Abstract
The apolipoprotein (apo) AI/CIII/AIV/AV cluster genes are expressed at different levels in the liver and intestine. The apoCIII enhancer, a common regulatory element, regulates the tissue-specific expression of apoAI, apoCIII, and apoAIV but not apoAV. To study this regulation at the chromatin level, the histone modifications and intergenic transcription in the human apoAI/CIII/AIV/AV cluster were investigated in HepG2 and Caco-2 cells and in the livers of transgenic mice carrying the human gene cluster constructs with or without the apoCIII enhancer. We found that both the promoters and the intergenic regions of the apoAI/CIII/AIV genes were hyperacetylated and formed an open subdomain that did not include the apoAV gene. Hepatic and intestinal intergenic transcripts were identified to transcribe bidirectionally with strand preferences along the cluster. The deletion of the apoCIII enhancer influenced both histone modification and intergenic transcription in the apoAI/CIII/AIV gene region. These results demonstrate that the apoCIII enhancer contributes to the maintenance of an active chromatin subdomain of the apoAI/CIII/AIV genes, but not apoAV.
The apolipoprotein (apo)3 AI/CIII/AIV/AV gene cluster includes some of the most important genes involved in lipid metabolism. Overexpression of apoAI or apoAIV increases cholesterol in plasma high density lipoprotein and protects against atherosclerosis (1). The apoCIII and apoAV genes have predominant but opposite roles in triglyceride homeostasis (2, 3). The aberrant expression of these apolipoprotein genes may lead to cardiovascular diseases, such as dislipidemia and atherosclerosis.
The human apoAI/CIII/AIV/AV gene cluster is located on chromosome 11q.23. The apoAI, apoCIII, and apoAIV genes are arranged in a 17-kb region in tandem, whereas apoAV is 28 kb downstream from the apoAIV gene (4, 5) (Fig. 1). The transcriptional direction of the apoCIII gene is opposite to that of the other three apo genes (6). The expression of the apoAI cluster genes displays distinct hepatic and intestinal specificity. ApoAI and apoCIII are expressed mainly in the liver and to a lesser extent in the intestine, whereas apoAIV is synthesized predominantly in the intestine and to a much lesser extent in the liver. ApoAV is only expressed in the liver (4). Previous reports have demonstrated that the expression of apoAI, apoCIII, and apoAIV in the liver and intestine is controlled by the apoCIII enhancer, which is located at the -890/-490 region of the apoCIII gene (7). Complex interactions between nuclear receptors bound to hormone response elements of the apoAI, apoCIII, and apoAIV promoters and SP1 on the apoCIII enhancer contribute to the tissue-specific expression of these apo genes (8). Recent results showed that similar nuclear receptors (HNF4, RORα, FXR, and PPARα) could also bind to the apoAV promoter as well as to other apo genes (9–11). However, our previous results from transgenic mice demonstrated that the apoCIII enhancer deletion had no obvious effect on the expression of apoAV (12).
FIGURE 1.
Schematic map of the human apoAI/CIII/AIV/AV gene cluster and the construct of transgenes. The 116-kb transgene fragment containing the apoAI gene cluster is shown. The sequence number is marked according to the GenBank™ registration sequence AC007707. The core apoCIII enhancer was deleted in the mutant transgenic mice. The angled arrows indicate transcriptional orientation of each apo gene.
As a key component of epigenetic regulation, histone modifications such as acetylation of histone H3 and H4 and methylation of histone H3 on Lys4 have been proven to correlate with the active chromatin structure. Histone acetylation and trimethylation of histone H3K4 (H3K4me3) usually denote active transcription, whereas dimethylated histone H3K4 (H3K4me2) has been characterized by modifications associated with a transcriptionally permissive chromatin (13–16). The local active chromatin modifications in the promoter region are necessary for recruiting the transcription initiation complex to activate transcription. Moreover, it has been shown that the intergenic regions between the locus control region (LCR) and the promoters of some gene loci are also hyperacetylated, forming an active chromatin subdomain (17, 18). This loose chromatin structure may facilitate an enhancer communicating with promoters at long range. Previous studies have also indicated that the extensively opened chromatin structure may be correlated with intergenic transcription (18–22), suggesting a potential relationship among remote regulatory elements, chromatin remodeling, and intergenic transcription (23). Because the LCR is a complex and long range enhancer, it is interesting to know how the 400-bp apoCIII enhancer regulates the liver- and intestine-specific expression of the apoAI, apoCIII, and apoAIV genes but not apoAV at the chromatin level.
In this study, we investigated chromatin modifications in the apoAI/CIII/AIV/AV gene cluster and the effect of the apoCIII enhancer in this process. We report that the apoAI/CIII/AIV genes form an extensively opened subdomain that does not include the apoAV gene, and the apoCIII enhancer regulates both the chromatin modification and the intergenic transcription of the apoAI/CIII/AIV region. Liver- and intestine-specific intergenic transcripts were transcribed bidirectionally with predominance from one strand, suggesting that there may be multiple transcription initiation sites in the chromatin template.
EXPERIMENTAL PROCEDURES
Cells—HepG2, HeLa, and K562 cells were cultured in Dulbecco's modified Eagle's medium (Hyclone, Logan, UT) supplemented with 10% fetal bovine serum (Hyclone). Caco-2 cells were maintained in modified Eagle's medium with nonessential amino acids (Hyclone) supplemented with 20% fetal bovine serum and 1% sodium pyruvate at 37 °C in a humidified incubator containing 5% CO2.
Mice—Human bacterial artificial chromosome (pBeloBAC 11 (286I4)) containing human apoAI/CIII/AIV/AV gene cluster (GenBank™ registration sequence AC007707 72491-188971) was screened and maintained in our laboratory. The apoCIII enhancer region-deleted (114824–115126 of AC007707) BAC clone was derived from the wild-type BAC clone 286I4 by temperature-sensitive shuttle vector-mediated homologous recombination (12) (Fig. 1). Intact transgenic lines carrying the 116-kb human apoAI/CIII/AIV/AV gene cluster and mutant transgenic lines carrying cluster with the apoCIII enhancer deletion were generated by BAC and maintained in the Peking Union Medical College animal facilities.
