficiently transferred [4Fe-4S] clusters to PsaC in a reaction that required contact between the two proteins. The NfuA-reconstituted PS I complexes had typical charge recombination kinetics from [FA/FB]- to P700+ and light-induced low-temperature EPR spectra. These results establish that cyanobacterial NfuA can act as a scaffolding protein for the insertion of [4Fe-4S] clusters into PsaC of PS I in vitro.
In the photosynthetic electron transport chains of cyanobacteria and chloroplasts of higher plants, iron-sulfur (Fe/S)4 clusters not only serve as the terminal electron-accepting cofactors in photosystem (PS) I but they also act as electron transfer cofactors in the cytochrome b6f complex and in soluble ferredoxin. PS I is a large, membrane-protein complex that catalyzes the light-induced transfer of electrons from plastocyanin or cytochrome c6 in the thylakoid lumen to ferredoxin or flavodoxin in the cytoplasm (1, 2). As indicated by the x-ray crystal structure, the PS I reaction center of Synechococcus elongatus contains 12 protein subunits, which bind 96 chlorophyll (Chl) a molecules, 22 β-carotenes, 2 phylloquinones, 3 [4Fe-4S] clusters, and 5 lipids (3). The PS I electron transport chain consists of P700 (Chl a—Chl a′ dimer), A0 (Chl a monomer), A1 (phylloquinone), and three [4Fe-4S] clusters, which are the terminal electron acceptors denoted FX, FA, and FB. PsaA and PsaB each provide two cysteine (Cys) ligands to the intrasubunit [4Fe-4S] FX cluster, and the FA and FB terminal electron acceptors are bound to a 9-kDa soluble protein, PsaC, which forms a ridge on the stromal surface of PS I with PsaD and PsaE (4). Our previous studies on PS I biogenesis have shown that the insertion of Fe/S clusters is a key step in its post-translational maturation (5, 6). The inability to assemble the FX cluster in a rubA mutant destabilizes PS I and prevents the association of PsaC, PsaD, and PsaE with the membrane-associated portion of the complex. Studies of psaC mutants with modified Cys ligands for FA and FB also point to the importance of Fe/S clusters in the biogenesis and stability of this complex (2).
The biosynthesis of Fe/S clusters is a complex and highly regulated metabolic process. Three major systems for Fe/S cluster biosynthesis have been identified in various organisms: the NIF, ISC, and SUF systems (7, 8). In cyanobacteria and in the chloroplasts of algae and higher plants, oxygen evolution accompanying water oxidation by PS II is one of the major processes associated with the light reactions. Perhaps for this reason, an oxygen-resistant system is essential for Fe/S cluster biogenesis in such organisms (see Ref. 9 for a review). The first evidence connecting the SUF system to PS I biogenesis was obtained through the characterization of pseudorevertants that had been isolated from site-specific psaC mutants of Synechocystis sp. PCC 6803 (10). These pseudorevertants were able to suppress the inability of certain PsaC variants to assemble Fe/S clusters when non-Cys ligands to FA and FB were present in vivo. Characterization of these intergenic suppressors identified the sufR gene, whose product was subsequently shown to encode a negative transcriptional regulator of the suf regulon (11, 12). In many cyanobacterial genomes the major components of the SUF system are organized as an operon, sufBCDS (9). Reverse genetics functional studies of genes for the SUF and ISC systems in Synechocystis sp. PCC 6803 and Synechococcus sp. PCC 7002 have shown that cyanobacteria primarily rely on the SUF system rather than the ISC system for the biogenesis and maintenance of Fe/S clusters (9).
Studies on the insertion of metalloclusters into the nitrogenase apoproteins in Azotobacter vinelandii have established the molecular scaffold model for Fe/S cluster biogenesis (7, 8). Together with NifS, a cysteine desulfurase that provides sulfide for Fe/S cluster assembly, NifU is known to participate in the assembly of nitrogenase metalloclusters (13). A. vinelandii NifU is a homodimer that carries two stable [2Fe-2S] clusters (14), and this protein is thought to provide a molecular scaffold for maturation of the nitrogenase metalloclusters (15). Sequence comparisons have revealed three domains in NifU proteins (8). All NifU proteins have nine highly conserved Cys residues: three Cys residues in the N-terminal domain, four Cys residues in the central domain, and two Cys residues in the C-terminal domain. Genetic and biochemical experiments suggest that transient Fe/S clusters are formed in both the N- and C-terminal domains of NifU. NifU-dependent activation of the Fe protein of nitrogenase (NifH) is lost if any of the Cys residues in the N- and C-terminal domains is replaced by alanine (16).
The genomes of all nitrogen-fixing cyanobacteria encode a homolog of the A. vinelandii nifU gene, but all cyanobacterial genomes additionally have a gene, nfuA, that encodes a small protein with strong sequence similarity to the C-terminal domain of NifU (see supplemental Fig. S1; for reviews, see Refs. 9 and 17). IscU, a protein with sequence similarity to the N-terminal domain of NifU, has been proposed to function as the scaffold protein in the ISC system (7, 8, 17). Because of their labile Fe/S clusters, both IscA and SufA have been proposed as alternate molecular scaffold proteins for the ISC or SUF systems (18, 19). However, a sufA iscA double mutant is viable and exhibits no detectable deficiencies in Fe/S proteins in cyanobacteria (20). In fact, the double deletion mutant grows at higher rates than does the wild-type under iron limitation and grows normally under growth conditions for which the wild-type undergoes chlorosis. Transcription analyses and physiological characterization of wild type and mutant strains lacking SufA, IscA, or both, suggest that SufA and IscA instead play regulatory roles in iron homeostasis, redox sensing, and sensing of the capacity of cells for Fe/S cluster biogenesis (20). In contrast, NfuA is absolutely required for cellular viability (20).
All cyanobacterial NfuA proteins contain a highly conserved CXXC motif that is presumably required for binding an Fe/S cluster. A previous study reported that NfuA (originally named SyNifU) from Synechocystis sp. PCC 6803 contained a labile [2Fe-2S] cluster and that this protein could act as scaffold protein in transferring its [2Fe-2S] cluster to a plant-type apoferredoxin (PetF) (21). Five NfuA-like proteins have been found in the Arabidopsis thaliana genome, and three of these proteins (Nfu1, Nfu2, and Nfu3) have been localized to plastids (22). Nfu2 displays the highest sequence similarity to cyanobacterial NfuA proteins and binds a labile [2Fe-2S] cluster in vitro (23). A reverse genetics approach has shown that Nfu2 plays an important role in Fe/S cluster biogenesis in plastids (23). A human NfuA-like protein, HIRIP5, has also been reported to carry a [4Fe-4S] cluster (24).
The goals of the current study were to clarify the Fe/S cluster-binding properties of cyanobacterial NfuA and to determine whether this protein might participate in the biogenesis of PS I. We report here our results from the biochemical and spectroscopic characterization of the NfuA protein from Synechococcus sp. PCC 7002. Non-denaturing gel electrophoresis indicates that cyanobacterial NfuA forms dimers and tetramers. Biochemical, EPR, and Mössbauer measurements show that cyanobacterial NfuA binds one [4Fe-4S] cluster per dimer. Finally, we demonstrate that the [4Fe-4S] clusters of NfuA can be rapidly and efficiently transferred to the PsaC apoprotein in vitro in a reaction that occurs without the release of Fe/S clusters into solution.
EXPERIMENTAL PROCEDURES
Construction of the nfuA Expression Plasmid—The nfuA gene (SYNPCC7002_A1413) was amplified by PCR from chromosomal DNA of Synechococcus sp. PCC 7002 using the primers 1nfuFN, 5′-TTCGTTCACCAAAAGACATATGGCATTAGC-3′ and 2nfuRB, 5′-CATGGCAAACCCCTAGGATCCTAAAGAAAGT-3′. The purified PCR product was digested with restriction enzymes NdeI and BamHI (sites are underlined above), and cloned into pET16b expression vector (Novagen, Madison, WI), which contains an ampicillin resistance gene, to produce plasmid pET16b-nfuA, which was verified by DNA sequencing (Penn State Nucleic Acid Facility). The product from pET16b-nfuA should produce a recombinant NfuA protein with a poly-His tag (His10) at its N terminus.
