Abstract
Nitration of tyrosine residues has been observed during various acute and chronic inflammatory diseases. However, the mechanism of tyrosine nitration and the nature of the proteins that become tyrosine nitrated during inflammation remain unclear. Here we show that eosinophils but not other cell types including neutrophils contain nitrotyrosine-positive proteins in specific granules. Furthermore, we demonstrate that the human eosinophil toxins, eosinophil peroxidase (EPO), major basic protein, eosinophil-derived neurotoxin (EDN) and eosinophil cationic protein (ECP), and the respective murine toxins, are post-translationally modified by nitration at tyrosine residues during cell maturation. High resolution affinity-mass spectrometry identified specific single nitration sites at Tyr349 in EPO and Tyr33 in both ECP and EDN. ECP and EDN crystal structures revealed and EPO structure modeling suggested that the nitrated tyrosine residues in the toxins are surface exposed. Studies in EPO-/-, gp91phox-/-, and NOS-/- mice revealed that tyrosine nitration of these toxins is mediated by EPO in the presence of hydrogen peroxide and minute amounts of NOx. Tyrosine nitration of eosinophil granule toxins occurs during maturation of eosinophils, independent of inflammation. These results provide evidence that post-translational tyrosine nitration is unique to eosinophils.
Human eosinophils are bone marrow-derived, non-dividing granulocytes of the innate immune system, which store the highly cationic proteins eosinophil peroxidase (EPO),3 major basic protein (MBP), eosinophil-derived neurotoxin (EDN), and eosinophil cationic protein (ECP) in secondary granules (1, 2). In rodents, eosinophil proteins with similar structure and activity have been identified (3-6). In response to allergen provocation or parasitic infection, expanded eosinophil populations from the bone marrow are selectively recruited to affected tissues (1). The release of cationic toxins from activated eosinophils by degranulation is regarded as a dominant effector function of these cells, mediating lysis of helminths and protozoae (7-10). The positive net charge conveys tight toxin binding to negatively charged cell surfaces where EPO causes oxidation of membrane components in the presence of hydrogen peroxide (H2O2) (8). MBP increases membrane permeability and perturbation following insertion of apolar residues into the membrane (11), whereas ECP creates membrane channels (12). Both EDN and ECP exert ribonuclease activity (13). Due to their unspecific binding to membranes and their cytolytic activities, eosinophil granule proteins also cause host tissue damage during parasite infections and inflammatory disorders such as allergic asthma (14-16).
Despite advances in elucidating the mechanisms of action for the eosinophil
granule proteins (1,
2), the cationic nature of
these proteins (pI values > 10) would predict electrostatic repulsion and
exclude interaction/cooperation of single granule proteins and among different
granule proteins. Because sequence data for human EPO, MBP, EDN, and ECP
reveal the presence of 4 to 16 tyrosine residues, and EPO can generate
3-nitrotyrosine (3NT) via oxidation of nitrite
(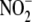 )
(17), we hypothesized that
selective post-translational 3NT formation in eosinophil granule proteins is
likely necessary to overcome such logistical issues.
)
(17), we hypothesized that
selective post-translational 3NT formation in eosinophil granule proteins is
likely necessary to overcome such logistical issues.
Here we show that all of the eosinophil granule proteins from both men and mouse are nitrated. We further identified specific single nitration sites, using high resolution affinity-mass spectrometry, in several of the human eosinophil granule proteins: Tyr349 in EPO and Tyr33 in both human ECP and EDN. Moreover, we demonstrated that Tyr nitration of the eosinophil granule proteins is exclusively mediated by EPO, in the presence of functional NADPH oxidase and minute amounts of NOx. EPO appears to nitrate itself via an autocatalytic mechanism.
EXPERIMENTAL PROCEDURES
Human Lung Tissue and Blood Samples—Cystic fibrosis (CF) lung tissue (Centre Hospitalier Lyon Sud, Pierre-Bénite, France) and healthy lung tissue (Klinik Schillerhöhe, Gerlingen, Germany) were cut in 1 × 1 × 1-cm blocks and frozen in liquid nitrogen. Frozen tissue blocks were cut into 5-6-μm sections and put on poly-l-lysine-coated glass slides for immunofluorescence. Blood samples were obtained from patients with helminth infections (Paul-Lechler-Krankenhaus, Tübingen, Germany), from healthy individuals (Institute of Medical Microbiology and Hygiene, Universitätsklinikum Tübingen, Germany), from patients with atopic rhinitis or asthma bronchiale (Swiss Institute of Allergy and Asthma Research (SIAF), Davos, Switzerland and Universitätshautklinik Tübingen, Germany), and from patients with chronic granulomatous disease (CGD) (Children's Hospital Universitätsklinikum Tübingen, Tübingen, Germany; Nachsorgeklinik Tannheim, Tannheim, Germany; University Clinics, Zürich, Switzerland). Cord blood was obtained from the Universitäts-Frauenklinik, Tübingen, Germany. Written informed consent was obtained from all patients or their parents and the investigations were approved by the ethics committee of the University of Tübingen and the respective ethics committees of the Hopitals Civils de Lyon, France, Nachsorgeklinik Tannheim, Germany, and the SIAF Davos, and University Clinics, Zürich, Switzerland.
Animals, Eosinophil Purification, and Granule Protein Preparation—Blood samples were taken from wild type IL-5 transgenic mice (18), IL-5-transgenic, homozygous null EPO (EPO-/-) mice (19), and from IL-5/gp91phox-/- mice, generated by mating wild type IL-5 transgenic C57BL/6J mice (Mayo Clinic, Arizona) with gp91phox-/- mice (The Jackson Laboratory, Bar Harbor, ME). Mice were housed in isolator cages that provided a pathogen-free environment. The mice were repeatedly investigated for microbial infections by bacterial culturing and serology and remained negative up to date. The hygienic status was repeatedly tested by a panel of common murine pathogens according to the FELASA recommendations of 2002. Additionally, eosinophils and, in turn, granules were purified (>98%) from IL-5 transgenic mice and IL-5/gp91phox-/- mice for granule protein preparations. Spleens were removed from transgenic animals (18), disassociated into a single cell suspension, and eosinophils were purified at >98% purity using standard purification methods (20). Purified eosinophils were lysed by suspension in 0.25 m sucrose and 600 units/ml heparin for 3 min. The free DNA was degraded with 4,000 units of DNase at 37 °C for 20 min. Granule pellets were obtained by centrifugation at 10,000 × g for 20 min at 4 °C. Eosinophil granules were re-suspended in 1 ml in 0.01 m HCl, pH 3, and disrupted by sonication (Sonifier, Branson, Schwäbisch Gmünd, Germany). The suspension was centrifuged at 5,000 × g for 10 min. The protein concentration in the supernantant was determined using the Pierce assay (Perbio Science GmbH, Bonn, Germany). Human granulocytes were isolated as described above and eosinophils were isolated by negative selection using CD 16 magnetic beads and the MACS system (Miltenyi Biotec GMBH, Bergisch Gladbach, Germany).
Localization of 3NT-positive Proteins in Human and Animal Tissues and Leukocyte Preparations by Indirect Immunofluorescence—The following antibodies were used: a polyclonal rabbit antibody or a monoclonal mouse IgG antibody against 3NT (Upstate Biotechnology, Lake Placid, NY) and a Cy2-conjugated goat anti-rabbit IgG or anti-mouse IgG antibody as second antibody. Different cell types were detected using Cy3-conjugated anti-mouse antibody (Dako, Hamburg, Germany) as second antibody and cell-specific monoclonal antibodies: human eosinophils (MBP, clone BMK-13; Monosan, Uden, The Nether-lands), T lymphocytes (anti-CD3, anti-CD45RO; Dako, Hamburg, Germany), neutrophils (neutrophil elastase; Dako), mast cells (C-kit 104D201), and monocytes (anti-CD14; Dako). Stained thin sections were examined using a Zeiss Axioplan microscope (Zeiss, Oberkochen, Germany); for controls, cells were stained without the respective first antibodies. Thirty thin sections of lung tissue from six CF patients and from three healthy individuals at a magnification of ×100 were investigated.