Chromatin Immunoprecipitation—ChIP assay for histone modification was carried out as described previously (24). Briefly, 5×107 cells of Caco-2, HepG2, and HeLa cells were cross-linked with 0.6% formaldehyde for 10 min at 37 °C. The livers of 4–5-week-old transgenic mice were isolated and disrupted separately in phosphate-buffered saline to prepare a single cell suspension. After passage through a 21-gauge needle, the cells were washed twice with cold phosphate-buffered saline and cross-linked immediately. The nuclei were sonicated to 100–500-bp fragments. The chromatin was immunoprecipitated using anti-acetylated histone H3 (06-599; Upstate), anti-dimethylated histone H3K4 (07-030; Upstate), anti-histone H3 (ab1791; Abcam), or rabbit IgG (sc-2027; Santa Cruz). The bound chromatin was diluted in parallel to the input DNA. The primers used in the ChIP assays are listed in Table 1.
TABLE 1.
Primers for ChIP assay
| Names | Primers (5′ → 3′) | Sequence on AC007707a |
|---|---|---|
| 1P-321s | GGTCCTGGCAATGTGGAACTT | 123420–123740 |
| 1P-321a | AGTAGTCTCCCTGGAATGCTGGT | |
| 3P-262s | GAGCCAGTCAGCTAGGAAGGAATGAG | 115349–115610 |
| 3P-262a | GGCAGGAGGGTTCTGACCTGTTTT | |
| 4P-290s | GGGGCAAAGTCCACATCTC | 109376–109665 |
| 4P-290a | GACCAAGGAGGCACAGCAT | |
| 5P-297s | GCCCGAAATCCTGTTACCC | 77621–77918 |
| 5P-297a | TTGCCTCTTCGCCCTCATT | |
| 5Pn-292s | CCTCACCCAGTTTCTCCC | 78279–78570 |
| 5Pn-292a | CTTCCCTTGCTTGATTCCA | |
| I31–188s | CGTAGGTTTGCCGTGAGATT | 119312–119499 |
| I31–188a | GCCTCTATTGGGAGACCTTGT | |
| I43–259s | GGGCAGAACTGAAGGACAG | 112801–113059 |
| I43–259a | GGGAGAAGCCAGTGGTAGAT | |
| I54–319s | CTGATTCCCAAGCATACATCC | 105282–105600 |
| I54–319a | AACCTCACAAGGAAAGAGCC | |
| I54–250s | GTGAGCGGGATGAGGAAGTCA | 105309–105060 |
| I54–250a | GTGGGAGGGATGTATGCTTGG | |
| I54–310s | GGGAATAGTTCTCGGGATGT | 93582–93891 |
| I54–310a | CCCACTAGATGAGGGTTTGT | |
| hGAPD-399s | ACCTTCCGTGCAGAAACCTC | |
| hGAPD-399 a | GGACCCTTACACGCTTGGAT | |
| mGAPD-151s | CCAATGTGTCCATCGTGGATCT | |
| mGAPD-151a | GTTGAAGTCGCAGGAGACAACC |
The start and end sequences of PCR product in GenBank™. The accession number is AC007707
Semi-quantitative PCR of input and bound chromatin was performed with <10 ng of DNA as a template in a total volume of 25 μl with the appropriate primer. PCR products were resolved on 1.5% agarose gels containing ethidium bromide and quantified by Gene Tool software (Syngene). All of the PCRs were performed at least three times and averaged.
RNA Isolation, RT-PCR, and Real Time PCR—HepG2, Caco-2, K562, and HeLa cells or the livers, the intestines, and the lungs of 4–5-week-old transgenic mice were used for extracting RNA. RNA isolation was performed by TRIzol (Invitrogen) according to the manufacturer's instructions. Isolated RNA was treated with RNase-free DNase I (Takara) to remove DNA contamination. The RNA was then reverse transcribed, and control reactions without avian myeloblastosis virus reverse transcriptase were performed as described according to the manufacturer's instructions (Promega).
Semi-quantitative RT-PCR was carried out on RNA samples from Caco-2, HepG2, HeLa, and K562 cells in parallel with identical control PCRs in increasing concentrations. Species-specific primers were designed in the exons of human apoAI, apoCIII, apoAIV, and apoAV genes. Primer pairs 3P, I31–188, I43–259, I54–310, and I54–319 used in the ChIP assays are listed in Table 1, and primer pairs I43–158, U5–224, and U5–324 are listed in Table 2. The intensity of the amplicon on the ethidium bromide-stained agarose gel was quantified with Gene Tools software (Syngene).
TABLE 2.
Additional Primers for measuring intergenic transcripts by RT-PCR and Real-time PCR
| Names | Primers (5′ → 3′) | Sequence on AC007707a |
|---|---|---|
| I43–158s | GGGCATCCCCAATACCAAAT | 113401–114000 |
| I43–158a | CACCCCAGGCAAATACCAGA | |
| U5p-244s | TGCGAACAGCCCAAATAC | 72589–72832 |
| U5p-244a | AGTGGCGAGAGGGAGAGA | |
| U5p-342s | GGCTAGGAGACATAGGGGGA | 73023–73364 |
| U5p-342a | GAGATAGTAGGGCGGGGAAA | |
| AI-173s | GGAAGTCGTCCAGGTAGGGC | 121930–122102 |
| AI-173a | TGACAACTGGGACAGCGTGA | |
| CIII-148s | GTGACCGATGGCTTCAGTTC | 118472–118619 |
| CIII-148a | ATGGATAGGCAGGTGGACTT | |
| AIV-291s | CACGTTCTGGTCGATCTTGG | 107185–107475 |
| AIV-291a | CTGAAGGAGGAGATTGGGAAG | |
| AV-476s | CTGGCGGAAAGCCTGAAGT | 76014–76419 |
| AV-476a | ACACGATGGATCTGATGGAGC | |
| I54–140s | GGACTTCCCCCTCCTCCTTAG | 108762–108902 |
| I54–140a | CAAACGACTCCACAGCCTGTTAC | |
| AV-76s | AGCTGGTGGGCTGGAATTT | 76461–76537 |
| AV-76a | GGCCACCTGCTCCATCAG | |
| CIII-62s | CAGCTTCATGCAGGGCTACA | 116511–116573 |
| CIII-62a | ACGCTGCTCAGTGCATCCT | |
| hGapdh-416s | CATCACCATCTTCCAGGAG | |
| hGapdh-416a | GGATGATGTTCTGAGAGCC | |
| mGapdh-105a | CATGGCCTTCCGTGTTCCTA | |
| mGapdh-105s | CCTGCTTCACCACCTTCTTGAT |
The start and end sequences of PCR product in GenBank™. The accession number is AC007707
Real time PCR was carried out on RNA samples from the livers and intestines of the intact and mutant apoAI gene cluster transgenic mice. Mouse glyceraldehyde-3-phosphate dehydrogenase acted as an internal control. DNA amplification was carried out in the Applied Biosystems 7500 Real-time PCR System. Thermocycle was initiated by incubating the mixtures at 95 °C for 10 min to denature genomic DNA and to activate AmpliTaq Gold® DNA polymerase. This was followed by 40 cycles of two steps of 95 °C for 15 s and 60 °C for 1 min. The fluorescence intensity was measured during the step at 60 °C. The primers used in real time PCR were I31–188, I43–158, I54–140, AV-76, CIII-62, and mGapdh-105 listed in Tables 1 and 2.