NfuA Overproduction and Purification—For overproduction of the NfuA protein, the pET16b-nfuA plasmid was transformed into Escherichia coli strain BL21(DE3). The overexpression of nfuA was induced by the addition of 0.1 mm isopropyl 1-thio-β-galactopyranoside to exponentially growing cells in LB medium at 37 °C. Cultures were grown for 4 h after addition of isopropyl 1-thio-β-galactopyranoside. Cells were harvested by centrifugation, and stored at -80 °C until required. For protein purification, cells were resuspended in 50 mm Tris-HCl buffer, pH 8.0, and lysed by passage through a chilled French pressure cell operated at 138 megapascal. The lysed cells were centrifuged at 12,000 × g. The soluble, cell-free extract was loaded onto a column packed with nickel-nitrilotriacetic acid resin (Qiagen, Valencia, CA), and washed with 5 column volumes of 50 mm Tris-HCl buffer at pH 8.0 containing 50 mm NaCl and 20 mm imidazole. NfuA was eluted with 50 mm Tris-HCl buffer at pH 8.0, containing 50 mm NaCl and 250 mm imidazole. The eluted NfuA fraction was immediately dialyzed against 50 mm Tris-HCl buffer, pH 8.0. The protein was concentrated by ultrafiltration with an Amicon concentrator equipped with a YM-3 membrane (membrane cut-off of 3 kDa) (Millipore, Billerica, MA).
Chemical Reconstitution of Fe/S Clusters in NfuA—Chemical reconstitution of Fe/S clusters in apo-NfuA was performed as described previously (5, 10, 25). Reconstitution reactions (50 ml) were routinely conducted at a protein concentration of 0.1 to 0.2 mg apo-NfuA ml-1. To prepare a sample for Mössbauer measurement, 4.5 mm 57Fe2+ was used in the reconstitution protocol. Reconstituted holo-NfuA was concentrated by ultrafiltration to 2.5 ml, and loaded onto a PD-10 desalting column (Amersham Biosciences) to remove excess iron, sulfide, and other reagents.
Protein Biochemistry—Quantitative amino acid analysis (Molecular Analysis Facility, University of Iowa) of NfuA was performed in triplicate to obtain a correction factor (0.669) for protein concentrations determined by the Coomassie Plus protein assay (Pierce) with bovine serum albumin as the standard. The protein concentration of an NfuA solution determined by quantitative amino acid analysis (3.20 ± 0.04 mg ml-1) was divided by the estimated protein concentration of the same protein solution from the dye-binding assay with bovine serum albumin as the standard (4.79 mg ml-1) to produce the correction factor 0.669. This correction factor was subsequently applied to all calculations in which protein concentrations were estimated by the dye binding assay.
Quantitation of Fe was performed with ferene (5,5′(3-(2-pyridyl)-1,2,4-triazine-5,6-diyl)-bis-2-furansulfonic-acid) as the detection reagent as described previously (26). A freshly prepared FeCl3 solution was used to produce a standard curve. The method for quantitation of sulfide has been described (27). A freshly prepared Na2S solution was used to produce the standard curve. All incubations were performed in an anaerobic chamber with an atmosphere of H2:CO2:N2 (10%:10%:80%, v/v) (Coy Laboratory Products, Grass Lake, MI).
Cross-linking and Alkylation—The methods for protein cross-linking with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) (28) and protein alkylation with iodoacetamide (29) have been described previously. Cross-linking and alkylation of apo-NfuA and holo-NfuA were performed in the anaerobic chamber to avoid oxidation of Cys residues.
Gel Electrophoresis—Methods for polyacrylamide gel electrophoresis in the presence of SDS-PAGE and immunoblotting have been described (5, 12). Native gel electrophoresis under anoxic conditions was performed in the anaerobic chamber. Non-denaturing gels (9% (w/v) acrylamide for the separating gel and 4% (w/v) acrylamide for the stacking gel) were prepared in the same way as for SDS-PAGE except that SDS was omitted from all buffers. All buffers were extensively sparged with N2 gas to remove oxygen. Apo-NfuA and holo-NfuA samples were prepared under anoxic conditions in loading buffer without SDS and 2-mercaptoethanol. Gels were stained for iron as described previously (30). The iron-stained gel was scanned, after which the gel was stained with Coomassie Blue.
Low-temperature X-band EPR Spectroscopy of Holo-NfuA—The method for low-temperature, X-band EPR spectroscopy has been described (31). Measurements were performed using an ECS-106 X-band spectrometer (Bruker Biospin, Billerica, MA) equipped with an ESR900 liquid helium cryostat and an ITC-4 temperature controller (Oxford Instruments, Billerica, MA). The microwave frequency was determined with a Hewlett-Packard 5340A frequency counter. The spectrometer parameters were: temperature, 16 K; microwave frequency, 9.47 GHz; receiver gain, 2 × 104; modulation amplitude, 10 gauss at 100 kHz. Spectra were obtained by averaging 10 scans. Holo-NfuA was prepared for EPR analysis by inclusion of 10 mm sodium dithionite, and 100 mm glycine at pH 10.0. The baseline was generated from a blank sample, which was prepared in the same manner as the NfuA samples except that buffer replaced the protein solution. The spectrum from the blank was subtracted from the spectrum of reduced NfuA.
Mössbauer Spectroscopy—The protocol for Mössbauer spectroscopy has been described (12). Spectra were recorded at a sample temperature of 4.2 K that was maintained by a liquid helium cryostat. The sample was kept inside an SVT-400 Dewar (Janis, Wilmington, MA) and a magnetic field of 53 millitesla was applied parallel to the γ-beam. The isomer shifts quoted are relative to the centroid of the spectrum of metallic Fe foil at room temperature. Data analysis was performed using the program WMOSS from WEB Research (Edina, MN).
PS I Reconstitution and Assay by Light-induced EPR Spectroscopy—The method for reconstitution of PS I complexes and the conditions for the light-induced EPR spectroscopy have been described (32). P700 FX-cores were prepared from purified PS I complexes of Synechococcus sp. PCC 7002 following the method described (25). Recombinant PsaC and PsaD were overproduced in E. coli and purified as described (33). For light-induced EPR spectroscopic studies of Fe/S cluster transfer from holo-NfuA to apo-PsaC, samples were prepared with 20 μm P700 FX-cores, 400 μm apo-PsaC, 400 μm PsaD, 840 μm holo-NfuA, 10 mm ascorbate, 10 μm 2,6-dichlorophenolindophenol (DCPIP), 4 mm dithiothreitol (DTT), and 0.04% (w/v) n-dodecyl-β-d-maltoside (DM), and frozen under low light. The baseline was recorded in the dark, and spectra were recorded before and after illumination with an argon ion laser operated at 2.5 watts in all-lines mode with a 3-fold beam expander to fill the resonator grid.
PS I Reconstitution and Assay by Room Temperature Optical Kinetic Spectroscopy in the Near-IR Region—Transient absorbance changes in the near-IR were measured for reconstituted PS I complexes with a laboratory-built, double-beam spectrophotometer as described (6, 31), except that the cuvettes used were 2 × 10 mm instead of 5 × 10 mm. The samples were prepared under anoxic conditions, with the following final concentrations: 1 μm P700-FX cores, 20 μm apo-PsaC, 20 μm PsaD, 42 μm holo-NfuA, 10 μm DCPIP, 10 mm ascorbate, 4 mm DTT, and 0.04% (w/v) DM. The concentrations of apo-PsaC, PsaD, and the P700 FX-cores were determined according to the extinction coefficients described previously (32, 33). The concentration of holo-NfuA was calculated from the [4Fe-4S] cluster content as described (25).
RESULTS
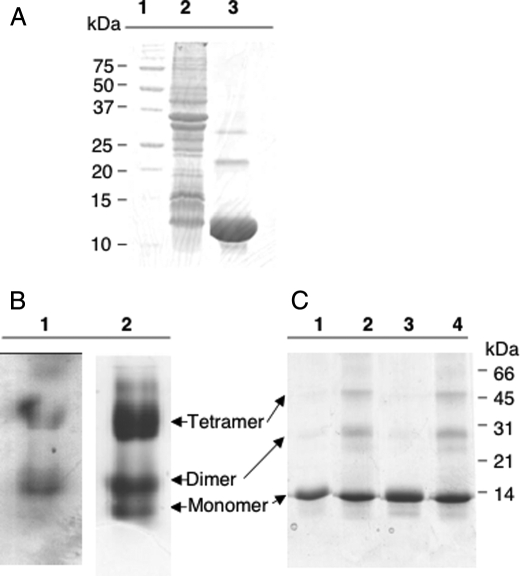
Overproduction and Purification of NfuA—The nfuA gene from Synechococcus sp. PCC 7002 was cloned into pET16b and overexpressed to produce a recombinant protein with a His10 tag at its N terminus. As determined by SDS-PAGE, purified NfuA had an apparent mass of ∼11 kDa (Fig. 1A, lane 3), which matched the predicted mass of 11.1 kDa (NfuA (8558 Da) + His10 tag (2538 Da) = 11.1 kDa). Apo-NfuA was purified from the soluble fraction of the whole cell lysate, which had a light brown color; this suggested that Fe/S clusters might be present in the NfuA protein. However, based upon the loss of the brownish color and a decrease in absorbance at about 420 nm, these Fe/S clusters were labile and were lost during the purification procedure. Therefore, it was necessary to reconstitute the Fe/S clusters of NfuA in vitro with inorganic reagents under anoxic conditions.