Blood smears from patients with atopic rhinitis, helminthic infection, and healthy individuals were stained with antibodies specific for MBP and 3NT, and with DAPI. Granulocytes and lymphocytes from human heparin blood were isolated by using Ficoll T 500 (Biochrom, Berlin, Germany). Cytospin samples were prepared and the cells were fixed on slides with methanol/acetone (50/50%) and stained with antibodies specific for MBP and 3NT, and with DAPI. For assessing the percentage of 3NT and MBP double stained cells to MBP single stained cells, five pictures (210 μm × 140 μm) of each slide were taken using the Zeiss Axio vision program.
To demonstrate localization of 3NT-positive proteins in human eosinophil granules, on-section immunolabeling (21) was used. Cells were fixed with 4% formaldehyde in phosphate-buffered saline, dehydrated in a graded series of ethanol at progressive lower temperature from 0 °C down to-40 °C, infiltrated with Lowicryl K11M and UV-polymerized at -40 °C. Thereafter, ultrathin sections were stained for immunofluorescence with a rabbit antibody against 3NT and a mouse antibody against MBP, detected by corresponding fluorochrome-labeled secondary goat antibodies (Jackson). Nuclei were counter-stained with DAPI and the samples visualized in a Zeiss Axiophot using ×63 (or ×100) oil immersion objectives.
In addition, for transmission electron microscopy ultrathin sections were labeled with a rabbit antibody against 3NT and protein A-10-nm gold complexes (gift from Dr. York-Dieter Stierhof, ZMBP Tübingen). These sections were finally stained with 1% aqueous uranyl acetate and lead citrate and analyzed in a Philips CM10 electron microscope at 60 kV using a 30-μm objective aperture.
Finally, for determination of granule sizes, eosinophils from IL-5 transgenic and IL-5 transgenic EPO-/- mice were isolated. For ultrastructural studies cells were fixed with 2.5% glutaraldehyde in phosphate-buffered saline and embedded in 2% agarose to cut small blocks of about 1 mm3 in size. After post-fixation with 1% osmium tetroxide in 100 mm PO4 buffer, pH 7.2, for 1 h on ice, these blocks were rinsed with aqua bidest, treated with 1% aqueous uranyl acetate for 1 h at 4°C, dehydrated through a graded series of ethanol at ambient temperature, infiltrated with Epon, and polymerized at 60 °C. Digital pictures were taken from 5 ultrathin sections of each sample and the granula diameter of 100 cells of each sample was measured using the Zeiss AxioVision Programm 4.6.3 (Zeiss).
Cord blood cells from three donors were collected in heparin and layered over Histopaque 1077 (Sigma). Mononuclear fractions were harvested after centrifugation at 250 × g for 30 min. The cells were washed in phosphate-buffered saline and incubated at 1 × 106 cells/ml in RPMI 1640 (Invitrogen) with 10% fetal calf serum with or without IL-5 (5 ng/ml) for 1 to 14 days. Every 6 days half of the medium was replaced with fresh medium. Cytospins of cell suspensions were prepared by centrifugation at 250 × g for 5 min onto a clean glass slide. The slides were air dried and stained with a monoclonal 3NT antibody and DAPI. Additionally, eosinophils from wild type IL-5 transgenic and IL-5 transgenic EPO-/- mice were double stained with 3NT antibodies and DAPI. Eosinophils from CGD patients were double stained with antibodies against 3NT, human MBP, and DAPI. De novo nitration of non-nitrated granula proteins from IL-5/gp91phox-/- mice was tested by incubation of the proteins with 100 μm KNO2 (Sigma) and 100 μm H2O2 (Sigma) in 20 mm sodium phosphate buffer, pH 7, for 5, 15, 30, and 60 s. Aliquots were spotted onto nitrocellulose membranes and 3NT-positive proteins were detected with a monoclonal antibody against 3NT.
Furthermore, blood smears from mice lacking nitric-oxide synthase (NOS) isoforms, specifically inducible (iNOS or type II) KO mice, neuronal (nNOS or type I), and endothelial (eNOS or type III) double KO mice and iNOS/eNOS/nNOS triple KO mice (22) were stained with antibodies specific for MBP, 3NT, and DAPI.
Identification of 3NT-positive Proteins by Immunoblotting and Matrix-assisted Laser Desorption Ionization Mass Spectrometry (MALDI-MS) in Different Cell Types—Crude protein solutions from immunobead purified eosinophils (102 cells) (23), Ficoll-purified neutrophils (107 cells), monocytes/lymphocytes (107 cells) (24), and 10 μg of purified MPO (25) were subjected to SDS-PAGE (200 V/75 mA). Samples were blotted on nitrocellulose (0.8 mA/cm2, 15 V) and skim milk-blocked blots were incubated with mouse anti-3NT IgG or mouse anti-EPO IgG (Mayo Clinic, Scottsdale, AZ). Bound antibodies were detected using appropriate peroxidase-conjugated secondary antibodies (Dako) using the chemiluminescent reagent Lumigen PS-3 (Amersham Biosciences) and the ECL Detection System (Amersham Biosciences). Detection of bound antibodies was achieved using x-ray film (Hyperfilm ECL, Amersham Biosciences) in the presence of molecular weight markers (Amersham Biosciences). The specificity of the primary 3NT-antibody was confirmed by preincubating the blots with 10 mm 3NT (Sigma). The specificity of the primary 3NT-antibody was further assessed by using the Upstate Biotechnology 3NT immunoblotting control. For identification of eosinophil proteins, target bands (60 and 15 kDa) were cut out from a non-fixed Coomassie-stained SDS-PAGE, subjected to tryptic digestion, and analyzed by MALDI-TOF-mass spectrometry (MS). In-gel tryptic digestion was performed as described (26) and modified as outlined below. Briefly, the protein band was excised from the gel, fully de-stained, and digested for 3 h with porcine trypsin (sequencing grade, modified; Promega, Heidelberg, Germany) at a concentration of 67 ng/μl in 25 mm ammonium bicarbonate, pH 8.1, at 37 °C. Prior to peptide mass mapping and sequencing of tryptic fragments by tandem mass spectrometry, the peptide mixture was extracted from the gel by two changes of 50% trifluoroacetic acid, 50% water followed by two changes of 50% trifluoroacetic acid, 50% acetonitrile. The combined extracts were vacuum-dried. The dried peptides were re-dissolved in 0.1% trifluoroacetic acid and purified using a ready-to-go pipette tip filled with C18 spherical silica reverse phase material (ZipTipC18™, Millipore GmbH, Schwalbach, Germany). Peptides were eluted with 10 μl of 50% methanol, 1% formic acid, and sequencing was performed by nanoelectrospray tandem mass spectrometry on a hybrid quadrupole orthogonal acceleration time-of-flight tandem mass spectrometer (Q-Tof, Micromass, Manchester, United Kingdom), equipped with a nanoflow electrospray ionization source. Gold-coated glass capillary nanoflow needles were obtained from Protana (Type Medium NanoES spray capillaries for the Micromass Q-Tof, Odense, Denmark). The needle was filled with 3 μl of the sample and subsequently opened by breaking the tapered end of the tip under a microscope. A stable spray was observed applying a needle voltage of 1200-1400 V, a back pressure of 2 p.s.i., and a source temperature of 40 °C. The estimated flow rate was 20-50 nl/min. For nanoflow electrospray ionization tandem MS experiments, fragmentation was achieved by collision with argon atoms. Q1 was set to the mass of interest, and optimized collision energy was applied. The integration time for the time-of-flight analyzer was 1 s with an interscan delay of 0.1 s. Data base searches (NCBInr, non-redundant protein data base) were done using the MASCOT software from Matrix Science (27).
Then, 10 μg of human EPO, MBP, EDN, and ECP (Mayo Clinic, Rochester, MN) was purified as previously described (28), and 10 μg of murine EPO, MBP, ribonuclease-like protein, and an eosinophil granule lysate from non-inflamed IL-5 transgenic mice (Mayo Clinic, Scottsdale, AZ), were subjected to Western blotting and stained with a 3NT-specific antibody.