Determination of Transcript Orientation—The orientation of the noncoding transcripts was identified as reported previously (25). Briefly, 2 μg of HepG2 RNA was reverse transcribed with strand-specific primers prior to PCR amplification with site-specific primers (Tables 1 and 2). PCR-generated cDNAs from the strand-specific analysis were separated on 1.8% agarose gels and then hybridized with 32P-labeled probes generated by the same sets of PCR primers from the BAC templates. Each band was quantified by Gene Tools software (Syngene). The apo BAC plasmid DNA was used as the control template to normalize the PCR efficiency of each primer set. Serial dilutions of the apo BAC plasmid DNA (80 ng to 0.08 pg) were assayed to determine the linear range for the PCR amplification. Because 0.008 ng of BAC template fell within the linear range of the RT-PCR signals from each primer, the RT-PCR signal levels were normalized to the PCR signal levels from 0.008 ng of apo BAC plasmid DNA generated with the same set of primers (C; BAC control) and were calculated in arbitrary units.
Statistical Analysis—To compare the results between the apoCIII enhancer deleted transgenic mice and the intact apoAI gene cluster transgenic mice, the histone modification levels were analyzed by Student's t test.
RESULTS
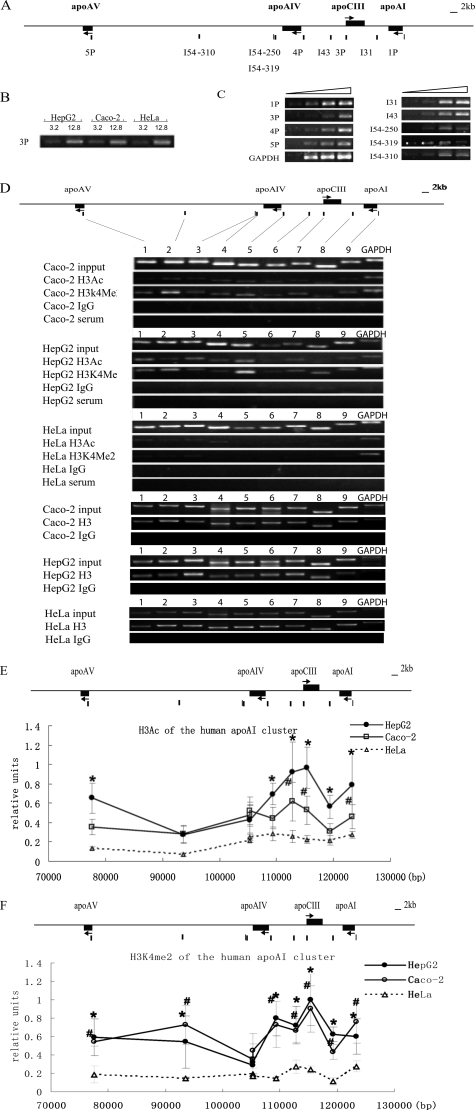
apoAI, apoCIII, apoAIV, but Not apoAV, Formed a Chromatin Subdomain with High Levels of Histone H3 Acetylation and H3K4 Dimethylation—The presence of histone H3 acetylation and histone H3K4 dimethylation in both the promoters and the intergenic regions in HepG2 and Caco-2 cells was detected by ChIP. The locations of the evaluated sites and primers are shown in the schematic map in Fig. 2A. The input DNA of three cell lines was normalized and verified by PCR gradient (Fig. 2B). All of the PCR primers used in this study were optimized to obtain appropriate PCR condition, in which the signals of PCR products were proportional to the amount of input DNA in the PCRs (Fig. 2C). HeLa cells without apo gene expression served as a negative control (Fig. 2D). Input DNA was used to calibrate the primer efficiency. Human histone H3 was used as an internal control (Fig. 2D).
FIGURE 2.
Histone H3 acetylation and H3K4 dimethylation pattern of the apoAI gene cluster in HepG2, Caco-2 and HeLa cells. A, the primer locations on the apoAI cluster. The primer pairs of 1P, 3P, 4P, and 5P represent the promoter sites of apoAI, apoCIII, apoAIV, and apoAV, respectively. The primer pairs of I54, I43, and I31 represent the sites in the intergenic regions. B, the concentrations of input DNA in different cells were adjusted to the same level and verified by PCR with the primer 3P. C, PCR conditions for each primer pair. PCR was performed with 4-fold serial dilution of input DNA. Each primer was in the linear amplification range. D, representative ethidium bromide-stained gels of ChIP assay are shown. Lanes 1–9 represent the PCR results from the sites of 5P, I54–310, I54–319, I54–250, 4P, I43, 3P, I31, and 1P on the apoAI cluster respectively. Total histone H3 served as an internal control, and rabbit serum and IgG were used as negative controls. The results are shown as the means ± S.D. of at least three independent PCRs. Compared with HeLa, p < 0.05 for difference from HepG2 values (*), and p < 0.05 for difference from Caco2 values (#). Ac, acetylated histone H3. Me, dimethylated histone H3K4. E, histone H3 acetylation pattern of the apoAI cluster in cells. F, histone H3K4 dimethylation pattern of the apoAI cluster in cells.