FIGURE 1.
Electrophoretic characterization of NfuA. A, lane 1, molecular mass markers whose masses are indicated at the left; lane 2, whole cell extract of E. coli cells overexpressing nfuA; lane 3, apo-NfuA purified by metal chelation chromatography. B, native gel electrophoresis of reconstituted NfuA under anoxic conditions. Lane 1, holo-NfuA stained for iron; lane 2, holo-NfuA stained with Coomassie Blue. C, SDS-PAGE analyses of holo-NfuA with and without alkylation and cross-linking with EDC. Lane 1, holo-NfuA; lane 2, holo-NfuA cross-linked with EDC; lane 3, holo-NfuA treated with iodoacetamide; lane 4, holo-NfuA cross-linked with EDC and then treated with iodoacetamide.
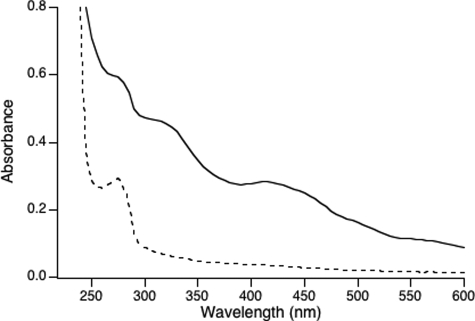
The absorption spectrum of chemically reconstituted holo-NfuA (Fig. 2, solid line) showed broad absorbance bands between 300 and 600 nm that are typical of sulfur → iron charge transfer bands of Fe/S proteins. The absence of maxima in the 450- and 550-nm regions suggested that holo-NfuA harbored [4Fe-4S] clusters. The stability of the Fe/S clusters in holo-NfuA was examined by exposing holo-NfuA to oxygen or 10 mm EDTA (data not shown). Exposure of holo-NfuA to air levels of oxygen led to a 50% decrease in absorption between 400 and 550 nm within 2 h, and EDTA treatment caused a 50% decrease in absorption after 1 h. These results indicated that the Fe/S cluster(s) in NfuA were sensitive to both oxygen and chelating agents. All subsequent manipulations with holo-NfuA were thus performed under anoxic conditions.
FIGURE 2.
UV-visible absorption spectra of apo-NfuA (dotted line) and holo-NfuA (solid line).
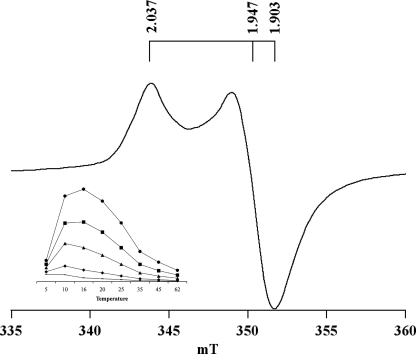
EPR Spectroscopy of Reconstituted NfuA—A low-temperature, X-band EPR spectrum of dithionite-reduced holo-NfuA is presented in Fig. 3. At 16 K and a microwave power of 40 milliwatts, NfuA had an axial spectrum with g values of 2.037, 1.947, and 1.903. The inset depicts the temperature dependence of the EPR signal. At constant microwave power, the intensity of the EPR signal was maximal at 10 to 16 K and decreased dramatically above 20 K. These spectroscopic properties suggested that chemically reconstituted holo-NfuA harbored [4Fe-4S]1+ cluster(s).
FIGURE 3.
Temperature and power dependence of EPR spectra of reconstituted holoNfuA. The EPR spectrum of holo-NfuA at 16 K and 40 milliwatts shows the axial line shape of an 4Fe-4S cluster with a low-field peak at g = 2.037 and a high-field trough at g = 1.903. Inset, temperature dependence (in Kelvins) of the holo-NfuA EPR signal at different microwave powers: 126 milliwatts (circles), 40 milliwatts (squares), 10 milliwatts (triangles), 1 milliwatt (diamonds), and 0.1 milliwatt (bars).
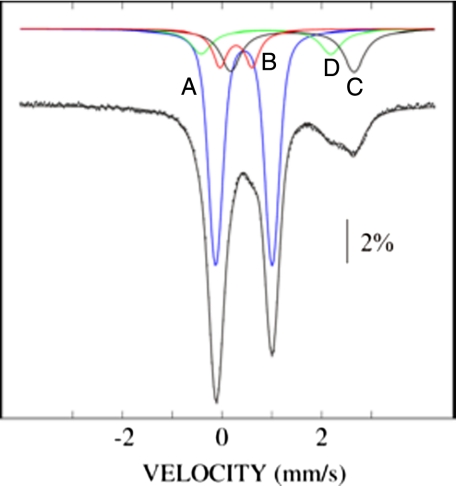
Mössbauer Spectroscopy of the Holo-NfuA—To obtain further evidence for the presence of [4Fe-4S] cluster(s) in holo-NfuA, the protein was reconstituted with 57Fe2+ and studied by Mössbauer spectroscopy. The spectrum of 57Fe-holo-NfuA was recorded at 4.2 K in a 53-millitesla external magnetic field oriented parallel to the γ-beam. The vertical bars in Fig. 4 are the Mössbauer spectrum of oxidized holo-NfuA, and the overlaid solid line shows a simulation of the experimental data. This spectrum was deconvoluted to assess the contributions from iron in different forms to the overall spectrum (Fig. 4, lines A–D). Line A represents the quadrupole doublet arising from [4Fe-4S]2+ clusters and has an isomer shift (δ) of 0.44 mm/s and a quadrupole splitting parameter (ΔEQ) of 1.13 mm/s. The deconvolution showed that 61% of the iron in the sample analyzed was contained within [4Fe-4S]2+ clusters. The presence of a shoulder at 0.6 mm/s is typical of [2Fe-2S]2+ clusters. Line B represents the iron quadrupole doublet attributed to [2Fe-2S]2+ clusters; it had δ = 0.28 mm/s and ΔEQ = 0.64 mm/s, and accounted for 11% of the iron in the reconstituted sample. The broad line at ∼2–3 mm/s is typical of high-spin Fe(II), and the broadness suggests the presence of several distinct Fe2+ species. Lines C and D are quadrupole doublet simulations with parameters typical of six-coordinate, N/O-ligated Fe2+ and four-coordinate, sulfur-ligated Fe2+ species; these account for 17 and 11% of the total iron, respectively. No signals were detected by low-temperature EPR spectroscopy of the identical oxidized holo-NfuA sample that was studied by Mössbauer spectroscopy (data not shown). These data indicate that most of the Fe/S clusters in holo-NfuA were [4Fe-4S] clusters, although the data suggests the presence of a small fraction of [2Fe-2S] clusters.
FIGURE 4.
Low-field Mössbauer spectra of 57Fe2+-reconsituted holo-Nfu at 4.2 K with an externally applied 53 millitesla magnetic field. The experimental data are indicated by the dotted line, and the solid overlaid line is the simulation of the experimental data. The lettered lines show the deconvoluted experimental data for iron in different forms: A, iron quadrupole doublet for [4Fe-4S]2+ clusters; B, iron quadrupole doublet for [2Fe-2S]2+ clusters; C and D, iron quadrupole doublets for six-coordinate Fe(II) and Fe(II) coordinated by four thiolate ligands, respectively.