Mass Spectrometric Identification of Tyr Nitration Structures—Purified EPO was proteolytically digested and the tryptic peptide mixture was separated by HPLC (column PepMap C18 (3 μm, 100 Å, 300-μm inner diameter × 5 cm; LC Packings/Dionex, Idstein, Germany). For the mobile phase a gradient of two solutions (A: 0.1% aqueous solution of HCOOH (Riedel-de Haën, Seelze, Germany); B: 0.1% solution of HCOOH in acetonitrile, 20:80, v/v) (ROTISOLV®, Carl Roth GmbH, Karlsruhe, Germany) was used with a binary pump flow of 100 μl/min, which was changed after splitting to 4-10 μl/min. ESI-MS analysis of nitrated EPO peptides was performed on an Agilent 1100 system and a Bruker Apex II FT-ICR mass spectrometer (Bruker Daltonics, Bremen) equipped with APOLLO II ESI source. The mass spectra were obtained by collecting 32-64 single scans. Experimental conditions were: capillary exit voltage (ΔCS), 45-70 V; setting of skimmer 1, 10; setting of skimmer 2, 7; RF amplitude, 500; ionization pulse time, 2500 ms; ionization delay time, 1 ms; excitation sweep pulse, 1.2 ms; excitation sweep attenuation 1, 2.16 dB. For ECP and EDN, mixtures of proteolytic peptides, carrying Tyr residues, were bound to a microaffinity column with immobilized 3NT antibodies. After washing, bound peptides were eluted by mild acidic treatment and the resulting nitrated peptides were identified by nano-electrospray Fourier transform ion cyclotron resonance mass spectrometry (nano-ESI-FTICR-MS). Prior to proteolytic digestion, cysteine residues of EPO, ECP, and EDN were derivatized (carbamidomethylated) using dithiothreitol and alkylated with iodacetamide.
Synthesis of Tyr-nitrated Peptides—The Tyr-nitrated EPO peptide, 333FGHTMLQPFMFRLDSQY(nitro)R350, and the respective non-nitrated peptide, FGHTMLQPFMFRLDSQYR, were synthesized on a semiautomated peptide synthesizer (EPS-221, Abimed, Langenfeld, Germany) with Biotin and a pentaglycine spacer coupled at the final N-terminal amino acid. All peptides were purified by preparative HPLC on a Grom-Sil ODS-4Me column and the exact mass ascertained by MALDI-MS analysis.
Structure Modeling—Molecular modeling of EPO was performed using the Swiss-MODEL (swissmodel.expasy.org//SWISS-MODEL.html, an automated comparative protein modeling server). The crystal structure of MPO (PDB code 1DNU) (29) was used as the target template for homology modeling and further comparative studies. To eliminate model bias, a round of energy minimization was carried out using the CNS package (30) after substituting Tyr349 with 3NT-Tyr349. The root mean square deviation between the EPO model and the template MPO prior to energy minimization and after energy minimization is 0.38 and 0.89 Å, respectively. For ECP, structure coordinates from Mohan et al. (35) (PDB code 1H1H) were used for nitro-Tyr modeling at the nitro-Tyr33 residue.
Cytotoxicity Assays—Confluent monolayers of the human alveolar type II cell line A549 (Deutsche Sammlung von Mikro-organismen und Zellkulturen, Braunschweig, Germany) were used for testing the toxicity of nitrated and non-nitrated eosinophil granule toxins. A549 cells were cultivated in Dulbecco's minimal essential medium, supplemented 1% penicillin/streptomycin and 10% fetal calf serum at 37 °C and 5% CO2 on chamber slides (BD Bioscience, Heidelberg, Germany). Non-nitrated recombinant ECP (rECP, Diagnostics Development, Uppsala, Sweden) and rEPO (GenWay, Bio-tech Inc., San Diego, CA) were nitrated using 100 μm KNO2 and 50 μm H2O2 in 20 mm sodium phosphate buffer, pH 7.2, for 10 min. The reaction was stopped with 50 μm catalase (Sigma). Nitration was confirmed with Western blotting (data not shown). A549 cells were treated with 20 μm rECP or 5 μm rEPO and the respected nitrated proteins for 24 h. Thereafter, cells were washed and the alive or dead status of the epithelial cells was determined using the Syto13/pro-pidium iodide viability test (Molecular Probes, Leiden, The Netherlands). Ten fluorescence micrographs of each sample were taken at a magnification of ×200 using the Axio Vision program (Zeiss, Oberkochen, Germany). The number of dead cells (red fluorescence) was determined as the percentage of total cells (green fluorescence).
Antibacterial Assay—Overnight cultures of Escherichia coli strain DH5α and Staphylococcus aureus strain ATCC35556 were washed twice and resuspended in 10 mm sodium phosphate buffer, pH 7.5, to a final concentration of 1.5 × 107 colony forming units/ml. The bacteria were treated with nitrated or non-nitrated 5 μm rECP or 5 μm rEPO for 24 h. The number of surviving bacteria (percentage of total colony forming units) was determined by plating serial 10-fold dilutions on Luria-Bertani (LB) agar.
BSA Nitration by Phorbol Myristate Acetate-stimulated Eosinophils—Human eosinophils from three healthy individuals were obtained as described above. The cells were incubated with 100 μg/ml bovine serum albumin (BSA) (Sigma) in the presence of 4 μg/ml phorbol myristate acetate (Sigma) for 30 min at 37 °C in a nitrite/nitrate-free phosphate-buffered saline buffer, pH 7.2. The suspension was centrifuged (1,000 × g for 10 min at 4 °C) and BSA was isolated from the supernatant using the MACS technology (Miltenyi), using albumin antibodies (DAKO). Nitrated BSA was detected by Western blotting using rabbit 3NT antibodies.
RESULTS
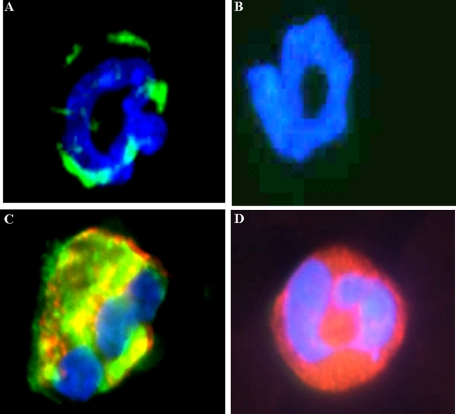
Identification of Post-translational Nitrated Cationic Proteins in Human and Mouse Eosinophil Secondary Granules—To identify 3NT-positive proteins, lung tissue sections of patients with CF that were also suffering from chronic bacterial lung infection/inflammation were stained with 3NT antibodies and cell markers specific for various cell types. Only eosinophils, i.e. cells staining with MBP, a representative eosinophil marker (2), stained positive for 3NT (Fig. 1, A-D), whereas monocytes, T lymphocytes, mast cells, and neutrophils were 3NT-negative (Fig. 1, E-L). In granulocyte preparations and blood smears from a patient with pronounced eosinophilia, 100% of purified eosinophils, positive for MBP, were 3NT-positive, whereas neutrophils and lymphocytes were completely 3NT-negative (Fig. 2A). Similar results were obtained in granulocyte preparations and blood smears from a patient with helmintic infection and blood smears from patients with allergic asthma (not shown). The results suggest that the inflammatory process leading to eosinophil activation in these patient groups is responsible for 3NT reactivity. Surprisingly, however, also in granulocyte preparations (Fig. 2B) and lung tissue sections of healthy individuals (Fig. 2C), eosinophils but not other cells stained positive for Tyr nitration. To avoid any activation of blood cells during manipulation, blood smears were taken. Also these cells from healthy individuals were 3NT-positive (Fig. 2D). These results indicate that nitration of eosinophil proteins is independent of activation events associated with inflammatory processes and occurs constitutively, because careful manipulation prevented cell activation as much as possible.
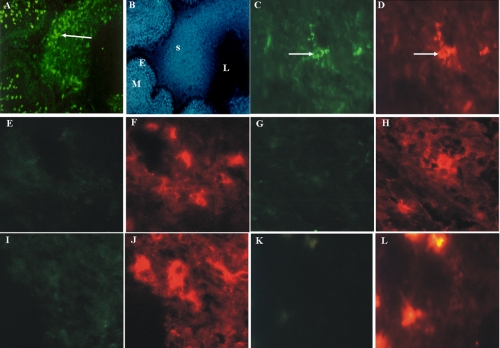
FIGURE 1.
Human tissue eosinophils from infected CF patients contain exclusively 3NT-positive residues. A and B, fluorescence micrograph of 3NT-positive cells in the lung mucosa (panel B, M) and in the sputum (panel B, S) filled bronchial lumen (panel B, L) of a patient with CF stained with a rabbit NT antibody (A) and DAPI (B)(panel B, E (epithelium)), original magnification: A and B, ×100. Double fluorescence micrographs of CF tissue sections stained with a rabbit antibody against 3NT (panels C, E, G, I, and K) and markers specific for eosinophils (D), T-lymphocytes (F), mast cells (H), monocytes (J), and neutrophils (L). Only eosinophils were 3NT-positive (panels C and D, arrows). Original magnification: panels C-L, ×400.
