Abstract
The tumor suppressor protein p53 is activated by distinct cellular stresses including radiation, hypoxia, type I interferon, and DNA/RNA virus infection. The transactivation domain of p53 contains a phosphorylation site at Ser20 whose modification stabilizes the binding of the transcriptional co-activator p300 and whose mutation in murine transgenics induces B-cell lymphoma. Although the checkpoint kinase CHK2 is implicated in promoting Ser20 site phosphorylation after irradiation, the enzyme that triggers this phosphorylation after DNA viral infection is undefined. Using human herpesvirus 6B (HHV-6B) as a virus that induces Ser20 site phosphorylation of p53 in T-cells, we sought to identify the kinase responsible for this virus-induced p53 modification. The p53 Ser20 kinase was fractionated and purified using cation, anion, and dye-ligand exchange chromatography. Mass spectrometry identified casein kinase 1 (CK1) and vaccinia-related kinase 1 (VRK1) as enzymes that coeluted with virus-induced Ser20 site kinase activity. Immunodepletion of CK1 but not VRK1 removed the kinase activity from the peak fraction, and bacterially expressed CK1 exhibited Ser20 site kinase activity equivalent to that of the virus-induced native CK1. CK1 modified p53 in a docking-dependent manner, which is similar to other known Ser20 site p53 kinases. Low levels of the CK1 inhibitor D4476 selectively inhibited HHV-6B-induced Ser20 site phosphorylation of p53. However, x-ray-induced Ser20 site phosphorylation of p53 was not blocked by D4476. These data highlight a central role for CK1 as the Ser20 site kinase for p53 in DNA virus-infected cells but also suggest that distinct stresses may selectively trigger different protein kinases to modify the transactivation domain of p53 at Ser20.
The tumor suppressor protein p53 is a key player in the survival or death decision that cells face after exposure to a variety of metabolic and genotoxic stresses (1). The transient accumulation and activation of p53 in response to various cellular stresses enables the protein to modulate the expression of numerous genes involved in cell cycle arrest, DNA repair, and/or apoptosis. The initiation of either transient cell cycle arrest and damage repair or apoptosis is dependent on the cell and damage type, the severity of damage, and the cellular microenvironment. Phosphorylation and acetylation events that control interactions between the transcription factor p53 and its negative regulators (Mdm2, COP1, and Pirh2) or co-activators (p300) are ultimately involved in modulating p53-dependent gene expression in response to cellular stress (2). In particular, phosphorylation at Thr18 within the N-terminal conserved BOX-I domain of p53 blocks the binding of Mdm2, whereas phosphorylation at Ser20, also within the BOX-I domain, enables the binding of p300 (3-5). Thus, phosphorylation in this transactivation domain serves to stimulate rather than inhibit p53 function. In addition, phosphorylation at Ser392 within the C terminus of p53 stimulates the sequence-specific DNA-binding function of p53 (6).
The generation of transgenic mice with phosphoacceptor site mutations (to alanine) at the key regulatory phosphoacceptor sites of Ser20 and Ser392 equivalents in murine p53 results in elevated cancer incidence. Mutation of Ser20 results in enhanced spontaneous B-cell lymphoma and attenuated damage-induced apoptosis in B-cells (7), whereas mutation of Ser392 results in enhanced UV-induced skin cancer or carcinogen-induced bladder cancer (8, 9). These biochemical and genetic results highlight the critical role that phosphorylation of p53 can play in modulating its tumor suppressor function and the likelihood that these phosphorylation events are “stress-” and/or cell type-specific. Presumably, the use of transgenic phosphomutated systems will further uncover the cell- and stress-specific function of these multiple covalent modifications.
Although members of the calcium calmodulin kinase superfamily, particularly the checkpoint kinases 1 and 2 (CHK1 and CHK2) and death-associated protein kinase 1 (DAPK-1), are genetic activators of the p53 pathway, other kinases have also been shown to phosphorylate and activate p53. For example, vaccinia-related kinase 1 (VRK1) and casein kinase 1 (CK1) have been reported to phosphorylate p53 at Thr18 (10), although the latter requires prior phosphorylation of p53 at Ser15 (11). Controversy remains as to which kinases are most important for the activation of p53 in response to distinct cellular stress. It is possible that the exact kinase(s) involved and the residue(s) modified are specific to both the cell and damage type, which would explain the slight disparities in the results reported to date, and provide a mechanism for a context-based cellular survival versus death decision to be made. Nevertheless, the identification and characterization of the kinases involved in the phosphorylation of p53 are essential to the further elucidation of the regulation of p53.
In addition to the well established activation of p53 by ionizing and nonionizing radiation, anoxia, hypoxia, and type I interferons, more recent research has shown that DNA and RNA viruses can activate the p53 response (12, 13). It has been previously shown that human herpesvirus 6B (HHV-6B) infection of HCT116 and MOLT-3 cancer cell lines induced p53 phosphorylation (at Ser15, Ser20, Ser33, and Ser392) and p53 accumulation in both nuclear and cytoplasmic fractions (14, 15). It has also been shown that HHV-6B-infected cells were mostly non-apoptotic, and it has been suggested that in this context, p53 phosphorylation and accumulation may act as a survival factor rather than cell death inducer. Thus, p53 is a central sensor of both DNA damage and immune system perturbation. The DNA virus-induced stress provides a novel model to examine the kinases that target the Ser20 site of p53.
The aim of this study was to identify the p53 Ser20 kinase that is activated after HHV-6B infection of MOLT-3 cells and to investigate whether the Ser20 kinase operates in a dual site docking-dependent manner, like members of the calcium calmodulin kinase superfamily. We show that the enzyme responsible for this virus-induced phosphorylation of p53 is the DNA damage-regulatory enzyme CK1. These data suggest a dominant role for CK1 in p53 activation after HHV-6B virus infection and highlight a specific kinase pathway for further integration into the antiviral sensing machinery of the cell.
EXPERIMENTAL PROCEDURES
Chemicals, Reagents, Recombinant Proteins, and Antibodies—All reagents were purchased from Sigma unless otherwise stated. The CK1 inhibitor D4476 was purchased from Merck. Overlapping synthetic 15-mer biotinylated peptides from the coding sequence of human p53 (see Table 1) (Mimotopes, Melbourne, Australia) were resuspended in DMSO to a concentration of 10 mm. Full-length p53 (FLp53)4 tetramers, recombinant DAPK-1 core domain, and recombinant GST-CK1δ kinase domain were purified as described previously (3, 11, 18). DO-1 and DO-12 antibodies to p53 were kindly provided by B. Vojtesek (Masaryk Memorial Cancer Institute, Brno, Czech Republic). Antibodies to p53 phosphorylated at Thr18 (P-p53 Thr18) and Ser392 (P-p53 Ser392) were described previously (16). 1F6 and VC1 antibodies to VRK1 were generated as described previously (17). P-p53 Ser15 antibody to p53 phosphorylated at Ser15 and an antibody against CK1 were purchased from Cell Signaling Technology (supplied by New England Biolabs, Hitchin, UK). P-p53 Ser20 antibody to p53 phosphorylated at Ser20 and antibodies against CK1α, CHK1, and CHK2 were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) (supplied by Insight Biotechnology, Wembley, UK). Antibodies to AMPKα1, AMPKα2, and CK2 were purchased from Upstate (Millipore, Watford, UK); an antibody to DAPK-1 was from BD Biosciences (Oxford, UK); and antibodies to DAPK-2 and DAPK-3 were from Abcam (Cambridge, UK). Rabbit anti-mouse or swine anti-rabbit secondary antibodies were obtained from Dako (Ely, UK), and donkey anti-goat secondary antibody was purchased from Santa Cruz Biotechnology.
