Abstract
In Saccharomyces cerevisiae, α-isopropylmalate (α-IPM), which is produced in mitochondria, must be exported to the cytosol where it is required for leucine biosynthesis. Recombinant and reconstituted mitochondrial oxalacetate carrier (Oac1p) efficiently transported α-IPM in addition to its known substrates oxalacetate, sulfate, and malonate and in contrast to other di- and tricarboxylate transporters as well as the previously proposed α-IPM transporter. Transport was saturable with a half-saturation constant of 75 ± 4 μm for α-IPM and 0.31 ± 0.04 mm for β-IPM and was inhibited by the substrates of Oac1p. Though not transported, α-ketoisocaproate, the immediate precursor of leucine in the biosynthetic pathway, inhibited Oac1p activity competitively. In contrast, leucine, α-ketoisovalerate, valine, and isoleucine neither inhibited nor were transported by Oac1p. Consistent with the function of Oac1p as an α-IPM transporter, cells lacking the gene for this carrier required leucine for optimal growth on fermentable carbon sources. Single deletions of other mitochondrial carrier genes or of LEU4, which is the only other enzyme that can provide the cytosol with α-IPM (in addition to Oac1p) exhibited no growth defect, whereas the double mutant ΔOAC1ΔLEU4 did not grow at all on fermentable substrates in the absence of leucine. The lack of growth of ΔOAC1ΔLEU4 cells was partially restored by adding the leucine biosynthetic cytosolic intermediates α-ketoisocaproate and α-IPM to these cells as well as by complementing them with one of the two unknown human mitochondrial carriers SLC25A34 and SLC25A35. Oac1p is important for leucine biosynthesis on fermentable carbon sources catalyzing the export of α-IPM, probably in exchange for oxalacetate.
The obligatory specific intermediate of leucine biosynthesis in Saccharomyces cerevisiae, α-isopropylmalate (α-IPM),2 is produced from KIV in the mitochondrial matrix by the action of two isopropylmalate synthetases, Leu9p3 and Leu4lp (the long isoform of Leu4p), and in the cytosol by the action of Leu4sp (the short isoform of Leu4p) (see Ref. 1 for a review). α-IPM, which is formed in mitochondria, must be exported to the cytosol because the subsequent two reactions of leucine biosynthesis producing KIC are catalyzed by two enzymes, IPM-isomerase and β-IPM dehydrogenase, which are exclusively cytosolic (2, 3).
It has been suggested that Fsf1p (YOR271c), the S. cerevisiae member of the sideroflexin family (4), might be the transporter for α-IPM across the mitochondrial membrane, because in all studied genomes the promoter of Fsf1p contains the consensus binding site for a specific regulator of leucine biosynthesis, Leu3p (5). However, no direct evidence was provided to support the ability of this protein to transport α-IPM. In the same study, it was claimed that the closest homologue with known function to Fsf1p is the citrate transporter from Rattus norvegicus (6), although Fsf1p and rat citrate transporter have a similarity of 20.7% and 12.6% identical amino acids.
Considering the metabolic significance of mitochondrial α-IPM transport, we sought to verify the hypothesis of Fsf1p involvement as well as the possible involvement of mitochondrial carrier family members (see Refs. 7, 8 for reviews) in the translocation of α-IPM across the mitochondrial membrane. In this study, we provide evidence that Oac1p, a member of the mitochondrial carrier family, is the transporter of α-IPM. Oac1p, overexpressed in Escherichia coli and reconstituted in phospholipid vesicles, was found to transport α-IPM across the liposomal membrane with high specificity and high affinity. Consistent with the function of Oac1p to export α-IPM from mitochondria to cytosol, the ΔOAC14 mutant showed a partial auxotrophy and the ΔOAC1ΔLEU4 double mutant a complete auxotrophy for leucine. The phenotype of ΔOAC1ΔLEU4 cells was partially rescued by adding leucine biosynthetic cytosolic intermediates to these cells as well as by complementing them with a plasmid containing one of the two unknown human mitochondrial carriers encoded by SLC25A34 and SLC25A35.
EXPERIMENTAL PROCEDURES
Materials—Radioactive compounds were supplied from Amersham Biosciences and PerkinElmer Life Science Products. α-IPM was obtained from Sigma and β-IPM from Wako.
Yeast Strains, Media, and Growth Conditions—BY4742 (wild-type) and single deletion yeast strains were provided by the EUROFAN resource center EUROSCARF (Frankfurt, Germany). Deleted genes of the S. cerevisiae strain BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) (9) were replaced by kanMX4, respectively. The double deletion strain ΔOAC1ΔLEU4 was constructed using the PCR-mediated gene disruption technique by replacing the LEU4 open reading frame with the hygromycin B resistance cassette (hphMX3) in the ΔOAC1 mutant strain (10). Deletion strains were verified by PCR. Each of the above-mentioned strains was transformed with empty pGREG505 plasmid (11) to obtain histidine and leucine prototrophy. Transformants were selected on SC medium without histidine. The WT and deletion strains were grown in rich medium containing 2% bactopeptone and 1% yeast extract, or SC medium (12). All media were supplemented with a fermentable or a nonfermentable carbon source (2% glucose, 2% galactose, 3% glycerol, 2% ethanol, or 2% dl-lactate). The final pH was adjusted to 4.5. Mitochondria were isolated by standard procedures from cells grown in SC medium containing a fermentable or a nonfermentable substrate until the early exponential phase (optical density between 1.0 and 1.5) was reached. The mitochondria were further purified on a discontinuous gradient (13).
Construction of Expression Plasmids—The coding sequences of CTP1, DIC1, FSF1, OAC1, ODC1, ODC2, and SFC1 were amplified from S. cerevisiae genomic DNA by PCR. Forward and reverse oligonucleotide primers were synthesized corresponding to the extremities of the coding sequences with additional NdeI and HindIII sites, respectively, except for ODC1 where additional NdeI and XhoI sites were used. The amplified products were cloned into the pMW7 expression vector. The OAC1-pRS416 plasmid was constructed by cloning a DNA fragment consisting of the OAC1 open reading frame, of 760-bp upstream and of 245-bp downstream of the open reading frame (amplified from S. cerevisiae genomic DNA by PCR using primers with additional HindIII and BamHI sites) into the low copy centromeric vector pRS416 (14). The SLC25A34-pYES2, SLC25A35-pYES2, and SLC25A10-pYES2 plasmids were constructed by cloning the coding sequences of SLC25A34, SLC25A35, and SLC25A10, respectively, into the yeast pYES2 expression vector (Invitrogen) under the control of the constitutive MIR1 promoter. The pMW7, pRS416, and pYES2 vectors, prepared as above, were transformed into E. coli DH5α cells. Transformants were selected on ampicillin (100 μg/ml) and screened by direct colony PCR and by restriction digestion of purified plasmids. The sequences of the inserts were verified.