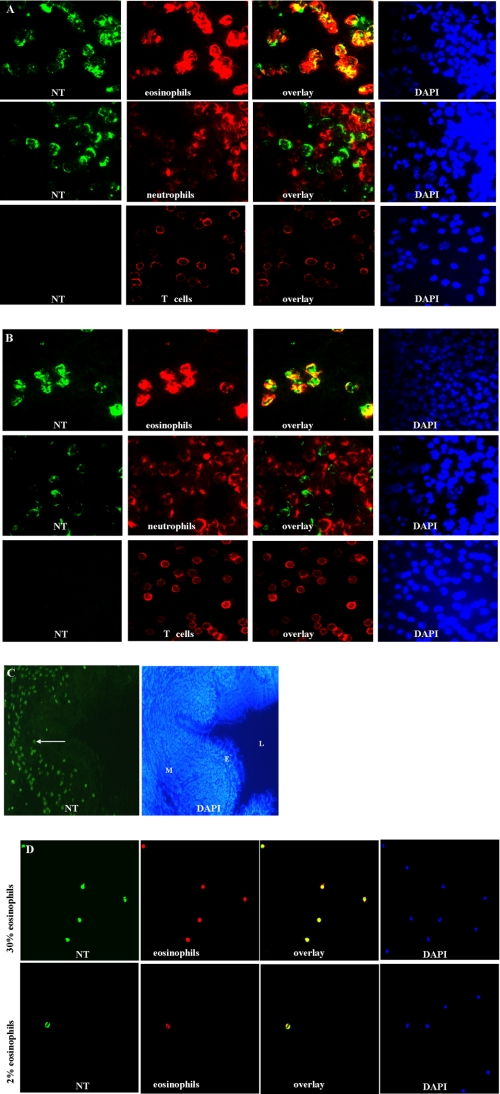
FIGURE 2.
Human blood eosinophils from inflamed and healthy individuals are exclusively 3NT-positive. Double fluorescence micrographs of granulocyte and lymphocyte preparations of a patient with atopic rhinitis (A) and a healthy individual (B) were stained with a rabbit antibody against 3NT (green) and cell-specific monoclonal antibodies against MBP (red) for eosinophils, neutrophil elastase for neutrophils, and CD3 for T-lymphocytes. Additionally, cells were stained with DAPI. The overlay shows perfect co-localization of MBP and 3NT in eosinophils but no co-localization of neutrohil elastase with 3NT or presence of 3NT in lymphocytes. C, in lung tissue of a healthy individual NT-positive cells are present in the lung mucosa (M)(L, lumen; E, epithelium). Original magnification: C, ×100. D, the overlay of double-stained blood smears of a patient with atopic rhinitis and a healthy individual show double staining exclusively for eosinophil cells. Original magnification: A, B, and D, ×400.
The eosinophil specificity of Tyr nitration was further substantiated by nitration pattern assessment of proteins derived from cell extracts of purified neutrophils (107 cells) or lymphocytes/monocytes (107 cells) from healthy individuals. In contrast to eosinophils (Fig. 3A, lane 1), which stained positively although a low number of cells was used (102 cells), no evidence of nitration was visible in neutrophils or lymphocytes/monocytes (Fig. 3A, lanes 2 and 3). We also tested for the presence of 3NT immunoreactivity in purified myeloperoxidase (MPO), an enzyme that is structurally (31) and enzymatically (17, 32) highly homologous to EPO. However, Western blotting did not show evidence of nitration, although 10 μg of protein was applied to the gel (Fig. 3A, lane 4), suggesting again that only the major cationic granule proteins from the eosinophils of humans and mice are nitrated.
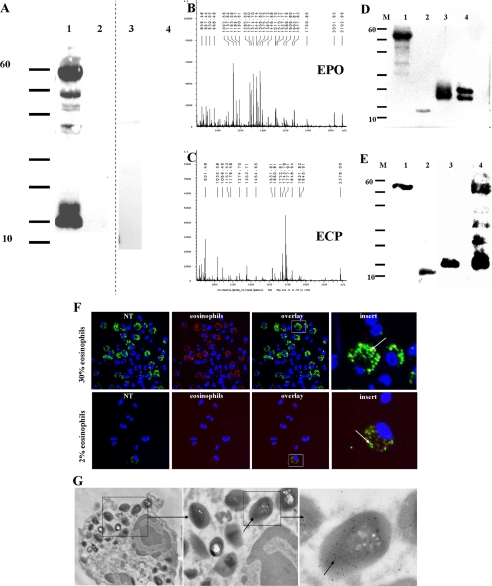
FIGURE 3.
Human and mouse eosinophils contain 3NT-positive EPO, MBP, EDN, and ECP. A, two Western blots of purified eosinophils (102 cells) (lane 1), purified neutrophils (107 cell, lane 2), or lymphocytes/monocytes (107 cells, lane 3) and myeloperoxidase (lane 4), stained with 3NT antibodies, are separated by a dotted line. Only eosinophil cell extracts were 3NT-positive. B and C, peptide mass fingerprint of the 60- and 15-kDa band of A, lane 1 shows EPO (60 kDa) and ECP (15 kDa). D, Western blot of purified human eosinophil granule proteins, stained with 3NT antibodies. Lane 1, EPO; lane 2, MBP; lane 3, ECP; lane 4, EDN. E, Western blot of eosinophil granule proteins from IL-5 transgenic mice, stained with 3NT antibodies. Lane 1, EPO; lane 2, MBP; lane 3, ribonuclease-like protein; lane 4, eosinophil granule lysate. F, immunofluorescence micrographs. Double staining with a rabbit antibody against 3NT and mouse antibody against MBP of ultrathin sections of granulocyte preparations from a atopic rhinitis patient and a healthy individual. The overlay shows double staining of MBP and 3NT within the granules of eosinophil cells (arrows, inserts). Original magnification: ×1000. G, immunogold labeling of ultrathin sections of a granulocyte preparation from a atopic rhinitis patient with a rabbit antibody against 3NT shows co-localization of granules and 3NT antibodies. Original magnification: left, ×8,900; middle and right, ×15,000.
The identities of the 3NT-containing proteins in eosinophils were determined by a proteomic approach using Western blot and MALDI-TOF analyses. Two 3NT-positive protein bands at 60 and 15 kDa from urea-solubilized human eosinophils of one patient, detectable by SDS-PAGE/Western blotting (Fig. 3A, lane 1), were identified as eosinophil secondary granule proteins upon trypsin digestion and MALDI-TOF analyses (Fig. 3, B and C). These results were corroborated by Western blot using 3NT antibodies and purified human granule proteins (i.e. EPO, MBP, ECP, and EDN) from several patients with eosinophilia (Fig. 3D). All four granule proteins stained positive for 3NT residues. Interestingly, only the heavy band, and not the light band (calculated molecular mass: 12.7 Da), of purified EPO was 3NT-positive. Significantly, Western blots of the respective mouse eosinophil granule proteins (i.e. EPO, MBP, and ribonuclease-like protein), isolated from non-inflamed IL-5 transgenic mice, demonstrated that each of these cationic secondary granule proteins were also 3NT-positive (Fig. 3E). The additional bands, seen in lane 4 of Fig. 2E are most probably digested peptides from the heavy band of EPO as also seen in Fig. 3, A and D (lanes 1). In lane 4 of Fig. 3E, the MBP band is present at a slightly higher molecular weight compared with lane 2, probably because the hydrophobic MBP aggregates with other murine granule proteins such as the ribonuclease-like protein. The murine granule lysate resembles very much the human eosinophil lysate in Fig. 3A, lane 1, strongly suggesting that only granule proteins are positive for 3NT. The silver stain of the respective SDS-PAGE gel revealed that many other proteins were present (see below, Fig. 6).
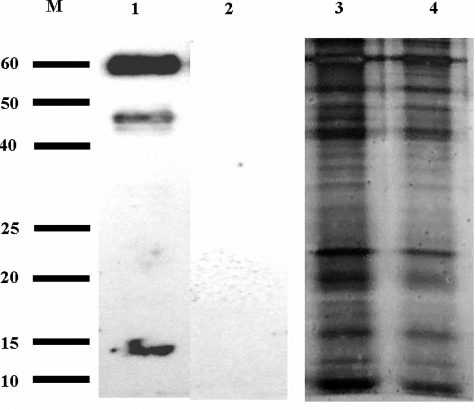
FIGURE 6.