TABLE 1.
p53 peptide amino acid sequences
Shown are overlapping synthetic 15-mer peptides from the coding sequence of human p53. All peptides were biotinylated at the N terminus with SGSG spacers and had a C terminal amide group.
| Peptide | Sequence |
|---|---|
| Peptide 1 | NRRPILTIITLEDSS |
| Peptide 2 | ILTIITLEDSSGNLL |
| Peptide 3 | LTIITLEDSSGNLLG |
| Peptide 4 | TIITLEDSSGNLLGR |
| Peptide 5 | IITLEDSSGNLLGRN |
| Peptide 6 | ITLEDSSGNLLGRNS |
| Peptide 7 | TLEDSSGNLLGRNSF |
| Peptide 8 | LEDSSGNLLGRNSFE |
| Peptide 9 | EDSSGNLLGRNSFEV |
| Peptide 10 | DSSGNLLGRNSFEVR |
| Peptide 11 | SSGNLLGRNSFEVRV |
| Peptide 12 | SGNLLGRNSFEVRVC |
| Peptide 13 | GNLLGRNSFEVRVCA |
| Peptide 14 | NLLGRNSFEVRVCAC |
| Peptide 15 | LLGRNSFEVRVCACP |
| Peptide 16 | LGRNSFEVRVCACPG |
| Peptide 17 | GRNSFEVRVCACPGR |
| Peptide 18 | RNSFEVRVCACPGRD |
Purification of the p53 Ser20 Kinase—The human T-cell acute lymphoblastic leukemia cell line, MOLT-3, was infected with HHV-6B strain PL-1 as previously described (14). High salt extracts were prepared from MOLT-3 cells infected with or without HHV-6B for 48 h. Briefly, cells were lysed in 20 mm Tris-HCl (pH 7.5), 0.5 m NaCl, 1 mm Na2EDTA, 1 mm EGTA, 1% (v/v) Triton X-100, 2.5 mm Na4P2O7, 1 mm β-glycerophosphate, 1 mm Na3VO4, and 1 μg/ml leupeptin. Lysates were treated with Benzonase nuclease (Merck), syringed, and collected after centrifugation at 12,000 rpm for 5 min at 4 °C. Fractionation of kinase activity from high salt lysates was performed using a Q-Sepharose Fast Flow anion exchanger with a linear gradient of 0-1 m KCl in a 10-column volume of 20 mm HEPES (pH 8.0), 10% (v/v) glycerol, 1 mm dithiothreitol (DTT), 1 mm benzamidine, and protease inhibitor mixture tablets (Roche Applied Science). The flow-through and 42 12-ml fractions were collected and assayed for their in vitro kinase activity toward FLp53 tetramers (see below), and positive fractions were pooled. Further fractionation of kinase activity from the positive fractions was performed using a Reactive Brown 10-agarose cation exchanger with a linear gradient of 10 mm to 2 m KCl in a 20-column volume of 25 mm HEPES (pH 7.5), 10% (v/v) glycerol, 0.02% (v/v) Triton X-100, 1 mm DTT, 1 mm benzamidine, and protease inhibitor mixture tablets. Flow-through and 77 1.3-ml fractions were collected and assayed as above, and positive fractions were pooled. A final fractionation of kinase activity was performed using a HiTrap SP-Sepharose HP cation exchanger (GE Healthcare) with a linear gradient of 10 mm to 2 m KCl in a 20-column volume of 25 mm HEPES (pH 7.5), 10% (v/v) glycerol, 0.02% (v/v) Triton X-100, 1 mm DTT, 1 mm benzamidine, and protease inhibitor mixture tablets. The flow-through and 100 1-ml fractions were collected and assayed as above.
In Vitro Kinase Assays—FLp53 tetramer in vitro kinase assays were described previously (18). Briefly, the kinase source (0.2-2 μl of lysate or fraction or flow-through, 0.2 μl of DAPK-1 core domain, or 1 μl of recombinant GST-CK1δ) was incubated with p53 substrate (100 ng of FLp53 tetramers) in a 20-μl reaction volume containing 25 mm HEPES (pH 7.5), 25 mm KCl, 1 mm MgCl2, 20 μm EDTA, 0.1 mm DTT, and 0.1 mm ATP. Reaction mixtures were incubated at 30 °C for 45 min, and reactions were terminated by the addition of 30 μl of SDS sample buffer containing 0.2 m DTT. Peptide kinase assays were performed by incubating p53 peptide fragments with the above described kinase reaction mixtures for 20 min at room temperature prior to the addition of the kinase source. p53 peptides fragments were added to a final concentration of 200 μm. Negative control reactions, lacking either the kinase source or the kinase substrate were included, along with DMSO solvent controls for peptide kinase assays.
CK1 Inhibitor Experiments—Mock-infected and HHV-6B-infected MOLT-3 cells were treated with 10-100 μm CK1 inhibitor D4476, concomitantly with infection, for 48 h. Alternatively, MOLT-3 cells were pretreated with 10-60 μm D4476 for 44 h before exposure (or sham exposure) to a 6-gray x-ray and further culture for 4 h. DMSO solvent controls were included, and cells were lysed as described previously (15).
Western Blotting—Lysates, flow-through, fractions, eluates, or kinase assay samples (10 μl) were resolved by SDS-PAGE through 10% (w/v) Tris-glycine gels and transferred onto nitrocellulose membranes (Hybond ECL; GE Healthcare). Membranes were probed with primary antibodies, followed by secondary antibodies conjugated to horseradish peroxidase. Bound antibody was detected by ECL.
Mass Spectrometry—Fractions from the HiTrap SP-Sepharose HP cation exchanger column (1 ml each) were concentrated ∼25-fold using Centricon YM10 centrifugal devices (Millipore). Flow-through and fractions (10 μl) were resolved by SDS-PAGE through a 4-12% (w/v) bis-tris gel (Invitrogen) and stained with colloidal blue (Invitrogen). Proteins enriched in the peak fraction (fraction 31) were excised from the gel, protein in-gel digestions were performed with 0.1 pmol/μl trypsin gold (Promega, Southampton, UK), and extracted peptides were resuspended in 2% (v/v) formic acid. Samples were analyzed by tandem mass spectrometry (MS/MS). Briefly, peptides were resolved in a gradient of 4-95% (v/v) MeCN using reverse phase high performance liquid chromatography, using the UltiMate 3000 system (Dionex, Camberley, UK). Samples were acquired on a 4000 Q TRAP® liquid chromatography/MS/MS system (Applied Biosystems, Warrington, UK). Data were analyzed using Analyst® software (Applied Biosystems).
Immunoprecipitation—Peak fractions from the HiTrap SP-Sepharose HP cation exchanger column (25 μl) were incubated with primary antibody (0.25-2 μl VC1 antibody to VRK1 or antibody to CK1α) overnight at 4 °C in immunoprecipitation buffer (0.1% (w/v) SDS, 1% (v/v) Nonidet P-40, 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA) in a final volume of 50 μl. Protein G-Sepharose Fast Flow beads (50 μl) were then added, and the reactions were incubated at 4 °C for a further 2 h. The flow-through was collected and assayed for in vitro kinase activity toward FLp53 tetramers, and the beads were washed six times in 100 μl of immunoprecipitation buffer. Bound protein was eluted after a 45-min incubation in 100 μl of elution buffer (SDS sample buffer containing 0.2 m DTT) at room temperature. Negative control reactions, where the antibody was omitted, were included.
