Abstract
P-selectin glycoprotein ligand-1 (PSGL-1) interacts with selectins to support leukocyte rolling along vascular wall. L- and P-selectin bind to N-terminal tyrosine sulfate residues and to core-2 O-glycans attached to Thr-57, whereas tyrosine sulfation is not required for E-selectin binding. PSGL-1 extracellular domain contains decameric repeats, which extend L- and P-selectin binding sites far above the plasma membrane. We hypothesized that decamers may play a role in regulating PSGL-1 interactions with selectins. Chinese hamster ovary cells expressing wild-type PSGL-1 or PSGL-1 molecules exhibiting deletion or substitution of decamers with the tandem repeats of platelet glycoprotein Ibα were compared in their ability to roll on selectins and to bind soluble L- or P-selectin. Deletion of decamers abrogated soluble L-selectin binding and cell rolling on L-selectin, whereas their substitution partially reversed these diminutions. P-selectin-dependent interactions with PSGL-1 were less affected by decamer deletion. Videomicroscopy analysis showed that decamers are required to stabilize L-selectin-dependent rolling. Importantly, adhesion assays performed on recombinant decamers demonstrated that they directly bind to E-selectin and promote slow rolling. Our results indicate that the role of decamers is to extend PSGL-1 N terminus far above the cell surface to support and stabilize leukocyte rolling on L- or P-selectin. In addition, they function as a cell adhesion receptor, which supports ∼80% of E-selectin-dependent rolling.
Selectins play a central role in regulating leukocyte migration into inflammatory lesions by mediating interactions between leukocytes, endothelial cells, and platelets (1, 2). L-selectin is expressed by most leukocytes, whereas P- and E-selectin expression is induced on activated platelets and/or endothelial cells (3). Early in inflammation (4), P-selectin supports leukocyte rolling by binding to its main ligand P-selectin glycoprotein ligand 1 (PSGL-1)2 (5). PSGL-1 is expressed on microvilli of most leukocytes. This homodimeric mucin-like glycoprotein (6, 7) also binds to L- and E-selectin (8, 9). Interactions between PSGL-1 and L-selectin promote rolling of free flowing leukocytes on leukocytes already adherent to the vascular wall (10). By interacting with PSGL-1 and other ligands, E-selectin supports leukocyte slow rolling along inflamed endothelium (11, 12).
Selectin binding to PSGL-1 N terminus has been extensively documented. By creating binding sites for P- and L-selectin, fucosylated core-2 O-glycans linked to Thr-57 and sulfated Tyr-46, -48, and -51 play a critical role in regulating PSGL-1 interactions with P- and L-selectin (13-18). E-selectin binding sites on PSGL-1 have not been completely characterized. Core-2 O-glycans linked to Thr-57 partially support E-selectin-dependent rolling, whereas tyrosine sulfate residues are not required (15, 19, 20). Flow adhesion assays performed on PSGL-1 T57A/μ chimera, in which Thr-57 was substituted by Ala, showed that an important part (∼75%) of E-selectin-mediated rolling is dependent on fucosylated carbohydrates distinct from core-2 O-glycans linked to Thr-57 (20). However, the involved structures have not been identified.
A large part of PSGL-1 extracellular domain is constituted by a mucin-like domain, which contains 14-16 decamers (7, 21, 22), the function of which has not been investigated yet. Decamers are characterized by repeated stretches of 10 amino acids containing numerous O-glycosylated threonines (∼30%) and prolines (10%), which elongate and strengthen the protein backbone and separate N-terminal selectin-binding sites from the cell membrane. We show here that deletion of decamers impairs L- and P-selectin-dependent interactions with PSGL-1. L-selectin-dependent rolling, in particular, is strongly affected. By performing rolling assays on recombinant decamers, we show for the first time that decamers directly interact with E-selectin, and that they support ∼80% of leukocyte rolling on E-selectin. However, they do not directly interact with L- or P-selectin. Taken together, our results show that both PSGL-1 decamers and N terminus mediate its interactions with L-, P-, and E-selectin. Moreover, decamers play a critical role in stabilizing L-selectin-dependent rolling and promoting slow rolling on E-selectin.
EXPERIMENTAL PROCEDURES
Antibodies and Chimeric Selectins—Monoclonal antibodies HECA-452 (anti-CLA, ATCC HB-11485) and CSLEX-1 (antisialyl Lewis x, ATCC HB-10135) were purified from hybridoma culture medium. Anti-PSGL-1 mAb PL2 was purchased from Coulter Immunotech (Marseille, France), and the PE-labeled KPL-1 was from BD Bioscience Pharmingen (Heidelberg, Germany). Isotypic mouse IgG1 control mAb, goat anti-mouse Ig-PE F(ab′)2, goat anti-human IgM-fluorescein isothiocyanate were purchased from DakoCytomation AG (Zug, Switzerland) and goat anti-human IgM from Caltag (Burlingame, CA). The hybridoma secreting PS5 mAb was generated in our laboratory by fusion of PX63Ag8 myeloma cells with Balb/c splenocytes that had been immunized with KG-1 cells (20). P-selectin/IgM heavy chain (P-selectin/μ) and L-selectin/μ chimeras were produced by stably transfected CHOdhFr- cells and used for immunostaining or cell rolling assays as described (8, 20, 23). Recombinant human P-selectin was purchased from R&D Systems (Abingdon, UK).
cDNA Constructs and Transfectants—Core-2 β1,6N-acetylglucosaminyltransferase (C2GlcNAcT-I) and α1,3-fucosyltransferase-VII (FucT-VII) cDNA sequences were inserted in pIRES ZeoSV vector, as described previously (18). PSGL-1 cDNA sequence (7) was inserted in pcDNA3 vector (Invitrogen).
PSGL-1/GPIbα (PSGL/GP) was obtained by introducing a BamHI and a KpnI restriction site before and after the decameric repeats of PSGL-1 and by exchanging the 450-bp sequence encoding PSGL-1 decamers (Ala-118 to Thr-267) by the sequence of platelet GPIbα macroglycopeptide (C variant, Ser-312 to Thr-461) (24). Restriction sites were introduced in PSGL-1 sequence using the QuikChange site-directed mutagenesis kit (Stratagene, Amsterdam, The Netherlands). Sequences of the forward and reverse primers used to generate a BamHI restriction site before the first decameric repeat were: 5′-CCT GTC CAC GGA TGG ATC CGC TAT GGA GAT AC-3′ and 5′-GTA TCT CCA TAG CGG ATC CAT CCG TGG ACA GG-3′. Primers used to generate a KpnI restriction site after the 15th decameric repeat were: 5′-CCA TGG AAC CTA CTG GTA CCA GAG GTC TGT TC-3′ and 5′-GAA CAG ACC TCT GGT ACC CGT AGG TTC CAT GG-3′. The macroglycopeptide region of human platelet GPIbα was amplified by PCR, using genomic DNA extracted from bone marrow mononuclear cells as template with the following primers: forward 5′-GCT AGG ATC CTC ATG GTC CAC TGC TTC TCT AGA C-3′ (containing a BamHI restriction site) and reverse 5′-CGA TGG TAC CGG TGG ATT CTA AGA GTG ATA CGG G-3′ (containing a KpnI restriction site). The amplification was conducted for 30 cycles at 62 °C using Pfu Turbo DNA polymerase (Stratagene). The 465-bp PCR product encoding the macroglycopeptide region of GPIbα was then digested with BamHI and KpnI and inserted in the restriction sites introduced by site-directed mutagenesis in PSGL-1 sequence.