NfuA Forms Oligomers That Carry One [4Fe-4S]2+ Cluster Per Dimer—To examine the oligomerization state of NfuA, apo-Nfu and reconstituted holo-NfuA were analyzed by native PAGE under anoxic conditions. After electrophoresis, two forms of holo-NfuA were detected by iron staining (Fig. 1B, lane 1); however, a third form of the protein was detected after staining with Coomassie Blue (Fig. 1B, lane 2). By comparing the electrophoretic migration of NfuA as a function of acrylamide concentration (data not shown), it was concluded that the three observed forms of NfuA were most likely monomeric apo-NfuA and dimeric and tetrameric holo-NfuA. As can been seen in Fig. 1B, the approximate ratio of these three NfuA forms was about 5:20:75, respectively. For apo-NfuA, two major bands, corresponding to monomeric (major) and dimeric (minor) forms, were resolved after native gel electrophoresis (supplemental Fig. S2A). Only a very faint band with the apparent migration of a tetramer was observed for apo-NfuA (supplemental Fig. S2A).
The oligomerization of apo-NfuA and holo-NfuA was studied further by cross-linking with the zero-length cross-linking reagent EDC and by using the alkylation reagent iodoacetamide, which was employed to prevent the formation of intermolecular disulfide bonds. As shown in Fig. 1A, lane 3, NfuA monomers, dimers, and trimers and/or tetramers were sometimes observed after SDS-PAGE of NfuA samples that had not been alkylated prior to SDS-PAGE. However, alkylation of the Cys residues with iodoacetamide prior to SDS-PAGE largely prevented the formation of these oligomers (Fig. 1C, lane 3). When holo-NfuA was subjected to cross-linking with EDC followed by alkylation, monomers, dimers, and tetramers were observed after SDS-PAGE (Fig. 1C, lane 4). NfuA dimers and tetramers were more abundant after EDC treatment of the holo-NfuA sample, but apo-Nfu dimers were also observed after EDC treatment (supplemental Fig. S2B). Based upon the results from native PAGE under anoxic conditions and from cross-linking of holo-NfuA with EDC, we concluded that both apo-NfuA and holo-NfuA could form dimers. However, in the presence of Fe/S clusters holo-NfuA occurred in solution as a mixture of dimers (minor form) and tetramers (major form).
Chemical analyses of holo-NfuA for non-heme iron and sulfide yielded ∼2.39 iron and ∼1.87 sulfides per NfuA monomer. Each NfuA polypeptide has only two conserved Cys residues that could serve as ligands to Fe/S clusters (supplemental Fig. S1); this suggests that a NfuA dimer is required to ligate an Fe/S cluster. Combining the chemical analyses with the results from EPR and Mössbauer spectroscopy, chemically reconstituted holo-NfuA contained ∼0.73 [4Fe-4S]2+ and ∼0.12 [2Fe-2S]2+ clusters per dimer. In summary, the data showed that holo-NfuA formed dimers and tetramers that predominantly harbored one [4Fe-4S]2+ cluster per homodimer.
Reconstitution of PS I Complexes by Cluster Transfer from Holo-NfuA to Apo-PsaC—Previous mutagenesis studies have shown that NfuA is essential in cyanobacteria, and other studies have suggested that NfuA might play an essential role in the assembly of Fe/S clusters (20). We next tested whether NfuA could transfer Fe/S clusters to apo-PsaC, which ligates two [4Fe-4S] clusters (1). In the presence of PsaD, Fe/S cluster transfer to apo-PsaC can be monitored by reconstitution of long-lived electron transfer in P700-FX cores (34). Optical kinetic spectroscopy can be used to monitor electron transfer in the reconstituted PS I complexes (35). In these measurements, a single turnover laser flash induces an absorbance increase at 820 nm, which arises from the P700+ Chl cation. The decay kinetics of this absorbance provides information about the electron acceptor from which the charge recombination occurs (35). Because binding of holo-PsaC to P700-FX cores is rapid, the recovery of long-lived charge separation on a laser flash provides a convenient and accurate assay for the insertion of the [4Fe-4S] clusters into PsaC.
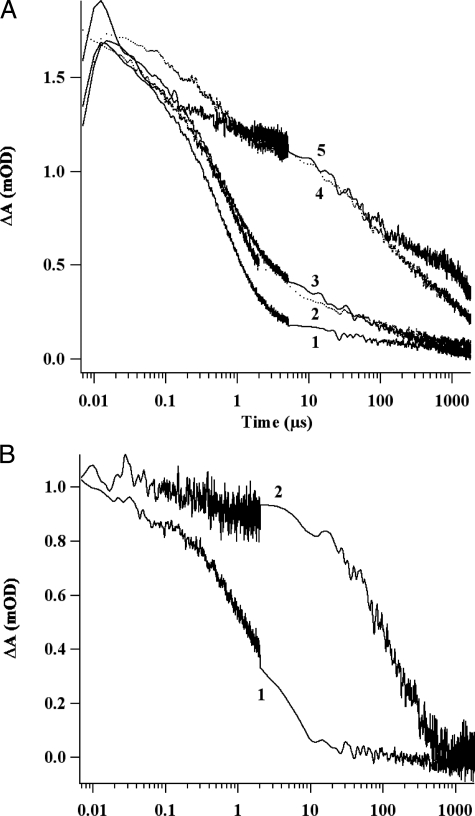
Fig. 5 compares the decay kinetics of the P700+ cation absorbance for P700-FX core complexes incubated with various combinations of PsaC, PsaD, and NfuA. As shown in Fig. 5A, trace 1, and supplemental Fig. S3A, four kinetic phases are observed in the decay of the 820-nm absorbance after P700-FX cores had been incubated with apo-PsaC and PsaD. The 51-μs phase was assigned to the backreaction of A1- with P700+; these represent PS I cores in which FX has been inadvertently denatured. The 643-μs and 4.3-ms phases were assigned to charge recombination between FX- and P700+. The remaining low-amplitude phase, which had a time constant of >100 ms, was sensitive to DCPIP concentration, allowing this phase to be assigned to electron donation from DCPIP to P700+ (data not shown). In PS I complexes showing this latter kinetic phase, electrons had been transferred to oxygen and thus were no longer available for charge recombination with P700+. Because PsaD is required for stable binding of PsaC to the stromal surface of PS I (33), very similar results were obtained when P700-FX cores were incubated with holo-PsaC in the absence of PsaD (Fig. 5A, trace 2).
FIGURE 5.
Flash-induced absorption changes for P700+ reduction at 820 nm. A, decay kinetics of samples containing: 1) 1 μm P700-FX cores, 20 μm apo-PsaC, 20 μm PsaD, 4 mm DTT, and 0.04% (w/v) DM; 2) 1 μm P700-FX cores, 20 μm holo-PsaC, 4 mm DTT, and 0.04% (w/v) DM; 3) 1 μm P700-FX cores, 42 μm holo-NfuA, 20 μm PsaD, 4 mm DTT, and 0.04% (w/v) DM; 4) 1 μm P700-FX cores, 20 μm holo-PsaC, 20 μm PsaD, 4 mm DTT, and 0.04% (w/v) DM; 5) 1 μm P700-FX cores, 42 μm holo-NfuA, 20 μm apo-PsaC, 20 μm PsaD, 4 mm DTT, and 0.04% (w/v) DM. After a 1-h incubation, 10 μm DCPIP and 10 mm ascorbate were added and flash-induced kinetics at 820 nm were measured. B, decay kinetics of samples containing: 1) 1 μm P700-FX cores, 42 μm holo-NfuA that had been boiled for 1 min, 20 μm apo-PsaC, 20 μm PsaD, 4 mm DTT, and 0.04% (w/v) DM; and 2) 1 μm P700-FX cores, 20 μm apo-PsaC, 20 μm PsaD, 4 mm DTT, and 0.04% (w/v) DM. Fe/S clusters were assembled by mixing 168 μm Fe2+ and 168 μm S2- and 4 mm DTT. Prior to addition of these reagents to the solution containing apo-PsaC, the Fe/S reagents were boiled for 1 min. After a 1-h incubation, 10 μm DCPIP and 4 mm ascorbate were added and flash-induced kinetics at 820 nm were measured. The discontinuity in the noise level is a consequence of fusing two kinetic traces taken at different time bases on the same laser flash.
When P700-FX cores were incubated with holo-PsaC and PsaD, electron transfer to FA and FB was restored and long-lived charge separation occurred (Fig. 5A, trace 4). The reconstituted PS I complexes also exhibited four kinetic phases, but a major new kinetic phase, corresponding to charge recombination between [FA/FB]- and P700+, was observed at ∼59 ms (Fig. 5A, trace 4; supplemental Fig. S3B). The two slowest kinetic phases (59 ms and 844 ms) accounted for ∼60% of the total absorbance change and were assigned to PS I complexes in which electrons had been transferred to FA/FB prior to charge recombination with P700+ or reduction of oxygen.