RESULTS
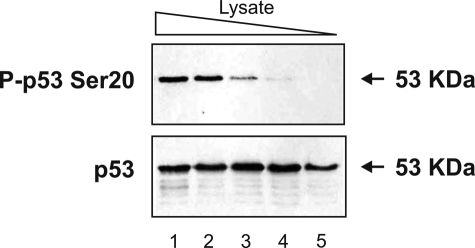
HHV-6B-infected MOLT-3 Cells Contain a Kinase That Displays Ser20 Site Activity toward p53 in Vitro—HHV-6B infection has previously been shown to induce Ser20 site phosphorylation of p53, although the kinase that mediates this stress-activated phosphorylation has not been identified (14). We set out to determine whether HHV-6B-infected MOLT-3 cell lysates were capable of mediating the phosphorylation of p53 at Ser20 in an in vitro assay. HHV-6B-infected MOLT-3 whole-cell lysates acted as the kinase source in an in vitro kinase assay using recombinant FLp53 tetramers as the substrate. Western blotting of the kinase assay products revealed that p53 was phosphorylated at Ser20 (Fig. 1, lanes 1-4) and that the amount of P-p53 Ser20 was proportional to the amount of kinase source used in the kinase assay. HHV-6B-infected MOLT-3 cell lysates therefore contain at least one kinase that is capable of mediating the phosphorylation of p53 at Ser20 in vitro.
FIGURE 1.
HHV-6B-infected MOLT-3 lysates display cell-free p53 Ser20 kinase activity. MOLT-3 cells were infected with HHV-6B for 48 h prior to lysis. Decreasing amounts of the whole-cell lysates (10-fold serial dilutions in lanes 1-5) acted as the kinase source in an in vitro kinase assay using recombinant FLp53 tetramers as the substrate. Kinase assay samples were subjected to Western blotting using P-p53 Ser20 (top) and DO-12 (bottom) antibodies to detect FLp53 phosphorylated at Ser20 and total FLp53, respectively.
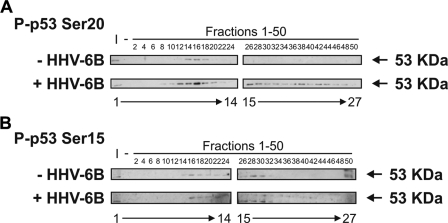
In order to identify the p53 Ser20 kinase, we subjected crude lysates to chromatographic fractionation to determine whether the kinase activity correlates with the known Ser20 site kinases, including CHK1/2 and DAPK-1. HHV-6B-infected and mock-infected MOLT-3 whole-cell lysates were applied onto a Q-Sepharose Fast Flow anion exchanger column, proteins were eluted in a linear gradient of 0-1 m KCl, and fractions were collected and assayed for kinase activity toward the Ser20 and Ser15 sites of FLp53 (Fig. 2). The induction of Ser20 site phosphorylation correlated with an increase in the specific activity of the Ser20 site kinase. The kinase eluted from fractions 14-18 in mock-infected cells (Fig. 2A, upper panels, lanes 9-11), whereas in HHV-6B-infected cells, the kinase eluted from fractions 10-20 (Fig. 2A, lower panels, lanes 7-12) and a broader elution peak from fractions 26-48 (lanes 15-26). By contrast, the induction of Ser15 site phosphorylation after HHV-6B infection did not exhibit the same qualitative increase in cell-free kinase activity (Fig. 2B). When these fractions containing the peak Ser20 site kinase activity were immunoblotted for the kinases known to phosphorylate p53 Ser20, none of the calcium calmodulin kinase superfamily members co-eluted precisely with the Ser20 site kinase activity (data not shown). Thus, we set up further chromatographic fractionations in order to enrich the HHV-6B-induced Ser20 site kinase activity sufficiently to identify candidate leads by identifying protein bands from SDS-PAGE gels using mass spectrometry (MS).
FIGURE 2.
p53 Ser20 kinase activity is elevated in HHV-6B-infected MOLT-3 cell lysates. Mock-infected (top) and HHV-6B-infected (bottom) MOLT-3 whole-cell lysates were subjected to chromatographic fractionation using a Q-Sepharose Fast Flow anion exchanger with a linear gradient of 0-1 m KCl. Even-numbered fractions (lanes 3-27) were assayed for in vitro kinase activity toward recombinant FLp53 tetramers, and Western blotting using P-p53 Ser20 and P-p53 Ser15 antibodies revealed phosphorylation at the Ser20 (A) and Ser15 (B) sites of FLp53. Input (I; lane 1) and negative (-; lane 2) controls were included and consisted of unfractionated whole-cell lysates and distilled H2O as the kinase source, respectively.
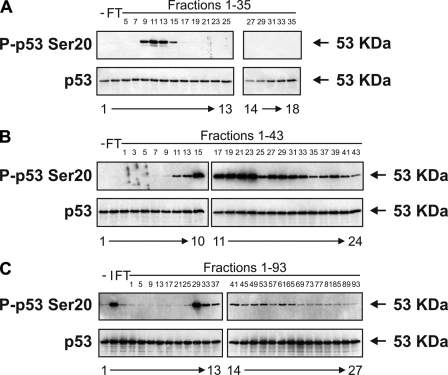
Purification of the p53 Ser20 Kinase—Large scaled up samples of HHV-6B-infected MOLT-3 whole-cell lysates were applied onto a Q-Sepharose Fast Flow anion exchanger column, proteins were eluted in a linear gradient of 0-1 m KCl, and fractions were collected and assayed for kinase activity toward the Ser20 site of FLp53 (Fig. 3A). Fractions 9-14 containing the most activity (Fig. 3A, lanes 5-8) were pooled and applied onto a Reactive Brown 10-agarose cation exchange column. Proteins were eluted in a linear gradient of 10 mm to 2 m KCl, and fractions were collected and assayed for kinase activity toward FLp53 Ser20, as described previously (Fig. 3B). Fractions 17-23 containing the first peak of activity (Fig. 3B, lanes 11-14) were pooled and applied onto a HiTrap SP-Sepharose HP cation exchanger column. Proteins were eluted in a linear gradient of 10 mm to 2 m KCl, and fractions were again collected and assayed for kinase activity toward FLp53 Ser20 (Fig. 3C). Fraction 29 displayed the most activity toward FLp53 Ser20 (Fig. 3C, lane 11).
FIGURE 3.
Purification of the virus-induced p53 Ser20 kinase. A, HHV-6B-infected MOLT-3 whole-cell lysates were subjected to chromatographic fractionation using a Q-Sepharose Fast Flow anion exchanger with a linear gradient of 0-1 m KCl. The column flow-through (FT; lane 2) and odd-numbered fractions (lanes 3-18) were assayed for in vitro kinase activity toward the Ser20 site of FLp53 tetramers, which was detected by Western blotting using P-p53 Ser20 (top) and DO-12 (bottom) antibodies. A negative control (-; lane 1), consisting of distilled H2O as the kinase source, was included. B, fractions 9-14 from A were pooled and subjected to further chromatographic fractionation using a Reactive Brown 10-agarose cation exchanger with a linear gradient of 0.01-2 m KCl. The column flow-through (lane 2) and odd-numbered fractions (lanes 3-24) were assayed for in vitro kinase activity toward the Ser20 site of FLp53 tetramers, which was detected by Western blotting using P-p53 Ser20 (top) and DO-12 (bottom) antibodies. A negative control (-; lane 1), consisting of distilled H2O as the kinase source, was included. C, fractions 17-23 from B were pooled and subjected to a final chromatographic fractionation using a HiTrap SP-Sepharose HP cation exchanger with a linear gradient of 0.01-2 m KCl. The column flow-through (lane 3) and every fourth fraction (lanes 4-27) were assayed for in vitro kinase activity toward the Ser20 site of FLp53 tetramers, which was detected by Western blotting using P-p53 Ser20 (top) and DO-12 (bottom) antibodies. Negative (-; lane 1) and input (I; lane 2) controls were included and consisted of distilled H2O and pooled fractions 17-23 from B as the kinase source, respectively.