PSGL-1 devoid of decameric repeats (PSGL/ΔDR) was obtained by amplifying the 351-bp 5′-sequence encoding the region located upstream of the decameric repeats with the forward primer: 5′-GCG GGA TCC AGC CAT GCC TCT GCA ACT CCT CCT-3′ and the reverse primer 5′-GCT GGA TCC ACT TAC CTG CTG AAT CCG TGG ACA G-3′ containing a splice donor site. The 408-bp 3′-sequence encoding PSGL-1 region located downstream of the decamers was amplified with the sense primer 5′-GCG GAA TTC CAG GTA CCA AAA GAG GTC TGT TC-3′ containing a splice acceptor site and the antisense primer 5′-CGT TCT AGA GAG CTA AGG GAG GAA GCT GTG-3′. Both PCR products were inserted in pCR-Blunt vector (Invitrogen), then subcloned and ligated in pcDNA3 vector (Invitrogen). The donor and acceptor splice sites were separated by a 73-bp non-coding sequence.
Wild-type PSGL-1/μ (WT PSGL/μ) chimera was generated by amplifying the sequence encoding the whole extracellular part of PSGL-1 (from Met-1 to Lys-308), as described (20). The cDNA sequence of PSGL/DR/μ chimera, encoding decamers (from Ala-118 to Thr-267) coupled to IgM heavy chain μ, was constructed by deleting WT PSGL/μ sequence encoding the propeptide and the sequence located upstream of the decameric repeats. A PvuII restriction site was introduced before PSGL-1 decamers by site-directed mutagenesis (Stratagene) using the following forward and reverse primers: 5′-GAA CCT GTC CAC GGA TCA GCT GGC TAT GGA GAT ACA GAC C-3′ and 5′-GGT CTG TAT CTC CAT AGC CAG CTG ATC CGT GGA CAG GTT C-3′. Because a PvuII restriction site is present at the beginning of the propeptide sequence, the propeptide and the N-terminal sequence (from Leu-20 to Ala-117) were deleted by PvuII digestion of WT PSGL/μ. PSGL/DR/μ chimera cDNA sequence was constructed by ligating the PvuII-digested DNA fragments, allowing the fusion of PSGL-1 signal sequence to the decamer sequence. PSGL/ΔDR/μ chimera was constructed by amplifying PSGL/ΔDR with the following primers: forward 5′-TCG CGA TAT CAA GCT TCT CGA GCC ACC ATG CCT CTG CAA CTC CTC-3′ and reverse 5′-TAT AGA TAT CAT CGA TAC CTG AGA TGT GGT CTG GGG C-3′, the latter introducing a ClaI restriction site and a splice donor site allowing the fusion of the amplified sequence to IgM heavy chain.
Cells and Transfections—CHOdhFr- cells (ATCC number: CRL 9096) expressing FucT-VII and C2GlcNAcT-I (18) were stably transfected with cDNAs encoding wild-type or mutant PSGL-1 using TransIT reagent (Mirus, Madison, WI) according to the manufacturer's protocol. Transfected CHO cells were cultured in minimal essential medium α containing 10% fetal bovine serum, hypoxanthine, and thymidine supplement (Invitrogen), 400 μg/ml G418 and 200 μg/ml Zeocin (Invitrogen). Individual clones expressing similar levels of the various forms of PSGL-1 and sLex were isolated by limiting dilution and screened by flow cytometry analysis using KPL-1 and CSLEX-1 mAbs.
Neutrophils were isolated from normal heparinized blood samples by Ficoll-Paque centrifugation, dextran sedimentation, and erythrocyte hypotonic lysis. K562-E-selectin cells (K562-E cells) and 300.19-L-selectin cells (300.19-L cells) were cultured in RPMI medium and CHO-P-selectin cells (CHO-P cells) in minimal essential medium α, containing 400 μg/ml G418 (Invitrogen) as described previously (20).
Immunophenotypic Analysis—PSGL-1, sLex, and CLA expressions were assessed by incubating cells with saturating concentrations of appropriate unlabeled mAbs, followed by PE-conjugated goat anti-mouse Ig. Cell staining with chimeric molecules was achieved by incubating chimeras with fluorescein isothiocyanate-conjugated rabbit anti-human IgM (20 μg/ml). The concentrations of P- and L-selectin/μ chimeras were measured by enzyme-linked immunosorbent assay (20). Chimeric proteins were suspended in RPMI 1640/1% fetal bovine serum, and binding specificity was assessed by its abrogation in the presence of 10 mm EDTA or anti-P- or -L-selectin blocking mAbs (WAPS 12.2 or LAM1-3). A total of 5000 cells was analyzed in each experiment. Flow cytometry was performed using a Cytomics™ FC 500 cytofluorometer (Beckman Coulter).
Epitopes present on chimeric proteins were identified by analyzing PSGL-1, sLex, and CLA expressions by flow cytometry. 10-μm polystyrene microspheres (2.5 × 107 microspheres/ml, Polysciences, Eppelheim, Germany) were coated overnight at 4 °C on a rotating wheel with goat anti-human IgM (Caltag), then blocked in phosphate-buffered saline/2% bovine serum albumin for 1 h and subsequently incubated with 2.5 μg of chimeric protein in 50 μl of phosphate-buffered saline (20). PSGL-1 epitopes were identified by incubating microspheres coated with captured chimeras with specific antibodies and PE-conjugated goat anti-mouse Ig. Immunostaining was analyzed by flow cytometry.
Cell Lysis and Western Blotting—106 CHO cells, expressing WT or mutant PSGL-1, were washed and lysed for 20 min on ice in 1 ml of lysis buffer (25 mm Tris-HCl, pH 7.6, containing 150 mm NaCl, Triton 1%, and protease inhibitors). After centrifugation of cell lysates, twice for 15 min at 2000 rpm at 4 °C, supernatants were boiled for 5 min in sample buffer containing 5% (v/v) β-mercaptoethanol and analyzed by electrophoresis on SDS-polyacrylamide gel and Western blotting. After transfer, nitrocellulose membranes (Bio-Rad Laboratories, Reinach, Switzerland) were incubated for 1 h in Tris-buffered saline-Tween 20 containing 5% (w/v) milk and then with the anti-PSGL-1 mAb PS5 (0.1 μg/ml), overnight at 4 °C. Recombinant decamers, PSGL/DR/μ chimera, were detected with PL2 mAb (0.5 μg/ml). mAb binding was revealed with horse radish peroxidase-conjugated sheep anti-mouse Ig (1/10,000) for 30 min at room temperature and chemiluminescence (ECL Plus, Amersham Biosciences). Extensive washes in Tris-buffered saline-Tween 20 (0.1% v/v) were performed between all steps.
Cell Rolling Assays—A laminar flow was generated in a parallel plate flow chamber (GlycoTech Corp., Rockville, MD) mounted on a glass coverslip (Polylabo SA, Plan-les-Ouates, Switzerland). Coverslips were coated with a confluent monolayer of transfected CHO cells or with recombinant P-selectin or L-selectin/μ chimera (0.25 μg) adsorbed on goat anti-human IgM heavy chain antibody (1 μgin50 μl of 0.1 m borate buffer, pH 8.0, on an area of 38 mm2) (20). Cells (0.5-1 × 106/ml in RPMI 1640/1% fetal bovine serum) were perfused through the chamber using a syringe pump (Harvard Apparatus, Indulab AG, Gams, Switzerland) under constant shear stress. Rolling cells were visualized using a phase contrast microscope (Leica Leitz DM IL, Renens, Switzerland) and a high resolution video camera (Sanyo CCD, Japan). Images were recorded on an S-VHS recorder (Panasonic MD830, Telecom Lausanne, Switzerland), and velocities were measured using a digital image analysis system (Mikado Software, GPL SA, Martigny, Switzerland) (23). Cell rolling interactions were analyzed from videotapes at 2-7 min of perfusion. Cell recruitment was counted in at least 24 microscopic fields of 0.25 mm2. Rolling interactions were analyzed when the interaction time was ≥1 s and when cell displacement during 20 s was ≥1 cell diameter. Rolling velocities illustrated in Fig. 4 were measured every 0.25 s by tracking individual cells in the flow direction within 0.25-mm2 microscopic fields. Cell displacements illustrated in Fig. 5 were measured every 0.05 s, over 1-s observation periods. The median velocity of tracked cells was included between percentiles 40 and 60 of the velocity of cell populations illustrated in Fig. 4D. The S.D. value of the median velocity of each tracked cell, an indicator of the variation of cell rolling velocity, was used to calculate the mean ± S.D. of each studied cell population. 156-236 independent determinations of frame-by-frame displacements were measured for each tested condition. In each assay, CHO-WT and mutant PSGL-1 cells expressed similar levels of PSGL-1.
