Abstract
The peptidoglycan glycosyltransferase (GT) module of class A penicillin-binding proteins (PBPs) and monofunctional GTs catalyze glycan chain elongation of the bacterial cell wall. These enzymes belong to the GT51 family, are characterized by five conserved motifs, and have some fold similarity with the phage λ lysozyme. In this work, we have systematically modified all the conserved amino acid residues of the GT module of Escherichia coli class A PBP1b by site-directed mutagenesis and determined their importance for the in vivo and in vitro activity and the thermostability of the protein. To get an insight into the GT active site of this paradigm enzyme, a model of PBP1b GT domain was constructed based on the available crystal structures (PDB codes 2OLV and 2OLU). The data show that in addition to the essential glutamate residues Glu233 of motif 1 and Glu290 of motif 3, the residues Phe237 and His240 of motif 1 and Gly264, Thr267, Gln271, and Lys274 of motif 2, all located in the catalytic cavity of the GT domain, are essential for the in vitro enzymatic activity of the PBP1b and for its in vivo functioning. Thus, the first three conserved motifs contain most of the residues that are required for the GT activity of the PBP1b. The residues Asp234, Phe237, His240, Thr267, and Gln271 are proposed to maintain the structure of the active site and the positioning of the catalytic Glu233.
Most bacteria are surrounded by cell wall peptidoglycan, a net-like polymer consisting of glycan strands made of alternating β-1,4-linked GlcNAc and N-acetylmuramic acid (MurNAc)2 residues cross-linked by peptides (1, 2). Peptidoglycan metabolism is linked to cell cycle and function to confer shape and osmotic stability to the cell (3–5). Its assembly is complex and requires the contribution of several proteins (polymerases, hydrolases, and skeletal proteins), which are believed to form protein assemblages specialized in cell elongation and division (3, 6, 7).
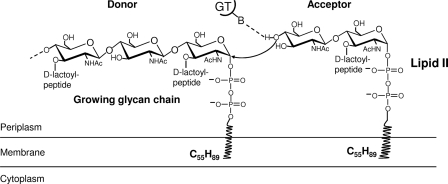
Multimodular penicillin-binding proteins (PBPs) are membrane-bound enzymes responsible for peptidoglycan polymerization (8, 9). They are essentially two-module proteins that belong either to class A or to class B, depending on the structure and the catalytic activity of their N-terminal module. The C-terminal penicillin-binding domain of both classes has a transpeptidase activity, catalyzing peptide cross-linking between two adjacent glycan chains. In class A, the N-terminal module is responsible for their glycosyltransferase (GT) activity. This module catalyzes the polymerization of uncross-linked glycan chains from a membrane bound intermediate called lipid II, the GlcNAc-MurNAc-(pentapeptide) linked to the undecaprenyl via a pyrophosphate bridge (Fig. 1). In class B, the N-terminal module appears to play a role in cell morphogenesis (10). Monofunctional enzymes similar to the GT module of class A PBPs also exist in some bacteria, but their exact role is still unknown (11).
FIGURE 1.
Mechanism of glycan chain elongation by peptidoglycan glycosyltransferases. The catalytic base (glutamate of motif 1) catalyzes the deprotonation of the GlcNAc 4-OH of lipid II, and the activated nucleophile then directly attacks the C1 of the lipid-linked MurNAc of the growing polysaccharide chain.
Crystal structures of the Staphylococcus aureus class A PBP2 in apo form (PDB code 2OLU) and in complex with moenomycin (PDB code 2OLV) and the GT module of Aquifex aeolicus PBP1a (PDB code 2O9O) reveal that the GT module has some similarity to phage λ lysozyme and shed light on its active site (12, 13). The available structural and biochemical data provide better understanding of the mechanism of glycan chain elongation by the glycosyltransferases (12–15). The essential glutamate of motif 1 functions as active site general base and deprotonates the GlcNAc 4-OH group of lipid II acting as an acceptor for nucleophilic attack on the reducing end of the growing glycan chain acting as a donor (16, 17) (Fig. 1). The reaction results in the formation of a β-1,4 glycosidic linkage with inversion of the configuration at the anomeric C1 carbon of MurNAc. The departure of the leaving diphospho-undecaprenyl group is probably assisted by anionic interaction with the second conserved glutamate in motif 3 via a divalent metal cation. The translocation of the new product from the acceptor to the donor site may be driven by the high affinity of the pyrophosphate for the positively charged donor site (12). Apart from S. aureus PBP2 (18), the activity of other GTs (16, 19, 20) was dependent on metal ions, but the exact role of bivalent ions in the reaction still need to be determined.
The multimodular class A PBP1b (Fig. 2) is the major bifunctional peptidoglycan synthase in Escherichia coli endowed with GT and transpeptidase activities (17, 21–23). It is an essential constituent of the peptidoglycan synthesis machineries involved in cell elongation and cell division (24–26), where its function is probably modulated by other proteins, such as FtsN, which stimulates its activity (27).