The ChIP results showed that the total histone H3 was distributed on the apoAI gene cluster with similar levels in all three cell lines. However, significant histone H3 acetylations were found in the promoters and the intergenic regions of the apoAI, apoCIII, and apoAIV genes, forming a hyperacetylated chromatin subdomain in both HepG2 and Caco-2 cells but not in HeLa cells (p < 0.05) (Fig. 2E). As to the apoAV gene, a high level of histone H3 acetylation was detected downstream from the transcription initiation site in HepG2, whereas a low level was detected in the apoAV nonexpressing Caco-2 cells. Furthermore, relatively low levels of the histone H3 acetylation were found in both cell lines at two intergenic regions: one site just downstream from apoAIV (I54–250 and I54–319) and a second in the middle of the apoAV-AIV region (I54–310). These results indicated that the apoAV gene was separated from the acetylated chromatin subdomain of the apoAI/CIII/AIV genes in the cluster (Fig. 2E).
H3K4me2 was found through the apoAI gene cluster in both HepG2 and Caco-2 cells (Fig. 2F). In addition to the identified hyperacetylation sites, the intergenic regions of apoAV-AIV in both HepG2 and Caco-2 cells and the apoAV promoter region in Caco-2 cells were also characterized by high H3K4me2 modifications. These results are consistent with the reports that histone H3K4me2 is present on active or inactive (poised) genes (15, 26, 27). Interestingly, a much lower histone H3K4me2 modification site was identified downstream from the apoAIV gene (I54–250 and I54–319), and this was similar to the pattern of the histone H3 acetylation. This extremely low H3K4me2 modification site appears to be a boundary between apoAV and the apoAI/CIII/AIV genes (Fig. 2F).
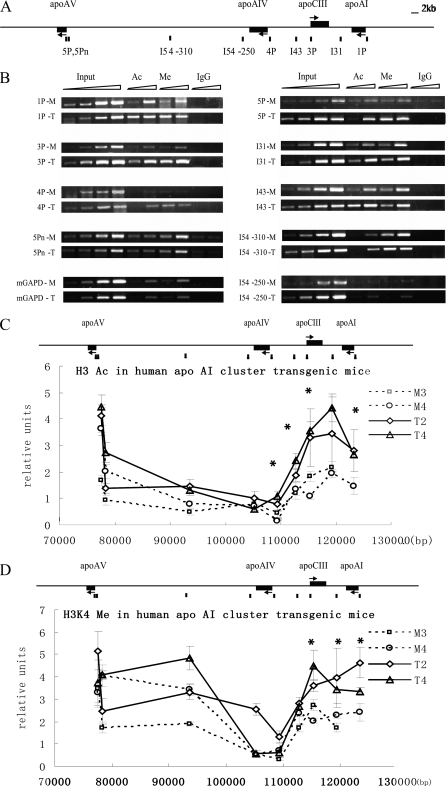
The apoCIII Enhancer Deletion Decreased Histone H3 Acetylation and H3K4 Dimethylation of the apoAI/CIII/AIV Genes but Not apoAV in Transgenic Mice—The results from cells showed that histone modification profiles of the apoAI gene cluster correlated well with its liver- and intestine-specific expression pattern. To test whether the apoCIII enhancer participates in regulating the chromatin modification, we performed ChIP assays on the liver of the apoAI gene cluster transgenic mice with low copy numbers (1 or 2 copies), including the two intact human apoAI gene cluster transgenic lines of T2 and T4 and the two apoCIII enhancer deleted transgenic lines of M3 and M4. The human apo genes exhibited an integrated position-independent and a closer approximation of copy number-dependent expression pattern in the intact apoAI cluster transgenic mice (12). It has been shown that the expression of apoAI, apoCIII, and apoAIV, but not of apoAV, was clearly decreased in the apoCIII enhancer deleted transgenic mouse lines (12). We compared the histone modification level in the liver between the intact (T2 and T4) and the mutant transgenic mouse lines (M3 and M4). As shown in Fig. 3C, high levels of histone H3 acetylation were restricted to the apoAI/CIII/AIV region, similar to that in HepG2 cells. One additional site (5Pn) located within 500 bp upstream from the apoAV transcription initiation site was analyzed (Fig. 3A). The result showed that the apoCIII enhancer deletion decreased the histone acetylation level in both the promoters and the intergenic regions of the apoAI/CIII/AIV genes (p < 0.05) but did not affect the modifications in the apoAV promoter (Fig. 3C).
FIGURE 3.
Acetylated histone H3 and dimethylated histone H3K4 pattern of the apoAI cluster in transgenic mice. A, the primer locations on the apoAI gene cluster. The primer pairs of 1P, 3P, 4P, and 5P and 5Pn represent the promoter sites of apoAI, apoCIII, apoAIV, and apoAV, respectively. The primer pairs of I54, I43, and I31 represent the sites in the intergenic region. B, representative ethidium bromide-stained gels are shown. PCR was performed with 4-fold serial dilution. The input DNA were 0.2, 0.8, 3.2, and 12.8 μl, respectively, and the DNA for antibody precipitation were 0.8 and 3.2 μl, respectively. The PCR products of the input in the liver of each transgenic mouse line were in the linear amplification range. Ac, acetylated histone H3. Me, dimethylated histone H3K4. T, the intact human apoAI gene cluster transgenic lines. M, the enhancer-deleted transgenic lines. C, histone H3 acetylation pattern of the apoAI cluster in transgenic mice. D, histone H3K4 dimethylation pattern of the apoAI cluster in transgenic mice. The results are shown as the means ± S.D. of at least three independent PCRs. The experiments were repeated with similar results in independent cell preparations. M3 and M4 represent the apoCIII enhancer deleted transgenic mice. T2 and T4 represent the intact human apoAI gene cluster transgenic mice. Compared with the apoAI gene cluster transgenic mice group, p < 0.05 (*).