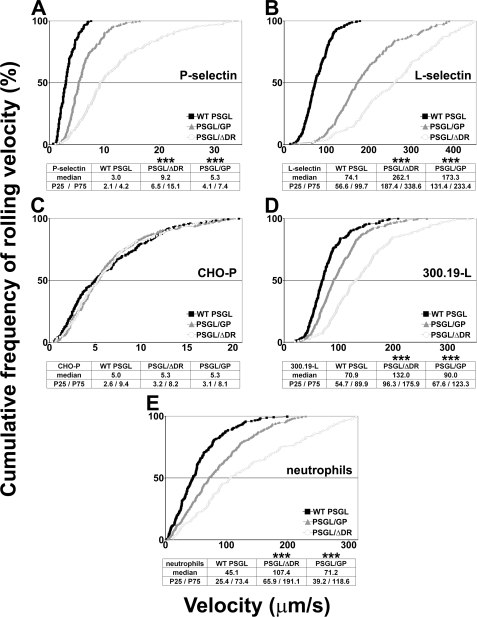
FIGURE 4.
Decamers are involved in regulating PSGL-1-dependent rolling velocities on P- and L-selectin. Rolling velocity of CHO transfectants perfused under a constant shear stress of 1.5 dyn/cm2 on recombinant P-selectin (A) or L-selectin/μ chimera (B). Rolling velocity of CHO-P-selectin cells (C), 300.19-L-selectin cells (D), or neutrophils (E) on CHO transfectants. Rolling velocities were assessed at 2-7 min of perfusion; curves represent 148-367 independent determinations of cell velocity within 3-4 independent experiments. *, statistically significant difference from CHO cells expressing WT PSGL-1 (***, p < 0.001).
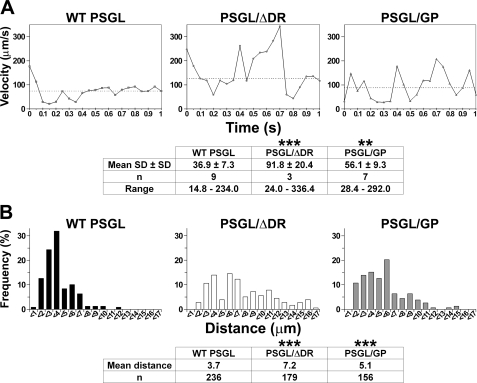
FIGURE 5.
Decamers are involved in stabilizing L-selectin-dependent rolling on PSGL-1. A, frame by frame rolling velocity of 300.19-L cells on CHO cells stably expressing WT PSGL, PSGL/GP, or PSGL/ΔDR. Each panel illustrates the rolling velocity of one representative cell tracked each 50 ms for 1 s on one transfectant. The median cell rolling velocity of represented cell was within percentiles 40 and 60 of velocity curves illustrated in Fig. 4D and is indicated by a dashed line. B, distribution of distances traveled by 300.19-L cells on CHO transfectants during successive 50-ms time lapses. Data are representative of 156-236 observations. *, statistically significant difference from CHO cells expressing WT PSGL-1 (**, p < 0.01; ***, p < 0.001).
Despite small differences in PSGL-1 expression, cell recruitment was not correlated to PSGL-1 expression levels. Several CHO-WT PSGL clones were compared in their ability to support CHO-P-selectin cell rolling. PE-labeled KPL-1 mAb was used to assess PSGL-1 expression; the mean fluorescence intensity of clone 1 was 2.8, clone 2 was 5.2, and clone 3 was 16.1. Cell recruitment/min/mm2 (mean ± S.E.) did not significantly differ among these clones: clone 1, 155 ± 10; clone 2, 180 ± 15; and clone 3, 181 ± 1. Similar observations were made with 300.19-L-selectin cells (clone 1, 1408 ± 76; clone 2, 1691 ± 86; and clone 3, 1657 ± 77). These observations suggest that the heterogeneity in PSGL-1 expression among tested clones does not significantly affect L- or P-selectin-dependent rolling on CHO-PSGL-1 cells, as long as a minimal level of glycosylated PSGL-1 is expressed at cell surface.
Statistical Analysis—Kruskal-Wallis nonparametric analysis of variance followed by Dunn's multiple comparisons test were used to determine statistical significance of difference between groups. p values < 0.05 were considered significant.
RESULTS
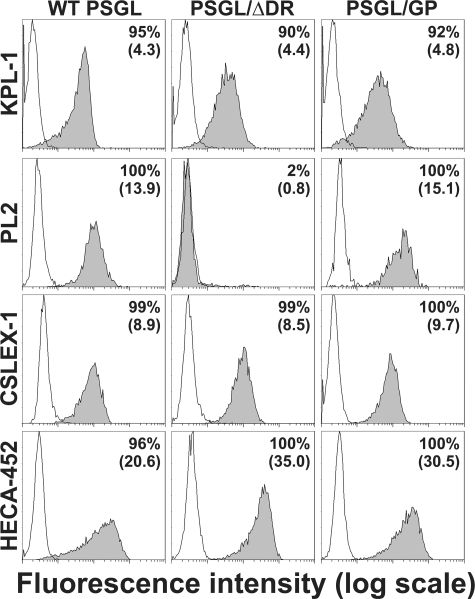
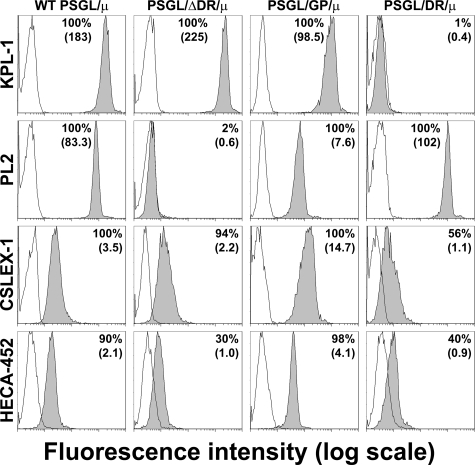
Wild-type and Mutant PSGL-1—cDNAs of WT and mutant PSGL-1 were expressed in CHO cells expressing C2GlcNAcT-I and FucT-VII (18, 20). They expressed similar levels of sLex and CLA (Fig. 1). PSGL-1 expression was examined by flow cytometry using 1) KPL-1 mAb, which binds to PSGL-1 N terminus, 2) PL2 mAb that reacts with the extracellular part of PSGL-1 containing decamers, 3) the anti-sLex mAb CSLEX-1, and 4) the anti-CLA mAb HECA-452 (Fig. 1). As expected, KPL-1 mAb bound to WT PSGL, PSGL/ΔDR, and PSGL/GP N terminus, whereas deletion of decamers in PSGL/ΔDR abrogated PL2 binding. Despite the absence of decamers, PL2 mAb reacted with PSGL/GP, suggesting that PL2 might also bind to amino acids located upstream of decamers. Reverse transcription-PCR analysis and cDNA sequencing confirmed the absence of decamers in CHO cells expressing PSGL/GP and PSGL/ΔDR (data not shown). Moreover, anti-CD42b mAb (Dako) strongly reacted with CHO cells expressing GPIbα, whereas PL2 mAb did not.