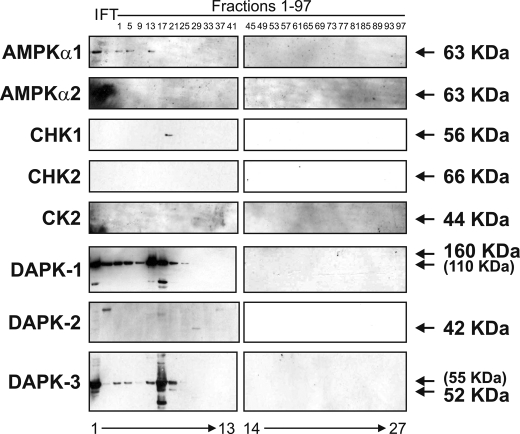
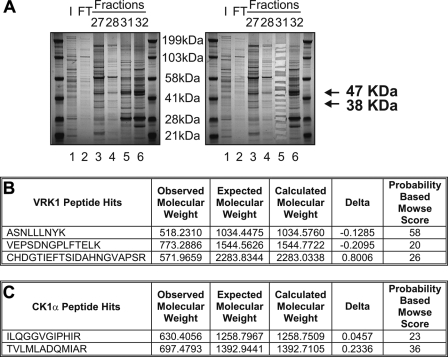
The p53 Ser20 Kinase Is CK1α—None of the calcium calmodulin kinase superfamily members co-eluted with the fractions from the HiTrap SP-Sepharose HP cation exchanger column that displayed the peak of p53 Ser20 kinase activity (Fig. 4, lane 10). As such, we resolved protein from the peak fractions by SDS-PAGE, and after staining with colloidal blue (Fig. 5A, left), we excised and processed the proteins that were enriched in the peak fraction (Fig. 5A, right, lane 5 versus lane 4 or 6) for analysis by MS in order to acquire candidate kinase leads. MS identified VRK1 (47 kDa) and CK1α (38 kDa) as candidate kinases that eluted in fraction 31 (Fig. 5, B and C), a fraction that displayed strong kinase activity toward the Ser20 site of FLp53.
FIGURE 4.
Calcium calmodulin kinases do not co-elute with virus-induced p53 Ser20 kinase activity. The flow-through (FT; lane 2) and every fourth fraction (lanes 3-27) from the HiTrap SP-Sepharose HP cation exchanger column were subjected to Western blotting with antibodies against known calcium calmodulin kinase superfamily members (AMPKα1, AMPKα2, CHK1, CHK2, CK2, DAPK-1, DAPK-2, and DAPK-3). An input control (I; lane 1), consisting of pooled fractions 17-23 from the Reactive Brown 10-agarose cation exchanger column, was included.
FIGURE 5.
Mass spectrometry identifies VRK1 and CK1α as candidate p53 Ser20 kinases. A, the flow-through (FT; lane 2) and concentrated fractions 27, 28, 31, and 32 (lanes 3-6) from the HiTrap SP-Sepharose HP cation exchanger column were resolved by SDS-PAGE and stained with colloidal blue (left). An input control (I; lane 1), consisting of pooled fractions 17-23 from the Reactive Brown 10-agarose cation exchanger column, was included. Proteins enriched in fraction 31 (lane 5) were excised from the gel (right) and processed for analysis by MS/MS. B and C, MS identified VRK1 (B) and CK1α (C) as candidate kinases.
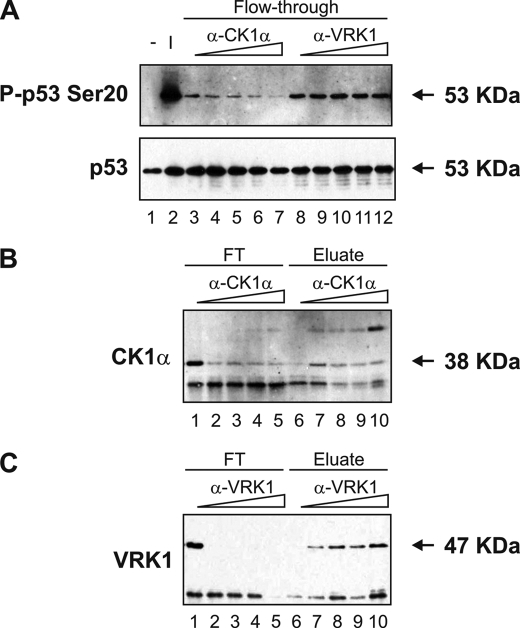
Western blotting confirmed that the elution profiles of CK1 or CK1α (Fig. 6, A and B) and VRK1 (Fig. 6C) matched the p53 Ser20 kinase activity profile. CK1 and CK1α eluted mostly in fraction 29 (Fig. 6, A and B, lanes 10 and 9, respectively), and VRK1 eluted in fractions 29-33 (Fig. 6C, lanes 10 and 11), suggesting that either kinase could be responsible for the observed phosphorylation of FLp53 at Ser20 in the peak fraction 29 (Fig. 3C). Immunoprecipitation studies revealed that the kinase activity toward FLp53 at Ser20 was attenuated after immunodepletion of CK1α (Fig. 7A, lanes 4-7 versus lane 3) and not after depletion of VRK1 (Fig. 7A, lanes 9-12 versus lane 8), implying that CK1α is the dominant p53 Ser20 kinase. This agrees with previous data showing that recombinant VRK1 does not exhibit Ser20 site kinase activity (10). However, this does rule out VRK1 as a co-purifying subunit of CK1. As controls, we affirmed that increasing the antibody titration did result in reduced CK1α (Fig. 7B, lanes 2-5 versus lane 1) and VRK1 (Fig. 7C, lanes 2-5 versus lane 1) in the immunodepleted samples as well as elevated CK1α (Fig. 7B, lanes 7-10 versus lane 6) and VRK1 (Fig. 7C, lanes 7-10 versus lane 6) in the immune pellet.
FIGURE 6.
CK1, CK1α, and VRK1 co-elute with p53 Ser20 kinase activity. The flow-through (FT; lanes 1 and 2) and every fourth fraction (lanes 2-26 or 3-27) from the HiTrap SP-Sepharose HP cation exchanger column were subjected to Western blotting with antibodies against CK1 (A), CK1α (B), or VRK1 (C). An input control (I; lane 1), consisting of pooled fractions 17-23 from the Reactive Brown 10-agarose cation exchanger column, was included in A and C.
FIGURE 7.
Immunodepletion of CK1α, but not VRK1, abolishes p53 Ser20 kinase activity. CK1α or VRK1 were immunoprecipitated from a peak fraction from the HiTrap SP-Sepharose HP cation exchanger column using increasing amounts (0.25-2 μl) of antibody to CK1α or VRK1 bound to Protein G-Sepharose Fast Flow beads. A, immunodepleted fractions, or flow-through (FT; lanes 3-12), were assayed for in vitro kinase activity toward the Ser20 site of FLp53 tetramers, which was detected by Western blotting using P-p53 Ser20(top) and DO-12 (bottom) antibodies. Negative (-; lane 1) and input (I; lane 2) controls were included and consisted of distilled H2O and the peak fraction as the kinase source, respectively. Fractions depleted of CK1α and VRK1 are shown in lanes 4-7 and lanes 9-12, respectively, and negative control reactions, where the antibody was omitted, are shown in lanes 3 and 8. B and C, immunodepleted fractions, or flow-through (lanes 1-5), and the corresponding immune pellet, or eluate (lanes 6-10), were subjected to Western blotting with antibodies to CK1α (B) or VRK1 (C). Fractions depleted of CK1α and VRK1 are shown in B and C, respectively, and negative control reactions, where the antibody was omitted from the immunoprecipitation reactions, are shown in lanes 1 and 6.