FIGURE 2.
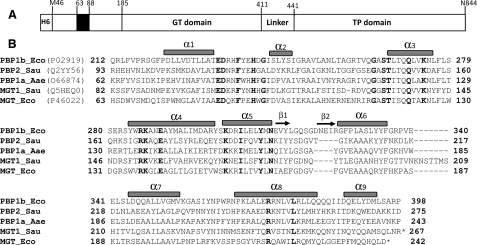
Organization of the multimodular PBP1b and structural model of its GT module. A, schematic representation of [H6]PBP1bγ. PBP1b has a cytoplasmic N-terminal tail (1–63 in PBP1bα; 46–63 in PBP1bγ) followed by a transmembrane anchor (64–87), an insertion of about 100 amino acids (88–185), and the GT (185–398) and transpeptidase (TP) (441–844) modules separated by a linker. B, sequence alignment (ClustalW) of the GT module of class A PBPs and monofunctional GTs. Eco, E. coli; Sau, S. aureus; Aae, A. aeolicus. The secondary structures of S. aureus PBP2 are shown on top of the sequences, and motifs 1–5 are underlined with the conserved residues in bold type. Numbers indicate the position of the first and last residues. The C-terminal end of monofunctional GT (MGT) is indicated by *.
E. coli PBP1b is the most studied class A PBP and exhibits the highest peptidoglycan polymerase activity in vitro (16, 17, 24, 25, 28). However, no tridimensional structure is available for this protein, and its active site is not well characterized. The crystal structure of the PBP1b ortholog, S. aureus PBP2 (PDB code 2OLU), is known (12), and the two proteins share high sequence similarity with each other. Based on the PBP2 structure, we have constructed a model of PBP1b GT domain to get an insight into its active site. The GT domains of the two PBPs, like almost all the proteins of the GT51 family (29), are characterized by five characteristic motifs containing several invariant amino acids, most of them located in the catalytic cavity of these enzymes (Figs. 2 and 3). We have modified all the conserved residues within these motifs and determined their importance for the function of the PBP1b both in vitro and in vivo.
FIGURE 3.
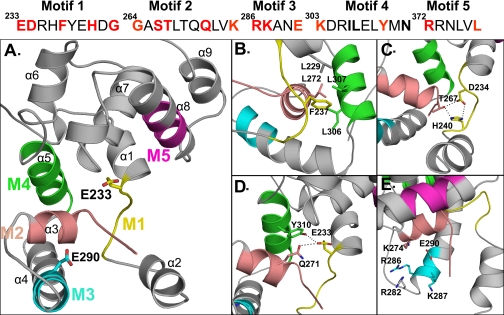
Structural model of PBP1b GT module (212–398) and close view into the catalytic site depicting the positions of the studied conserved residues in sticks. A, motif 1 (M1) is shown in yellow, motif 2 (M2) is shown in salmon, motif 3 (M3) is shown in cyan, motif 4 (M4) is shown in green, and motif 5 (M5) is shown in magenta. The catalytic Glu233 and the Glu290 residues are shown in sticks. B, Phe237 is part of a hydrophobic cluster. C, interaction between Asp234 and His240 of motif 1 and Thr267 of motif 2. D, Gln271 and Tyr310 are within interacting distance of the catalytic Glu233. E, positively charged residues within the donor site close to the Glu290 are shown. The figures were generated with the help of the PyMOL software.
EXPERIMENTAL PROCEDURES
Construction of the Mutants—Single amino acid mutations were introduced into the ponB gene using the QuikChange site-directed mutagenesis kit (Stratagene) as described previously (17). Briefly, the primers (supplemental Table S1) were used to PCR-amplify the pDML920C plasmid containing the 1300-bp NcoI-PstI sequence encoding the His tag (Met46–Gln423) PBP1b GT module. After DNA sequencing to check for the desired mutation and absence of any errors, the 840-bp NheI-SacII or 300-bp SacII-PstI fragments were exchanged with the corresponding unmodified segments of the expression plasmid pDML924 carrying ponB gene encoding the His tag PBP1bγ (Met46–Asn844) (17) and sequenced. Supplemental Table S1 shows the pDML924 derivatives (E to N) allowing the production of PBP1b mutants.
Expression and Purification of the His Tag PBP1bγ and the Mutants—E. coli BL21 (DE3)/pDML924-E to R transformants were grown at 37 °C in Luria Bertani (LB) medium containing 50 μg of kanamycin/ml. When the absorbance at 600 nm reached a value of 0.6, the cultures were supplemented with 1 mm IPTG and grown for two additional hours. The cells were disrupted and fractionated. The membrane-bound PBPs were solubilized in 25 mm Tris-HCl, pH 7.5, 1 m NaCl, and l % CHAPS and purified on Ni2+-Sepharose column (HisTrap HP from GE healthcare) as described previously (17).