FIGURE 1.
Expression of PSGL-1, sLex, and CLA by CHO-PSGL-1 transfectants. Immunostaining of CHO cells stably expressing C2GlcNAcT-I, FucT-VII, and WT or mutant PSGL-1 with isotype-matched control mAb (white histograms) or with the anti-PSGL-1 (KPL-1 or PL2), anti-sLex (CSLEX-1), or anti-CLA (HECA-452) mAbs (gray histograms). The proportion of positive cells and mean fluorescence intensity are indicated on each histogram.
WT PSGL and PSGL/GP (with substituted decamers) migrated with similar apparent molecular masses on SDS-polyacrylamide gel (∼121 kDa and ∼130 kDa, respectively, data not shown). In contrast, PSGL/ΔDR (without decamers) was much smaller (∼48 kDa).
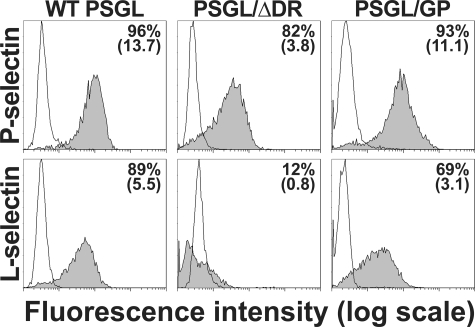
PSGL-1 Decamers Are Required For Optimal Binding of L-selectin—The involvement of decamers in regulating P- or L-selectin interactions with PSGL-1 was first examined by comparing P- and L-selectin/μ chimera binding to CHO cells expressing WT PSGL, PSGL/GP, or PSGL/ΔDR.
P- and L-selectin/μ chimeras efficiently bound to CHO cells expressing WT PSGL (P- versus L-selectin/μ binding (mean fluorescence intensity ± S.E.): 13.0 ± 0.7 versus 6.9 ± 1.4, (Fig. 2). Decamer deletion more strongly affected L- than P-selectin binding. L-selectin/μ binding to CHO-PSGL/ΔDR was almost abrogated (90% of inhibition), whereas P-selectin/μ binding was diminished by 70%. GPIbα macroglycopeptide substitution for decamers almost restored P-selectin/μ binding (18% of decrease compared with WT PSGL), whereas L-selectin binding remained diminished (55% of decrease).
FIGURE 2.
Role of PSGL-1 decamers in regulating P- (upper panel) and L-selectin (lower panel) binding to CHO transfectants. Binding of P- and L-selectin/μ chimeras (gray histograms) was abolished by 10 mm EDTA (white histograms). The proportion of positive cells and mean fluorescence intensity are indicated on each panel. The figure illustrates results from one representative experiment out of two. Chimeric selectins did not significantly bind (<2%) to mock-transfected CHO cells (not illustrated).
PSGL-1 Decamers Regulate L- and P-selectin-dependent Rolling—The involvement of decamers in regulating PSGL-1-dependent rolling on P- or L-selectin was assessed under a constant shear stress of 1.5 dyn/cm2 (Fig. 3). The ability of CHO cells expressing WT or mutant PSGL-1 to roll on L- or P-selectin was compared side by side (Fig. 3, A and B). Additional assays were performed in the reverse setting to examine L- or P-selectin-dependent rolling on WT or mutant PSGL-1 (Fig. 3, C and D). These experiments mimic the interactions of flowing leukocytes with adherent leukocytes, activated platelets or inflamed endothelium.
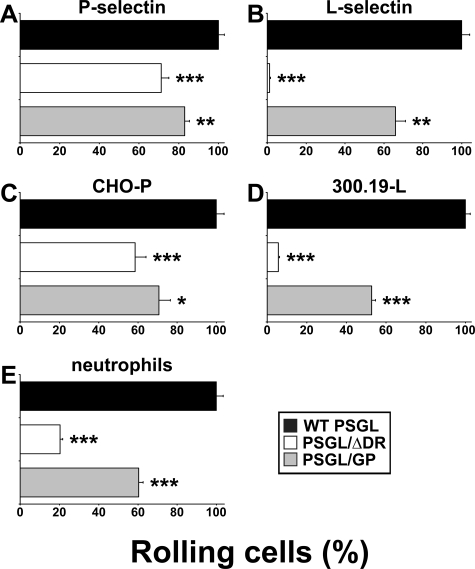
FIGURE 3.
Decamers are involved in regulating P- and L-selectin-dependent recruitment on PSGL-1. CHO transfectants (0.5 × 106/ml) were perfused under a constant shear stress of 1.5 dyn/cm2 on recombinant P-selectin (A) or L-selectin/μ chimera adsorbed on glass coverslips coated with goat anti-human IgM (B). C, CHO-P-selectin cells; D, 300.19-L-selectin cells; or E, neutrophils were perfused under a constant shear stress of 1.5 dyn/cm2 on confluent monolayers of CHO transfectants. Cell recruitment was assessed by videomicroscopy. Results represent the mean ± S.E. of 3-4 independent experiments. Error bars represent the S.E. *, represents statistically significant difference from CHO cells expressing WT PSGL-1 (*, p < 0.05; **, p < 0.01; and ***, p < 0.001).
Transfectants strongly differed in their rolling capacity. CHO-WT PSGL cells were more efficiently recruited on P-selectin (mean number of rolling cells/min/mm2 ± S.E.: 163 ± 5) than CHO-PSGL/ΔDR cells (116 ± 6; 38% inhibition, p < 0.001). Replacement of PSGL-1 decamers by GPIbα macroglycopeptide partially restored cell rolling (135 ± 4; 17% inhibition, p < 0.01). Similar results were obtained in the reverse setting, when CHO-P cells were perfused on a monolayer of CHO transfectants (Fig. 3C). The mean number of rolling cells/min/mm2 ± S.E. on CHO-WT PSGL was 226 ± 9. The deletion of decamers reduced rolling by 41% (137 ± 13, p < 0.001), whereas substitution of decamers by GPIbα tandem repeats did not fully restore CHO cell rolling (153 ± 13; 29% inhibition, p < 0.05). The strong decrease in cell recruitment observed in the absence of decamers was not due to a difference in PSGL-1 expression, as CHO-PSGL/ΔDR cell recruitment was compared with that of CHO-WT PSGL cells expressing the same level of cell surface PSGL-1.
Cell rolling on L-selectin/μ was strongly affected by decamer deletion (Fig. 3B). CHO-PSGL/ΔDR were poorly recruited on L-selectin/μ chimera compared with CHO-WT PSGL cells (mean number of rolling cells/min/mm2 ± S.E.: 161 ± 8 versus 1 ± 1; 99% inhibition, p < 0.001). The replacement of decamers by GPIbα macroglycopeptide partially restored CHO-PSGL/GP cell recruitment on L-selectin (109 ± 10; 34% inhibition, p < 0.01). Similar results were obtained in the reverse setting (Fig. 3D). 300.19-L cells were strongly recruited on CHO-WT PSGL (1683 ± 42 cells/min/mm2). In contrast, L-selectin-dependent cell recruitment was almost abrogated on CHO-PSGL/ΔDR (95 ± 9; 94% inhibition, p < 0.001). The replacement of PSGL-1 decamers by GPIbα macroglycopeptide partially restored cell rolling (886 ± 32; 47% inhibition, p < 0.001).