Eosinophils from patients with chronic granulomatosis and IL-5/gp91phox-/- mice do not express tyrosine-nitrated granules toxins. Western blotting (lanes 1 and 2) and silver-stained SDS gel (lanes 3 and 4) of equal concentrations of eosinophil secondary granule proteins from IL-5 transgenic (lane 1 and 3) and IL-5/gp91phox-/- (lanes 2 and 4) mice. M, molecular weight markers.
To rigorously confirm our notion that 3NT-positive proteins of eosinophils are exclusively present in granules, ultrathin sections from granulocyte preparations both from a patient with atopic rhinitis and from a healthy individual were stained with antibodies against 3NT and MBP. Immunofluorescence (Fig. 3F) revealed co-localization within eosinophils granules but not outside of granules and immunogold labeling showed 3NT labeling within the granules (Fig. 3G).
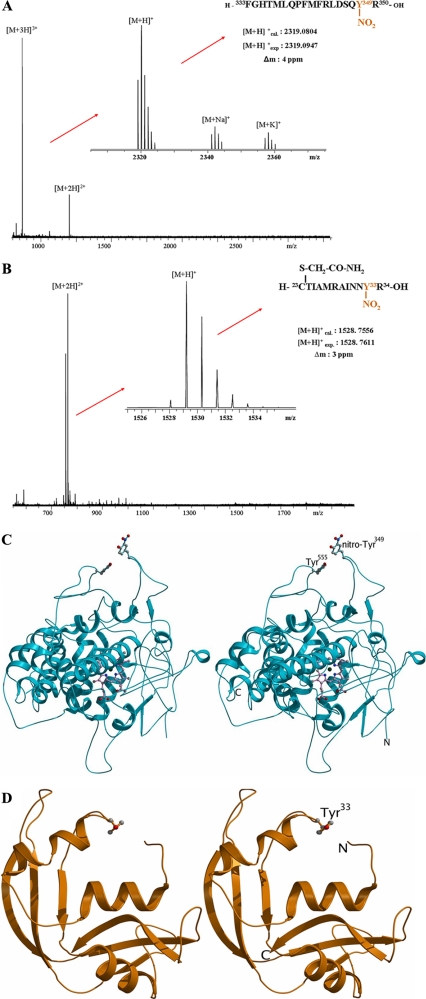
EPO and ECP/EDN Carry a Single, Surface-exposed Nitrotyrosine Residue at Tyr349 and Tyr33, Respectively—Identification of the specific Tyr nitration sites in highly purified human EPO were obtained after trypsin proteolysis and separation of the tryptic peptides mixture by HPLC followed by ESI-MS analysis. Tyr nitration sites in ECP and EDN were obtained by a combination of affinity proteomics and high resolution nano-ESI-FTICR-MS (33). Both methods provided the unequivocal identification of single nitration sites in EPO by the isolated tryptic peptide fragment (333FGHTMLQPFMFRLDSQY(NO2)R350) nitrated at Tyr349 (Fig. 4A), and in ECP by the tryptic fragment (23CTIAMRAINNY(NO2)R34) nitrated at Tyr33 (Fig. 4B), whereas the nitration site in EDN with high sequence homology to ECP was identified at Tyr33 by the affinity-isolated thermolysin peptide fragment (29VINNY(NO2)QRRCKNQNTF43) (data not shown). Sequence coverage values obtained for the proteomic identification were ∼38% for EPO, which may be possibly explained by its large molecular mass, possible multiple missed cleavages (e.g. at Arg344 and Arg27), and different glycosylation sites. Six of 13 Tyr residues were covered in mature EPO. In ECP, a sequence coverage of 67% was obtained, including all unmodified tyrosine residues. These results showed specific nitration at single, specific Tyr residues in eosinophil proteins, detectable at low levels.
FIGURE 4.
EPO carries a single, surface-exposed, 3NT residue at Tyr349, which mediates binding of EPO to positively charged surfaces. A, high resolution ESI-FT-ICR-mass spectrum of the tryptic EPO peptide 472-489-NO2. Upon deconvolution of the 3-fold charged ion, sodium and potassium adducts can be observed in addition to [M + H]+. B, high resolution ESI-FT-ICR-mass spectrum of the tryptic ECP peptide CTIAMRAINNY(NO2). Upon deconvolution of the 3-fold charged ion, sodium and potassium adducts can be observed in addition to [M + H]+. C, stereo diagram representation of an EPO model, using structural data of MPO as the target template for homology modeling. Tyr349 is represented as a ball-and-stick model. D, stereo diagram representation of ECP. Tyr33 is represented as a ball-and-stick model.
The crystal structure of the related myeloid-specific peroxidase MPO (PDB code 1DNU) (29) was used as a template for homology modeling and further comparative studies to assess the spatial orientation of the nitrated Tyr349 residue in EPO. EPO and MPO share ∼71% sequence identity (31), suggesting that both enzymes have similar tertiary structures. This comparison showed that non-nitrated Tyr349 is present in a highly flexible loop region (∼23 amino acids in length) in a weak, parallel stacking interaction (34) with Tyr555 (Fig. 4C). However, upon nitration of Tyr349, the flexibility of this loop is lost due to steric hindrance and the net electronegativity of the nitrated Tyr349 residue likely prevents the stacking interactions with Tyr555, forcing the nitro group of Tyr349 to be permanently exposed to the surface of the molecule. The spatial orientation of the nitro-Tyr residues in ECP and EDN were deduced via a similar strategy based on the available crystal structure for these proteins (35, 36). These assessments showed that Tyr33 residues of ECP and EDN were both surface exposed (Fig. 4D), whereas the other three Tyr residues of these granule proteins are instead oriented toward the inner structure of the molecule (Table 1). Interestingly, the nitro group on Tyr33 was not found when ECP and EDN crystal structures were analyzed previously, because recombinant material was used to generate crystals of these molecules. Dot blots of recombinant ECP (rECP) and rEPO were 3NT-negative when stained with the 3NT antibody (data not shown).
TABLE 1.
Solvent accessibility and contacts for the four tyrosine residues in ECP
Calculated using the program Areaimol from CCP4 software suite (60).
| Tyrosine residue position | Solvent accessibility | Contact surface area |
|---|---|---|
| 33 | 101.2 | 30.1 |
| 98 | 6.8 | 1.9 |
| 107 | 8.6 | 2.5 |
| 122 | 57.2 | 17.4 |
| Nitro-Tyr33 | 136.9 | 39.9 |
Tyr Nitration of Eosinophil Granule Proteins Is Mediated by EPO in the Presence of H2O2 and NO—Previous studies by Hazen and colleagues demonstrated that in the presence of H2O2 and a source of nitrite EPO specifically nitrates Tyr residues in proteins (17). We therefore examined the 3NT status of eosinophil granule proteins from EPO knock-out mice (19) to determine whether nitration of Tyr was a function of EPO activity. Immunofluorescence microscopy of 3NT antibody-stained eosinophils from wild type and EPO-/- mice was used to assess the EPO dependence of Tyr nitration. The analysis showed that in contrast to eosinophils from wild type mice (Fig. 5A), 3NT immunostaining was not detectable in eosinophils from EPO-/- animals (Fig. 5B), suggesting that mouse EPO catalyzes the oxidative nitration of Tyr residues.
FIGURE 5.
Tyrosine nitration of eosinophil granule toxins is mediated by EPO in the presence of H2O2 and a nitrite source. Fluorescence micrographs of eosinophils from a wild type mouse (A) and an interleukin 5-transgenic, homozygous null EPO (EPO-/-) mouse (B), from an otherwise healthy human individual (C) and a CGD patient stained for 3NT, green; MBP, red; and DAPI, blue (D) Original magnification: ×1000.
The linkage between Tyr nitration and EPO-mediated activities was further substantiated by 3NT staining of eosinophils from patients with CGD. These patients lack NADPH oxidase activity and hence, do not produce intracellular H2O2 (37), a required substrate for EPO-mediated nitration events. Thus, our expectation was that the lack of intracellular H2O2 would prevent the nitration of eosinophil granule proteins in CGD patients. Indeed, immunofluorescence microscopy, following dual immunofluorescence staining with antibodies for the secondary granule protein MBP and 3NT residues, showed that whereas eosinophils of normal individuals were 3NT-positive (Fig. 5C), eosinophils from patients with CGD were negative for 3NT residues (Fig. 5D).