Bacterial Expression and Purification of Recombinant Proteins—Ctp1p, Dic1p, Fsf1p, Oac1p, Odc1p, Odc2p, and Sfc1p were overproduced as inclusion bodies in the cytosol of E. coli, as previously described (15); host cells were E. coli CO214[DE3] (16). Control cultures with empty vector were processed in parallel. Inclusion bodies were purified on a sucrose density gradient (15) and washed at 4 °C first with TE buffer (10 mm Tris-HCl, 1 mm EDTA, pH 7.0), then twice with a buffer containing Triton X-114 (3%, w/v), 1 mm EDTA, and 10 mm PIPES, pH 7.0, and once again with TE buffer. Proteins were solubilized in sarkosyl (w/v), as described (17, 18). Small residues were removed by centrifugation (258,000 × g for 20 min at 4 °C).
Reconstitution into Liposomes and Transport Assays—The recombinant proteins in sarkosyl were reconstituted into liposomes in the presence of the indicated substrate, as previously described (19). The external substrate was removed from proteoliposomes on Sephadex G-75 columns pre-equilibrated with 50 mm NaCl and 10 mm PIPES at pH 7.0. Reconstituted antiport activity was determined by measuring the uptake (forward exchange) or the efflux (backward exchange) of the indicated radioactive substrate in exchange for cold substrate (19). For backward-exchange measurements, the proteoliposomes containing 2 mm internal sulfate were prelabeled immediately after reconstitution by carrier-mediated exchange equilibration with 5 μm [35S]sulfate for 40 min. The external substrate was removed from proteoliposomes on a Sephadex G-75 column. Transport was started by adding labeled substrate (forward exchange) and cold substrate (backward exchange). When measuring the activity of carrier proteins catalyzing uniport besides antiport (e.g. Oac1p), external substrate was removed by exclusion chromatography in the presence of a reversible inhibitor (50 μm p-hydroxymercuribenzoate) to avoid efflux of internal substrate, and transport was started by adding combined substrate and 0.5 mm DTE. In some experiments, uniport activity was measured by adding radioactive substrate to proteoliposomes containing 10 mm NaCl and no substrate. In all cases, transport was terminated by addition of 30 mm pyridoxal 5′-phosphate and 10 mm bathophenanthroline according to the “inhibitor-stop” method (19). In control samples, the inhibitors were added at the beginning together with the external substrate. The assay temperature was 25 °C. All transport measurements were carried out in the presence of 10 mm PIPES at pH 7.0 in the internal and external compartments. Finally, the external substrate was removed, and radioactivity in the liposomes was measured (19). The experimental values were corrected by subtracting control values. In forward exchange kinetic measurements, the initial transport rate was calculated from the radioactivity taken up by the proteoliposomes after 1 min in the initial linear range of substrate uptake. In backward exchange experiments, the decrease in radioactivity inside the liposomes was fitted to the equation: α = 100 (1 - e-kt), where α is the percentage of isotopic equilibrium (20). The rates were expressed as apparent velocities, i.e. the product of k and the substrate concentration inside the liposomes, and are directly proportional to the actual transport rates (19–21).
Other Methods—The amount of recombinant protein was estimated on Coomassie Blue-stained SDS-PAGE gels by the Bio-Rad GS-700 Imaging Densitometer equipped with the Bio-Rad Multi-Analyst software using bovine serum albumin as standard. The amount of protein incorporated into liposomes was measured as described (22) and was about 25% of the protein added to the reconstitution mixture. The homology model of Oac1p was built based upon the structure of the bovine ADP/ATP carrier as described (23, 24). An anaerobic Oxoid jar was used to grow yeast strains in anaerobiosis.
RESULTS
α-IPM Is Transported by Oac1p—Recombinant Fsf1p, the previously proposed α-IPM transporter (5), was reconstituted into liposomes, and its ability to transport α-IPM was tested using reconstituted liposomes. Because α-IPM is not available in radioactive form, its transport was tested by measuring the exchange of 1 mm externally added labeled compounds for 20 mm α-IPM within proteoliposomes. No uptake whatsoever of malate, malonate, phosphate, sulfate, citrate, oxoglutarate, KIC, leucine, ADP, or ATP was observed with internal α-IPM, nor an uptake of any of these compounds into proteoliposomes containing the same (compound) (data not shown).
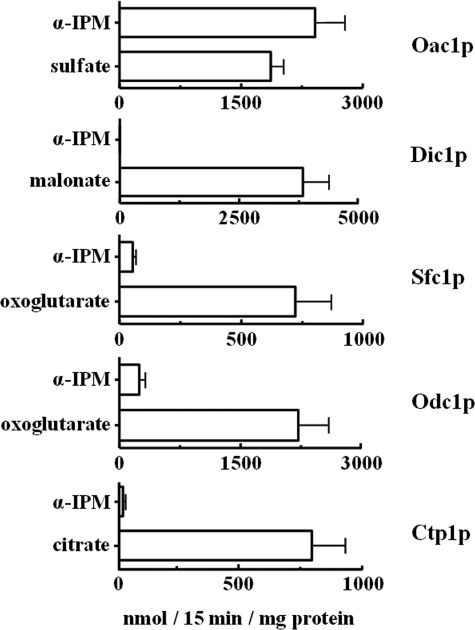
Given that α-IPM is a dicarboxylate analogue, the ability of yeast mitochondrial dicarboxylate carriers to transport α-IPM was investigated. The citrate carrier, Ctp1p, was also tested for its ability to transport α-IPM because it is known to transport citrate in exchange for malate (25). In these experiments, a labeled compound known to be transported by the investigated carrier ([35S]sulfate for Oac1p, [14C]malonate for Dic1p, [14C]oxoglutarate for Sfc1p and Odc1p, and [14C]citrate for Ctp1p) was added to reconstituted liposomes preloaded with 10 mm α-IPM or 10 mm of the same substrate used externally (Fig. 1). In the case of Oac1p, the activity of the [35S]sulfate/α-IPM exchange was very high and comparable to that of the sulfate/sulfate exchange (Fig. 1). In contrast, the activities of [14C]malonate/α-IPM via Dic1p, [14C]oxoglutarate/α-IPM via Sfc1p and Odc1p, and [14C]citrate/α-IPM via Cpt1p were very low or absent. Negligible activity of the [14C]oxoglutarate/α-IPM exchange was also found in liposomes reconstituted with Odc2p (data not shown). These results demonstrate that among the yeast carriers investigated, only Oac1p transports α-IPM efficiently.
FIGURE 1.
Ability of S. cerevisiae di- and tricarboxylate mitochondrial carriers to transport α-IPM. Liposomes were reconstituted with recombinant Oac1p, Dic1p, Sfc1p, Odc1p, or Ctp1p and preloaded internally with the indicated substrates (concentration, 10 mm). Transport was started by adding 0.1 mm [35S]sulfate (Oac1p), 0.1 mm [14C]malonate (Dic1p), 0.1 mm [14C]oxoglutarate (Sfc1p and Odc1p), or 0.1 mm [14C]citrate (Ctp1p). The reaction time was 15 min. Data are means ± S.D. of at least three independent experiments.