Additional experiments were performed with neutrophils to examine whether observations made with transfectants were reproducible with physiological cells (Fig. 3E). Neutrophil rolling was strongly decreased on CHO-PSGL/ΔDR (158 ± 10 rolling cells/min/mm2; 80% inhibition, p < 0.001) compared with that observed on CHO-WT PSGL cells (778 ± 26). It was only partially restored on CHO-PSGL/GP cells (470 ± 17; 40% inhibition, p < 0.001).
Decamers Are Involved in Regulating Cell Rolling Velocity on L- and P-selectin—CHO-PSGL/ΔDR rolled much faster than CHO-WT PSGL cells on P-selectin (median: 9.2 versus 3.0 μm/s, Fig. 4A) and L-selectin (262.1 μm/s versus 74.1 μm/s, Fig. 4B). PSGL-1 decamer substitution by GPIbα tandem repeats slowed down CHO cell rolling velocities on L-selectin (median: 173.3 μm/s, Fig. 4B). However, they remained higher than that of CHO-WT PSGL cells. Similarly, CHO-PSGL/GP rolled at intermediate velocities on P-selectin (5.3 μm/s, Fig. 4A).
In the reverse setting, 300.19-L cells rolled slower on CHO-WT PSGL (median: 70.9 μm/s) than on CHO-PSGL/ΔDR cells (median: 132.0 μm/s, Fig. 4D). Intermediate velocities were measured on CHO-PSGL/GP (median: 90.0 μm/s). Rolling velocities of CHO-P cells were slow on all transfectants and did not differ significantly (median: 5.1 μm/s on CHO-WT PSGL versus 5.3 μm/s on CHO-PSGL/GP and CHO-PSGL/ΔDR, Fig. 4C).
Neutrophils rolled much faster on CHO-PSGL/ΔDR (median: 107.4 μm/s, Fig. 4E) than on CHO-WT PSGL cells (median: 45.1 μm/s); intermediate velocities were measured on CHO-PSGL/GP (median: 71.2 μm/s).
PSGL-1 Decamers Stabilize L-selectin-dependent Cell Displacements—The stability of L-selectin-dependent rolling velocities on PSGL-1 was assessed by tracking 300.19-L cells every 50 ms for 1 s. In Fig. 5A, each peak corresponds to an acceleration, whereas each valley indicates a deceleration. The dashed line indicates the median velocity, which was included within percentiles 40 and 60 of the velocity curves illustrated in Fig. 4D. The stability of rolling velocity is represented by the mean S.D. ± S.D. of tracked cell rolling velocities (20). Cell displacements were very unstable in the absence of decamers; intermediate values were measured on CHO-PSGL/GP cells (mean S.D. ± S.D. on CHO-WT PSGL: 36.9 ± 7.3 μm/s versus 56.1 ± 9.3 μm/s on CHO-PSGL/GP, p < 0.01 versus 91.8 ± 20.4 μm/s on CHO-PSGL/ΔDR, p < 0.001).
The distribution of travel distances illustrated in Fig. 5B was assessed by measuring cell displacements within successive 50-ms periods (156-236 determinations). Rolling stability was correlated with shorter rolling distances (70% of 300.19-L cells rolled <4 μm within 50-ms periods on CHO-WT PSGL). Longer rolling distances were observed in the absence of decamers. Substitution of decamers by GPIbα macroglycopeptide poorly stabilized cell displacements. Thus, 37 and 15% of 300.19-L cells rolled >8 μm within 50-ms periods on CHO-PSGL/ΔDR and CHO-PSGL/GP, respectively, whereas only 3% did so on CHO-WT PSGL (Fig. 5B).
PSGL-1 Decamers Play a Key Role in Supporting E-selectin-dependent Rolling—Additional experiments were performed to examine the contribution of PSGL-1 N terminus versus decamers in supporting selectin-dependent rolling. WT and mutant PSGL-1/μ chimeras were produced by CHO cells. WT PSGL/μ and PSGL/GP/μ migrated with similar molecular mass (∼145 and ∼150 kDa, respectively), whereas PSGL/ΔDR/μ and PSGL/DR/μ chimeras migrated as bands of ∼82 and ∼130 kDa, respectively. As expected, PSGL/DR/μ chimera, which expresses recombinant decamers, did not react with the anti-PSGL-1 mAb PS5, whereas PL2 that reacts with decamers did not bind to PSGL/ΔDR/μ chimera (data not shown).
WT PSGL/μ, PSGL/GP/μ, PSGL/ΔDR/μ, and PSGL/DR/μ chimeras were coated on polystyrene microspheres to examine KPL-1, PL2, CSLEX-1, and HECA-452 epitopes by flow cytometry (Fig. 6). WT PSGL/μ, PSGL/GP/μ, and PSGL/ΔDR/μ chimeras exhibited a similar reactivity with KPL-1 mAb. As expected, KPL-1 did not bind to PSGL/DR/μ chimera, which does not express PSGL-1 N terminus. On the other hand, PL2 epitope was present on all chimeras, except PSGL/ΔDR/μ. CSLEX-1 and HECA-452 mAbs reacted with the four PSGL-1/μ chimeras.
FIGURE 6.
Immunophenotypic analysis of PSGL-1, sLex, and CLA expression by PSGL-1 chimeras. The proportion of positive microspheres and mean fluorescence intensity are indicated on each histogram. White histograms: isotype-matched control immunoglobulin. The figure illustrates results from one representative experiment out of three.
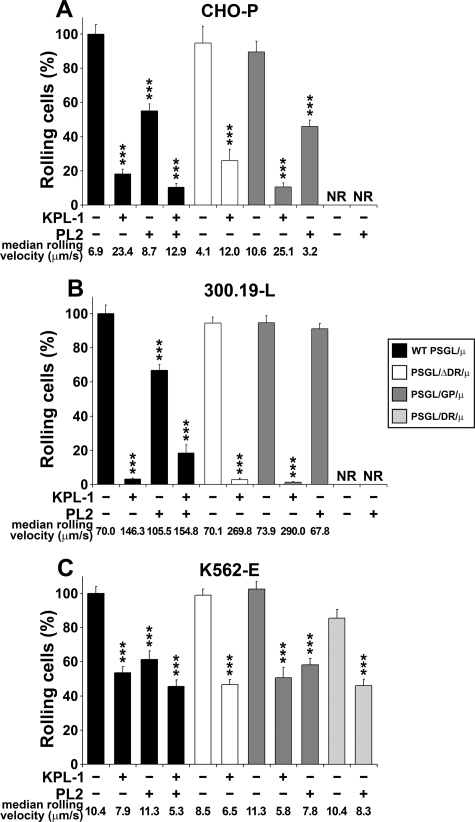
WT PSGL/μ, PSGL/GP/μ, PSGL/ΔDR/μ, and PSGL/DR/μ chimeras were adsorbed on glass coverslips to perform flow adhesion assays. CHO-P cells were similarly recruited on WT PSGL/μ, PSGL/GP/μ, or PSGL/ΔDR/μ chimeras (mean number of rolling cells/min/mm2 ± S.E.: 179 ± 18 versus 150 ± 3 versus 177 ± 10, respectively, Fig. 7A). In contrast, they did not roll on recombinant decamers, PSGL/DR/μ (Fig. 7A). KPL-1 inhibited by >75% P-selectin-dependent rolling on WT PSGL/μ, PSGL/GP/μ, and PSGL/ΔDR/μ chimeras, whereas PL2 had a partial inhibitory effect (∼40% inhibition, Fig. 7A). PSGL/μ chimeras treatment with KPL-1 mAb destabilized and strongly accelerated CHO-P cell rolling velocity (Fig. 7A). The median rolling velocity increased from 6.9 to 23.4 μm/s on WT PSGL/μ (n = 158, p < 0.001), from 10.6 to 25.1 μm/s on PSGL/GP/μ (n = 296, p < 0.001), and from 4.1 to 12.0 μm/s on PSGL/ΔDR/μ chimeras (n = 203, p < 0.001).