Evidence of EPO/H2O2-dependent nitration of eosinophil granule proteins is not limited to human patients but was also observed in mice. In particular, we compared the nitration pattern observed among eosinophil granule proteins isolated from the peripheral blood eosinophils of IL-5 transgenic mice (18) and compared this pattern of Tyr nitration among granule proteins that were isolated from peripheral blood eosinophils of IL-5 transgenic mice that were also deficient in a gene resulting in X-linked CGD (gp91phox-/-) (37). Specifically, Western blots of equal concentrations of eosinophil secondary granule proteins from IL-5 transgenic mice versus IL-5 transgenic animals, which were also gp91phox-/-, revealed, that only proteins from IL-5 transgenic mice were 3NT-positive (Fig. 6).
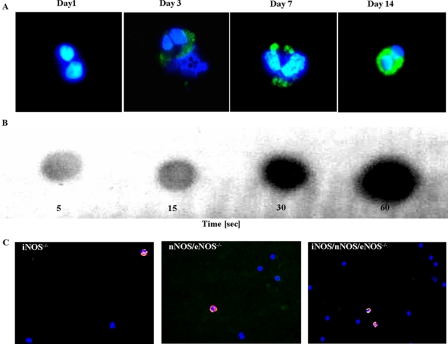
Ex vivo differentiation of cord blood-derived hematopoietic stem cells also provides additional evidence for the EPO-dependent nitration of eosinophil granule proteins. Specifically, we determined if a linear relationship exists between the kinetic appearance of 3NT and the rapid rise (maximal at day 8 (31)) of EPO expression (i.e. increase in EPO transcript prevalence). That is, given our demonstration that EPO is responsible for Tyr nitration of secondary granule proteins, 3NT-immunoreactivity should be visible during this period. Indeed, umbilical cord stem cells stimulated for 2 weeks with the eosinophil agonist cytokine IL-5 (38) showed that initially 3NT-negative cord blood cells became 3NT-positive at day 3 (Fig. 7A). Thereafter, 3NT-immunoreactivity increased significantly and was maximal at day 7.
FIGURE 7.
Tyrosine nitration of eosinophil granule toxins from human umbilical cord stem cells, IL-5/gp91phox-/- mice, and mice lacking nitric-oxide synthases. A, cord blood eosinophils after in vitro culture at days 1, 3, 7, and 14 stained for 3NT (green). Original magnification: ×1000. 3NT-negative cord blood cells became 3NT-positive at day 3. B, rapid tyrosine nitration in vitro is dependent on hydrogen peroxide and nitrite. Dot blot shows autonitration of IL-5/gp91phox-/- mouse granula toxins at 5, 10, 15, and 30 s. C, blood smears from mice lacking iNOS (left), nNOS and eNOS (middle), and iNOS/eNOS/nNOS (right), stained with antibodies specific for MBP (red), 3NT (green), and with DAPI. Original magnification: ×1000.
To further support our notion that tyrosine nitration of eosinophil granule toxins is mediated by the enzymatic action of EPO, de novo nitration of non-nitrated toxins was assessed by addition of H2O2 and nitrate to a granule extract from gp91phox-/- mice in vitro. Dot blots revealed 3NT immunoreactivity within 5 s (Fig. 7B).
To investigate the possibility that NOS isoforms provide a source for tyrosine nitration, we investigated eosinophils of mice that lack different isoforms of NOS. Our results show that even iNOS/eNOS/nNOS triple KO mice were positive for 3NT in eosinophil granule proteins (Fig. 7C), suggesting that minute amounts of plasma NOx are sufficient for tyrosine nitration in these animals and that eNOS, which has been detected in eosinophils granule membranes, is sufficient for tyrosine nitration in wild type animals and humans. Taken together, these data demonstrate that nitration of Tyr residues in eosinophil granule proteins is mediated by the enzymatic activity of EPO in the presence of H2O2 and NOx.
Biological Function of Nitrated Eosinophil Granule Proteins Versus Non-nitrated Eosinophil Granule Proteins—First, we investigated whether eosinophils would nitrate other proteins. When we incubated stimulated human eosinophils with BSA, nitrated BSA was detectable by Western blotting using rabbit 3NT antibodies (Fig. 8A). To compare the cytotoxic activities of nitrated versus non-nitrated eosinophil granule proteins, we incubated monolayers of the respiratory cell line A549 with either of the granule preparations. However, we did not observe a statistical difference in cytotoxicity between nitrated versus non-nitrated eosinophil granule extract preparations (Fig. 8B). Similarly, no difference in cytotoxicity was observed between non-nitrated rEPO or rECP and their nitrated counterparts (Fig. 8B). Furthermore, nitration had no impact on the bactericidal activities of non-nitrated rEPO or rECP and their nitrated counterparts when the bacterial pathogens S. aureus and E. coli were investigated (Fig. 8, C-F). Finally, we tested the question whether the size of eosinophil granules differs when granules are filled with nitrated or non-nitrated toxins. Again, we did not find any significant difference in the granule diameters in eosinophils, expressing nitrated or non-nitrated toxins using transmission electron microscopy (Fig. 8, G-I).
FIGURE 8.
Biological function of nitrated eosinophil granule proteins versus non-nitrated eosinophil granule proteins. A and B, cytotoxic activities of nitrated (3NT+) versus non-nitrated (3NT-) eosinophil granule proteins and nitrated rEPO or rECP versus their non-nitrated counterparts were assayed in incubations on monolayers of A549 cells. After a 24-h incubation, the alive or dead status of the epithelial cells was determined using immunofluorescence. C-F, the bactericidal activities of non-nitrated rEPO or rECP and their nitrated counterparts were tested in 24-h incubations with S. aureus or E. coli. Thereafter bacterial numbers were detemined by a plate assay. G-I, the size of eosinophil granules, filled with nitrated or non-nitrated toxins, was determined using transmission electron microscopy preparations of the respective eosinophils.
DISCUSSION
This study provides substantial evidence demonstrating that EPO uniquely mediated the nitration of the tyrosine residues of secondary granule proteins in mature resting eosinophils from both humans and mice.
(i) Antibodies specific for 3NT residues and the eosinophil cell marker MBP demonstrated that blood and tissue eosinophils from healthy individuals, as well as those from patients with inflammatory diseases, e.g. CF or atopic rhinitis, contained nitrated proteins. This observation was also true of eosinophils isolated from wild type mice and, furthermore, in both species, the nitrated eosinophil-derived proteins were restricted to the abundant proteins stored with EPO in the secondary granules of eosinophils. Interestingly, the presence of nitrated proteins among leukocytes tested was specific for eosinophils. Despite the significant structural identity between EPO and MPO (the peroxidase contained in neutrophils and monocytes/macrophages) (31, 39), and the fact that MPO has been shown to be capable of nitrating tyrosine residues (40-42), although to a lower extend (32), we were unable to detect 3NT residues in leukocytes other than eosinophils or in MPO itself, suggesting that the endogenous tyrosine nitration of eosinophil secondary granule proteins is uniquely mediated by EPO.
(ii) The nitration of eosinophil granule proteins did not occur in EPO knock-out mice and was absent in patients with CGD who lack NADPH oxidase activity and, in turn, the ability to produce the intracellular H2O2 needed to mediate EPO-dependent oxidative nitration.
(iii) Uncommitted human cord blood hematopoietic stem cells were 3NT-negative but became positive for 3NT approximately 3 days after in vitro exposure to the eosinophil agonist cytokine IL-5, in parallel with the rise of EPO mRNA transcripts (31).
(iv) De novo nitration of non-nitrated toxins by addition of H2O2 and nitrate to a granule extract from gp91phox-/- mice resulted in 3NT immunoreactivity within 5 s.
(v) The observation that the eosinophil granule proteins are already nitrotyrosine-positive in resting blood eosinophils from healthy individuals suggests that EPO, and not inflammatory responses leading to the production of superoxide anion and nitric oxide generating the nitrating species peroxinitrate (43, 44), is responsible for the observed protein nitration. This result is corroborated by other studies (e.g. Ref. 45) that showed that New Zealand White mice, an inbred mouse strain with a spontaneous deficiency of EPO (46), did not display 3NT immunoreactivity after ovalbumin challenge despite increased NO and superoxide anion production. Moreover, our demonstration that eosinophils are exclusively associated with 3NT-containing proteins (i.e. these proteins are absent in other leukocytes that generate reactive oxygen species and contain NOS isoforms (i.e. neutrophils, lymphocytes, and monocytes/macrophages)), strongly argues against a tyrosine nitrating mechanism involving peroxinitrate (47).