When P700-FX cores were incubated with apo-PsaC, PsaD, and holo-NfuA, electron transfer to FA/FB was also restored, resulting in long-lived charge separation (Fig. 5A, trace 5). These reconstituted PS I complexes exhibited four kinetic phases, and their decay kinetics were very similar to those of P700-FX cores that had been reconstituted with holo-PsaC and PsaD (compare Fig. 5A, traces 4 and 5, and supplemental Fig. S3C). The two slowest kinetic phases together accounted for ∼53% of the total absorbance change and were assigned to PS I complexes in which the electron had been transferred to FA/FB prior to charge recombination with P700+ or reduction of oxygen. In other similar experiments, these slow kinetic phases accounted for as much as 60–70% of the total absorbance change. These results clearly showed that the [4Fe-4S] clusters could be transferred from holo-NfuA to the FA and FB sites in apo-PsaC. In these experiments, the molar ratio of [4Fe-4S] clusters to PsaC was 2.1:1, and much higher molar ratios (5-, 10-, and 20-fold) of [4Fe-4S] clusters to PsaC did not significantly improve the reconstitution as measured by charge recombination from [FA/FB]- to P700+ (data not shown). Thus, cluster transfer from holo-NfuA to apo-PsaC appeared to be nearly stoichiometric. As shown in Fig. 5B (trace 1), the ability of holo-NfuA to transfer Fe/S clusters to PsaC was completely abolished when holo-NfuA was denatured by boiling for 1 min. However, when the reagents for chemical reconstitution of Fe/S proteins were boiled prior to mixing with apo-PsaC, PsaD, and P700-FX cores, reconstitution of the PS I complexes still occurred (Fig. 5B, trace 2). This experiment clearly suggested that interactions between holo-NfuA and apo-PsaC were required during transfer of Fe/S clusters from holo-NfuA to apo-PsaC.
The standard reconstitution buffer for cluster transfer from holo-NfuA to apo-PsaC contained 4 mm DTT. DTT could be acting as a reductant, a thiol reagent, or both in the cluster-transfer reaction. When DTT was omitted from the reaction mixture, very little (<12%) charge recombination occurred from electron carriers beyond FX (data not shown); thus, the Fe/S clusters were not transferred from holo-NfuA to apo-PsaC. As described above, the charge recombination between [FA/FB]- and P700+ accounted for >55% of the total absorption change in the presence of 4 mm DTT. However, DTT could be replaced by the non-thiol reductant, tris(2-carboxyethyl)phosphine. When the cluster transfer from holo-NfuA to apo-PsaC was performed in the presence of 1 mm tris(2-carboxyethyl)phosphine, the 121-ms phase, which was assigned to charge recombination between [FA/FB]- and P700+, accounted for ∼44% of the total absorbance change (data not shown). Presently it is not clear whether DTT and tris(2-carboxyethyl)phosphine increase the efficiency of the reconstitution reaction by acting as reducing agents for the Fe/S clusters or whether these reducing agents improve the efficiency of cluster transfer by reducing disulfide bonds in apo-PsaC that interfere with cluster transfer, or both. However, because cluster transfer occurs in the presence of the non-thiol reductant, tris(2-carboxyethyl)phosphine, thiol reagents are clearly not required for cluster transfer from holo-NfuA to apo-PsaC.
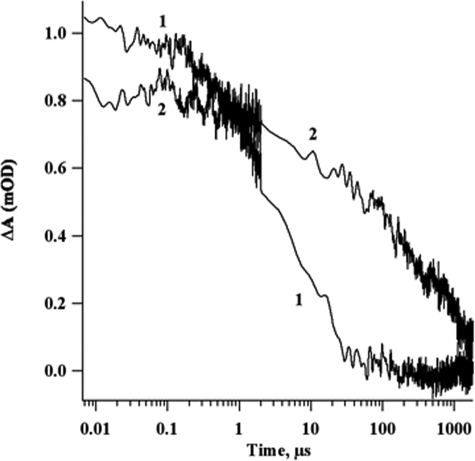
Holo-NfuA Rapidly and Efficiently Transfers Fe/S Clusters Directly to Apo-PsaC Without Cluster Release—The relative efficiencies of Fe/S cluster insertion into apo-PsaC by holo-NfuA and by chemical reconstitution were compared in the following experiment. As shown in Fig. 6 (trace 2), PS I complexes were reconstituted with P700-FX cores, PsaD, apo-PsaC, and holo-NfuA. The decay kinetics of P700+ were measured at 820 nm after an incubation of only 10 min, which was approximately the minimal time to mix the reconstitution reagents in the glove box and transfer the reaction mixture to the spectrophotometer. In this experiment, the ∼30-ms phase corresponding to electron transfer from [FA/FB]- to P700+ accounted for ∼56% of the total absorbance change. This result showed that holo-NfuA rapidly transferred [4Fe-4S] clusters to apo-PsaC. Increasing the incubation time (30 min, 1 h, 2 h, and 16 h) did not lead to a significant increase in cluster transfer (data not shown).
FIGURE 6.
Flash-induced absorption changes for P700+ reduction at 820 nm. Decay kinetics of 1) 1 μm P700-FX cores, 20 μm apo-PsaC, 20 μm PsaD, 4 mm DTT, 0.04% (w/v) DM, 84 μm Fe2+, 84 μm sulfide, 10 μm DCPIP, and 10 mm ascorbate following a 1-h incubation; 2) 1 μm P700-FX cores, 42 μm holo-Nfu, 20 μm apo-PsaC, 20 μm PsaD, 0.04% (w/v) DM, and 4 mm DTT. After a 10-min incubation, 10 μm DCPIP and 10 mm ascorbate were added and flash-induced kinetics at 820 nm were measured. The discontinuity in the noise level is a consequence of fusing two kinetic traces taken at different time bases on the same laser flash.
Dos Santos et al. (16) compared the activation of the nitrogenase iron-protein, NifH, by A. vinelandii NifU and by chemical reconstitution. A similar experiment was devised to show that holo-NfuA transfers clusters to apo-PsaC more efficiently than chemical reconstitution. In this experiment, 84 μm Fe2+ and 84 μm sulfide were added to the standard reconstitution reaction mixture in place of 42 μm holo-NfuA. After a 1-h incubation, no charge recombination from [FA/FB]- to P700+ was detected for the chemical reconstitution (Fig. 6, trace 1). Increasing the incubation time did not substantially increase the reconstitution (data not shown). This experiment showed that free Fe2+, sulfide, and DTT were unable to reconstitute apo-PsaC as rapidly and efficiently as an equivalent concentration of holo-NfuA. This experiment further suggested that the cluster transfer reaction was probably not due to the release of Fe/S clusters from holo-NfuA as free iron and sulfide that subsequently assembled Fe/S clusters on apo-PsaC.