FIGURE 7.
Decamers participate in E-selectin-dependent interaction with PSGL-1. CHO-P-selectin (A), 300.19-L-selectin (B), or K562-E-selectin (C) cells (106/ml) were perfused under a constant shear stress of 1.0 dyn/cm2 on WT or mutant PSGL-1/μ chimeras. KPL-1 and PL2 mAbs were added alone or in combination as indicated. Results are representative of three independent experiments. *, statistically significant difference in rolling cells (%) on chimeras treated with KPL-1 and/or PL2 mAbs compared with rolling cells observed on chimeras treated with isotypic control mAb (***, p < 0.001; NR, no rolling on PSGL/DR/μ).
The number of 300.19-L cells that rolled on WT PSGL/μ, PSGL/GP/μ, or PSGL/ΔDR/μ chimeras was similar (mean number of rolling cells/min/mm2 ± S.E.: 1507 ± 114 versus 1451 ± 67 and 1354 ± 116, respectively, Fig. 7B). Cell rolling was almost abolished by KPL-1 mAb (>97% inhibition), and the median velocity of residual rolling cells was strongly increased on WT PSGL/μ (from 70.0 to 146.3 μm/s; n = 156, p < 0.001), on PSGL/ΔDR/μ (from 70.1 to 269.8 μm/s n = 130, p < 0.001), and on PSGL/GP/μ (from 73.9 to 290.0 μm/s, n = 203, p < 0.001). Treatment of WT PSGL/μ with PL2 mAb accelerated 300.19-L cell rolling velocity (from 70.0 to 105.5 μm/s; n = 236, p < 0.001). In contrast, it did not affect 300.19-L cell rolling velocity on PSGL/ΔDR/μ (from 70.1 to 63.1 μm/s, n = 352, data not shown) or PSGL/GP/μ (from 73.9 to 67.8 μm/s, n = 314). As observed with CHO-P cells, PSGL/DR/μ chimera did not support L-selectin-dependent rolling (Fig. 7B).
K562-E cell recruitment on WT PSGL/μ, PSGL/GP/μ, or PSGL/ΔDR/μ chimeras did not significantly differ (mean number of rolling cells/min/mm2: 933 ± 81 versus 940 ± 112 versus 942 ± 171, respectively, Fig. 7C). In contrast to CHO-P and 300.19-L cells, K562-E cells were efficiently recruited on PSGL/DR/μ chimera (805 ± 85 cells/min/mm2, Fig. 7C). K562-E cell recruitment on PSGL/DR/μ was slightly lower (∼14% diminution) than that observed on chimeras exhibiting a conserved PSGL-1 N-terminal sequence. Interestingly, chimera pretreatment with KPL-1 mAb slowed down K562-E cell rolling velocity on WT PSGL/μ (from 10.4 to 7.9 μm/s, n = 297, p < 0.001) and PSGL/GP/μ (from 11.3 to 5.8 μm/s, n = 294, p < 0.001), whereas it did slightly affect cell rolling velocity on PSGL/ΔDR/μ chimera (from 8.5 to 6.5 μm/s, n = 158, p < 0.01). PL2 mAb did not significantly affect K562-E cell rolling velocity on WT PSGL/μ chimera (Fig. 7C). On the other hand, it slightly decreased cell rolling velocity on PSGL/DR/μ (from 10.4 to 8.3 μm/s, n = 205, p < 0.01).
It should be noted that, because the lengths and structures of chimeric molecules (Fig. 7) and those of the extracellular domain of cell membrane PSGL-1 (Fig. 3) strongly differ, quantitative results obtained in Figs. 3 and 7 cannot be compared side by side (the length of cell membrane PSGL-1 is ∼3-fold shorter than that of PSGL-1/μ chimera bound to coverslip through a goat anti-human IgM antibody and PSGL-1/ΔDR expressed by CHO cells is ∼5.5-fold shorter than PSGL-1/ΔDR/μ). The sole aim of experiments performed with chimeras was to identify PSGL-1 domains that contribute to support selectin-dependent rolling.
DISCUSSION
Selectins and PSGL-1 play a critical role in regulating leukocyte migration into inflammatory lesions (1). The involvement of core-2 O-glycans attached to PSGL-1 Thr-57 in regulating L- and P-selectin-dependent rolling has been clearly demonstrated (14, 15, 18, 20, 25). On the other hand, the role of decamers in regulating selectin-dependent rolling has not been previously examined. Data presented here lead to the following conclusions: 1) A primary function of decamers is to extend PSGL-1 N terminus away from the cell surface to present it to flowing leukocytes/activated platelets, inflamed endothelium/adherent platelets, or leukocytes. They enhance the ability of PSGL-1 N terminus to support L- and P-selectin-dependent rolling. In addition, they play a critical role in stabilizing L-selectin-dependent rolling interactions. 2) Decamers directly interact with E-selectin and function as an adhesion receptor. In the absence of PSGL-1 N terminus, they support ∼80% of E-selectin-dependent rolling. This suggests that they play a major role in mediating E-selectin-dependent rolling.
PSGL-1 decamers are rich in threonine residues and contain at least 50 potential O-glycosylation sites. Glycosylation of decamers contributes to almost two-thirds of PSGL-1 molecular weight. The extensive O-glycosylation (7) and high proline content of decamers confer a highly extended structure to PSGL-1, which was revealed by electron microscopy analysis (26). The rod-like structure of the mucin-like domain positions PSGL-1 N terminus far above cell surface and restricts the flexibility of the peptide backbone. This suggests that a major function of the mucin domain is to provide a rigid stalk to present PSGL-1 N terminus to flowing leukocytes/platelets, adherent platelets/leukocytes, or inflamed endothelium. This hypothesis was examined by comparing rolling interactions of CHO cells expressing WT PSGL-1, a PSGL-1 form devoid of decamers (PSGL/ΔDR) or a chimeric molecule in which decamers were replaced by the mucin domain of GPIbα (PSGL/GP) (24). CHO-PSGL/ΔDR cells suboptimally supported P-selectin-dependent rolling and very inefficiently L-selectin-dependent rolling (Fig. 3). Interestingly, CHO-PSGL/GP supported P-selectin-dependent rolling almost as efficiently as CHO-WT PSGL cells. This implies that a major function of decamers is to provide a stalk of sufficient length to extend PSGL-1 N terminus away from the leukocyte cell surface to support PSGL-1 interactions with P-selectin. On the other hand, CHO-PSGL/GP cells suboptimally supported L-selectin-mediated rolling, suggesting that PSGL-1 length is not the sole parameter that promotes L-selectin binding. Other biophysical mechanisms that regulate the formation of bonds between L-selectin and PSGL-1 may play a role.