At the same time, the histone H3K4 dimethylation pattern of the apoAI gene cluster in transgenic mice was similar to that in HepG2 cells. The histone H3K4 dimethylation level in the apoAI-apoCIII region, but not in the apoAV promoter region, was also decreased (p < 0.05) in the apoCIII enhancer deleted transgenic mice (Fig. 3D). Taken together, these results demonstrate that the apoCIII enhancer mediates the extensive histone modification of the apoAI/CIII/AIV genes in vivo but not that of the apoAV gene.
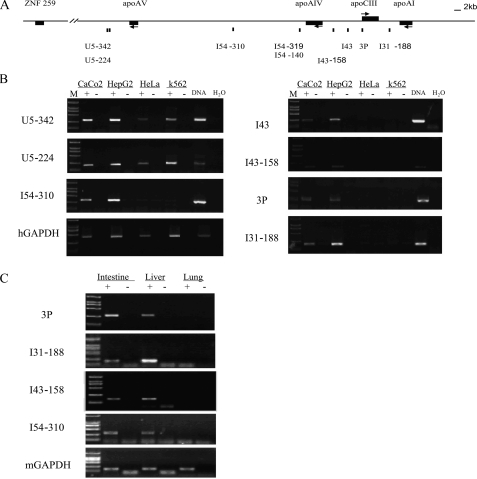
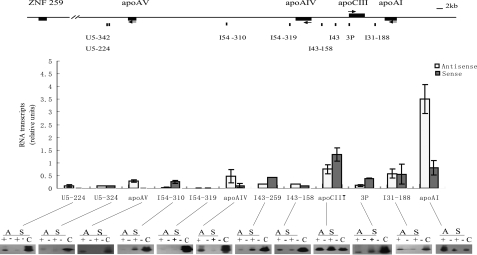
The apoCIII Enhancer Deletion Decreased the Expression of Intergenic Transcripts of the apoAI/CIII/AIV Genes in Transgenic Mice—The studies from gene loci with LCR have shown that there are correlations between the intergenic transcription and the extensively hyperacetylated chromatin domain (19, 28). Using RT-PCR and real time PCR, hepatic and intestinal specific intergenic transcripts were identified on the apoAI/CIII/AIV/AV gene cluster in HepG2 and Caco2 cells, as well as in the livers and the intestines of transgenic mice (Fig. 4). The intergenic transcripts at 2 and 2.5 kb downstream from the apoAV gene (U5–342 and U5–224) were identified even in apo gene nonexpressing HeLa and K562 cells, suggesting that the intergenic transcription beyond the apoAI gene cluster has no hepatic and intestinal specificity. In addition to intergenic transcripts in the apoCIII-AI, apoAIV-CIII, and apo AV-AIV regions, we also identified one in the apoCIII promoter. Furthermore, the expression pattern of intergenic transcripts seemed to be in accordance with the distribution of dimethylated histone H3K4 in the apoAI gene cluster.
FIGURE 4.
Tissue-specific intergenic transcripts throughout the human apoAI cluster. A, schematic map of the apoAI locus indicating the sites assayed. The primer pairs of 3P represent the site at the apoCIII promoter. The primer pairs of I54, I43, and I31 represent the sites in the intergenic region of apoAV-AIV, apoAIV-CIII, and apoCIII-AI, respectively. The primer pairs of U5 represent the site in the intergenic region between apoAV and ZNF259. B, intergenic transcripts of the apoAI gene cluster were expressed specifically in HepG2 and Caco-2 cells. C, intergenic transcripts of the apoAI cluster were expressed specifically in the liver and the intestine of transgenic mice. + and - represent the reverse transcription with or without avian myeloblastosis virus reverse transcriptase. The ladder for DL2000 marker is 100, 250, 500, 750, 1000, and 2000 bp, respectively (low to high).
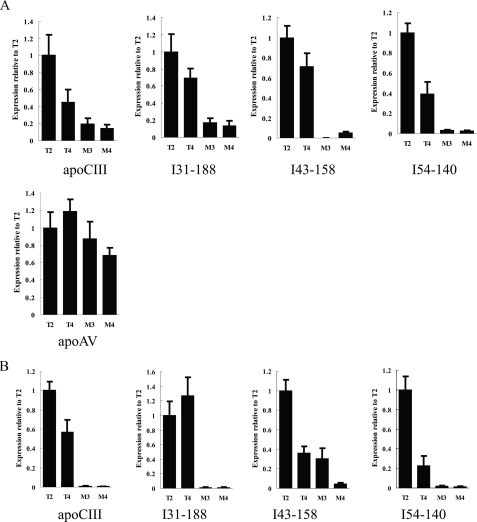
To determine the role of the apoCIII enhancer in intergenic transcription, we compared the levels of intergenic transcripts in livers and intestines of the intact and mutant apoAI cluster transgenic mice (Fig. 5). It has been shown, by RNase protection assays, that the transcription level of apoCIII, but not apoAV, was lowered in the apoCIII enhancer deleted transgenic lines, M3 and M4 (12). In this study, we further confirmed these results by real time PCR. More importantly, we found that the expression levels of intergenic transcripts I31–188, I43–158, and I54–140 (located just downstream from the apoAIV gene) were much lower in the liver and intestine from the mutant lines than those from the intact lines (Fig. 5). These results indicate that the apoCIII enhancer also regulates the intergenic transcription in the apoAI/CIII/AIV region.
FIGURE 5.
Comparing the levels of intergenic transcripts between the mutant and the intact transgenic mice. A, intergenic transcripts in the liver of transgenic mice. B, intergenic transcripts in the intestine of transgenic mice. Mouse glyceraldehyde-3-phosphate dehydrogenase served as an internal control, and the results in the histogram were normalized to the expression of transgenic mouse line T2. M3 and M4 represent the apoCIII enhancer-deleted transgenic mice. T2 and T4 represent the intact human apoAI gene cluster transgenic mice.