Complementation Assay—The in vivo activity of the PBP1b mutants was assayed by their ability to complement for the deletion of PBP1b and the thermosensitive PBP1a in E. coli strain EJ801 at 42 °C (30). EJ801 was transformed with the plasmids pDML924-E to R at 30 °C. The transformants were first grown at 30 °C in LB medium containing 30 μg of kanamycin/ml. The preculture was then diluted in 20 ml of fresh LB medium in the absence and the presence of IPTG to an A600 nm value of 0.1–0.3, and the absorbance of the culture was monitored for 4 h at 42 °C.
Glycosyltransferase Assay—The [14C]meso-diaminopimelic acid (A2pm)-labeled C55 lipid II intermediate (N-acetylglucosaminyl-N-acetylmuramoyl(l-Ala-γ-d-Glu-(l)-meso-A2pm-(l)-d-Ala-d-Ala)-pyrophosphate-undecaprenol) was prepared essentially as described previously (31).
In vitro peptidoglycan polymerization was performed by incubation of the [14C]lipid II (0.5–5 μm; 0.126 μCi/nmol) and the His tag PBP1bγ or the mutants (20 nm to 1 μm) in 50 mm Tris-HCl, pH 7.5; 0.046% CHAPS; 33 mm NaCl; 10 mm MgCl2; 0.5% decyl polyethylene glycol; 12.5% 1-octanol; 25% dimethyl sulfoxide (DMSO) for 15–60 min at 30 °C. The reaction products were separated by an overnight chromatography on Whatman No. 1 filter paper in isobutyric acid:1 m ammonium hydroxide (5:3; V/V) and analyzed as described previously (17).
Interaction of the PBP1b Mutants with Fluorescent Ampicillin or [14C]Benzylpenicillin—[14C]Benzylpenicillin (54 nCi nmol-1) was from Amersham Biosciences. Fluoresceyl-labeled ampicillin was prepared as described previously (32). The interaction between PBP1b and β-lactams was analyzed using the same procedure as described previously (17, 19).
Thermostability of the PBP1b Mutants—Purified PBP1b and PBP1b mutants (5 μm final concentration) were incubated at 60 °C for various times and then labeled with fluoresceyl-ampicillin (10-4 m final concentration) in 25 mm Tris-HCl, pH 7.5, 0.5 m NaCl, and 0.7% CHAPS (buffer A) for 5 min at 30 °C. The samples were analyzed by SDS-PAGE, and the acyl-enzyme complex formed was detected with a Molecular Imager scanner and analyzed with the Quantity One software (Bio-Rad).
PBP1b GT Domain Modeling—A model of E. coli PBP1b GT domain was constructed based on S. aureus PBP2 crystallographic structure (Protein Data Bank code 2OLV) (12). From residue 185 to residue 400 (PBP1b numbering), S. aureus PBP2 side chains have been replaced by the equivalent ones of E. coli PBP1b and have been reoriented so as to avoid steric clashes. Side chains positions have not been modified when the side chains are common to both enzymes. Residues 255–263 have not been modeled as the structure of equivalent residues in S. aureus PBP2 is unknown, like residues 320–323, which are not present in S. aureus PBP2. The model was energy-minimized with the program Insight II® (BIOSYM Technologies, San Diego, CA; Accelrys Software Inc.) in a two-step process, on a all-atom framework, thus taking all the hydrogen atoms into account. In the first step, all the Cα atoms were fixed, and the rest of the protein was subject to a geometry optimization. In the second step, the geometry optimization was performed without restraint.
RESULTS
Model of the GT Domain of E. coli PBP1b—Alignment of the E. coli PBP1b GT module sequence, starting from residues 212–398, with that of S. aureus PBP2 showed 39% of identity and 65% of similarity. This high degree of conservation suggests that their three-dimensional structures must also be well conserved. A model of E. coli PBP1b GT domain based on the crystal structure of S. aureus PBP2 (PDB code 2OLV) (12) was constructed (see “Experimental Procedures”). The fold adopted by the 212–398 polypeptide model with the five conserved motifs is shown in Fig. 3A.
At least 18 amino acid residues within these motifs are highly or strictly conserved (Fig. 2B). In the E. coli PBP1b GT domain, four of them (Glu233, Asp234, Glu290, and Tyr310) were analyzed previously (17). The 14 additional conserved residues in the PBP1b were modified by site-directed mutagenesis, and the mutants were characterized (Table 1).
TABLE 1.