To test whether NOS isoforms are necessary to generate NO for the EPO-triggered tyrosine nitration, we investigated eosinophils of mice that lack different isoforms of NOS, i.e. nNOS, eNOS, and iNOS types. eNOS and iNOS isoforms have been observed in rat peritoneal eosinophils where they are strongly expressed and localized in cytoplasmic granules (48). In human peripheral blood eosinophils, iNOS (49) and eNOS (50) have been detected. Our results that show also that triple NOS KO mice were positive for 3NT in eosinophil granule proteins reveals that minute amounts of plasma NOx are sufficient for tyrosine nitration. This notion stems from the fact that plasma NOx levels were ∼60, 20, 10, and 3 μmol/liter in wild type, single NOs KO, double NOS KO, and triple NOS KO mice, respectively (22). These minute NOx amounts may be derived from NOS-independent mechanisms, including NOx metabolizing intestinal bacteria, myoglobin turnover of NO from NOx (51), or catalase NO production from hydroxylamine (52). These data indicate that eNOS is sufficient for tyrosine nitration in wild type animals and humans.
Regarding the reaction mechanism, several possibilities exist: intracellular NO, produced by NOS isoforms, is oxidized by EPO in the presence of H2O2 to the nitrosonium ion NO+, which nitrosylates the phenol ring of the tyrosine residue (electrophilic substitution) with subsequent oxidation to nitrotyrosin by H2O2. Alternatively, intracellular NO is oxidized to NO2, and further to the nitronium ion by H2O2, which then nitrates the phenol ring.
The findings that stimulated human eosinophils nitrate BSA is in line with many reports that tyrosine nitration occurs in inflamed animal tissues (53) and in the inflammatory cells of patients (54-56). Here we show that activated eosinophils can generate 3NT-positive albumin. Thus, eosinophil granule proteins may also actively modify other endogenous proteins thereby influencing inflammatory pathways. For instance, in an investigation of human bronchoalveolar lavage fluid, catalase activity was found to be reduced in patients with asthma by up to 50% relative to healthy controls and this reduction was linked to protein oxidation and nitrotyrosine modification (56). Similarly, inactivation of human manganese-superoxide dismutase by peroxynitrite is caused by exclusive nitration of Tyr34 to 3NT (57).
It is intriguing to speculate that the biophysical consequences of nitration facilitate the ability of the cationic eosinophil granule proteins, to interact/aggregate with one another despite their cationic character. Such interactions are mainly pH-dependent, because the pK values of ortho-nitrophenol (7.21) (58) and 3NT (7.0) would allow dissociation of the 3NT group only at neutral but not at acidic pH. Consequently, in the acidic granules of eosinophils (pH 5.1) (59), EPO, ECP, and EDN should not interact with each other via the 3NT group. This notion is supported by our results regarding the measurement of granule diameters packed with nitrated or non-nitrated toxins, where we did not observe a statistical difference between the preparations.
On the other hand, toxin interaction would rapidly occur upon degranulation into an environment with neutral pH. The rise in pH may thus increase local concentrations of granule protein at sites of deposition (e.g. the surface of invading parasites). Further studies are needed to unravel the biological effects of tyrosine nitration of eosinophil toxins.
In summary, we show that EPO constitutively catalyzes post-translational nitration of eosinophil granule proteins, a novel mechanism that may have significant consequences concerning the innate immune defense toward parasites and possibly for host tissue destruction during parasite infection and disorders characterized by eosinophil cell activation such as allergic asthma.
Acknowledgments
We thank Peter Döller and Martin Schaller for providing eosinophil-enriched blood samples from their patients and Claudia Gerber, Roland Dopfer, and Taifun Guengoer for providing blood smears from CGD patients. In addition, we thank Godehart Friedel for providing healthy human lung tissue, Hans-Jörg Bühring and Christian Sommerhoff for antibodies against mast cells, Sena Sezen and Arthur Burnett for blood smears from NOS1/NOS3 double KO mice, and Ulrike Schleicher and Christian Bogdan for blood smears from NOS2 KO mice. We also thank Stefan Stevanovic and Alfred Nordheim for assistance with MALDI-MS analyses, Diethelm Wallwiener for a sample of human cord blood, and Frank Schreiber, Ulrich Nagel, and Gerald B. Pier for discussions on the manuscript.
This work was supported in part by grants from the Deutsche Forschungs-gemeinschaft, Bonn, Germany, and the European Union (to M. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: EPO, eosinophil peroxidase; rEPO, recombinant EPO; CGD, chronic granulomatous disease; CF, cystic fibrosis; ECP, eosinophil cationic protein; rECP, recombinant eosinophil peroxidase; EDN, eosinophil-derived neurotoxin; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; MS, mass spectrometry; MBP, major basic protein; MPO, myeloperoxidase; nano-ESI-FTICR-MS, nano-electrospray Fourier transform ion cyclotron resonance mass spectrometry; NOS, nitric-oxide synthase; iNOS, inducible nitric-oxide synthase; nNOS neuronal nitric-oxide synthase; eNOS, endothelial nitric-oxide synthase; 3NT, 3-nitrotyrosine; IL, interleukin; KO, knock-out; HPLC, high pressure liquid chromatography; BSA, bovine serum albumin; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Giembycz, M. A., and Lindsay, M. A. (1999) Pharmacol. Rev. 51 213-340 [PubMed] [Google Scholar]
- 2.Gleich, G. J., Adolphson, C. R., and Leiferman, K. M. (1993) Annu. Rev. Med. 44 85-101 [DOI] [PubMed] [Google Scholar]
- 3.Larson, K. A., Horton, M. A., Madden, B. J., Gleich, G. J., Lee, N. A., and Lee, J. J. (1995) J. Immunol. 155 3002-3012 [PubMed] [Google Scholar]
- 4.Horton, M. A., Larson, K. A., Lee, J. J., and Lee, N. A. (1996) J. Leukocyte Biol. 60 285-294 [DOI] [PubMed] [Google Scholar]
- 5.Macias, M. P., Welch, K. C., Denzler, K. L., Larson, K. A., Lee, N. A., and Lee, J. J. (2000) J. Leukocyte Biol. 67 567-576 [DOI] [PubMed] [Google Scholar]
- 6.Cormier, S. A., Larson, K. A., Yuan, S., Mitchell, T. L., Lindenberger, K., Carrigan, P., Lee, N. A., and Lee, J. J. (2001) Mamm. Genome 12 352-361 [DOI] [PubMed] [Google Scholar]
- 7.Butterworth, A. E. (1984) Adv. Parasitol. 23 143-235 [DOI] [PubMed] [Google Scholar]
- 8.Nogueira, N. M., Klebanoff, S. J., and Cohn, Z. A. (1982) J. Immunol. 128 1705-1708 [PubMed] [Google Scholar]
- 9.Villalta, F., Pankratz, H. S., and Kierszenbaum, F. (1987) J. Protozool. 34 285-290 [DOI] [PubMed] [Google Scholar]
- 10.Molina, H. A., Kierszenbaum, F., Hamann, K. J., and Gleich, G. J. (1988) Am. J. Trop. Med. Hyg. 38 327-334 [DOI] [PubMed] [Google Scholar]