Intergenic Transcripts Were of Biorientation with Dominating in One Strand—We used strand-specific RT-PCR combined with Southern blot to determine the orientation of these intergenic transcripts (Fig. 6). The BAC 286I4 containing the human apoAI gene cluster was used to normalize the cDNA quantity. Compared with the apoAI and apoCIII expression, the levels of intergenic transcripts were much lower (Fig. 6). Intriguingly, these transcripts could be detected from both strands. Each region analyzed showed a strand preference for transcription from one strand or the other, suggesting that these transcripts may have originated from multiple sites. We found that the exons of apoCIII and apoAI genes were also transcribed from both strands but predominated from the coding strand (Fig. 6).
FIGURE 6.
Bidirectional transcriptions along the apoAI gene locus with strand preferences. Each RT reaction was performed using a sense or an antisense specific primer from the relevant amplimer set (as in Fig. 4). Strand-specific RT reactions were followed with PCRs corresponding to the primer sites indicated above. The results were quantified by Southern blot hybridization with unique sequence probes and calculated. The results are shown as the means ± S.D. of at least three independent PCRs. Representative data for the transcriptional orientation are shown at the bottom of the histogram. Amplification products are shown in the presence (+) and absence (-) of reverse transcriptase, and the polarity of the initial strand-specific RT primer, antisense (A) or sense (S), is indicated. The control PCR was optimized into linear amplification range, and 0.008 ng of apo BAC plasmid DNA was used as the template (C) for determining the relative levels of antisense and sense transcripts along the apoAI gene cluster.
DISCUSSION
In this study, we investigated the role of the apoCIII enhancer in regulating the liver- and intestine-specific expression of apoAI, apoCIII, and apoAIV but not apoAV, at the chromatin level. Our results indicate that the apoCIII enhancer participates in modulating the histone modification and the intergenic transcription of the apoAI/CIII/AIV chromatin subdomain instead of regulating these apo genes separately. Meanwhile, the apoAV gene is not included in the apoAI/CIII/AIV subdomain.
It is well established that the transcriptional regulation of the enhancer is mediated by the specific interaction between regulatory elements and transcription factors, as well as by the interactions among transcription factors. In the apoAI cluster, there are some important nuclear factors, such as HNF4 and Sp1, binding to the enhancer and each promoter of the apo genes. The apoAV promoter could bind similar nuclear receptors as other apo genes do in the cluster (9–11). Furthermore, our results showed that the apoAV promoter activity increased about five times when the promoter was close to the apoCIII enhancer in vitro (data not shown). All of these observations suggest that the apoCIII enhancer may regulate the expression of apoAV. However, our previous data demonstrated that the apoCIII enhancer deletion had no obvious effect on the apoAV gene expression in transgenic mice (12). In this study ChIP analysis showed that, in the apo gene expressing HepG2 and Caco-2 cells, broad histone H3 hyperacetylations were found in both the promoters and the intergenic regions of apoAI/CIII/AIV, and this modification did not extend to the apoAV gene, indicating that the apoAV gene was separated from the apoAI/CIII/AIV chromatin subdomain (Fig. 2). The extensively acetylated subdomain in the apoAI/CIII/AIV region may make the chromatin flexible, thus providing the opportunity for interactions between the apoCIII enhancer and the apoAI and apoAIV promoters (29). CBP/p300, the cofactor of HNF4 and Sp1, exhibits histone acetyltransferase activity, suggesting that the apoCIII enhancer could specifically recruit factors to achieve histone acetylation (30–32). By comparing the histone acetylations of the apoAI gene cluster in the liver of transgenic mice, we found that these modifications were decreased significantly in the promoters and the intergenic regions of the apoAI/CIII/AIV genes, but not of the apoAV gene region, after apoCIII enhancer deletion (Fig. 3). These results demonstrate that the apoCIII enhancer regulates the transcription of apoAI, apoCIII, and apoAIV, partially by maintaining an extensive histone modification pattern. The communications between the apoCIII enhancer and the promoters are conducted by physical interactions between protein-protein interactions and protein-DNA interactions, possibly via a looping mechanism (8, 33, 34). Using a modified chromosome conformation capture method, we determined the spatial organization of the apoAI gene cluster and did not detect an interaction between the apoAV promoter and the apoCIII enhancer.4 This confirmed the ChIP results that the apoAV gene was not included in the apoCIII enhancer-maintained chromatin subdomain.
Note that although the apoCIII enhancer deletion dramatically reduced the expression of the apoAI, apoCIII, and apoAIV gene (12), it lowered the histone H3 acetylation and H3K4 dimethylation level significantly but less markedly (one-half to one-third). This indicates that other factors may participate in the chromatin opening process. Developmental associated intergenic transcription and extensively opened chromatin structure have been observed in the globin gene cluster (19). It has been verified in the human growth hormone locus that the intergenic transcription in LCR plays a direct role in the hGH-N gene expression (25). In this study, intergenic transcripts of the apoAI gene cluster were detected specifically in the hepatic and intestinal cells, suggesting that they may participate in regulating the expression of the apo genes. Our data indicated that the distribution of intergenic transcripts was in accordance with the regions marked with H3K4me2. This result supports the idea that histone methylations may be involved in a process of transcriptional elongation (35). It has been shown that Set1, a H3K4 methylase, is also a component of the RNA pol II elongation complex (36). Thus, the transcriptional elongation process may provide more opportunities for transcription factors to bind to the intergenic region then to mediate histone modifications and open the chromatin broadly. Consistent with the ChIP data, real time PCR results showed that the deletion of the apoCIII enhancer clearly down-regulated the intergenic transcripts level in the apoAI/CIII/AIV region, confirming that the apoCIII enhancer regulates the whole apoAI/CIII/AIV chromatin subdomain.
We found that the intergenic transcription in the apoAI/CIII/AIV gene region was bidirectional, dominating from one strand. These results were consistent with the observation from human growth hormone that the transcription in the LCR region was also bidirectional (25). This indicates that the promoters of apo genes are not the only sites for starting transcription. Previous data showed that the apoCIII enhancer had the potential to initiate transcription (37). In this study we identified a transcript at the apoCIII promoter. In the β-globin gene cluster, it has been reported that the intergenic transcription and histone acetylation is dependent on the HS2 enhancer (22). The HS2 enhancer complex tracks along the intervening DNA, synthesizing short, polyadenylated, intergenic RNAs to ultimately loop with the ε-globin promoter (38). Because enhancers and promoters are active sites undergoing various chromatin modifications (39), the transcription initiation complex, including the transcription factors and RNApolII, may get access to multiple sites along an “open” apoAI gene cluster. Thus, the apoCIII enhancer deletion may reduce the recruitment of transcription factors to the chromatin domain. This would then reduce the initiation of intergenic transcription. It also reduces the histone modifications relative to chromatin opening.