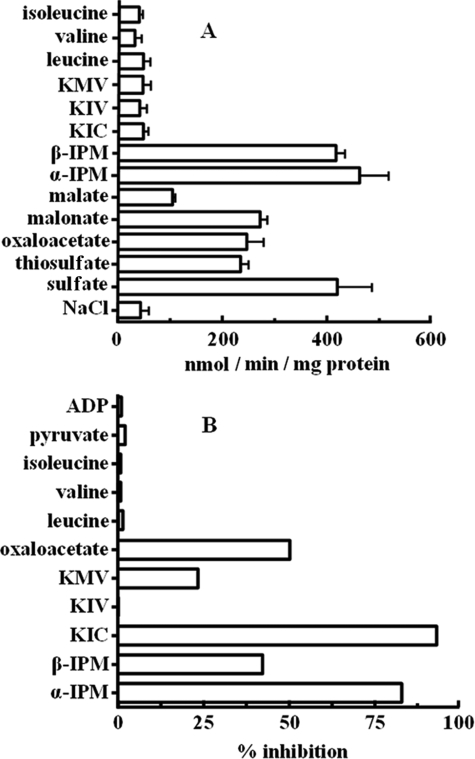
The substrate specificity of reconstituted Oac1p was examined in detail by measuring the uptake of [35S]sulfate into proteoliposomes preloaded with various potential substrates (Fig. 2A). The highest rates of [35S]sulfate uptake into proteoliposomes were observed with internal α-IPM, β-IPM, and sulfate. In agreement with previously published results (26), reconstituted Oac1p also exchanged [35S]sulfate for internal thiosulfate, oxalacetate, and malonate and, to a much lesser extent, malate. The uptake of [35S]sulfate was low in the presence of internal KIC, KIV, KMV, leucine, valine, isoleucine, and (not shown) pyruvate, succinate, fumarate, oxoglutarate, phosphate, citrate, glutamate, glutamine, lysine, carnitine, ATP, and GTP. Residual activity in the presence of these substrates was approximately the same as the activity observed in the presence of NaCl, in line with the reported slow uniport mode of Oac1p transport (26). In addition, the [35S]sulfate/sulfate exchange mediated by Oac1p was strongly inhibited by external addition of α-IPM and KIC (Fig. 2B). β-IPM, KMV, and oxalacetate were less effective. In contrast, no inhibition was observed with KIV, leucine, valine, isoleucine, pyruvate, or ADP.
FIGURE 2.
Specificity of α-IPM transport catalyzed by Oac1p. A, dependence of Oac1p activity on internal substrate. Liposomes reconstituted with Oac1p were preloaded internally with various substrates (concentration, 10 mm). Transport was started by addition of 70 μm [35S]sulfate and terminated after 1 min. Values are means ± S.D. of at least three independent experiments. B, inhibition of the rate of [35S]sulfate/sulfate exchange by external substrates. Liposomes reconstituted with Oac1p were preloaded internally with 10 mm sulfate. Transport was started by adding 70 μm [35S]sulfate and stopped after 1 min. External substrates (concentration, 0.7 mm) were added together with [35S]sulfate. The extents of inhibition (%) from a representative experiment are reported.
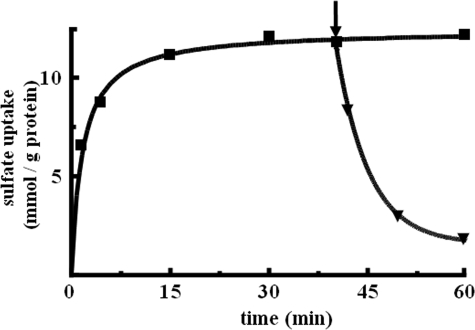
Kinetic Characteristics of Reconstituted [35S]Sulfate/α-IPM Exchange—The kinetics of the uptake of 0.7 mm [35S]sulfate by proteoliposomes in exchange for internal α-IPM are shown in Fig. 3. The exchange followed first-order kinetics (rate constant, 0.38 min-1; initial rate, 4.5 mmol/min/g protein), isotopic equilibrium being approached exponentially. The addition of 10 mm unlabeled α-IPM after incubation for 40 min, when radioactive uptake by proteoliposomes had almost reached equilibrium, caused an extensive efflux of radiolabeled sulfate. This efflux shows that the [35S]sulfate taken up by proteoliposomes is released in exchange for externally added α-IPM. Therefore, α-IPM is transported by reconstituted Oac1p not only when present inside liposomes but also when added externally. The kinetic constants of the reconstituted [35S]sulfate/α-IPM exchange were determined by measuring the initial transport rate at various external [35S]sulfate concentrations, in the presence of a constant saturating internal concentration of 10 mm α-IPM (forward exchange). The half-saturation constant (Km) and specific activity (Vmax) values for the [35S]sulfate/α-IPM exchange at 25 °C were 0.69 ± 0.07 mm and 7.0 ± 1.6 mmol/min/g protein, respectively. KIC and KMV inhibited [35S]sulfate/α-IPM exchange competitively by increasing the apparent Km without changing the Vmax of [35S]sulfate uptake (data not shown). The inhibition constants (Ki) of KIC and KMV were 91 ± 14 μm and 1.2 ± 0.2 mm, respectively.
FIGURE 3.
Time course of reconstituted [35S]sulfate/α-IPM exchange and efflux of [35S]sulfate from proteoliposomes after addition of an excess of unlabeled α-IPM. [35S]sulfate (0.7 mm) was added to liposomes reconstituted with Oac1p and preloaded internally with 10 mm α-IPM. The arrow indicates the addition of 10 mm nonradioactive α-IPM.
To determine the Km for transport of nonradioactive α-IPM, β-IPM, and oxalacetate, we applied the backward exchange method (19), which has been used before in the kinetic analysis of other mitochondrial carriers (20–22). Intraliposomal sulfate was radiolabeled, and export of [35S]sulfate was initiated by adding different concentrations of nonradioactive α-IPM, β-IPM, or oxalacetate. From a series of experiments of this type, a Km of 75 ± 4 μm for α-IPM, of 0.27 ± 0.04 mm for oxalacetate, and of 0.31 ± 0.04 mm for β-IPM was calculated. To verify that the backward exchange technique is applicable also to reconstituted Oac1p, the Km value for external sulfate was measured by the backward exchange procedure in the presence of internal [35S]sulfate and external sulfate ranging from 0.02 to 3 mm. The values obtained from five of these experiments (Km = 0.56 ± 0.08 mm) are consistent with those from the forward exchange determinations (see above). Therefore, α-IPM is a good substrate for reconstituted Oac1p. The half-saturation constant of α-IPM was 3.6-, 4.1-, and 7.5-fold lower than that of β-IPM, oxalacetate, and sulfate, respectively.