Properties of E. coli PBP1b mutants The kinetic parameters were determined, with lipid II as substrate. In vivo activities of the mutants were determined by complementation assay at 42 °C with and without IPTG induction in ponA (ts) ponB E. coli EJ801 double mutant. The kinetic parameters values are the mean of three to five separate experiments. + indicates that the mutant is active or stable; –, the mutant is not active or instable; ±, the mutant has intermediate activity or not less stable. ND, not determined because of low activity or instability of the protein.
| In vitro activity | Penicillin binding/thermostability | kcat | Km | kcat/Km | GT activity | |
|---|---|---|---|---|---|---|
| s–1 | μm | m–1 s–1 | % | |||
| PBP1b wild-typea | + | +/+ | 70 ± 13 | 1.8 ± 0.8 | 39,000 | 100% |
| I. E233Qa | – | +/+ | ND | ND | ND | 0.2 |
| D234Na,b | – | +/± | 9.4 ± 1.8 | 1.68 ± 0.65 | 5600 | 14 |
| F237A | – | +/+ | – | – | – | 0 |
| H240A | – | +/+ | – | – | – | 0 |
| H240Q | ± | +/+ | 5.2 ± 1.24 | 1.8 ± 0.21 | 4700 | 7 |
| G242A | +c | +/+ | 2.9 ± 1 | 1.6 ± 0.9 | 1800 | 4 |
| II. G264L | – | +/+ | – | – | – | 0 |
| S266A | + | +/+ | 7.6 ± 2 | 1.2 ± 0.67 | 6300 | 11 |
| T267Ab | – | +/± | – | – | – | 0 |
| Q271A | – | +/+ | – | – | – | 0 |
| K274A | – | +/+ | – | – | – | 0 |
| III. R286Ad | – | ×/× | × | × | × | × |
| K287A | + | +/+ | 44 ± 10 | 1.2 ± 0.6 | 36,500 | 63 |
| E290Qa | ± | +/+ | 1.44 ± 0.4 | 0.4 ± 0.2 | 3600 | 2 |
| IV. K303Ab | – | +/– | ND | ND | ND | – |
| Y310Fa,d | – | ×/× | × | × | × | – |
| N312A | + | +/+ | 11 ± 3.6 | 1.2 ± 0.94 | 9200 | 15 |
| V. R372A | + | +/+ | 13.4 ± 4 | 1.1 ± 0.63 | 12,200 | 19 |
| L377Gb | + | +/± | ND | ND | ND | + |
From Ref. 17
These PBP1b mutants were expressed only at lower temperature or at lower level at 37 °C when compared with the wild-type
Active only after induction with IPTG
These mutants were not expressed at all (×)
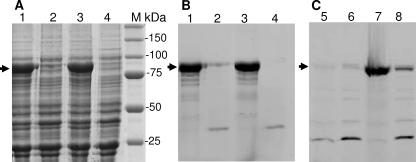
Expression, Purification, and Properties of the PBP1b Mutants—All the PBP1b mutants were expressed and purified in the same conditions as for the wild type, except that PBP1bT267A could only be expressed at 18 °C instead of 37 °C, and the PBP1b R286A was not produced even at 18 °C (Fig. 4). The PBP1b mutants were characterized in terms of penicillin binding activity, thermostability, in vivo activity by complementation, and GT activity using lipid II as substrate. The expression levels (2–3 mg of protein/liter of culture) and the stability of the mutants, unless otherwise mentioned, were similar to those of the wild-type protein. The penicillin binding capacity of the native PBP1b was reduced by 50% after 8–10 min of preincubation at 60 °C. The acylation rate constants of the PBP1b mutants by β-lactams were comparable with those of the native PBP1b (500–700 m-1 s-1 for [14C]benzylpenicillin or 20–30 m-1 s-1 for fluoresceyl-ampicillin). The results of the complementation experiments with and without induction were similar unless otherwise indicated.
FIGURE 4.
Expression and fluoresceyl-ampicillin labeling of some PBP1b mutants. A and B, Coomassie Blue staining (A) and fluoresceyl-ampicillin (50 μm) labeling (B) of PBPs after SDS-PAGE loaded with equivalent amount of membrane extract from cells expressing PBP1b mutants at 37 °C. Lane 1, F237A; lane 2, T267A; lane 3, Q271A; lane 4, R286A; M = molecular mass standard. C, fluorescence detection of fluoresceyl-ampicillin labeled PBPs at 30 °C. In lanes 5 and 6, the expression of the PBP1b R286A was realized at 18 and 37 °C, respectively. In lanes 7 and 8, the expression of PBP1b T267A was realized at 18 and 37 °C, respectively (no overexpression of R286A was visible after Coomassie Blue staining, not shown).
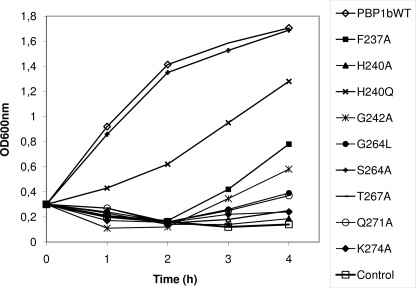
Mutations in Motif 1 (233EDRHFYEHDG242), F237A, H240Q, or H240A and G242A—Motif 1 is located in a loop between the helices α1 and α2 and contains five conserved residues, Glu233, Asp234, Phe237, His240, and Gly242 (Figs. 2 and 3A). The residues, Phe237, His240, and Gly242 were modified to Ala. In addition, His240 was modified to Gln. The thermostability and the penicillin binding activity of the four PBP1b mutants were similar to those of the wild type. The PBP1b F237A and H240A were inactive in vivo as assayed by complementation using the ponA (ts) ponB EJ801 E. coli strain producing a thermosensitive PBP1a and depleted of PBP1b (see “Experimental Procedures”), whereas PBP1b H240Q was partially functional (Fig. 5), and G242A was only active in the presence of IPTG (data not shown). The GT activity of PBP1b F237A and H240A was completely abolished. The glycan polymerase activities of PBP1b H240Q and G242A were 8- and 22-fold lower than that of the wild type with efficiencies of 4700 m-1 s-1 and 1800 m-1 s-1, respectively, when compared with 39,000 m-1 s-1 for the wild type (Table 1).