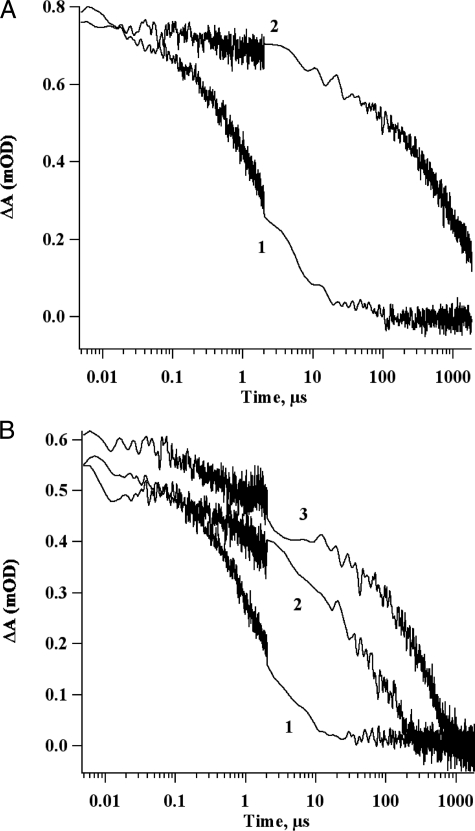
To demonstrate conclusively that Fe/S clusters or their constituents were not being released from holo-NfuA, an ultrafiltration experiment was performed. Two solutions were subjected to ultrafiltration over a membrane with a 3-kDa cut-off: holo-NfuA treated with 4 mm DTT; and a solution containing 84 μm Fe2+, 84 μm sulfide, and 4 mm DTT. (This solution was incubated for 4 h to allow the formation of Fe/S clusters prior to the ultrafiltration.) As shown by the long-lived charge separation observed in Fig. 7A (trace 2), the retentate from the holo-NfuA solution was able to reconstitute electron transfer to FA/FB when mixed with apo-PsaC, PsaD, and P700-FX cores. However, the filtrate did not reconstitute electron transfer to FA/FB (Fig. 7A, trace 1). In the case of the solution containing Fe, sulfide, and DTT, half of the solution was passed through the membrane (filtrate) and half was retained as the “retentate.” Both solutions were similarly effective in reconstituting electron transfer to FA/FB when the solutions were mixed with apo-PsaC, PsaD, and P700-FX cores (Fig. 7B, traces 2 and 3). This result indicates that Fe/S clusters assembled from Fe2+, sulfide, and DTT could readily pass through the membrane filter and were available to reconstitute the two [4Fe-4S] clusters in apo-PsaC. However, the failure to observe any reconstitution of electron transfer when the filtrate from the solution containing holo-NfuA was mixed with apo-PsaC, PsaD, and P700-FX cores (Fig. 7A, trace 1) indicated that no significant amount of iron, sulfide, and/or Fe/S clusters were released from holo-NfuA in the presence of DTT. Chemical analyses of the filtrate fraction showed the free iron concentration was ∼2 μm and the sulfide concentration was ∼33 μm. This experiment implied that cluster transfer from holo-NfuA to apo-PsaC required a direct interaction between the two proteins.
FIGURE 7.
Decay kinetics of the P700+ flash-induced absorption change at 820. A, decay kinetics of 1 μm P700-FX cores, 20 μm apo-PsaC, 20 μm PsaD, 0.04% (w/v) DM and filtrate (1) or retentate (2) from ultrafiltration of 42 μm holo-NfuA. The reconstitution mixture was incubated for 1 h, 10 μm DCPIP and 10 mm ascorbate were added prior to measurement. B, 1 μm P700-FX cores, 20 μm apo-PsaC, 20 μm PsaD, 0.04% (w/v) DM and no addition (1), filtrate (2), or retentate (3) from ultrafiltration of chemically assembled Fe/S clusters. The reconstitution mixture was incubated for 1 h, 10 μm DCPIP and 10 mm ascorbate were added, and flash-induced kinetics at 820 nm were then measured. The discontinuity in the noise level is a consequence of fusing two kinetic traces taken at different time bases on the same laser flash.
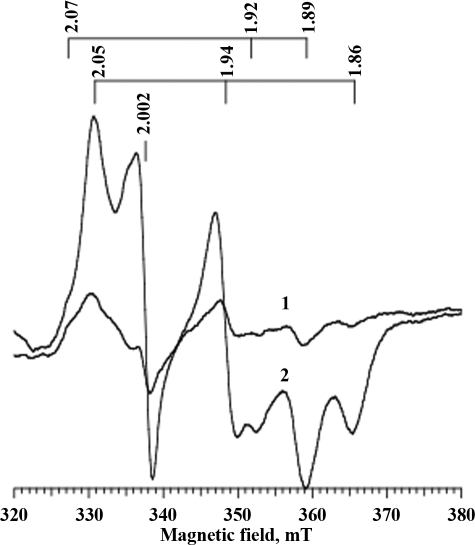
Light-induced EPR Spectra of PS I Complexes Reconstituted with Holo-NfuA—The FA and FB clusters of PsaC have characteristic EPR resonances that can easily be detected when PS I complexes are illuminated at low temperature (36). As shown in Fig. 8 (trace 1), P700-FX cores that were frozen at low light intensity exhibited only weak EPR signals after illumination at 16 K. However, P700-FX cores that had been reconstituted with holo-NfuA, apo-PsaC, and PsaD had the characteristic EPR resonances of reduced FA and FB when the complexes were illuminated at low temperature (Fig. 8, trace 2). Consistent with the results from optical kinetic spectroscopy, this result showed that holo-NfuA was able to transfer [4Fe-4S] clusters to apo-PsaC and that both of these clusters could be photoreduced.
FIGURE 8.
Light-induced EPR spectra of P700-FX cores and PS I complexes reconstituted with P700-FX cores, PsaD, apo-PsaC, and holo-NfuA proteins. Comparison of EPR spectra for P700-FX cores (trace 1) and reconstituted PS I (trace 2) after both had been illuminated at 20 K. The PS I sample was reconstituted from 20 μm FX cores, 840 μm holo-NfuA, 400 μm apo-PsaC, 400 μm PsaD, 10 mm DCPIP, 10 μm ascorbate, 0.04% (w/v) DM, and 4 mm DTT. The g values for the FA cluster are shown above traces 2.05, 1.94, and 1.86; the g values for the FB cluster are 2.07, 1.92, and 1.89, and the P700+ radical is shown at g = 2.002.
DISCUSSION
The NifU/Nfu Family in Cyanobacteria—Through studies in various organisms from all three kingdoms of life, three major systems for Fe/S cluster biogenesis are now recognized: the NIF, ISC, and SUF systems (7–9). Although these systems may have different biochemical mechanisms for Fe/S cluster assembly and are differently regulated, each system apparently requires a cysteine desulfurase and the participation of a “scaffold” protein for the assembly and transfer of Fe/S clusters. The NifU protein family is generally believed to play a central role in Fe/S cluster formation and transfer in the NIF system, which functions in the maturation of nitrogenase (7, 8, 16). It has been proposed that this scaffold protein provides an intermediate site for the assembly of Fe/S clusters or Fe/S cluster precursors, and there is strong biochemical and genetic evidence that NifU can transfer Fe/S clusters to NifH, the iron-protein of nitrogenase (7, 8).
Although all cyanobacteria perform oxygenic photosynthesis, the cyanobacteria can further be divided into two major groups on the basis of their ability to perform nitrogen fixation. The genomes of all nitrogen-fixing cyanobacteria encode a nifU gene that is clustered together with other nif genes. Cyanobacterial NifU proteins are highly similar (∼49% identity and 63% similarity) to A. vinelandii NifU, and they form a distinctive clade of sequences within the NifU/NfuA sequences (supplemental Fig. S4).
All cyanobacterial genomes contain a single nfuA gene, and NfuA proteins, which have strong sequence similarity to the C terminus of NifU, also form a discrete clade in phylogenetic analyses (supplemental Fig. S4). Because efforts to inactivate nfuA inevitably led to the generation of meridiploids, we previously reported that NfuA is essential for viability in cyanobacteria (20). Because nfuA was essential even in cells for which the psaAB genes had been deleted, it is clear that NfuA is required for functions that extend beyond the assembly of PS I (see below). NfuA-like proteins have recently been identified in several non-photosynthetic organisms, including E. coli (37), A. vinelandii (38), Saccharomyces cerevisiae (39), and humans (24). In contrast to the situation in cyanobacteria, an S. cerevisiae nfu1 mutant had no obvious growth defect (39). In oxygenic photosynthetic cyanobacteria and the chloroplasts of higher plants, the SUF system is the major machinery for Fe/S cluster assembly (9). NfuA-like proteins have been suggested to be involved in Fe/S cluster biogenesis in the chloroplasts of A. thaliana (22). Because several proteins of the ISC system are not essential for Fe/S biogenesis in cyanobacteria, it is reasonable to conclude that NfuA functions together with the SUF machinery in Fe/S cluster assembly. Interestingly, wild type A. vinelandii can grow at an oxygen concentration twice that of the atmosphere, but an nfuA mutant was unable to grow under these conditions (38). This observation is consistent with the idea that NfuA might participate in an Fe/S cluster biogenesis pathway that is relatively insensitive to oxygen.
The genomes of most nitrogen-fixing cyanobacteria also contain a third member of the NifU/Nfu family of proteins, which we designate here as NfuB (supplemental Fig. S4). Members of the NfuB clade have sequence similarity to the C-terminal domain of NifU, have Rieske domains at their C termini, have an N-terminal extension not found in NfuA (supplemental Fig. S1), and are typically clustered with genes for the maturation of nickel-iron uptake hydrogenases in cyanobacteria and other bacteria. The N- and C-terminal extensions for NfuB may specify its unique functionality and substrate recognition. Based upon these observations, we hypothesize that NfuB plays a specialized role in the insertion of Fe/S clusters in uptake hydrogenases.