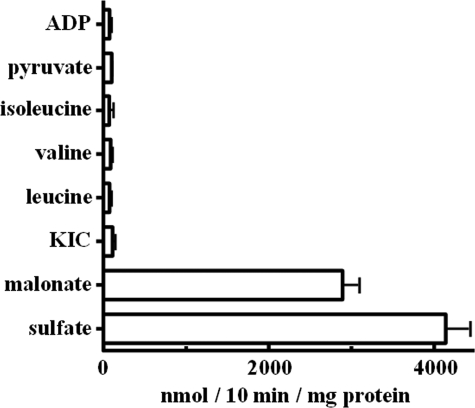
KIC, Leucine, Isoleucine, and Valine Are Not Substrates of Oac1p—In another set of experiments, the ability of Oac1p to transport KIC, leucine, isoleucine, and valine in reconstituted liposomes was tested in homo-exchange (i.e. the same substrate inside and outside) experiments. Using external and internal substrate concentrations of 1 and 10 mm, respectively, virtually no homo-exchange of KIC, leucine, valine, isoleucine, pyruvate, or ADP was observed even after 10 min of incubation (Fig. 4). By contrast, reconstituted Oac1p catalyzed active sulfate/sulfate and malonate/malonate exchanges (i.e. homo-exchanges of substrates known to be transported by this carrier) (Fig. 4).
FIGURE 4.
Homo-exchanges of various substrates in proteoliposomes containing recombinant Oac1p. Transport was initiated by adding radioactive substrate (final concentration, 1 mm) to proteoliposomes preloaded internally with the same substrate (concentration, 10 mm). The reaction was terminated after 10 min. Values are the means ± S.D. of at least three independent experiments.
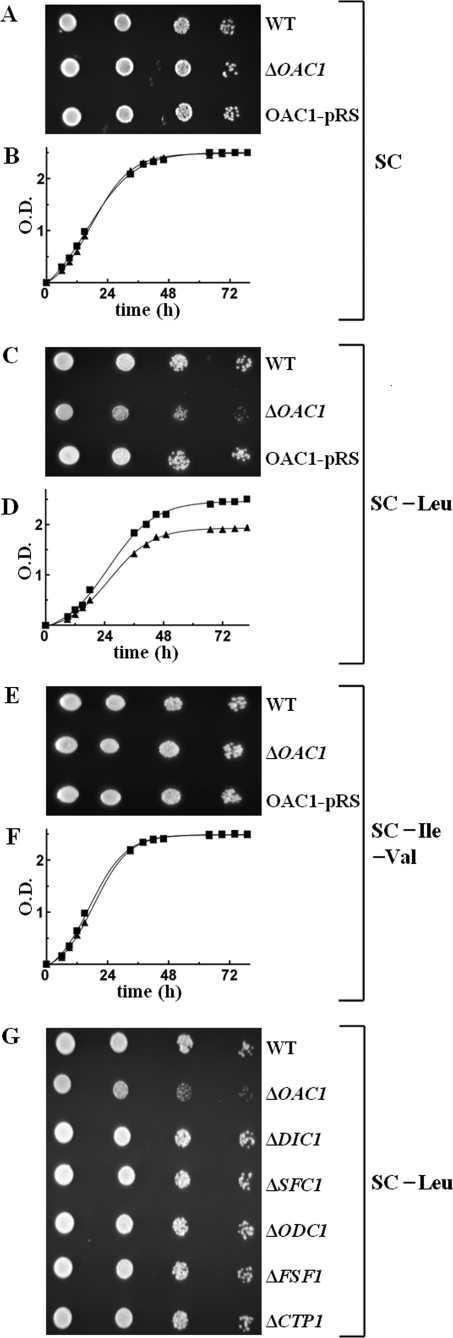
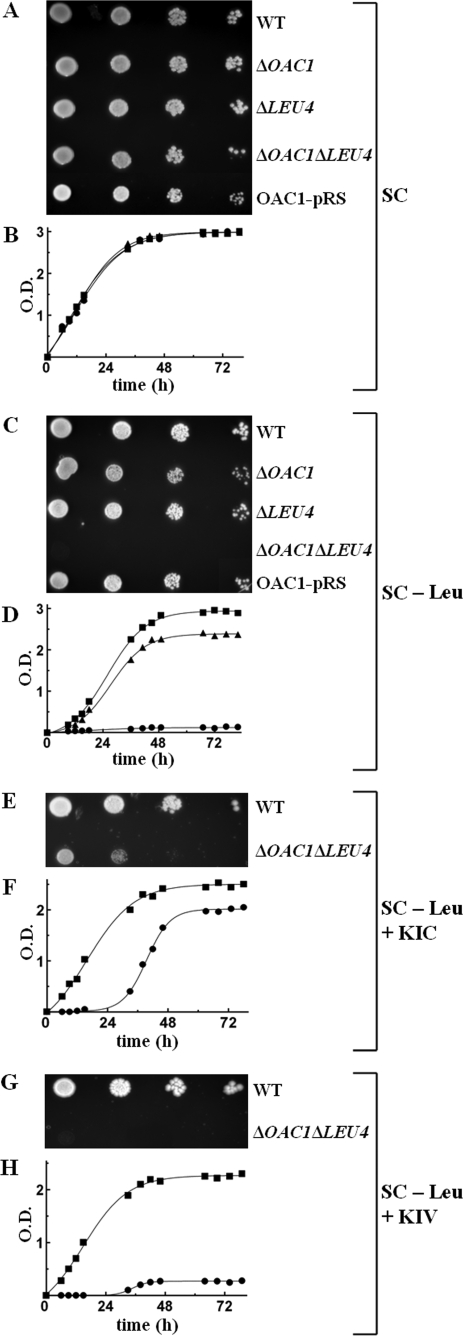
Effect of Deleting OAC1 and/or LEU4 on the Growth of S. cerevisiae Cells in the Absence of Leucine—Having established the ability of Oac1p to transport α-IPM by means of in vitro assays, the ΔOAC1 strain was tested for its ability to grow in the absence of leucine. On SC medium supplemented with glucose or (not shown) galactose, yeast cells lacking OAC1 exhibited a slight but definite growth defect in the absence of leucine compared with WT cells (Fig. 5, C and D versus A and B in the presence of leucine). The observed defect of ΔOAC1 cell growth was corrected by complementing the deletion strain with the OAC1-pRS416 plasmid containing the gene for the mitochondrial α-IPM carrier (Fig. 5C), demonstrating that the difference in phenotype is the result of the absence of Oac1p and not a secondary effect. Notably, ΔOAC1 cells grew similarly to the WT cells when both isoleucine and valine, instead of leucine, were subtracted from the SC medium (Fig. 5, E and F). Furthermore, the partial auxotrophy of ΔOAC1 cells for leucine was specific, because single deletions of dicarboxylate carrier genes (ΔDIC1, ΔSFC1, ΔODC1, and (data not shown) ΔODC2) as well as of ΔFSF1 and ΔCTP1 exhibited no growth defect (Fig. 5G). Because cytosolic α-IPM can be provided by both Oac1p (see “Results” above) and cytosolic Leu4p (1), we investigated the effect of deleting the OAC1 and LEU4 genes on the growth of S. cerevisiae cells. The double mutant ΔOAC1ΔLEU4 did not grow at all in leucine-less SC medium supplemented with glucose or (not shown) with galactose (Fig. 6, C and D versus A and B in the presence of leucine); this severe phenotype was fully restored by complementation with the OAC1-pRS416 plasmid (Fig. 6C). Of note, there was no growth defect of the single mutant ΔLEU4, because these cells grew in glucose-supplemented SC medium without leucine similarly to the WT cells and more than the single mutant ΔOAC1 (Fig. 6C). These findings indicate that on fermentable carbon sources, the role of Oac1p in producing cytosolic α-IPM is more significant than that of Leu4p. Moreover, the immediate precursor of leucine, KIC, added at a high concentration (10 mm) to ΔOAC1ΔLEU4 cells, partially restored their lack of growth (Fig. 6, E and F). Also, the addition of 10 mm α-IPM to solid or liquid leucine-less SC medium containing ΔOAC1ΔLEU4 cells partially rescued their lack of growth (not shown). Despite limitations in the ability of KIC and α-IPM to cross the cell plasma membrane, these results show that the complete auxotrophy of the double mutant for leucine is caused by lack of leucine biosynthesis cytosolic intermediates. As a control, growth of the double mutant was not restored by 10 mm KIV because of the deletion of LEU4 (Fig. 6, G and H).