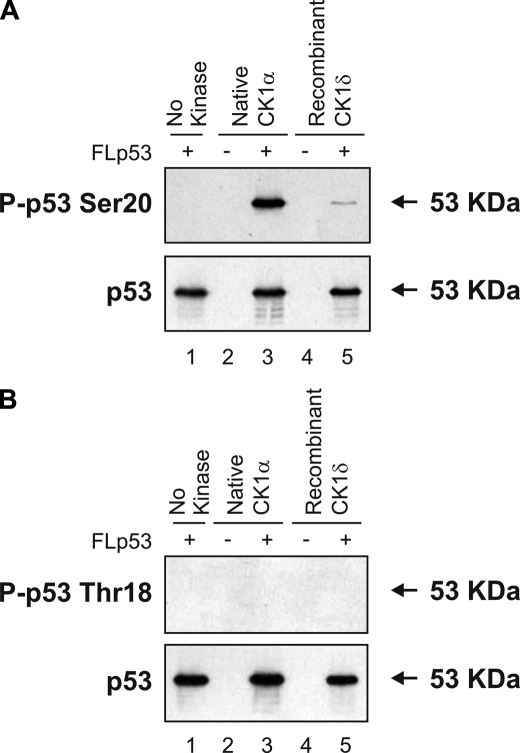
Together, these results demonstrate that CK1α is the most likely cellular virus-induced Ser20 site kinase. However, we still set up assays to compare recombinant CK1δ kinase domain purified from Escherichia coli with the native enzyme purified from MOLT-3 cells in order to determine whether the native enzyme has the same properties as the recombinant CK1. Recombinant CK1δ was able to mimic the native enzyme and catalyze Ser20 site phosphorylation of FLp53 tetramers in vitro (Fig. 8A, lane 5 versus lane 3). This finding is consistent with the original data on CK1δ, which showed that mutation of p53 at Ser20 is sufficient to block completely p53 phosphorylation (11). We could not detect p53 Thr18 phosphorylation induced by native or recombinant CK1 using a well validated Thr18-phosphospecific monoclonal antibody (Fig. 8B, lanes 3 and 5), which might be expected, since recombinant CK1δ-mediated phosphorylation at Thr18 requires a prior phosphorylation of p53 at Ser15 by phosphoinositide 3-kinases (11).
FIGURE 8.
Native and recombinant CK1 phosphorylate p53 at Ser20. Native CK1α purified from HHV-6B-infected MOLT-3 cells (lanes 2 and 3) or recombinant GST-CK1δ kinase domain purified from E. coli (lanes 4 and 5) was assayed for in vitro kinase activity toward the Ser20 (A) and Thr18 (B) sites of FLp53 tetramers, which was detected by Western blotting using P-p53 Ser20 (A, top), P-p53 Thr18 (B, top), and DO-12 (A and B, bottom) antibodies. Negative control reactions, lacking either the kinase source (lane 1) or the kinase substrate (lanes 2 and 4) were included.
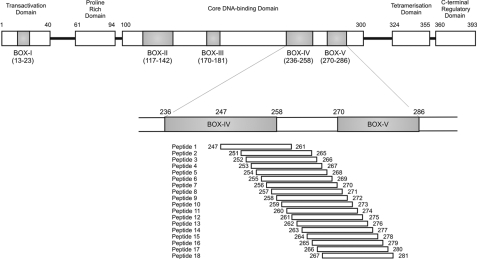
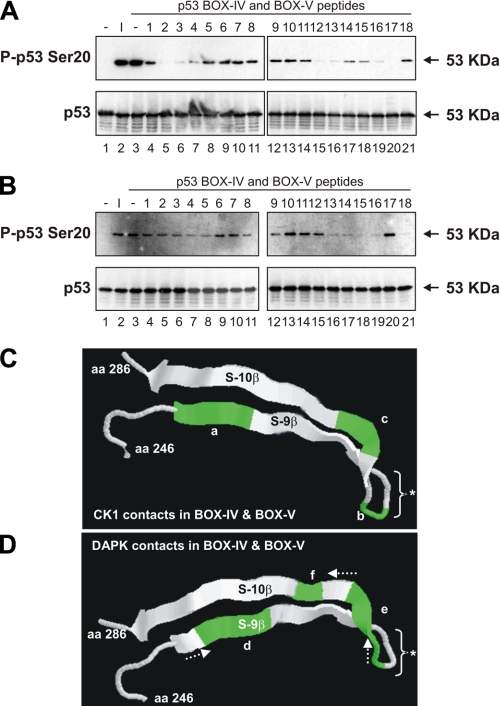
The Phosphorylation of p53 at Ser20 by Native CK1α Is Dependent on Interactions with the BOX-IV and BOX-V Domains of p53—All enzymes reported to modify the Thr18 or Ser20 sites within the BOX-I domain of p53 to date also interact with the multiprotein docking site in the BOX-V domain of p53 (Fig. 9). The three-dimensional proximity of the BOX-V motif to the BOX-I motif in tetrameric p53 is not known. However, synthetic peptide fragments from the BOX-V domain of p53 can alter in trans the BOX-I Ser20 site kinase activity of the respective enzymes (18, 19). Kinase assays were performed using peptides designed to the BOX-IV and BOX-V domains of the coding sequence of human p53 (Fig. 9 and Table 1) in order to monitor peptide stimulation or inhibition of kinase function toward FLp53 tetramers (Fig. 10). Several peptides (peptides 2-4, 12 and 13, and 16 and 17) had an inhibitory effect on the kinase activity of CK1α toward the Ser20 site of FLp53 tetramers (Fig. 10A, lanes 5-7, 15 and 16, and 19 and 20). This implies that residues 251-253, 261-262, and 265-266 of p53 encompass docking sites in the BOX-IV and BOX-V domains that are crucial for the Ser20 site activity of CK1α. The location of these key docking sites within the localized three-dimensional structure of p53 is shown in Fig. 10C.
FIGURE 9.
Location of the synthetic peptides within the coding sequence of p53. The domain structure of p53 (393 amino acids) is represented, including the conserved domains, labeled with the indicated amino acids flanking BOX-I through BOX-V. Overlapping synthetic 15-mer peptides were designed to the BOX-IV and BOX-V domains of the coding sequence of human p53. See Table 1 for peptide sequences.
FIGURE 10.
The BOX-IV and BOX-V domains of p53 act as multiprotein docking sites necessary for the p53 Ser20 kinase activity of CK1α and DAPK-1. A and B, native CK1α purified from HHV-6B-infected MOLT-3 cells (A) or DAPK-1 core domain (B) were assayed for in vitro kinase activity toward the Ser20 site of FLp53 tetramers in the presence of p53 BOX-IV and BOX-V peptides (lanes 4-21) or a DMSO solvent control (lane 3). Western blotting using P-p53 Ser20 (top) and DO-12 (bottom) antibodies revealed FLp53 phosphorylated at Ser20 and total FLp53, respectively. Negative control reactions (-; lane 1) lacking the kinase source and positive or input controls (I; lane 2) with the kinase source were included. C and D, location of key CK1α (C) or DAPK-1 (D) docking sites within the BOX-IV and BOX-V domains of the three-dimensional structure of p53 that are necessary for the p53 Ser20 kinase activity of either CK1α (C) or DAPK-1 (D). Highlighted docking regions encompass residues 251-253 (a), 261 and 262 (b), 265 and 266 (c), 253 and 254 (d), 262-265 (e), and 267 (f). The arrows indicate the shift in docking site preferences of DAPK-1 versus CK1α. *, the conformationally flexible monoclonal antibody epitope, which can be exposed or cryptic, depending upon p53 conformation. aa, amino acids.
Distinct Residues in the BOX-IV and BOX-V Domains of p53 Are Important for the Interaction of p53 with Different Proteins—We evaluated whether DAPK-1 core domain has a similar or overlapping sensitivity to the p53 BOX-IV and BOX-V peptides as native CK1α. Several peptides (peptides 4 and 5, 13-16, and 18) had an inhibitory effect on the kinase activity of DAPK-1 core toward the Ser20 site of FLp53 tetramers (Fig. 10B, lanes 7 and 8, 16-19, and 21). This implies that residues 253-254, 262-265, and 267 in the BOX-IV and BOX-V domains of p53 are important for the Ser20 site activity of DAPK-1 core. The location of these key docking sites within the three-dimensional structure of p53 is shown in Fig. 10D. These data indicate that at least two distinct kinases have an overlapping, but nonidentical, interaction with the BOX-IV and BOX-V domains of p53.