From the ChIP data, it is shown that H3K4me2 was enriched in the region with low H3 acetylation level, supporting the previous observation that H3K4me2 was the epigenetic marker for euchromatin (40). It was enriched broadly across active or poised chromatin (15, 27). Interestingly, we also identified an extremely low local H3K4me2 modification site downstream from the apoAIV gene. Note that the histone H3 acetylation level in the intergenic region of apoAV-AIV is low. This forms a relatively sharp edge of the apoAI/CIII/AIV chromatin subdomain, which suggests that a specific chromatin structure keeps the apoCIII enhancer from regulating the apoAV gene expression. Congruent with the histone modification pattern, a site characterized by palindromic AT-rich repeats has been identified downstream from the apoAIV gene. This cruciform structure, formed by self-folding, could mediate translocation (41, 42). We speculate that the special structure near the 3′ end of the apoAIV gene may insulate the apoCIII enhancer from the apoAV gene or hamper the tracking process from the apoCIII enhancer.
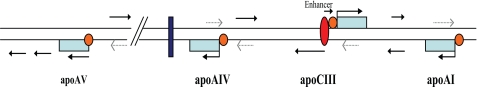
Collectively, the apoCIII enhancer is involved in maintaining an active chromatin subdomain in the apoAI/CIII/AIV region. However, the apoAV gene is separated from this broadly opened chromatin subdomain and thus cannot be regulated by the apoCIII enhancer (Fig. 7). This study provides the chromatin level evidence that the apoCIII enhancer regulates the expression of the apoAI, apoCIII, and apoAIV genes but not apoAV. These findings shed new light on the mechanisms whereby the tissue-specific expression of the apoAI gene cluster is regulated and may provide new options for therapeutic intervention in dyslipidemia.
FIGURE 7.
A model for transcriptional regulation of the apoAI gene cluster by the apoCIII enhancer. The apoCIII enhancer controls the expression of the apoAI, apoCIII, and apoAIV genes by maintaining an extensively opened chromatin structure. The tracking process of intergenic transcription plays an important role in mediating the enhancer's function. The apoAV gene is separated from the extensively opened apoAI/CIII/AIV subdomain and not regulated by the apoCIII enhancer. Specific chromatin structure downstream from the apoAIV gene may insulate the apoAV gene from the apoCIII enhancer. The red ellipse represents the complex on the apoCIII enhancer. The orange circle represents the transcription complex on promoters. The arrows indicate the transcriptional orientation. The solid arrows indicate the predominant transcriptional orientation, whereas the dashed arrows mark the opposite strand transcription. The blue box represents a potential insulator, where both the histone H3 acetylation and H3K4 dimethylation level were much lower.
Acknowledgments
We thank Dr. Rod Becher, Dr. Hui-dong Shi, Dr. Kristen Taylor, and Angel Surdin for proofreading the manuscript.
This work was supported by the National Natural Science Foundation of China Grants 30500297 and 30721063, National Basic Research Program of China Grants 2005CB522402 and 2006CB503801, and National High-tech Research and Development Program of China Grant 2006AA02A406. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: apo, apolipoprotein; H3K4me2, dimethylated histone H3 lysine 4; ChIP, chromatin immunoprecipitation; LCR, locus control region; RT, reverse transcription.
Y.-J. Li, Y.-S. Wei, L.-J. Di, X.-H. Fu, D.-L. Hao, Z. Xue, H. Gong, Z.-Q. Zhang, D.-P. Liu, and C.-C. Liang, unpublished data.
References
- 1.Rong, J. X., and Fisher, E. A. (2000) Ann. Med. 32 642-651 [DOI] [PubMed] [Google Scholar]
- 2.van Dijk, K. W., Rensen, P. C., Voshol, P. J., and Havekes, L. M. (2004) Curr. Opin. Lipidol. 15 239-246 [DOI] [PubMed] [Google Scholar]
- 3.Jakel, H., Nowak, M., Helleboid-Chapman, A., Fruchart-Najib, J., and Fruchart, J. C. (2006) Ann. Med. 38 2-10 [DOI] [PubMed] [Google Scholar]
- 4.Haddad, I. A., Ordovas, J. M., Fitzpatrick, T., and Karathanasis, S. K. (1986) J. Biol. Chem. 261 13268-13277 [PubMed] [Google Scholar]
- 5.Pennacchio, L. A., Olivier, M., Hubacek, J. A., Cohen, J. C., Cox, D. R., Fruchart, J. C., Krauss, R. M., and Rubin, E. M. (2001) Science 294 169-173 [DOI] [PubMed] [Google Scholar]
- 6.Zannis, V. I., Cole, F. S., Jackson, C. L., Kurnit, D. M., and Karathanasis, S. K. (1985) Biochemistry 24 4450-4455 [DOI] [PubMed] [Google Scholar]
- 7.Zannis, V. I., Kan, H. Y., Kritis, A., Zanni, E. E., and Kardassis, D. (2001) Curr. Opin. Lipidol. 12 181-207 [DOI] [PubMed] [Google Scholar]
- 8.Kardassis, D., Falvey, E., Tsantili, P., Hadzopoulou-Cladaras, M., and Zannis V. I. (2002) Biochemistry 41 1217-1228 [DOI] [PubMed] [Google Scholar]
- 9.Prieur, X., Coste, H., and Rodriguez, J. C. (2003) J. Biol. Chem. 278 25468-25480 [DOI] [PubMed] [Google Scholar]