FIGURE 5.
The ΔOAC1 mutant is partially auxotrophic for leucine. A, C, and E, 4-fold serial dilutions of WT and ΔOAC1 cells and ΔOAC1 cells transformed with the OAC1-pRS416 plasmid (OAC1-pRS) were spotted on solid glucose-supplemented SC (A), glucose-supplemented SC without leucine (C), or glucose-supplemented SC medium without isoleucine and valine (E). B, D, and F, WT and ΔOAC1 cells were inoculated in liquid glucose-supplemented SC (B), glucose-supplemented SC without leucine (D), or glucose-supplemented SC medium without isoleucine and valine (F). The values of optical density at 600 nm refer to cell cultures after the indicated times of growth. (▪), WT cells; (▴), ΔOAC1 cells. G, growth behavior of various mutants in the absence of leucine. 4-fold serial dilutions of WT, ΔOAC1, ΔDIC1, ΔSFC1, ΔODC1, ΔFSF1, and ΔCTP1 cells were spotted on solid glucose-supplemented SC medium without leucine.
FIGURE 6.
The ΔOAC1ΔLEU4 double mutant is auxotrophic for leucine. A and C, 4-fold serial dilutions of WT, ΔOAC1, ΔLEU4, and ΔOAC1ΔLEU4 cells and ΔOAC1ΔLEU4 cells transformed with the OAC1-pRS416 plasmid (OAC1-pRS) were spotted on solid glucose-supplemented SC medium with leucine (A) or without leucine (C). B and D, WT, ΔOAC1, and ΔOAC1ΔLEU4 cells were inoculated in liquid glucose-supplemented SC medium with leucine (B) or without leucine (D). The values of optical density at 600 nm refer to cell cultures after the indicated times of growth. (▪), WT cells; (▴), ΔOAC1 cells; (•), ΔOAC1ΔLEU4 cells. E–H, effect of the addition of KIC and KIV on the growth of ΔOAC1ΔLEU4 cells in the absence of leucine. E and G, 4-fold serial dilutions of WT and ΔOAC1ΔLEU4 cells were spotted on solid glucose-supplemented leucine-less SC medium containing 10 mm KIC (E) or 10 mm KIV (G). F and H, WT and ΔOAC1ΔLEU4 cells were inoculated in liquid glucose-supplemented leucine-less SC medium containing 10 mm KIC (F) or 10 mm KIV (H). (▪), WT cells; (•), ΔOAC1ΔLEU4 cells.
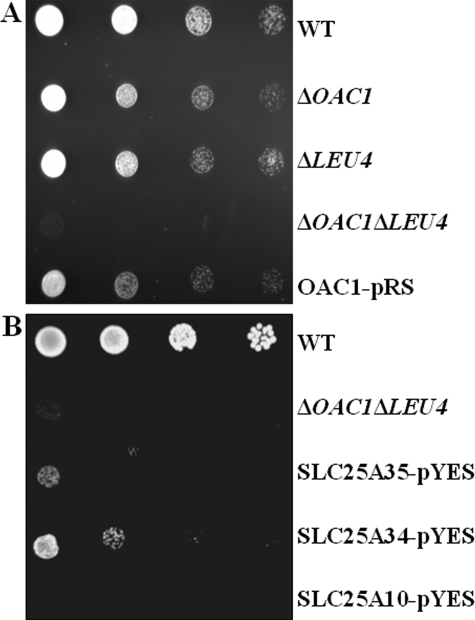
It has been speculated that in anaerobiosis and under glucose repression, the mitochondrial form of Leu4p might be unstable or nonfunctional, and the cytosolic form takes over (1). If so, under anaerobic conditions, one would expect a less significant role of Oac1p in leucine biosynthesis. The results presented in Fig. 7A show that in anaerobiosis as well, ΔOAC1 cells exhibited a partial auxotrophy and ΔOAC1ΔLEU4 cells a complete auxotrophy for leucine in contrast to WT cells and (not shown) the deleted cells in the presence of leucine.
FIGURE 7.
A, growth behavior of ΔOAC1, ΔLEU4, and ΔOAC1ΔLEU4 in anaerobiosis. 4-fold serial dilutions of WT, ΔOAC1, ΔLEU4, and ΔOAC1ΔLEU4 cells and ΔOAC1ΔLEU4 cells transformed with the OAC1-pRS416 plasmid (OAC1-pRS) were spotted on glucose-supplemented SC medium without leucine and incubated in anaerobiosis. B, effect of complementing ΔOAC1ΔLEU4 cells with the human SLC25A34 and SLC25A35 proteins on leucine auxotrophy. 4-fold serial dilutions of WT and ΔOAC1ΔLEU4 cells and ΔOAC1ΔLEU4 cells transformed with the SLC25A34-pYES, SLC25A35-pYES, or SLC25A10-pYES plasmid (SLC25A34-pYES, SLC25A35-pYES, or SLC25A10-pYES, respectively) were spotted on glucose-supplemented SC medium without leucine.