FIGURE 5.
In vivo activity of PBP1b mutants modified in motifs 1 and 2 by complementation assays at 42 °C using the ponA(TS) ponB E. coli strain EJ801. WT, wild type.
Mutations in Motif 2 (264GASTLTQQLVK274), G264L, S266A, T267A, Q271A, and K274A—We propose that motif 2 extends to Lys274 since the nature (Lys/Arg) of the residue at this position is highly conserved among the hundreds of class A PBPs/monofunctional GTs (CAZy (carbohydrate-active enzymes) data base), and the three-dimensional structures of the GT domain show that it may play an important role in the binding of the substrate (12, 13). Motif 2 contains five conserved residues starting before the α3 helix (Gly264, Ser266, and Thr267) and extends through the whole helix (Gln271 and Lys274) (Figs. 2B and 3A). To test their importance in the function of PBP1b, the mutations G264L, S266A, T267A, Q271A, and K274A were performed. The PBP1b G264L, S266A, Q271A, and K274A mutants were expressed in stable form at 37 °C and behave as the wild-type protein. They all bound penicillin, but their GT activity was completely abolished except that of PBP1b S266A, which had a 6-fold reduced GT activity with an efficiency of 6300 m-1 s-1 (Table 1) and was active in the complementation assay (Fig. 5). PBP1b G264L, Q271A, and K274A do not support the growth of E. coli EJ801 ponA (ts) ponB at the nonpermissive temperature (Fig. 5). The PBP1b T267A was not expressed at 37 °C, but when the expression was tested at 18 °C for 16 h, the protein was produced (Fig. 4). The purified PBP1b T267A was stable and bound penicillin (Fig. 4) but was devoid of GT activity and was inactive in the complementation assay at 42 °C (Fig. 5). The ability of the PBP1b T267A to bind penicillin after incubation at 60 °C was similar to the wild type (50% labeling after 8–10 min at 60 °C), suggesting that the mutation had not affected the overall stability and the structure of the protein (other than the GT active site) but perhaps affected the folding process of the protein, which was restored at lower temperature.
Mutations in Motif 3 (286RKANE290), R286A and K287A—Motif 3 is located at the beginning of the α4 helix and contains three conserved residues, Arg286, Lys287, and Glu290 (Figs. 2B and 3A). Arg286 and Lys287 were modified to Ala. The PBP1b R286A mutant was not detected in induced cells using the same condition as for the wild type. Its expression was also tested at 18 °C for 3–16 h, but no protein was produced (Fig. 4). This mutation may have affected the expression or the folding and stability of the protein. Moreover, when the plasmid encoding the PBP1b R286A was used in the complementation assay, the ponA (ts) ponB EJ801 strain was not able to grow at 42 °C. By contrast, the behavior of the PBP1b K287A was similar to the nonmutated protein with respect to the level of production, thermostability, susceptibility to penicillin, glycan chain elongation from lipid II (kcat/Km 36,500 m-1 s-1) (Table 1), and in vivo activity.
Mutations in Motif 4 (303KDRILELYMN312), K303A and N312A—Motif 4 holds within the entire α5 helix and contains four conserved residues, Lys303, Ile306, Tyr310, and Asn312 (Figs. 2B and 3A). Lys303 and Asn312 were modified to Ala. The PBP1b K303A mutant was produced at a 3-fold lower level than the wild type. The K303A mutant from membrane preparations bound penicillin, but the CHAPS-solubilized protein underwent spontaneous precipitation during purification, making its in vitro characterization difficult. This mutation may have affected the stability of the protein. The plasmid, which encoded the PBP1b K303A mutant, was not able to rescue the strain EJ801 from lysis at the nonpermissive temperature, showing that the protein was not functional in vivo. The PBP1b N312A behaves as the wild type in terms of expression level, stability, and penicillin binding. This mutant has 4-fold lower GT activity than the wild type with an efficiency of 9200 m-1 s-1 (Table 1). E. coli EJ801 transformants expressing the PBP1b N312A grew at 42 °C, indicating that the mutant was active in vivo.