- 10.Prieur, X., Schaap, F. G., Coste, H., and Rodriguez, J. C. (2005) Mol. Endocrinol. 19 3107-3125 [DOI] [PubMed] [Google Scholar]
- 11.Genoux, A., Dehondt, H., Helleboid-Chapman, A., Duhem, C., Hum, D. W., Martin, G., Pennacchio, L. A., Staels, B., Fruchart-Najib, J., and Fruchart, J. C. (2005) Arterioscler. Thromb. Vasc. Biol. 25 1186-1192 [DOI] [PubMed] [Google Scholar]
- 12.Gao, J., Wei, Y., Huang, Y., Liu, D., Liu, G., Wu, M., Wu, L., Zhang, Q., Zhang, Z., Zhang, R., and Liang, C. (2005) J. Biol. Chem. 280 12559-12566 [DOI] [PubMed] [Google Scholar]
- 13.Bulger, M., Schubeler, D., Bender, M. A., Hamilton, J., Farrell, C. M., Hardison, R. C., and Groudine, M. (2003) Mol. Cell Biol. 23 5234-5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adachi, S., and Rothenberg, E. V. (2005) Nucleic Acids Res. 33 3200-3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider, R., Bannister, A. J., Myers, F. A., Thorne, A. W., Crane-Robinson, C., and Kouzarides, T. (2004) Nat. Cell Biol. 6 73-77 [DOI] [PubMed] [Google Scholar]
- 16.Bottardi, S., Aumont, A., Grosveld, F., and Milot, E. (2003) Blood 102 3989-3997 [DOI] [PubMed] [Google Scholar]
- 17.Forsberg, E. C., Downs, K. M., Christensen, H. M., Im, H., Nuzzi, P. A., and Bresnick, E. H. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 14494-14499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elefant, F., Cooke, N. E., and Liebhaber, S. A. (2000) J. Biol. Chem. 275 13827-13834 [DOI] [PubMed] [Google Scholar]
- 19.Gribnau, J., Diderich, K., Pruzina, S., Calzolari, R., and Fraser, P. (2000) Mol. Cell 5 377-386 [DOI] [PubMed] [Google Scholar]
- 20.Kim, A., and Dean, A. (2003) Mol. Cell Biol. 23 8099-8109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cajiao, I., Zhang, A., Yoo, E. J., Cooke, N. E., and Liebhaber, S. A. (2004) EMBO J. 23 3854-3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, A., Zhao, H., Ifrim, I., and Dean, A. (2007) Mol. Cell Biol. 27 2980-2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Y. J., Fu, X. H., Liu, D. P., and Liang, C. C. (2004) Int. J. Biochem. Cell Biol. 36 1411-1423 [DOI] [PubMed] [Google Scholar]
- 24.Fu, X. H., Liu, D. P., Tang, X. B., Liu, G., Lv, X., Li, Y. J., and Liang, C. C. (2005) Exp. Cell Res. 309 174-184 [DOI] [PubMed] [Google Scholar]
- 25.Ho, Y., Elefant, F., Liebhaber, S. A., and Cooke, N. E. (2006) Mol. Cell 23 365-375 [DOI] [PubMed] [Google Scholar]
- 26.Sims, R. J., and Reinberg, D. (2006) Genes Dev. 20 2779-2786 [DOI] [PubMed] [Google Scholar]
- 27.Bernstein, B. E., Kamal, M., Lindblad-Toh, K., Bekiranov, S., Bailey, D. K., Huebert, D. J., McMahon, S., Karlsson, E. K., Kulbokas, E. J., Gingeras, T. R., Schreiber, S. L., and Lander, E. S. (2005) Cell 120 169-181 [DOI] [PubMed] [Google Scholar]
- 28.Gerber, M., and Shilatifard, A. (2003) J. Biol. Chem. 278 26303-26306 [DOI] [PubMed] [Google Scholar]
- 29.Li, Q., Barkess, G., and Qian, H. (2006) Trends Genet. 22 197-202 [DOI] [PubMed] [Google Scholar]
- 30.Suzuki, T., Kimura, A., Nagai, R., and Horikoshi, M. (2000) Genes Cells 5 29-41 [DOI] [PubMed] [Google Scholar]
- 31.Krumm, A., Madisen, L., Yang, X. J., Goodman, R., Nakatani, Y., and Groudine, M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 13501-13506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soutoglou, E., Viollet, B., Vaxillaire, M., Yaniv, M., Pontoglio, M., and Talianidis, I. (2001) EMBO J. 20 1984-1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F., and de Laat, W. (2002)Mol. Cell 10(6), 1453-1465 [DOI] [PubMed] [Google Scholar]
- 34.Carter, D., Chakalova, L., Osborne, C. S., Dai, Y. F., and Fraser, P. (2002) Nat. Genet. 32 623-626 [DOI] [PubMed] [Google Scholar]
- 35.Schubeler, D., MacAlpine, D. M., Scalzo, D., Wirbelauer, C., Kooperberg, C., van Leeuwen, F., Gottschling, D. E., O'Neill, L. P., Turner, B. M., Delrow, J., Bell, S. P., and Groudine, M. (2004) Genes Dev. 18 1263-1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng, H. H., Robert, F., Young, R. A., and Struhl, K. (2003) Mol. Cell 11 709-719 [DOI] [PubMed] [Google Scholar]
- 37.Kan, H. Y., Georgopoulos, S., and Zannis, V. (2000) J. Biol. Chem. 275 30423-30431 [DOI] [PubMed] [Google Scholar]
- 38.Zhu, X., Ling, J., Zhang, L., Pi, W., Wu, M., and Tuan, D. (2007) Nucleic Acids Res. 35 5532-5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mito, Y., Henikoff, J. G., and Henikoff, S. (2007) Science 315 1408-1411 [DOI] [PubMed] [Google Scholar]
- 40.Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., Schreiber, S. L., Mellor, J., and Kouzarides, T. (2002) Nature 419 407-411 [DOI] [PubMed] [Google Scholar]
- 41.Kurahashi, H., and Emanuel, B. S. (2001) Hum. Mol. Genet. 10 2605-2617 [DOI] [PubMed] [Google Scholar]
- 42.Kurahashi, H., Inagaki, H., Yamada, K., Ohye, T., Taniguchi, M., Emanuel, B. S., and Toda, T. (2004) J. Biol. Chem. 279 35377-35383 [DOI] [PMC free article] [PubMed] [Google Scholar]