Homologues of Oac1p in Other Organisms—Data base searches identified several protein sequences that are, or may be, orthologues of Oac1p. These sequences include: in fungi, XP_448721.1 from Candida glabrata (80% of identical amino acids with Oac1p); XP_001647483.1 from Vanderwaltozyma polyspora (80%); XP_452102.1 from Kluyveromyces lactis (69%); XP_001383163.2 from Pichia stipitis (52%); XP_458551.1 from Debaryomyces hansenii (52%); XP_962817.1 from Neurospora crassa (51%); NP_593169.1 from Schizosaccharomyces pombe (49%); XP_001824075.1 from Aspergillus oryzae (50%); and XP_001259461.1 from Neosartorya fischeri (47%); in metazoa, NP_001102503.1 from R. norvegicus (33%); NP_082324.1 from Mus musculus (34%); XP_001504869.1 from Equus caballus (34%); NP_001092917.1 from Danio rerio (37%); XP_001631510.1 from Nematostella vectensis (36%); XP_972193.1 from Tribolium castaneum (35%); NP_610933.1 from Drosophila melanogaster (35%); XP_001606805.1 from Nasonia vitripennis (36%); XP_789671.1 from Strongylocentrotus purpuratus (35%); and NP_493694. B0432.4 from Caenorhabditis elegans (33%); and in mycetozoa, XP_645658.1 from Dictyostelium discoideum (36%). In plants, the degree of homology between Oac1p and the closest proteins is too low to identify any true orthologue. The closest human relatives of Oac1p are two unknown proteins, encoded by the SLC25A34 and SLC25A35 genes (27), which share 49% identical amino acids, have the characteristics of the mitochondrial carrier protein family, and display 33 and 32% identical amino acids and 51 and 48% homology, respectively, with Oac1p. To functionally identify these proteins, we first attempted to characterize them upon heterologous expression and reconstitution into liposomes. The SLC25A34 protein was not expressed in E. coli. Moreover, the recombinant SLC25A35 protein showed neither homo-exchange activity with sulfate, malonate, malate, and KIC nor [35S]sulfate/oxalacetate hetero-exchange, under the same experimental conditions used for Oac1p, perhaps because of the inability to renature and/or reconstitute it functionally. Therefore, an alternative approach allowing identification of mitochondrial carriers (28, 29) was used. As reported above, the yeast ΔOAC1ΔLEU4 double mutant is growth-restricted without leucine, a phenotype explained by its inability to transport mitochondrially synthesized α-IPM to the cytosol where it is converted to leucine. Thus, expression of a mitochondrial carrier protein that recognizes α-IPM as a substrate should mitigate leucine auxotrophy of the double mutant. The SLC25A34 protein and, to a lesser extent, the SLC25A35 protein expressed in ΔOAC1ΔLEU4 cells via the yeast vector pYES2 partially restored the capacity of the ΔOAC1ΔLEU4 strain to grow on glucose-supplemented leucine-less SC medium (Fig. 7B), indicating that both proteins can export α-IPM from mitochondria to cytosol leading to the production of leucine. By contrast, when the ΔOAC1ΔLEU4 cells were transformed with a plasmid carrying the human dicarboxylate carrier cDNA (SLC25A10), or with the empty vector (not shown), no growth restoration was observed (Fig. 7B).
DISCUSSION
In a previous study, Oac1p was shown to be the yeast mitochondrial transporter for oxalacetate and sulfate (26). Oac1p transports oxalacetate into the yeast mitochondrial matrix with protons or by exchange with other substrates and operates with the same transport properties in vivo and in reconstituted liposomes (26). In the same study, it was suggested that the role of Oac1p is to take up into mitochondrial oxalacetate produced by pyruvate carboxylase, which in S. cerevisiae is only cytoplasmic. Later, by means of DNA microarray analysis, Boer et al. (30) showed that OAC1 gene expression is downregulated by leucine because the gene promoter contains a binding site for Leu3p, the transcriptional activator of leucine biosynthesis. Indeed, our investigation by Western blots has demonstrated a leucine-induced decrease in mitochondrial Oac1p content of yeast cells grown on both fermentable and nonfermentable carbon sources.5 Boer et al. (30) suggested that Oac1p may have a role in nitrogen metabolism because its substrate, oxalacetate, in mitochondria may produce pyruvate (needed for the first step in leucine and valine biosynthesis) and/or increase the level of free CoA that inhibits α-IPM synthase (1).
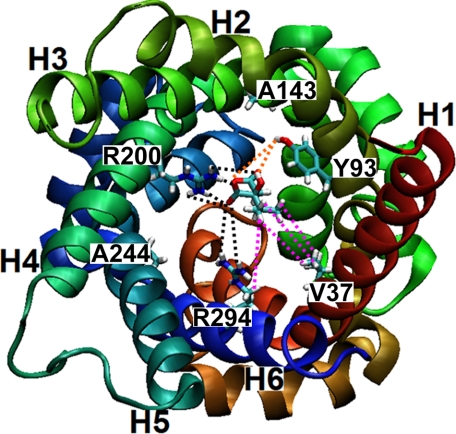
The transport measurements presented in this study demonstrate that α-IPM is transported efficiently by recombinant and reconstituted Oac1p. The lower half-saturation constant of α-IPM for Oac1p can be rationalized on the basis of the homology model of Oac1p built by comparison with the crystallographic structure of the ADP/ATP carrier (31) and on the basis of the common substrate binding site in mitochondrial carriers proposed by Robinson and Kunji (32). This site is formed by the residues of the three even-numbered transmembrane α-helices (corresponding to Tyr-93, Arg-200, and Arg-294 in Oac1p) which protrude into the internal cavity of the carrier at the midpoint of the membrane. As shown in Fig. 8, α-IPM docked in this site may bind not only with Arg-200, Arg-294, and Tyr-93 but also with the side chain of Val-37, which is the residue of the first transmembrane α-helix protruding into the cavity at the same height of Arg-200, Arg-294, and Tyr-93. It may also be that Val-37 together with Leu-97, Ala-90, Ile-48, and Phe-150 form a hydrophobic pocket surrounding the isopropylic chain of α-IPM. It should be noted that Leu-97 and Ala-90 belong to the second transmembrane α-helix immediately above and below Tyr-93, and Ile-48 and Phe-150 belong to the lower portions of the first and third transmembrane α-helices, which are kinked toward the center of the internal cavity of the carrier in the homology model of Oac1p. We hypothesize that the hydrophobic interactions between α-IPM and Val-37 or the hydrophobic pocket would allow a better fit of this substrate with Oac1p in the transition state, as compared with other carrier substrates, providing energy for the conformational changes required for substrate translocation through the protein (32–34). Interestingly, the immediate precursor of leucine biosynthesis, KIC, is an effective competitive inhibitor of Oac1p. Though not transported because of the requirement of two carboxylate groups (as in oxalacetate) or two negative charges (as in sulfate) for transport, KIC has high affinity for the substrate binding site because of its hydrophobic portion and its α-ketonic group, which may react with Arg-200. The importance of the α-ketonic group in the binding of KIC to Oac1p is supported by the finding that leucine, which differs from KIC for the presence of an amino group in place of the keto group, is unable to inhibit Oac1p activity.
FIGURE 8.