Under hydrodynamic flow conditions, bonds form rapidly between selectins and their ligands and initiate cell tethering. In the flow direction, shear forces induce the dissociation of bonds at the back of the cell, while new bonds are formed at the front, resulting in cell rolling. A minimum level of fluid shear stress is required to sustain rolling interactions (27, 28). Below the threshold level, fewer cells tether; they roll faster and begin to detach (28). Shear forces decrease L-selectin-PSGL-1 off-rates (catch bonds) between this threshold level and an optimal shear value, enabling the generation of long-lived tethers, which increases rolling stability and decreases rolling velocity (29, 30). Above optimal shear stress, the increase in off-rates (slip bonds) accelerates rolling velocity and decreases rolling stability (30). Thus, L-selectin-mediated rolling is controlled by force-dependent alterations of bond lifetimes below and above a shear optimum. Protrusion of PSGL-1 N terminus into the bloodstream might be an important mechanism by which hydrodynamic flow could modulate PSGL-1 interactions. In the absence of decamers, shear forces generated on L-selectin-PSGL-1 N terminus could not be sufficient to promote efficient leukocyte rolling and to stabilize cell displacement. This may explain why 300.19-L cells rolled at high velocities, unstably traveling much longer distances on CHO-PSGL/ΔDR than on CHO-WT PSGL cells (Fig. 5). Substitution of PSGL-1 decamers by GPIbα was not sufficient to completely recover rolling stability. This may be explained by differences in the biophysical properties of WT PSGL and PSGL/GP, in particular in their ability to form catch bonds promoting rolling stability.
By providing sufficient length and rigidity to PSGL-1, decamers play a critical role in stabilizing rolling interactions. Tandem repetition of decameric repeats has been an evolutionary mechanism used to change the biophysical properties of PSGL-1 by increasing the size of the mucin-like domain (21). A consequence is the exposure of PSGL-1 N terminus to shear forces, promoting its rapid binding to selectins and prompt initiation of leukocyte rolling at site of inflammation, without the need of exogenous modulators. The control of the lifetime of PSGL-1-selectin bonds by a threshold of fluid shear may also be important to prevent inappropriate leukocyte aggregation of flowing leukocytes exposed to low shear forces (31). A polymorphism in the number of decameric repeats has been observed in humans and other mammals (21, 22). Variants with 16, 15, and 14 decameric repeats have been reported in the Caucasian population at frequencies of ∼85, ∼14, and ∼1%, respectively. Interestingly, an association has been disclosed between expression of smaller alleles (B and C) and lower risk of developing cerebrovascular diseases (32). On the other hand, no correlation was observed with coronary heart disease or deep venous thrombosis (33-35).
Several parallels exist between PSGL-1 and GPIbα. Both molecules exhibit a mucin-like domain that separates the ligand binding region from the cell membrane. PSGL-1 and GPIbα mucin-like regions are highly glycosylated and sialylated, rich in serine, threonine, and proline residues, and contain repeated tandem sequences with a variable number of repeats that account for polymorphism (21, 22, 24). PSGL-1, like GPIbα, has N-terminal tyrosine sulfate residues that are important for ligand binding (15, 18, 36). Importantly, PSGL-1 and selectins, like GPIbα and its ligand the von Willebrand factor, require shear forces for binding. Furthermore, similarities are observed in the kinetics of PSGL-1 and GPIbα binding to their ligands. As described for PSGL-1 and selectins, catch-slip transitional bonds between GPIbα and von Willebrand factor regulate platelet adhesion (37, 38). GPIbα interactions with von Willebrand factor mediate flow-enhanced platelet adhesion on subendothelium of disrupted arterial vessels, whereas platelets do not aggregate under low shear stress of sluggish flows. Interestingly, both PSGL-1 and GPIbα can interact with P-selectin (39). However, PSGL-1 and GPIbα differ by several aspects in their interactions with their ligands. Core-2 O-glycosylation and fucosylation of PSGL-1 and the presence of calcium is required to support PSGL-1 interactions with selectins, whereas GPIbα interaction with von Willebrand factor requires neither. Like GPIbα macroglycopeptide, decamers function as a rigid spacer extending the ligand-binding site far above the plasma membrane to support leukocyte rolling on L- or P-selectin. Importantly, we show here for the first time that decamers also function as an adhesion receptor by directly interacting with E-selectin and by cooperating with PSGL-1 N terminus in supporting E-selectin-dependent rolling (Fig. 7C).
The mucin domain can function as a rigid stalk that separates the ligand-binding site from the plasma membrane, as observed for GPIbα, the human low density lipoprotein receptor (40) or fractalkine (41). However, it can also directly support adhesive interactions, as described for CD34, glycosylation-dependent cell adhesion molecule-1 (GlyCAM-1) or mucosal vascular addressin cell adhesion molecule-1 (MadCAM-1). These L-selectin ligands are highly fucosylated, sialylated, and sulfated (42). The sulfation of O-glycans allows the preferential interaction of these sialomucins with L-selectin (43). In contrast, the mucin-like domain of PSGL-1 is not sulfated. This might explain why L- and P-selectin do not interact with decamers, whereas they preferentially bind to N-terminal tyrosine sulfate residues and core-2 O-glycans linked to Thr-57. Because PSGL-1 sulfation is not required for E-selectin binding (19), E-selectin can interact with both the N-terminal selectin-binding site and with decamers (Fig. 7C). In the absence of PSGL-1 N terminus, E-selectin-dependent cell recruitment is decreased by only ∼14% indicating that decamers play a major role in supporting E-selectin-dependent cell rolling. Interestingly, KPL-1 mAb binding to PSGL-1 N terminus partially inhibits E-selectin binding and slows down E-selectin-dependent rolling, whereas it strongly accelerates L- and P-selectin-dependent rolling and decreases cell recruitment. These results suggest that both PSGL-1 N terminus and decamers interact with E-selectin and promote slow rolling on E-selectin.
Taken together, data reported here show for the first time that PSGL-1 decamers play a crucial role in regulating leukocyte rolling on L-, P-, and E-selectin. They provide a sufficient length and an appropriate structure to PSGL-1 to interact with L- and P-selectin and to stabilize leukocyte rolling on L-selectin. Finally, PSGL-1 N terminus and decamers directly interact with E-selectin and promote slow rolling.
Acknowledgments
We thank Jacqueline Cvetanovic for careful reading of the manuscript.
This work was supported by the Swiss National Foundation for Scientific Research (Grant 3200B0-105593). The authors have no conflict of interest disclosures. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PSGL-1, P-selectin glycoprotein ligand-1; PSGL/DR, PSGL-1 decameric repeats; GP, glycoprotein Ibα; CHO, chinese hamster ovary cells; WT, wild-type; PSGL/GP, PSGL-1/GPIbα; PSGL/ΔDR, PSGL-1/ΔDR; sLex, sialyl Lewis x; CLA, cutaneous lymphocyte antigen; C2GlcNAcT-I, core-2 β1,6N-acetylglucosaminyltransferase; FucT-VII, α1,3-fucosyltransferase-VII; mAb, monoclonal antibody; PE, phycoerythrin.