Mutations in Motif 5 (372RRNLVL377), R372A and L377G—Motif 5 is located in the middle of the α8 helix and contains two conserved residues, Arg372 and Leu377, which were modified to Ala and Gly, respectively (Figs. 2 and 3A). The thermostability of PBP1b R372A and its affinity for penicillin were similar to those of the wild type. The mutant R372A was active in vivo and catalyzed glycan chain polymerization in vitro with an efficiency of 12,200 m-1 s-1 (Table 1). The expression level (0.3 mg/l) and thermostability (50% labeling after 4 min at 60 °C) of the PBP1b L377G were lower than those of the wild-type PBP1b. The PBP1b L377G mutant was active in vivo in the complementation assay and had GT activity in vitro with lipid II as substrate (kinetic parameters not determined). The PBP1b mutants, which retained some activity (Table 1), all have their kcat values affected to different extents, but there was almost no change in the Km values.
DISCUSSION
PBP1b is the major peptidoglycan polymerase in E. coli.Itisa bifunctional enzyme catalyzing both glycan chain elongation and peptide cross-linking. Here we have studied the structure-function relationship and the effect of point mutations within the PBP1b GT domain on the peptidoglycan polymerase activity of the PBP1b.
The glycosyltransferase domain of the PBP1b catalyzes the transfer of the lipid linked growing glycan chain, which acts as the glycosyl donor, to the lipid II precursor, which functions as the glycosyl acceptor (12, 14) (Fig. 1). The catalytic base Glu233 catalyzes the deprotonation of the GlcNAc 4-OH of lipid II, and the activated nucleophile then directly attacks the C1 of the lipid-linked MurNAc of the growing polysaccharide chain (SN2-like mechanism). In addition to the essential dicarboxylic acid residue, the GT module contains several other conserved residues, most of them clustered in the five conserved motifs. The importance of these residues for the function of the PBP1b was analyzed both in vitro and in vivo, and their possible role is discussed below. The results presented here show a nice correlation between the GT activity in vitro and the in vivo activity of the mutants.
A three-dimensional structural model of E. coli PBP1b GT domain was constructed using the x-ray structure of S. aureus PBP2 (12). By similarity, the motif 1 containing the essential Glu233 is located in a loop at the end of the α1 helix and just before the flexible region, which encompasses the α2 helix (Fig. 3A). Therefore, the structure of the loop, which also contains Asp234, Phe237, and His240, must be very important for the correct positioning of the glutamate and the activity of the enzyme. The side chain of Phe237 is part of a hydrophobic cluster that contains Tyr238 of motif 1, Leu272 of motif 2, Ile306 and Leu307 of motif 4, and Leu229 on the α1 helix (Fig. 3B). This hydrophobic cluster is likely to play an important role in the maintenance of the structure of the motif 1 loop. In addition, the side chains of Asp234 and His240 of motif 1 and Thr267 of motif 2 interact with each other (Fig. 3C), and therefore, the mutation of one of these residues may destabilize the loop that holds Glu233, resulting in the loss of activity. Consistently, the PBP1b mutants D234N, F237A, H240A, and T267A were all not functional in vivo, and only PBP1b D234N had 13% of the wild-type GT activity in vitro (17). Note that although the PBP1b H240A was completely inactive, the mutant H240Q, the activity of which was reduced about 8-fold, was partially functional in the complementation assay. These results show that the glutamine can replace the histidine and suggest that the hydrogen bonds network of the triad His-Asp-Thr (Fig. 3C) is partially maintained in the PBP1b H240Q mutant and is thus important for the activity.
According to the model, the Glu233 side chain (motif 1) interacts directly with that of Gln271 (motif 2) and the hydroxyl group of Tyr310 (motif 4) (Fig. 3D). The PBP1b Q271A mutant was completely inactive in vitro and in vivo, suggesting that this substitution also had an effect on the position of the glutamate. In the case of Y310F, the modified protein could not be isolated, probably because Tyr310 may also play a role in the folding process and stability of the protein (17).
Gly242 and Gly264 are located in the flexible region upstream and downstream of the α2 helix. PBP1b G242A mutant has low activity, and PBP1b G264L mutant was inactive, suggesting that the flexibility of the region may be important for the activity of the enzyme and that the mutations may have caused local conformational change or steric effect.
Residues Lys274 (motif 2), Arg286, and Lys287 (motif 3), together with Arg282 (residues equivalent to Lys155, Arg167, Lys168, and Lys163 in S. aureus), create a positively charged pocket in the donor site (Fig. 3E) and may play a role in the positioning of the negatively charged pyrophosphate of the donor substrate (12). Below this region lays a hydrophobic surface, which likely interacts with the membrane and where the lipid moiety would bind. Among the positively charged residues, Lys274 seems to play a major role in the interaction with the substrate as the modification of this residue into Ala completely abolished the in vivo and in vitro functions of the protein. The PBP1b R286A mutant was not expressed, and its exact role cannot be determined. However, the equivalent residue in A. aeolicus (Arg136) was shown to be important for enzymatic activity (15), and therefore, Arg286 may also play similar function in PBP1b. Lys287 seems not to be important since the K287A mutant retained high GT activity and was functional in vivo.