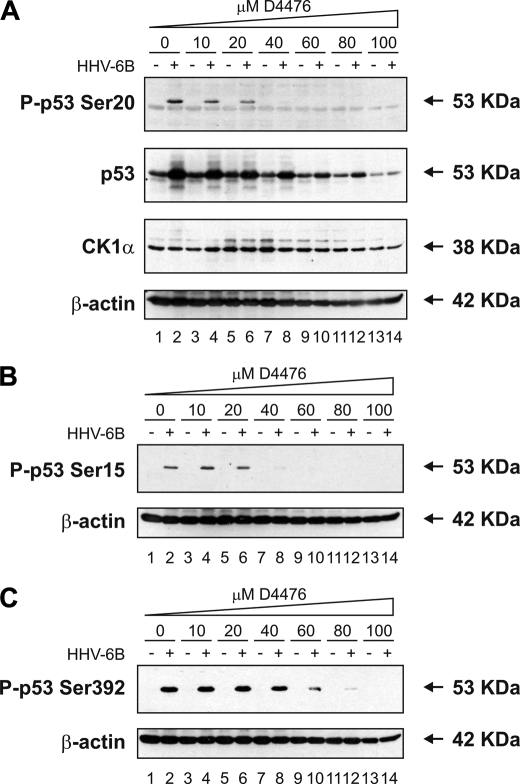
CK1 Inhibitors Attenuate p53 Induction by HHV-6B Infection—In order to obtain independent genetic evidence that CK1 is the major Ser20 site kinase for p53 in HHV-6B-infected cells, we needed to inhibit the enzyme to determine whether p53 induction would be reduced. The MOLT-3 cells are well known to be difficult to transfect using small interfering RNA, so we could not deplete endogenous CK1 using this approach. However, there are three chemical inhibitors of CK1 that can be used, one of which (D4476) shows a more pronounced specificity for CK1 (31). Upon increasing addition of D4476 to HHV-6B-infected MOLT-3 cells, there was a dose-dependent reduction in Ser20 site phosphorylation (Fig. 11A, top, lanes 4-14 versus lane 2). This reduction was especially pronounced from a concentration of 20 μm D4476 and higher (Fig. 11A, top, lanes 6-14 versus lane 2). This inhibition of p53 Ser20 site phosphorylation did not result in loss of CK1 enzyme (Fig. 11A, third panel, lanes 4-14 versus lane 2); however, there was an increase in a higher molecular weight adduct at the 20 μm concentration of D4476 irrespective of HHV-6B infection status (Fig. 11A, third panel, lanes 5 and 6 versus lanes 1 and 2).
FIGURE 11.
A CK1 inhibitor attenuates Ser20 site phosphorylation of p53 and p53 induction mediated by HHV-6B infection. A-C, MOLT-3 cells were infected with (even-numbered lanes) or without (odd-numbered lanes) HHV-6B for 48 h in the presence of increasing concentrations (10-100 μm) of the CK1 inhibitor D4476 (lanes 3-14) or a DMSO solvent control (lanes 1 and 2). Cell lysates were examined by Western blotting with antibodies against the indicated proteins.
Although previous studies were unable to show a reduction in HHV-6B-induced phosphorylation of p53 at Ser15, Ser20, or Ser392 using chemical inhibitors designed to CK2, double-stranded RNA-activated protein kinase, or p38 mitogen-activated protein kinase (15), we evaluated whether CK1 inhibition of Ser20 site phosphorylation could be uncoupled from HHV-6B-induced phosphorylation at other sites. At the concentrations of D4766 where we began to observe reductions in Ser20 site phosphorylation (10 and 20 μm), there was no reduction detected in HHV-6B induction of Ser15 or Ser392 phosphorylation of p53 (Fig. 11, B and C, top, lanes 4 and 6 versus lane 2). However, from a concentration of 40 μm D4476 and higher, there was a complete inhibition of HHV-6B-induced Ser15 site phosphorylation (Fig. 11B, top, lanes 8-14) but not Ser392 site phosphorylation (Fig. 11C, top, lane 8). Only at concentrations of 60 μm D4476 or higher did we see inhibition of Ser392 site phosphorylation of p53 (Fig. 11C, top, lanes 10-14 versus lane 2). Together, these data suggest that CK1 may not only be the Ser20 site kinase after virus infection but that there might be a degree of cross-talk between CK1 and the enzymes that mediate p53 phosphorylation at Ser15 and Ser392.
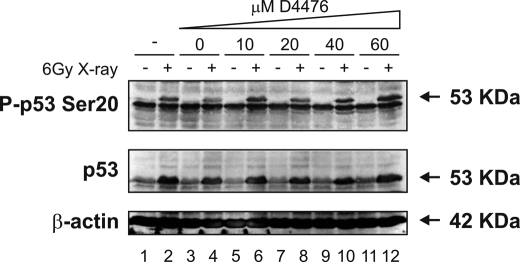
CK1 Inhibitors Do Not Attenuate p53 Induction by X-ray Treatment—We next set out to determine whether CK1 is a global Ser20 site kinase for p53 in response to a wide range of stresses or whether CK1 acts specifically in response to viral infection. MOLT-3 cells were therefore treated with and without a 6-gray x-ray in the presence of increasing amounts of the CK1 inhibitor D4476. Although x-rays induced the phosphorylation of p53 at Ser20 along with an increase in p53 levels (Fig. 12, even-numbered lanes versus odd-numbered lanes), increasing the addition of D4476 did not attenuate either the Ser20 site phosphorylation or the induction of p53 (Fig. 12, lanes 6-12 versus lanes 2 and 4). The data therefore suggest that CK1 is a virus-specific Ser20 site kinase for p53 and that other kinases are responsible for mediating the phosphorylation of p53 at Ser20 after distinct stresses, such as x-rays.
FIGURE 12.
A CK1 inhibitor does not attenuate Ser20 site phosphorylation of p53 and p53 induction mediated by treatment with x-rays. MOLT-3 cells were treated with (even-numbered lanes) or without (odd-numbered lanes) 6-gray x-ray and cultured for 4 h after an initial 44-h pretreatment with increasing concentrations (10-60 μm) of the CK1 inhibitor D4476 (lanes 5-12), a DMSO solvent control (lanes 3 and 4), or a culture medium control (lanes 1 and 2). Cell lysates were examined by Western blotting with antibodies against the indicated proteins.
DISCUSSION
The p53 tetramer is subject to a complex set of diverse post-translational modifications that are induced in a coordinated manner by stress-activated pathways. These covalent adducts include phosphorylation, ubiquitination, acetylation, additional ubiquitin-like modifications, and methylation. These modifications occur within different domains of the p53 protein and result in distinct effects both on the tetrameric conformation and the protein interaction network of p53, and they ultimately affect the specific activity of p53. These post-translational modifications are catalyzed by a range of enzymes, including p300, UBCH5/Mdm2, cyclin-dependent kinases, CK2, and CHK2. A feature of these enzymatic modifications is that they occur through enzyme docking to multiple linear peptide interaction sites within the p53 tetramer (2). Presumably, p53 has therefore evolved these unstructured and flexible motifs common to regulatory enzymes in order to coordinate multiple stress-activated signaling events.