References
- 1.Ley, K. (2003) Trends Mol. Med. 9 263-268 [DOI] [PubMed] [Google Scholar]
- 2.Lowe, J. B. (2003) Curr. Opin. Cell Biol. 15 531-538 [DOI] [PubMed] [Google Scholar]
- 3.McEver, R. P. (2002) Curr. Opin. Cell Biol. 14 581-586 [DOI] [PubMed] [Google Scholar]
- 4.Ley, K., Bullard, D. C., Arbonés, M. L., Bosse, R., Vestweber, D., Tedder, T. F., and Beaudet, A. L. (1995) J. Exp. Med. 181 669-675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norman, K. E., Moore, K. L., McEver, R. P., and Ley, K. (1996) Blood 86 4417-4421 [PubMed] [Google Scholar]
- 6.Moore, K., Stults, N., Diaz, S., Smith, D., Cummings, R., Varki, A., and McEver, R. (1992) J. Cell Biol. 118 445-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sako, D., Chang, X.-J., Barone, K. M., Vachino, G., White, H. M., Shaw, G., Veldman, G. M., Bean, K. M., Ahern, T. J., Furie, B., Cumming, D. A., and Larsen, G. R. (1993) Cell 75 1179-1186 [DOI] [PubMed] [Google Scholar]
- 8.Spertini, O., Cordey, A.-S., Monai, N., Giuffré, L., and Schapira, M. (1996) J. Cell Biol. 135 523-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goetz, D. J., Greif, D. M., Ding, H., Camphausen, R. T., Howes, S., Comess, K. M., Snapp, K. R., Kansas, G. S., and Luscinskas, F. W. (1997) J. Cell Biol. 137 509-519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sperandio, M., Smith, M. L., Forlow, S. B., Olson, T. S., Xia, L., McEver, R. P., and Ley, K. (2003) J. Exp. Med. 197 1355-1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norman, K. E., Katopodis, A. G., Thoma, G., Kolbinger, F., Hicks, A. E., Cotter, M. J., Pockley, A. G., and Hellewell, P. G. (2000) Blood 96 3585-3591 [PubMed] [Google Scholar]
- 12.Hidalgo, A., Peired, A. J., Wild, M. K., Vestweber, D., and Frenette, P. S. (2007) Immunity 26 477-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, F., Wilkins, P. P., Crawley, S., Weinstein, J., Cummings, R. D., and McEver, R. P. (1996) J. Biol. Chem. 271 3255-3264 [PubMed] [Google Scholar]
- 14.Ramachandran, V., Nollert, M. U., Qiu, H. Y., Liu, W. J., Cummings, R. D., Zhu, C., and McEver, R. P. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3771-13776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somers, W. S., Tang, J., Shaw, G. D., and Camphausen, R. T. (2000) Cell 103 467-479 [DOI] [PubMed] [Google Scholar]
- 16.Leppanen, A., White, S. P., Helin, J., McEver, R. P., and Cummings, R. D. (2000) J. Biol. Chem. 275 39569-39578 [DOI] [PubMed] [Google Scholar]
- 17.Leppanen, A., Yago, T., Otto, V. I., McEver, R. P., and Cummings, R. D. (2003) J. Biol. Chem. 278 26391-26400 [DOI] [PubMed] [Google Scholar]
- 18.Bernimoulin, M. P., Zeng, X. L., Abbal, C., Giraud, S., Martinez, M., Michielin, O., Schapira, M., and Spertini, O. (2003) J. Biol. Chem. 278 37-47 [DOI] [PubMed] [Google Scholar]
- 19.Pouyani, T., and Seed, B. (1995) Cell 83 333-343 [DOI] [PubMed] [Google Scholar]
- 20.Martinez, M., Joffraud, M., Giraud, S., Baisse, B., Bernimoulin, M. P., Schapira, M., and Spertini, O. (2005) J. Biol. Chem. 280 5378-5390 [DOI] [PubMed] [Google Scholar]
- 21.Baisse, B., Galisson, F., Giraud, S., Schapira, M., and Spertini, O. (2007) BMC Evol. Biol. 7 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afshar-Kharghan, V., Diz-Kucukkaya, R., Ludwig, E. H., Marian, A. J., and Lopez, J. A. (2001) Blood 97 3306-3307 [DOI] [PubMed] [Google Scholar]
- 23.Xie, X., Rivier, A.-S., Zakrzewicz, A., Bernimoulin, M., Zeng, X.-L., Wessel, H. P., Schapira, M., and Spertini, O. (2000) J. Biol. Chem. 275 34818-34825 [DOI] [PubMed] [Google Scholar]
- 24.Lopez, J. A., Ludwig, E. H., and McCarthy, B. J. (1992) J. Biol. Chem. 267 10055-10061 [PubMed] [Google Scholar]
- 25.Liu, W., Ramachandran, V., Kang, J., Kishimoto, T. K., Cummings, R. D., and McEver, R. P. (1998) J. Biol. Chem. 273 7078-7087 [DOI] [PubMed] [Google Scholar]
- 26.Li, F., Erickson, H. P., James, J. A., Moore, K. L., Cummings, R. D., and McEver, R. P. (1996) J. Biol. Chem. 271 6342-6348 [DOI] [PubMed] [Google Scholar]
- 27.Finger, E. B., Puri, K. D., Alon, R., Lawrence, M. B., von Andrian, U. H., and Springer, T. A. (1996) Nature 379 266-269 [DOI] [PubMed] [Google Scholar]
- 28.Lawrence, M. B., Kansas, G. S., Kunkel, E. J., and Ley, K. (1997) J. Cell Biol. 136 717-727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarangapani, K. K., Yago, T., Klopocki, A. G., Lawrence, M. B., Fieger, C. B., Rosen, S. D., McEver, R. P., and Zhu, C. (2004) J. Biol. Chem. 279 2291-2298 [DOI] [PubMed] [Google Scholar]
- 30.Yago, T., Wu, J., Wey, C. D., Klopocki, A. G., Zhu, C., and McEver, R. P. (2004) J. Cell Biol. 166 913-923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu, C., Lou, J., and McEver, R. P. (2005) Biorheology 42 443-462 [PubMed] [Google Scholar]
- 32.Lozano, M. L., Gonzalez-Conejero, R., Corral, J., Rivera, J., Iniesta, J. A., Martinez, C., and Vicente, V. (2001) Br. J. Haematol. 115 969-976 [DOI] [PubMed] [Google Scholar]
- 33.Bugert, P., Vosberg, M., Entelmann, M., Jahn, J., Katus, H. A., and Kluter, H. (2004) Clin. Chem. Lab. Med. 42 997-1004 [DOI] [PubMed] [Google Scholar]
- 34.Tregouet, D. A., Barbaux, S., Poirier, O., Blankenberg, S., Bickel, C., Escolano, S., Rupprecht, H. J., Meyer, J., Cambien, F., and Tiret, L. (2003) Ann. Hum. Genet. 67 504-511 [DOI] [PubMed] [Google Scholar]
- 35.Ozben, B., Diz-Kucukkaya, R., Bilge, A. K., Hancer, V. S., and Oncul, A. (2007) J. Thromb. Thrombolysis 23 181-187 [DOI] [PubMed] [Google Scholar]
- 36.Marchese, P., Murata, M., Mazzucato, M., Pradella, P., De Marco, L., Ware, J., and Ruggeri, Z. M. (1995) J. Biol. Chem. 270 9571-9578 [DOI] [PubMed] [Google Scholar]
- 37.Doggett, T. A., Girdhar, G., Lawshe, A., Schmidtke, D. W., Laurenzi, I. J., Diamond, S. L., and Diacovo, T. G. (2002) Biophys. J. 83 194-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doggett, T. A., Girdhar, G., Lawshe, A., Miller, J. L., Laurenzi, I. J., Diamond, S. L., and Diacovo, T. G. (2003) Blood 102 152-160 [DOI] [PubMed] [Google Scholar]
- 39.Romo, G. M., Dong, J. F., Schade, A. J., Gardiner, E. E., Kansas, G. S., Li, C. Q., McIntire, L. V., Berndt, M. C., and Lopez, J. A. (1999) J. Exp. Med. 190 803-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein, J. L., Brown, M. S., Anderson, R. G., Russell, D. W., and Schneider, W. J. (1985) Annu. Rev. Cell Biol. 1 1-39 [DOI] [PubMed] [Google Scholar]
- 41.Fong, A. M., Erickson, H. P., Zachariah, J. P., Poon, S., Schamberg, N. J., Imai, T., and Patel, D. D. (2000) J. Biol. Chem. 275 3781-3786 [DOI] [PubMed] [Google Scholar]
- 42.Rosen, S. D., and Bertozzi, C. R. (1994) Curr. Opin. Cell Biol. 6 663-673 [DOI] [PubMed] [Google Scholar]
- 43.Fukuda, M., Hiraoka, N., and Yeh, J. C. (1999) J. Cell Biol. 147 467-470 [DOI] [PMC free article] [PubMed] [Google Scholar]