The PBP1b R372A retained 20% of GT activity in vitro and was active in vivo. The equivalent residue to Arg372 in S. aureus PBP2 (Arg249) was proposed to stabilize the deprotonated form of the catalytic glutamate (12). A mutant of the equivalent residue (R218A) in A. aeolicus PBP1a seems to be more affected as it retained only 10% of GT activity. Its in vivo activity is not known. However, under certain conditions (more enzyme), good turnover by A. aeolicus PBP1a was observed (15). One explanation is that the role of Arg372 in the PBP1b mutant or Arg218 in the PBP1a mutant may be taken over by the amino group of a lysine downstream of helix α7, which is not strictly conserved but is well positioned and could fold into the active site upon substrate binding: Lys355 in E. coli PBP1b R372A mutant and Lys200 in A. aeolicus PBP1a R218A mutant.
In contrast to E. coli PBP1b mutants F237A, H240A, and T267A, which were completely inactive in vivo and in vitro even when high protein concentrations (20–50 times more than the wild type) were used in vitro to monitor their activity, the mutants of the equivalent residues (F87A, H90A, and T117A) in A. aeolicus PBP1a GT domain retain residual GT activity in vitro (5–10%) (13, 15). These results suggest that some local variation may exist between different GT domains. However, the general tendency of the effects of mutations in E. coli PBP1b and A. aeolicus PBP1a is similar and shows that most of the conserved residues of motifs 1, 2, and 3 play an important role in the activity of their GT domains.
Consistent with the location of the first three conserved motifs in the catalytic cavity of the GT domain, most of the mutations in motifs 1 and 2 and to a lesser extent motif 3 severely affected the function of the PBP1b; only 2 of 12 mutants (S266A in motif 2 and K287A in motif 3) were fully active in vivo, and these residues are probably dispensable. Except from D234N (motif 1), T267A (motif 2), and R286A (motif 3), whose thermostabilities were affected to different extents when compared with the wild type, all the mutants were thermostable. On the contrary, in terms of in vitro GT activity, six mutants (F237A, H240A, G264L, T267A, Q271A, and K274A) were completely inactive, and D234N, G242A, and Glu290 have only residual GT activities estimated at 13, 4.6, and 2% of the wild-type activity, respectively.
In conclusion, motifs 1, 2, and 3 are important for the GT activity of the PBP1b. The residues Asp234, Phe237, His240, Thr267, and Gln271 are proposed to maintain the structure of the active site and the positioning of the catalytic Glu233 and may interact with the sugar moiety of the substrates as suggested (12). Residue Lys274, located in a positively charged pocket of the donor site, is involved in the positioning of the pyrophosphate of the growing glycan chain. Motifs 4 and 5 seem less critical for the activity and may be involved mainly in maintaining the overall structure of the GT domain.
Supplementary Material
Acknowledgments
We thank Astrid Zervozen for the preparation of fluoresceyl-labeled ampicillin.
This work was supported by grants from the European Commission within the “EUR-INTAFAR” (Inhibition of New Targets for Fighting Antibiotic Resistance) (LSHM-CT-2004-512138) network, the Belgian programme of Interuniversity Poles of Attraction (IAP) initiated by the Federal Office for Scientific Technical and Cultural Affairs (IAP Number 6/19), the Actions de Recherche Concertées (Grant 03/08-297), and the Fonds National de la Recherche Scientifique (Fonds de la Recherche Fondamentale Collective d'initiative des chercheurs (FRFC) Numbers 2.4506.08 and 2.4511.06). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental table.
Footnotes
The abbreviations used are: MurNAc, N-acetylmuramic acid; PBP, penicillin-binding protein; GT, glycosyltransferase; IPTG, isopropyl-1-thio-β-d-galactopyranoside; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate; PDB, Protein Data Bank.