Top view from the cytosolic side of the structural homology model of Oac1p highlighting the carrier substrate binding site. Transmembrane α-helices are colored as follows: H1, red; H2, tan; H3, green; H4, emerald green; H5, cyan; and H6, blue. The residues (V37, Y93, A143, R200, A244, and R294) of the six transmembrane α-helices protruding into the cavity at the height of the binding site and the substrate α-isopropylmalate are shown in stick representation using the VMD color code: cyan, C-atoms; red, O-atoms; white, H-atoms; and blue, N-atoms. Possible interactions between the substrate and residues (V37, Y93, R200, and R294) are highlighted as follows: electrostatic/polar (black dashed lines), hydrophobic (purple dashed lines), and via bridging water (orange dashed lines).
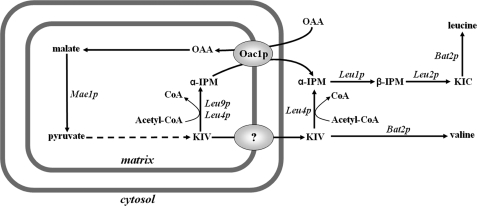
Because α-IPM is produced in mitochondria by Leu9p and Leu4lp and is required in the cytosol for the synthesis of leucine, our data indicate that a primary function of Oac1p is to catalyze the export of α-IPM in exchange for oxalacetate. Inside mitochondria, oxalacetate can be converted (by malate dehydrogenase and malic enzyme, which is also up-regulated by Leu3p (1)) into pyruvate from which the synthesis of leucine begins. In addition, the observed inhibition of Oac1p activity by KIC shows that the important step in leucine biosynthesis mediated by Oac1p is subjected to regulatory mechanisms operating not only at the transcriptional level (30) but at the protein level as well. Our conclusion that Oac1p is important for the export of α-IPM from mitochondria is supported by the growth defect of ΔOAC1 in leucine-less SC medium supplemented with glucose. It is worthwhile mentioning that all the tested S. cerevisiae strains deleted of other genes, including the ΔLEU4 and ΔFSF1 strains, do not exhibit any growth defect in the same medium. The crucial role of Oac1p in leucine biosynthesis is further demonstrated by the complete auxotrophy of the double mutant ΔOAC1ΔLEU4 for leucine under the same experimental conditions. This finding is in agreement with the current view that α-IPM is produced from KIV both in mitochondria, by Leu9p and the mitochondrial isoform of Leu4p, and in the cytosol by the cytosolic isoform of Leu4p (see Fig. 9). This metabolic compartmentalization requires export of α-IPM from mitochondria via Oac1p (this study) as well as export of KIV via a still unknown transporter. It should be stressed that the complete auxotrophy of the double mutant for leucine shows that Oac1p is the only transporter that exports α-IPM in significant amounts. Furthermore, the complete lack of growth of ΔOAC1ΔLEU4 cells in the absence of leucine is partially rescued by the addition of high concentrations of KIC to the cells and to a lesser extent by the addition of α-IPM. Notably, KIV, which also may partially cross the plasma membrane of cells, has no effect on the growth of the double mutant because it cannot be converted into α-IPM because of the lack of cytosolic Leu4p. The significance of Oac1p for leucine biosynthesis on nonfermentable carbon sources is less than that on fermentable substrates, because ΔOAC1 does not exhibit any growth defect on lactate-supplemented SC medium without leucine,6 and ΔLEU4 displays a marked growth defect under these conditions6 (see also Ref. 35). Moreover, the expression of OAC1 and LEU9 was shown by microarray analysis to be down-regulated on nonfermentable carbon sources compared with glucose and galactose media (36) and to decrease during the diauxic shift (37), whereas the expression of LEU4 behaved in the opposite manner (36, 37). Consistently, we found that the amount of Oac1p detected by immunoreaction with a specific antibody is considerably lower on lactate, glycerol, or ethanol (by about 56%) than on glucose.7 It may be that the greater number of mitochondria on nonfermentable substrates compensates the shortage of Oac1p and Leu9p by supplying a sufficient amount of KIV (to be converted into α-IPM by Leu4sp) to the cytosol.
FIGURE 9.
Scheme showing metabolic compartmentalization and the role of mitochondrial Oac1p in leucine biosynthesis in yeast. The question mark indicates a still unknown transporter that catalyzes the exit of KIV from mitochondria. Bat2p, cytosolic branched-chain amino acid aminotransferase; Leu1p, isopropylmalate isomerase; Leu2p, β-isopropylmalate dehydrogenase; Mae1p, malic enzyme; OAA, oxalacetate.
There is little doubt that the closest homologues found in several species of fungi are orthologues of Oac1p in view of the high percentage (>47%) of shared amino acids. The closest human relatives to Oac1p, i.e. the two unknown mitochondrial carriers encoded by SLC25A34 and SLC25A35, display a significantly high degree of identity and homology with Oac1p. In addition, the six residues of the SLC25A34 and SLC25A35 transmembrane α-helices protruding into the carrier cavity at the height of the proposed common substrate binding site (32) are identical to each other and strikingly similar to the corresponding residues in Oac1p; in this respect, they differ from Oac1p only by a valine in place of Ala-244 and a cysteine in place of Val-37. Our finding that the SLC25A34 and SLC25A35 proteins partially rescued the phenotype of the ΔOAC1ΔLEU4 double mutant in the absence of leucine indicates their ability to transport α-IPM. However, given that humans do not synthesize leucine, it is likely that α-IPM is not the physiological substrate of SLC25A34 and SLC25A35, which remains to be determined.
This work was supported by grants from Ministero dell'Università e della Ricerca (MUR), the Center of Excellence in Genomics (CEGBA) and Apulia Region, a contract from the European Commission (LSHM-CT-2004-503116), and University local funds. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: α-IPM, α-isopropylmalate; Ctp1p, citrate transport protein; Dic1p, dicarboxylate carrier; Fsf1p, fungal sideroflexin 1; β-IPM, β-isopropylmalate; KIC, α-ketoisocaproate; KIV, α-ketoisovalerate; KMV, α-keto-β-methylvalerate; Leu3p, transcription factor that regulates genes involved in branched-chain amino acid biosynthesis; Leu4p, α-isopropylmalate synthase I; Leu9p, α-isopropylmalate synthase II; Oac1p, oxalacetate carrier; Odc1p, oxodicarboxylate carrier, isoform 1; Odc2p, oxodicarboxylate carrier, isoform 2; Sfc1p, succinate-fumarate carrier; WT, wild-type; PIPES, 1,4-piperazinediethanesulfonic acid.
Protein nomenclature is from the Saccharomyces Genome Database (SGD).
Gene nomenclature is from the Saccharomyces Genome Database (SGD).