Properties of the Iron-Sulfur Clusters in NfuA—NfuA is similar in sequence to the C-terminal domain of NifU; this domain contains a highly conserved CXXC motif that likely provides the ligands to coordinate a [4Fe-4S] cluster (supplemental Fig. S1). The functional importance of these two Cys residues has recently been established for both E. coli NfuA (formerly YhgI) and A. vinelandii NfuA, which like the human NfuA-like protein carry a [4Fe-4S] cluster (24, 37, 38). However, two NfuA orthologs, SyNifU from Synechocystis sp. PCC 6803 (21) and Nfu2 from A. thaliana (22), have been reported to harbor labile [2Fe-2S] clusters. In both cases the evidence for the presence of [2Fe-2S] clusters was only based on the UV-visible absorption properties of the proteins, and results from more sensitive and definitive methods, such as EPR or Mössbauer spectroscopy, were not reported. The EPR and Mössbauer spectroscopic studies reported here clearly showed that recombinant, chemically reconstituted holo-NfuA contains one [4Fe-4S] cluster per NfuA dimer. The [4Fe-4S] clusters of reconstituted NfuA were sensitive to both oxygen and chelating agents, and this lability might have contributed to the differences between the results presented here and those of previous studies. Only [4Fe-4S] clusters were detected in holo-NfuA by EPR spectroscopy; however, Mössbauer spectroscopy suggested the presence of a small population of [2Fe-2S] clusters. Four ligands are required to ligate [4Fe-4S] and [2Fe-2S] clusters stably, and our studies showed that NfuA apparently forms dimers and tetramers to provide four Cys residues (see Fig. 1, B and C, supplemental Fig. S2, A and B). Thus, it is reasonable to assume that dimeric NfuA ligates one [4Fe-4S] cluster via two Cys residues provided by each subunit. On the basis of NifS-mediated cluster assembly in A. vinelandii NifU, Smith et al. (40) had suggested that the C-terminal domain of NifU bound one [4Fe-4S] cluster per monomer. However, our results showed that only dimeric and tetrameric NfuA bound Fe/S clusters (Fig. 1B). This result is similar to more recent studies on the A. vinelandii NfuA protein, which showed that one [4Fe-4S] cluster is bound per NfuA dimer (38).
NfuA and PS I Biogenesis—The A. thaliana genome contains five NfuA-like homologs, and three of these homologs (AtNfu1, -2, and -3) have been localized to chloroplasts (22). By T-DNA insertional mutagenesis, it has been reported that interruption of the AtNfu2 gene resulted in the A. thaliana mutant lines with reduced PS I content and activity, which mostly seemed to be due to the loss of PsaC and other PS I subunits (23, 41). Using holo-NfuA and apo-PsaC to reconstitute PS I complexes, we show here that holo-NfuA can rapidly transfer [4Fe-4S] clusters to the PsaC apoproteins with very high efficiency (Figs. 5 and 6). The functional properties of the transferred Fe/S clusters in PsaC from NfuA could be verified by optical kinetic spectroscopy (Figs. 5, 6, 7), and by the light-induced EPR signals characteristic of the [4Fe-4S]1+ clusters in FA and FB (Fig. 8). Based on results of the titration with holo-NfuA, a 2-fold molar excess of [4Fe-4S] clusters (i.e. a 2.1:1 molar ratio of holo-NfuA to apo-PsaC) was sufficient for rapid and efficient cluster transfer to apo-PsaC. The results showed that Fe/S cluster transfer from holo-NfuA to apo-PsaC was nearly stoichiometric and was essentially complete in less than 10 min (Fig. 6). This result is similar to results obtained for the larger NfuA proteins of A. vinelandii and E. coli, which could rapidly and nearly stoichiometrically transfer a [4Fe-4S] cluster to apo-aconitase (37, 38). Our results indicated that intact [4Fe-4S] clusters were transferred from holo-NfuA to apo-PsaC. In studies on the activation of the apoprotein (NifH) of nitrogenase with NifU, it was found that Fe2+ and sulfide only reconstituted ∼11% of the intrasubunit [4Fe-4S] cluster of dimeric apo-NifH. In contrast about 85% of the NifH Fe/S clusters could be inserted when NifU was used as the Fe/S cluster donor (15). Because PS I accounts for about 25% of the iron in cyanobacterial cells (42), the transfer of [4Fe-4S] clusters to apo-PsaC could be one of the major processes mediated by holo-NfuA in cyanobacteria.
It was recently shown that holo-NfuA from E. coli and A. vinelandii can transfer a [4Fe-4S] cluster to apo-aconitase to reactivate the active site of this enzyme (37, 38). Interestingly, these NfuA proteins contain an additional N-terminal domain, which resembles an A-type scaffold domain, and, unlike the results reported here, the C-terminal domain alone was not active in cluster transfer (37). E. coli nfuA null mutants had oxidative stress and iron starvation phenotypes, which suggested that NfuA participates in both ISC- and SUF-dependent Fe/S cluster biogenesis. The nfuA gene is found in all cyanobacterial genomes, is essential for viability, and cannot be inactivated in strains in which the psaAB genes have been deleted (20). Thus, it is highly likely that NfuA is involved in Fe/S cluster transfer to proteins that are not involved in photosynthesis. Moreover, its product can transfer [2Fe-2S] clusters to apo-(PetF)-ferredoxin (21, 23) and [4Fe-4S] clusters to PsaC. Therefore, we propose that NfuA plays a central and ubiquitous role in Fe/S cluster biogenesis and iron homeostasis in cyanobacteria, and related proteins probably play similar roles in E. coli and many other organisms.
It is possible that redundant pathways for Fe/S cluster insertion into cyanobacterial proteins exist. Picciocchi et al. (43) recently showed that cyanobacteria have a monothiol glutaredoxin, which ligates one intrasubunit [2Fe-2S] cluster per dimer by single cysteines on each subunit in combination with two glutathione ligands. The SyGrx3p-like protein of Synechocystis sp. PCC 6803, the product of open reading frame slr1846, can complement the Fe/S cluster biogenesis defect of a yeast mitochondrial mutant lacking ScGrx5p (44). Another recent study suggested that two chloroplast-targeted monothiol glutaredoxins, GrxS14 and GrxS16, transfer their [2Fe-2S] clusters to chloroplast apo-ferrodoxin (45). Finally, recent studies suggest that IscU can harbor either two [2Fe-2S] clusters or one [4Fe-4S] cluster but only the latter can be transferred to apoaconitase to activate this enzyme (46, 47). Given the wide range of Fe/S proteins that exist in biological systems and the importance of these proteins to cellular metabolism and physiology, it is not surprising that nature has apparently devised multiple pathways to produce these essential prosthetic groups.
Supplementary Material
This work was supported by U. S. Dept. of Agriculture Contract 2005-35318-15284 (to J. H. G. and D. A. B.), National Science Foundation Grant MCB-0077586 (to D. A. B. and J. H. G.), and “Center for the Study of Biometals in Health and Disease” Grant 417-12Hy TSF (to C. K.). This work was also supported, in part, under a grant with the Pennsylvania Dept. of Health using Tobacco Settlement Funds. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: Fe/S, iron-sulfur; Chl, chlorophyll; EPR, electron paramagnetic resonance; ISC, iron sulfur cluster; NIF, nitrogen fixation; P700-FX cores, PS I complexes from which PsaC, PsaD, and PsaE have been removed by treatment with a strong chaotropic agent; PS, photosystem; SUF, sulfur utilization factor; EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride; DCPIP, 2,6-dichlorophenolindophenol; DTT, dithiothreitol; DM, n-dodecyl-β-d-maltoside.