References
- 1.van Heijenoort, J. (2001) Glycobiology 11 25R-36R [DOI] [PubMed] [Google Scholar]
- 2.Vollmer, W., Blanot, D., and de Pedro, M. A. (2008) FEMS Microbiol. Rev. 32 149-167 [DOI] [PubMed] [Google Scholar]
- 3.Vollmer, W., and Höltje, J. V. (2001) Curr. Opin. Microbiol. 4 625-633 [DOI] [PubMed] [Google Scholar]
- 4.Nanninga, N. (1991) Mol. Microbiol. 5 791-795 [DOI] [PubMed] [Google Scholar]
- 5.Goehring, N. W., and Beckwith, J. (2005) Curr. Biol. 15 R514-526 [DOI] [PubMed] [Google Scholar]
- 6.Höltje, J. V. (1998) Microbiol. Mol. Biol. Rev. 62 181-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Blaauwen, T., de Pedro, M. A., Nguyen-Distèche, M., and Ayala, J. A. (2008) FEMS Microbiol. Rev. 32 321-344 [DOI] [PubMed] [Google Scholar]
- 8.Goffin, C., and Ghuysen, J. M. (1998) Microbiol. Mol. Biol. Rev. 62 1079-1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauvage, E., Kerff, F., Terrak, M., Ayala, J. A., and Charlier, P. (2008) FEMS Microbiol. Rev. 32 234-258 [DOI] [PubMed] [Google Scholar]
- 10.Marrec-Fairley, M., Piette, A., Gallet, X., Brasseur, R., Hara, H., Fraipont, C., Ghuysen, J. M., and Nguyen-Distèche, M. (2000) Mol. Microbiol. 37 1019-1031 [DOI] [PubMed] [Google Scholar]
- 11.Spratt, B. G., Zhou, J., Taylor, M., and Merrick, M. J. (1996) Mol. Microbiol. 19 639-640 [DOI] [PubMed] [Google Scholar]
- 12.Lovering, A. L., de Castro, L. H., Lim, D., and Strynadka, N. C. (2007) Science 315 1402-1405 [DOI] [PubMed] [Google Scholar]
- 13.Yuan, Y., Barrett, D., Zhang, Y., Kahne, D., Sliz, P., and Walker, S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5348-5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perlstein, D. L., Zhang, Y., Wang, T. S., Kahne, D. E., and Walker, S. (2007) J. Am. Chem. Soc. 129 12674-12675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett, D., Wang, T. S., Yuan, Y., Zhang, Y., Kahne, D., and Walker, S. (2007) J. Biol. Chem. 282 31964-31971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz, B., Markwalder, J. A., Seitz, S. P., Wang, Y., and Stein, R. L. (2002) Biochemistry 41 12552-12561 [DOI] [PubMed] [Google Scholar]
- 17.Terrak, M., Ghosh, T. K., van Heijenoort, J., Van Beeumen, J., Lampilas, M., Aszodi, J., Ayala, J. A., Ghuysen, J. M., and Nguyen-Distèche, M. (1999) Mol. Microbiol. 34 350-364 [DOI] [PubMed] [Google Scholar]
- 18.Barrett, D., Leimkuhler, C., Chen, L., Walker, D., Kahne, D., and Walker, S. (2005) J. Bacteriol. 187 2215-2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zawadzka-Skomial, J., Markiewicz, Z., Nguyen-Distèche, M., Devreese, B., Frère, J. M., and Terrak, M. (2006) J. Bacteriol. 188 1875-1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terrak, M., and Nguyen-Distèche, M. (2006) J. Bacteriol. 188 2528-2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamaki, S., Nakajima, S., and Matsuhashi, M. (1977) Proc. Natl. Acad. Sci. U. S. A. 74 5472-5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki, H., Nishimura, Y., and Hirota, Y. (1978) Proc. Natl. Acad. Sci. U. S. A. 75 664-668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki, H., van Heijenoort, Y., Tamura, T., Mizoguchi, J., Hirota, Y., and van Heijenoort, J. (1980) FEBS Lett. 110 245-249 [DOI] [PubMed] [Google Scholar]
- 24.Bertsche, U., Breukink, E., Kast, T., and Vollmer, W. (2005) J. Biol. Chem. 280 38096-38101 [DOI] [PubMed] [Google Scholar]
- 25.Bertsche, U., Kast, T., Wolf, B., Fraipont, C., Aarsman, M. E., Kannenberg, K., von Rechenberg, M., Nguyen-Distèche, M., den Blaauwen, T., Höltje, J. V., and Vollmer, W. (2006) Mol. Microbiol. 61 675-690 [DOI] [PubMed] [Google Scholar]
- 26.Vollmer, W., von Rechenberg, M., and Höltje, J. V. (1999) J. Biol. Chem. 274 6726-6734 [DOI] [PubMed] [Google Scholar]
- 27.Müller, P., Ewers, C., Bertsche, U., Anstett, M., Kallis, T., Breukink, E., Fraipont, C., Terrak, M., Nguyen-Distèche, M., and Vollmer, W. (2007) J. Biol. Chem. 282 36394-36402 [DOI] [PubMed] [Google Scholar]
- 28.Barrett, D. S., Chen, L., Litterman, N. K., and Walker, S. (2004) Biochemistry 43 12375-12381 [DOI] [PubMed] [Google Scholar]
- 29.Coutinho, P. M., Deleury, E., Davies, G. J., and Henrissat, B. (2003) J. Mol. Biol. 328 307-317 [DOI] [PubMed] [Google Scholar]
- 30.Hara, H., and Suzuki, H. (1984) FEBS Lett. 168 155-160 [DOI] [PubMed] [Google Scholar]
- 31.van Heijenoort, Y., Gomez, M., Derrien, M., Ayala, J., and van Heijenoort, J. (1992) J. Bacteriol. 174 3549-3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakaye, B., Damblon, C., Jamin, M., Galleni, M., Lepage, S., Joris, B., Marchand-Brynaert, J., Frydrych, C., and Frère, J. M. (1994) Biochem. J. 300 141-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.