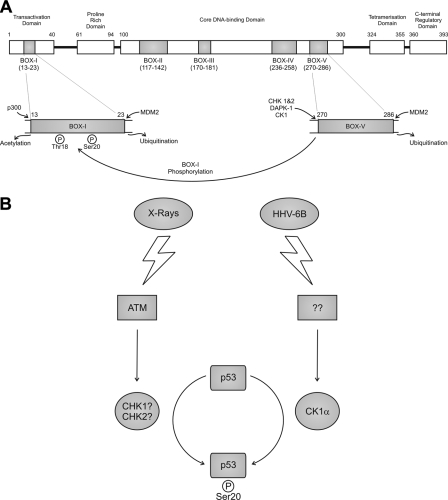
Since phosphorylation of the Ser20 site of p53 plays a dominant role in stabilizing p300 and mediating DNA-dependent acetylation of p53, as well as B-cell lymphoma suppression, it is of interest to define the environmental stresses and the kinases that mediate this phosphorylation. Studies originally highlighted that CHK1 and CHK2 were the major Ser20 kinases for p53, although this has been questioned (20, 21). Subsequent work has shown that members of the calcium calmodulin kinase superfamily, including CHK1, CHK2, AMPK (AMP-activated protein kinase), DAPK-1, DAPK-3, and DRAK-1 (DAPK-related 1) can phosphorylate the Thr18 and/or Ser20 residues of p53 in a cell-free assay (18). These kinases, including CHK1/2, modify p53 in a dual site docking-dependent manner. This shows that phosphorylation events within the BOX-I domain of p53 are dependent on kinase docking interactions within the conformationally flexible BOX-V domain of p53 (model depicted in Fig. 13A). Similarly, recent work has also shown that the ubiquitination of p53 by Mdm2 is regulated by a related dual site mechanism. Stimulation of the ubiquitin ligase function of Mdm2 requires the interaction of Mdm2 both with the BOX-I domain of p53 and with a ubiquitination signal found within the BOX-V domain of p53 (22). It is therefore becoming increasingly apparent that the BOX-V domain of p53 is a key multiprotein docking region, equivalent to the BOX-I domain, with both activating kinases and negative regulators potentially competing for binding to overlapping motifs (Fig. 13A).
FIGURE 13.
Multiprotein docking site and p53 activation models. A, the interaction of CK1 and calcium calmodulin kinases with the multiprotein docking sites in the BOX-IV and BOX-V domains of p53 allows their subsequent phosphorylation of Ser20 and/or Thr18 sites within the BOX-I domain of p53. The domain structure of p53 (393 amino acids) is represented, including the conserved domains, labeled with the indicated amino acids flanking BOX-I through BOX-V. The BOX-I domain from amino acid 13 to 23 contains the Thr18 and Ser20 phosphorylation sites and the N-terminal docking sites for p300 and MDM2 that are required for acetylation and ubiquitination of p53, respectively. The BOX-V domain from amino acid 270 to 286 contains the protein kinase and MDM2 docking sites that are required for phosphorylation and ubiquitination, respectively. B, different kinase signaling pathways link the two distinct stresses of DNA damage and viral infection to p53 Ser20 site phosphorylation and activation. p53 is activated by distinct stresses, including DNA damage, metabolic stress, oncogene activation, and virus infection. CHK1 and/or CHK2 have been implicated as the most likely Ser20 site kinases in response to DNA damage (18, 20), but this has been questioned using gene knock-out technologies (21). Thus, the identity of the DNA damage-induced Ser20 site kinase is not necessarily evident. By contrast, this current study demonstrates that DNA virus-induced Ser20 site phosphorylation of p53 is not mediated by CHK1/2 but rather by CK1. These data support the model that the Ser20 site phosphorylation of p53 is triggered by distinct stress-responsive signaling cascades, and future analysis will be required to determine the identity of the enzymes that mediate stress-induced phosphorylation of p53 at this site.
Although DNA damage induced by radiation has been the most well characterized stress that activates p53, perhaps more biologically intrinsic stresses have also been reported that have not been as well dissected. These include hypoxia, interferon-signaling, and DNA or RNA virus infection. A range of DNA or RNA viruses can induce p53-dependent apoptosis in some cell types (23, 24). Further, type I interferons can activate p53-dependent processes in animal models (12). Whether these interferon signaling effects occur in dendritic cells and/or macrophages and how this alters immune system integrity in either normal or cancer cells remains to be determined. The link between p53 and interferon or virus responses is particularly intriguing, since a role for p53 in immunity might have been the physiological selection pressure under which the p53 pathway appeared during evolution in chordates. This would have occurred evolutionarily to maintain host survival during viral or pathogen infection. Indeed, some tumor viruses have evolved proteins that bind to and inactivate or degrade p53, thus permitting virus propagation. Understanding the kinase signaling pathways that cells use to activate p53 after virus infection should provide fundamental insights into the physiology of the p53 response to immune system stresses. Although there is a range of RNA and DNA viruses that have been reported to activate p53-dependent processes, we have used HHV-6B as a model. This virus is thought to be the most ancient, founding member of the HHV family of viruses and has the unusual property of inducing growth arrest rather than apoptosis in infected T-cells (14, 25).
HHV-6B has previously been reported to induce phosphorylation of p53 at Ser15, Ser20, Ser33, and Ser392 (14, 15). Phosphorylation at Thr18 after viral infection cannot be detected in this system (data not shown). Of these sites, the Ser20 and Ser392 phosphoacceptor site mutations have been made in animal transgenic models and result in stress-induced or cell-specific tumor-prone phenotypes (7-9). The Ser20 phosphoacceptor site has appeared rather late in evolution, since it is found only in mammals. The phosphorylation of p53 at Ser20 has the most striking effect in stabilizing the binding of p300 (4), which in turn stimulates the DNA-dependent acetylation of p53 (3, 26). Interestingly, although the Ser20 site is confined to mammals, certain fish species have evolved a negatively charged amino acid at this position, suggesting that other vertebrates also exploit this site to anchor p300. Defining which kinase pathway(s) targets the Ser20 site after viral infection will help us to identify potential evolutionarily core signaling pathways that have maintained mammalian host survival under the natural selection imposed by virus infection.
In this report, we identify CK1α as the dominant enzyme that catalyzes Ser20 site phosphorylation of p53 after viral infection. This conclusion is based on (i) the purification of the virus-induced Ser20 site kinase away from the known Ser20 site kinases, including CHK1/2 and DAPK-1, (ii) the immunodepletion of p53 Ser20 kinase activity from infected cell lysates, and (iii) the ability of a relatively specific CK1 inhibitor to attenuate virus-induced Ser20 site phosphorylation in cells. Like all enzymes reported to modify the Thr18 or Ser20 sites of p53 to date, CK1α also interacts with the multiprotein docking site in the BOX-V domain of p53 (model illustrated in Fig. 13A). Further, despite the fact that CK1 can catalyze p53 phosphorylation at Thr18 (27), with a prior DNA damage-inducible modification at Ser15 by phosphoinositide 3-kinases, the original characterization of CK1δ specificity in targeting the transactivation domain of p53 showed that mutation of the Ser20 site resulted in the most pronounced block of p53 phosphorylation (11).
The function of CK1 in virus signaling was only recently realized, in part, due to the availability of relatively specific inhibitors of CK1. In particular, CK1 was implicated in mediating NSP5-dependent rotavirus RNA replication (28) and in mediating hepatitis C virus NS5A protein hyperphosphorylation (29, 30). Whether cells exploit CK1 to modify p53 after infection by RNA viruses or other proapoptotic DNA viruses remains to be established. In addition, whether cells exploit CHK1/2 after virus infection to target p53 remains to be determined; it may be a possibility if DNA replication and genome checkpoints sense and signal to the ataxia telangiectasia-mutated and Rad3-related (ATM/ATR)-CHK1/2 axis. Further, the function of p53 after HHV-6B infection remains to be determined, and the changes in gene expression induced by p53 need to be characterized. Presumably the virus-induced Ser20 site phosphorylation will alter p300 binding and related p300 co-activated transcriptional targets. However, we do not know whether this will facilitate virus survival and replication or whether it will facilitate host T-cell responses and organism survival.