References
- 1.Golbeck, J. H. (1994) in The Molecular Biology of Cyanobacteria (Bryant, D. A., ed) pp. 319-360, Kluwer Academic Press, Dordrecht, The Netherlands
- 2.Golbeck, J. H. (2006) Photosystem I, The Light-driven Plastocyanin:Ferredoxin Oxidoreductase, Springer, Dordrecht, The Netherlands
- 3.Jordan, P., Fromme, P., Witt, H. T., Klukas, O., Saenger, W., and Krauss, N. (2001) Nature 411 909-917 [DOI] [PubMed] [Google Scholar]
- 4.Antonkine, M. L., Jordan, P., Fromme, P., Krauss, N., Golbeck, J. H., and Stehlik, D. (2003) J. Mol. Biol. 327 671-697 [DOI] [PubMed] [Google Scholar]
- 5.Shen, G., Zhao, J., Reimer, S. K., Antonkine, M. L., Cai, Q., Weiland, S. M., Golbeck, J. H., and Bryant, D. A. (2002) J. Biol. Chem. 277 20343-20354 [DOI] [PubMed] [Google Scholar]
- 6.Shen, G., Antonkine, M. L., van der Est, A., Vassiliev, I. R., Brettel, K., Bittl, R., Zech, S. G., Zhao, J., Stehlik, D., Bryant, D. A., and Golbeck, J. H. (2002) J. Biol. Chem. 277 20355-20366 [DOI] [PubMed] [Google Scholar]
- 7.Frazzon, J., and Dean, D. R. (2003) Curr. Opin. Chem. Biol. 7 166-167 [DOI] [PubMed] [Google Scholar]
- 8.Johnson, D., Dean, D. R., Smith, A. D., and Johnson, M. K. (2005) Annu. Rev. Biochem. 74 247-281 [DOI] [PubMed] [Google Scholar]
- 9.Shen, G., and Golbeck, J. H. (2006) in Photosystem I, The Light-driven Plastocyanin:Ferredoxin Oxidoreductase (Golbeck, J. H., ed) pp. 529-547, Springer, Dordrecht, The Netherlands
- 10.Yu, J., Shen, G., Wang, T., Bryant, D. A., Golbeck, J. H., and McIntosh, L. (2003) J. Bacteriol. 185 3878-3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, T., Shen, G., Balasubramanian, R., McIntosh, L., Bryant, D. A., and Golbeck, J. H. (2004) J. Bacteriol. 186 956-967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen, G., Balasubramanian, R., Wang, T., Wu, Y., Hoffart, L. M., Krebs, C., Bryant, D. A., and Golbeck, J. H. (2007) J. Biol. Chem. 282 31909-31919 [DOI] [PubMed] [Google Scholar]
- 13.Dean, D. R., Bolin, J. T., and Zheng, L. (1993) J. Bacteriol. 175 6737-6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, W., Jack, R. F., Morgan, T. V., Dean, D. R., and Johnson, M. K. (1994) Biochemistry 33 13455-13463 [DOI] [PubMed] [Google Scholar]
- 15.Yuvaniyama, P., Agar, J. N., Cash, V. L., Johnson, M. K., and Dean, D. R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 599-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dos Santos, P. C., Smith, A. D., Frazzon, J., Cash, V. L., Johnson, M. K., and Dean, D. R. (2004) J. Biol. Chem. 279 19705-19711 [DOI] [PubMed] [Google Scholar]
- 17.Mühlenhoff, U., and Lill, R. (2000) Biochim. Biophys. Acta 1459 370-382 [DOI] [PubMed] [Google Scholar]
- 18.Wollenberg, M., Berndt, C., Bill, E., Schwenn, J. D., and Seidler, A. (2003) Eur. J. Biochem. 270 1662-1671 [DOI] [PubMed] [Google Scholar]
- 19.Wu, S. P., and Cowan, J. A. (2003) Biochemistry 42 5784-5791 [DOI] [PubMed] [Google Scholar]
- 20.Balasubramanian, R., Shen, G., Bryant, D. A., and Golbeck, J. H. (2006) J. Bacteriol. 188 3182-3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishio, K., and Nakai, M. (2000) J. Biol. Chem. 275 22615-22618 [DOI] [PubMed] [Google Scholar]
- 22.Leon, S., Touraine, B., Ribot, C., Briat, J. F., and Lobreaux, S. (2003) Biochem. J. 371 823-830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Touraine, B., Boutin, J.-P., Marion-Poll, A., Brat, J.-F., Peltier, G., and Lobréaux, S. (2004) Plant J. 40 101-111 [DOI] [PubMed] [Google Scholar]
- 24.Tong, W. H., Jameson, G. N., Huynh, B. H., and Roualt, T. A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9762-9767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, N., Warren, P. V., Golbeck, J. H., Frank, G., Zuber, H., and Bryant, D. A. (1991) Biochim. Biophys. Acta 1059 215-235 [DOI] [PubMed] [Google Scholar]
- 26.Beinert, H. (1978) Methods Enzymol. 54 435-445 [DOI] [PubMed] [Google Scholar]
- 27.Golbeck, J. H., Lien, S., and San Pietro, A. (1976) Biochem. Biophys. Res. Commun. 71 452-458 [DOI] [PubMed] [Google Scholar]
- 28.Grabarek, Z., and Gergely, J. (1990) Anal. Biochem. 185 131-135 [DOI] [PubMed] [Google Scholar]
- 29.Crankshaw, M. W., and Grant, G. A. (1996) in Current Protocols in Protein Science (Coligan, J. E., ed) pp. 2218-2235, John Wiley & Sons, Inc., Hoboken, NJ
- 30.Kuo, C. F., and Fridovich, I. (1988) Anal. Biochem. 170 183-185 [DOI] [PubMed] [Google Scholar]
- 31.Heinnickel, M., Shen, G., Agalarov, R., and Golbeck, J. H. (2005) Biochemistry 44 9950-9960 [DOI] [PubMed] [Google Scholar]
- 32.Golbeck, J. H., Mehari, T., Parrett, K. G., and Ikegami, I. (1988) FEBS Lett. 240 9-14 [Google Scholar]
- 33.Li, N., Zhao, J., Warren, P. V., Warden, J. T., Bryant, D. A., and Golbeck, J. H. (1991) Biochemistry 30 7863-7872 [DOI] [PubMed] [Google Scholar]
- 34.Golbeck, J. H. (1995) in CRC Handbook of Organic Photochemistry and Photobiology (Song, P. S., and Horspool, W. M., eds) pp. 1407-1419, CRC Press, Boca Raton, FL
- 35.Vassiliev, I. R., Jung, Y. S., Mamedov, M. D., Semenov, A., and Golbeck, J. H. (1997) Biophys. J. 72 301-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bearden, A. J., and Malkin, R. (1976) Biochim. Biophys. Acta 430 538-547 [DOI] [PubMed] [Google Scholar]
- 37.Angelini, S., Gerez, C., Ollagnier-de Choudens, S., Sanakis, Y., Fontecave, M., Barras, F., and Py, B. (2008) J. Biol. Chem. 283 14084-14091 [DOI] [PubMed] [Google Scholar]
- 38.Bandyopadhyay, S., Naik, S. G., O'Carroll, I. P., Huynh, B.-H., Dean, D. R., Johnson, M. K., and Dos Santos, P. C. (2008) J. Biol. Chem. 283 14092-14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schilke, B., Voisine, C., Beinert, H., and Craig, E. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 10206-10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, A. D., Jameson, G. N., Dos Santos, P. C., Agar, J. N., Naik, S., Krebs, C., Frazzon, J., Dean, D. R., Huynh, B. H., and Johnson, M. K. (2005) Biochemistry 44 12955-12969 [DOI] [PubMed] [Google Scholar]
- 41.Yabe, T., Morimoto, K., Kikuchi, S., Nishio, K., Terashima, I., and Nakai, M. (2004) Plant Cell 16 993-1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keren, N., Aurora, R., and Pakrasi, H. B. (2004) Plant Physiol. 135 1666-1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picciocchi, A., Saguez, C., Bussac, A., Cassier-Chauvat, C., and Chauvat, F. (2007) Biochemistry 46 15018-15026 [DOI] [PubMed] [Google Scholar]
- 44.Molina-Navarro, M., Casas, M., Piedrafita, L., Belli, G., and Herrero, E. (2006) FEBS Lett. 580 2273-2280 [DOI] [PubMed] [Google Scholar]
- 45.Bandyopadhyay, S., Gama, F., Molina-Navarro, M. M., Gualberto, J. M., Claxton, R., Naik, S. G., Huynh, B.-H., Herrero, E., Jacquot, J. P., Johnson, M. K., and Rouhier, N. (2008) EMBO J. 27 1122-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandramouli, K., Uniciuleac, M. C., Naik, S., Dean, D. R., Huynh, B.-H., and Johnson, M. K. (2007) Biochemistry 46 6804-6811 [DOI] [PubMed] [Google Scholar]
- 47.Uniciuleac, M. C., Chandramouli, K., Naik, S., Mayer, S., Huynh, B.-H., Johnson, M. K., and Dean, D. R. (2007) Biochemistry 46 6812-6821 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.