C. M. T. Marobbio, G. Giannuzzi, and F. Palmieri, unpublished observations.
C. M. T. Marobbio, E. Paradies, and F. Palmieri, unpublished observations.
C. M. T. Marobbio, G. Giannuzzi, and F. Palmieri, unpublished observations.
References
- 1.Kohlhaw, G. B. (2003) Mol. Biol. Rev. 67 1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan, E., Tracy, J. W., and Kohlhaw, G. B. (1973) J. Bacteriol. 116 222-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar, A., Agarwal, S., Heyman, J. A., Matson, S., Heidtman, M., Piccirillo, S., Umansky, L., Drawid, A., Jansen, R., Liu, Y., Cheung, K. H., Miller, P., Gerstein, M., Roeder, G. S., and Snyder, M. (2002) Genes Dev. 16 707-719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miotto, G., Tessaro, S., Adami Rotta, G., and Bonatto, D. (2007) Fungal Genet. Biol. 44 740-753 [DOI] [PubMed] [Google Scholar]
- 5.Kovaleva, G. Y., Bazykin, G. A., Brudno, M., and Gelfand, M. S. (2006) J. Bioinform. Comput. Biol. 4 981-998 [DOI] [PubMed] [Google Scholar]
- 6.Kaplan, R. S., Mayor, J. A., and Wood, D. O. (1993) J. Biol. Chem. 268 13682-13690 [PubMed] [Google Scholar]
- 7.Palmieri, F. (2004) Pflugers Arch., Eur. J. Physiol. 447 689-709 [DOI] [PubMed] [Google Scholar]
- 8.Palmieri, F., Agrimi, G., Blanco, E., Castegna, A., Di Noia, M. A., Iacobazzi, V., Lasorsa, F. M., Marobbio, C. M. T., Palmieri, L., Scarcia, P., Todisco, S., Vozza, A., and Walker, J. (2006) Biochim. Biophys. Acta 1757 1249-1262 [DOI] [PubMed] [Google Scholar]
- 9.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., and Boeke, J. D. (1998) Yeast 14 115-132 [DOI] [PubMed] [Google Scholar]
- 10.Goldstein, A. L., and McCusker, J. H. (1999) Yeast 15 1541-1553 [DOI] [PubMed] [Google Scholar]
- 11.Jansen, G., Wu, C., Schade, B., Thomas, D. Y., and Whiteway, M. (2005) Gene 344 43-51 [DOI] [PubMed] [Google Scholar]
- 12.Sherman, F. (1991) Methods Enzymol. 194 3-21 [DOI] [PubMed] [Google Scholar]
- 13.Meisinger, C., Sommer, T., and Pfanner, N. (2000) Anal. Biochem. 287 339-342 [DOI] [PubMed] [Google Scholar]
- 14.Sikorski, R. S., and Hieter, P. (1989) Genetics 122 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiermonte, G., Walker, J. E., and Palmieri, F. (1993) Biochem. J. 294 293-299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marobbio, C. M. T., Agrimi, G., Lasorsa, F. M., and Palmieri, F. (2003) EMBO J. 22 5975-5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmieri, L., Palmieri, F., Runswick, M., and Walker, J. E. (1996) FEBS Lett. 399 299-302 [DOI] [PubMed] [Google Scholar]
- 18.Palmieri, L., Pardo, B., Lasorsa, F. M., del Arco, A., Kobayashi, K., Iijima, M., Runswick, M. J., Walker, J. E., Saheki, T., Satrustegui, J., and Palmieri, F. (2001) EMBO J. 20 5060-5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmieri, F., Indiveri, C., Bisaccia, F., and Iacobazzi, V. (1995) Methods Enzymol. 260 349-369 [DOI] [PubMed] [Google Scholar]
- 20.Schroers, A., Kramer, R., and Wohlrab, H. (1997) J. Biol. Chem. 272 10558-10564 [DOI] [PubMed] [Google Scholar]
- 21.Palmieri, L., Lasorsa, F. M., De Palma, A., Palmieri, F., Runswick, M. J., and Walker, J. E. (1997) FEBS Lett. 417 114-118 [DOI] [PubMed] [Google Scholar]
- 22.Fiermonte, G., Dolce, V., and Palmieri, F. (1998) J. Biol. Chem. 273 22782-22787 [DOI] [PubMed] [Google Scholar]
- 23.Cappello, A. R., Curcio, R., Miniero, D. V., Stipani, I., Robinson, A. J., Kunji, E. R. S., and Palmieri, F. (2006) J. Mol. Biol. 363 51-62 [DOI] [PubMed] [Google Scholar]
- 24.Giangregorio, N., Tonazzi, A., Indiveri, C., and Palmieri, F. (2007) Biochim. Biophys. Acta 1767 1331-1339 [DOI] [PubMed] [Google Scholar]
- 25.Kaplan, R. S., Mayor, J. A., Gremse, D. A., and Wood, D. O. (1995) J. Biol. Chem. 270 4108-4114 [DOI] [PubMed] [Google Scholar]
- 26.Palmieri, L., Vozza, A., Agrimi, G., De Marco, V., Runswick, M. J., Palmieri, F., and Walker, J. E. (1999) J. Biol. Chem. 274 22184-22190 [DOI] [PubMed] [Google Scholar]
- 27.Haitina, T., Lindblom, J., Renström, T., and Fredriksson, R. (2006) Genomics 88 779-790 [DOI] [PubMed] [Google Scholar]
- 28.Catoni, E., Schwab, R., Hilpert, M., Desimone, M., Schwacke, R., Flugge, U.-I., Schumacher, K., and Frommer, W. B. (2003) FEBS Lett. 534 87-92 [DOI] [PubMed] [Google Scholar]
- 29.Hoyos, M. E., Palmieri, L., Wertin, T., Arrigoni, R., Polacco, J. C., and Palmieri, F. (2003) Plant J. 33 1027-1035 [DOI] [PubMed] [Google Scholar]
- 30.Boer, V. M., Daran, J.-M., Almering, M. J. H., de Winde, J. H., and Pronk, J. T. (2005) FEMS Yeast Res. 5 885-897 [DOI] [PubMed] [Google Scholar]
- 31.Pebay-Peyroula, E., Dahout-Gonzalez, C., Kahn, R., Trézéguet, V., Lauquin, G. J.-M., and Brandolin, G. (2003) Nature 426 39-44 [DOI] [PubMed] [Google Scholar]
- 32.Robinson, A. J., and Kunji, E. R. S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2617-2622-2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klingenberg, M. (2005) Biochemistry 44 8563-8570 [DOI] [PubMed] [Google Scholar]
- 34.Cappello, A. R., Miniero, D. V., Curcio, R., Ludovico, A., Daddabbo, L., Stipani, I., Robinson, A. J., Kunji, E. R. S., and Palmieri, F. (2007) J. Mol. Biol. 369 400-412 [DOI] [PubMed] [Google Scholar]
- 35.Casalone, E., Barberio, C., Cavalieri, D., and Polsinelli, M. (2000) Yeast 16 539-545 [DOI] [PubMed] [Google Scholar]
- 36.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., and Brown, P. O. (2000) Mol. Biol. Cell 11 4241-4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeRisi, J. L., Iyer, V. R., and Brown, P. O. (1997) Science 278 680-686 [DOI] [PubMed] [Google Scholar]