Although we identified a novel role for CK1α in linking the p53 checkpoint pathway to the cellular response to DNA virus infection, our studies suggest that distinct kinases are responsible for mediating the phosphorylation of p53 at Ser20 after different stresses (model depicted in Fig. 13B). For example, Ser20 site phosphorylation in response to irradiation damage has been historically attributed to CHK1/2. It therefore appears that distinct stresses lead to the activation of different p53-activating kinases, which presumably enables cells to adopt appropriate stress-specific responses.
In conclusion, the enzymes that coordinately modify p53 protein after cell stress form a relatively large family of enzymes, including kinases, acetyltransferase, and ubiquitin conjugation enzymes. Although irradiation damage has been the most well characterized stress-activating system for p53, in part because anti-cancer agents often function by inducing DNA damage, more physiological stresses have not been as well characterized. In particular, the ability of cells to respond to DNA or RNA virus infection by switching on p53 activation pathways provides an opportunity to identify signaling pathways that might shed light on how viruses target the immune system. In this report, we provide evidence suggesting that CK1 is a major enzyme that targets the transactivation domain of p53 by phosphorylation after HHV-6B infection. Our work highlights CK1 as a component of an intriguing pathway that warrants further investigation in order to understand the interaction between viruses and the mammalian immune system.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FLp53, full-length p53; GST, glutathione S-transferase; P-p53, phosphorylated p53; DTT, dithiothreitol; bis-tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; MS, mass spectrometry; MS/MS, tandem mass spectrometry.
References
- 1.Levine, A. J., Hu, W., and Feng, Z. (2006) Cell Death Differ. 13 1027-1036 [DOI] [PubMed] [Google Scholar]
- 2.Hupp, T. R., and Walkinshaw, M. (2007) Nat. Struct. Mol. Biol. 14 885-887 [DOI] [PubMed] [Google Scholar]
- 3.Dornan, D., Shimizu, H., Perkins, N. D., and Hupp, T. R. (2003) J. Biol. Chem. 278 13431-13441 [DOI] [PubMed] [Google Scholar]
- 4.Polley, S., Guha, S., Roy, N. S., Kar, S., Sakaguchi, K., Chuman, Y., Swaminathan, V., Kundu, T., and Roy, S. (2008) J. Mol. Biol. 376 8-12 [DOI] [PubMed] [Google Scholar]
- 5.Teufel, D. P., Freund, S. M., Bycroft, M., and Fersht, A. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7009-7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hupp, T. R., Sparks, A., and Lane, D. P. (1995) Cell 83 237-245 [DOI] [PubMed] [Google Scholar]
- 7.MacPherson, D., Kim, J., Kim, T., Rhee, B. K., Van Oostrom, C. T., DiTullio, R. A., Venere, M., Halazonetis, T. D., Bronson, R., De Vries, A., Fleming, M., and Jacks, T. (2004) EMBO J. 23 3689-3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruins, W., Zwart, E., Attardi, L. D., Iwakuma, T., Hoogervorst, E. M., Beems, R. B., Miranda, B., van Oostrom, C. T., van den Berg, J., van den Aardweg, G. J., Lozano, G., van Steeg, H., Jacks, T., and de Vries, A. (2004) Mol. Cell Biol. 24 8884-8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruins, W., Jonker, M. J., Bruning, O., Pennings, J. L., Schaap, M. M., Hoogervorst, E. M., van Steeg, H., Breit, T. M., and de Vries, A. (2007) Carcinogenesis 28 1814-1823 [DOI] [PubMed] [Google Scholar]
- 10.Vega, F. M., Sevilla, A., and Lazo, P. A. (2004) Mol. Cell Biol. 24 10366-10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumaz, N., Milne, D. M., and Meek, D. W. (1999) FEBS Lett. 463 312-316 [DOI] [PubMed] [Google Scholar]
- 12.Takaoka, A., Hayakawa, S., Yanai, H., Stoiber, D., Negishi, H., Kikuchi, H., Sasaki, S., Imai, K., Shibue, T., Honda, K., and Taniguchi, T. (2003) Nature 424 516-523 [DOI] [PubMed] [Google Scholar]
- 13.Boutell, C., and Everett, R. D. (2004) J. Virol. 78 8068-8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oster, B., Bundgaard, B., and Hollsberg, P. (2005) J. Virol. 79 1961-1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oster, B., Bundgaard, B., Hupp, T. R., and Hollsberg, P. (2008) J. Gen. Virol. 89 87-96 [DOI] [PubMed] [Google Scholar]
- 16.Craig, A. L., Bray, S. E., Finlan, L. E., Kernohan, N. M., and Hupp, T. R. (2003) Methods Mol. Biol. 234 171-202 [DOI] [PubMed] [Google Scholar]
- 17.Valbuena, A., Lopez-Sanchez, I., Vega, F. M., Sevilla, A., Sanz-Garcia, M., Blanco, S., and Lazo, P. A. (2007) Arch. Biochem. Biophys. 465 219-226 [DOI] [PubMed] [Google Scholar]
- 18.Craig, A. L., Chrystal, J. A., Fraser, J. A., Sphyris, N., Lin, Y., Harrison, B. J., Scott, M. T., Dornreiter, I., and Hupp, T. R. (2007) Mol. Cell Biol. 27 3542-3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig, A., Scott, M., Burch, L., Smith, G., Ball, K., and Hupp, T. (2003) EMBO Rep. 4 787-792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shieh, S. Y., Ahn, J., Tamai, K., Taya, Y., and Prives, C. (2000) Genes Dev. 14 289-300 [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn, J., Urist, M., and Prives, C. (2003) J. Biol. Chem. 278 20480-20489 [DOI] [PubMed] [Google Scholar]
- 22.Wallace, M., Worrall, E., Pettersson, S., Hupp, T. R., and Ball, K. L. (2006) Mol. Cell 23 251-263 [DOI] [PubMed] [Google Scholar]
- 23.Ping-Yuan, L., Hung-Jen, L., Meng-Jiun, L., Feng-Ling, Y., Hsue-Yin, H., Jeng-Woei, L., and Wen-Ling, S. (2006) Apoptosis 11 2179-2193 [DOI] [PubMed] [Google Scholar]
- 24.Sun, Y., and Leaman, D. W. (2005) J. Biol. Chem. 280 15561-15568 [DOI] [PubMed] [Google Scholar]
- 25.Pedersen, S. M., and Hollsberg, P. (2006) Virology 356 1-3 [DOI] [PubMed] [Google Scholar]
- 26.Dornan, D., Shimizu, H., Burch, L., Smith, A. J., and Hupp, T. R. (2003) Mol. Cell Biol. 23 8846-8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alsheich-Bartok, O., Haupt, S., Alkalay-Snir, I., Saito, S., Appella, E., and Haupt, Y. (2008) Oncogene 27 3653-3661 [DOI] [PubMed] [Google Scholar]
- 28.Campagna, M., Budini, M., Arnoldi, F., Desselberger, U., Allende, J. E., and Burrone, O. R. (2007) J. Gen. Virol. 88 2800-2810 [DOI] [PubMed] [Google Scholar]
- 29.Quintavalle, M., Sambucini, S., Summa, V., Orsatti, L., Talamo, F., De Francesco, R., and Neddermann, P. (2007) J. Biol. Chem. 282 5536-5544 [DOI] [PubMed] [Google Scholar]
- 30.Quintavalle, M., Sambucini, S., Di Pietro, C., De Francesco, R., and Neddermann, P. (2006) J. Virol. 80 11305-11312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rena, G, Bain, J., Elliot, M., and Cohen, P. (2004) EMBO Rep. 5 60-65 [DOI] [PMC free article] [PubMed] [Google Scholar]