- 11.Wasmoen, T. L., Bell, M. P., Loegering, D. A., Gleich, G. J., Prendergast, F. G., and McKean, D. J. (1988) J. Biol. Chem. 263 12559-12563 [PubMed] [Google Scholar]
- 12.Young, J. D., Peterson, C. G., Venge, P., and Cohn, Z. A. (1986) Nature 321 613-616 [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg, H. F., and Dyer, K. D. (1995) J. Biol. Chem. 270 30234. [PubMed] [Google Scholar]
- 14.Molina, H. A., and Kierszenbaum, F. (1989) Immunology 66 289-295 [PMC free article] [PubMed] [Google Scholar]
- 15.Motojima, S., Frigas, E., Loegering, D. A., and Gleich, G. J. (1989) Am. Rev. Respir. Dis. 139 801-805 [DOI] [PubMed] [Google Scholar]
- 16.Wang, J., and Slungaard, A. (2006) Arch. Biochem. Biophys. 445 256-260 [DOI] [PubMed] [Google Scholar]
- 17.Wu, W., Chen, Y., and Hazen, S. L. (1999) J. Biol. Chem. 274 25933-25944 [DOI] [PubMed] [Google Scholar]
- 18.Lee, N. A., McGarry, M. P., Larson, K. A., Horton, M. A., Kristensen, A. B., and Lee, J. J. (1997) J. Immunol. 158 1332-1344 [PubMed] [Google Scholar]
- 19.Denzler, K. L., Borchers, M. T., Crosby, J. R., Cieslewicz, G., Hines, E. M., Justice, J. P., Cormier, S. A., Lindenberger, K. A., Song, W., Wu, W., Hazen, S. L., Gleich, G. J., Lee, J. J., and Lee, N. A. (2001) J. Immunol. 167 1672-1682 [DOI] [PubMed] [Google Scholar]
- 20.Pero, R. S., Borchers, M. T., Spicher, K., Ochkur, S. I., Sikora, L., Rao, S. P., Abdala-Valencia, H., O'Neill, K. R., Shen, H., McGarry, M. P., Lee, N. A., Cook-Mills, J. M., Sriramarao, P., Simon, M. I., Birnbaumer, L., and Lee, J. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 4371-4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz, H., and Humbel, B. M. (2007) Meth. Mol. Biol. 369 229-256 [DOI] [PubMed] [Google Scholar]
- 22.Morishita, T., Tsutsui, M., Shimokawa, H., Sabanai, K., Tasaki, H., Suda, O., Nakata, S., Tanimoto, A., Wang, K. Y., Ueta, Y., Sasaguri, Y., Nakashima, Y., and Yanagihara, N. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10616-10621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansel, T. T., De Vries, I., Iff, T., Rihs, S., Wandzilak, M., Betz, S., Blaser, K., and Walker, C. (1991) J. Immunol. Meth. 145 105-110 [DOI] [PubMed] [Google Scholar]
- 24.Metcalf, J. A., Gallin, J. I., Nauseef, W. M., and Root, R. K. (1986) Laboratory Manual of Neutrophil Function, pp. 1-200, Raven Press, New York
- 25.Goldstein, W., and Doring, G. (1986) Am. Rev. Respir. Dis 134 49-56 [DOI] [PubMed] [Google Scholar]
- 26.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996) Anal. Chem. 68 850-858 [DOI] [PubMed] [Google Scholar]
- 27.Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. (1999) Electrophoresis 20 3551-3567 [DOI] [PubMed] [Google Scholar]
- 28.Abu-Ghazaleh, R. I., Dunnette, S. L., Loegering, D. A., Checkel, J. L., Kita, H., Thomas, L. L., and Gleich, G. J. (1992) J. Leukocyte Biol. 52 611-618 [DOI] [PubMed] [Google Scholar]
- 29.Fiedler, T. J., Davey, C. A., and Fenna, R. E. (2000) J. Biol. Chem. 275 11964-11971 [DOI] [PubMed] [Google Scholar]
- 30.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 31.Ten, R. M., Pease, L. R., McKean, D. J., Bell, M. P., and Gleich, G. J. (1989) J. Exp. Med. 169 1757-1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang, Q., and Hurst, J. K. (1997) J. Biol. Chem. 272 32767-32772 [DOI] [PubMed] [Google Scholar]
- 33.Macht, M., Marquardt, A., Deininger, S. O., Damoc, E., Kohlmann, M., and Przybylski, M. (2004) Anal. Biochem. 378 1102-1111 [DOI] [PubMed] [Google Scholar]
- 34.McGaughey, G. B., Gagne, M., and Rappe, A. K. (1998) J. Biol. Chem. 273 15458-15463 [DOI] [PubMed] [Google Scholar]
- 35.Mohan, C. G., Boix, E., Evans, H. R., Nikolovski, Z., Nogues, M. V., Cuchillo, C. M., and Acharya, K. R. (2002) Biochemistry 41 12100-12106 [DOI] [PubMed] [Google Scholar]
- 36.Leonidas, D. D., Boix, E., Prill, R., Suzuki, M., Turton, R., Minson, K., Swaminathan, G. J., Youle, R. J., and Acharya, K. R. (2001) J. Biol. Chem. 276 15009-15017 [DOI] [PubMed] [Google Scholar]
- 37.Pollock, J. D., Williams, D. A., Gifford, M. A., Li, L. L., Du, X., Fisherman, J., Orkin, S. H., Doerschuk, C. M., and Dinauer, M. C. (1995) Nat. Genet. 9 202-209 [DOI] [PubMed] [Google Scholar]
- 38.Popken-Harris, P., Checkel, J., Loegering, D., Madden, B., Springett, M., Kephart, G., and Gleich, G. J. (1998) Blood 92 623-631 [PubMed] [Google Scholar]
- 39.Furtmuller, P. G., Zederbauer, M., Jantschko, W., Helm, J., Bogner, M., Jakopitsch, C., and Obinger, C. (2006) Arch. Biochem. Biophys. 445 199-213 [DOI] [PubMed] [Google Scholar]
- 40.Sampson, J. B., Ye, Y., Rosen, H., and Beckman, J. S. (1998) Arch. Biochem. Biophys. 356 207-213 [DOI] [PubMed] [Google Scholar]
- 41.Eiserich, J. P., Hristova, M., Cross, C. E., Jones, A. D., Freeman, B. A., Halliwell, B., and van den Vliet, A. (1998) Nature 391 393-397 [DOI] [PubMed] [Google Scholar]
- 42.van den Vliet, A., Eiserich, J. P., Halliwell, B., and Cross, C. E. (1997) J. Biol. Chem. 272 7617-7625 [DOI] [PubMed] [Google Scholar]
- 43.Beckman, J. S., and Koppenol, W. H. (1996) Am. J. Physiol. 271 C1424-C1437 [DOI] [PubMed] [Google Scholar]
- 44.Ischiropoulos, H. (1998) Arch. Biochem. Biophys. 356 1-11 [DOI] [PubMed] [Google Scholar]
- 45.Duguet, A., Iijima, H., Eum, S. Y., Hamid, Q., and Eidelman, D. H. (2001) Am. J. Respir. Crit. Care Med. 164 1119-1126 [DOI] [PubMed] [Google Scholar]
- 46.Ohmori, J., Tokunaga, H., Ezaki, T., Maruyama, H., and Nawa, Y. (1996) Int. Arch. Allergy Immunol. 111 30-35 [DOI] [PubMed] [Google Scholar]
- 47.Pfeiffer, S., Lass, A., Schmidt, K., and Mayer, B. (2001) FASEB J. 15 2355-2364 [DOI] [PubMed] [Google Scholar]
- 48.Zanardo, R. C., Costa, E., Ferreira, H. H., Antunes, E., Martins, A. R., Murad, F., and De Nucci, G. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 14111-14114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.del Pozo, V., de Arruda-Chaves, E., de Andres, B., Cardaba, B., Lopez-Farre, A., Gallardo, S., Cortegano, I., Vidarte, L., Jurado, A., Sastre, J., Palomino, P., and Lahoz, C. (1997) J. Immunol. 158 859-864 [PubMed] [Google Scholar]
- 50.Kobzik, L., Imrich, A., Massaro, A., and Drazen, J. M. (1997) Am. J. Respir. Crit. Care Med. 155 A60. [DOI] [PubMed] [Google Scholar]
- 51.Koizumi, C., and Brown, W. D. (1971) J. Food Sci. 36 1105-1109 [Google Scholar]
- 52.Nicholls, P. (1964) Biochem. J. 90 331-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aulak, K. S., Miyagi, M., Yan, L., West, K. A., Massillon, D., Crabb, J. W., and Stuehr, D. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12056-12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saleh, D., Barnes, P. J., and Giaid, A. (1997) Am. J. Respir. Crit. Care Med. 155 1763-1769 [DOI] [PubMed] [Google Scholar]
- 55.MacPherson, J. C., Comhair, S. A., Erzurum, S. C., Klein, D. F., Lipscomb, M. F., Kavuru, M. S., Samoszuk, M. K., and Hazen, S. L. (2001) J. Immunol. 166 5763-5772 [DOI] [PubMed] [Google Scholar]
- 56.Ghosh, S., Janocha, A. J., Aronica, M. A., Swaidani, S., Comhair, S. A. A., Xu, W., Zheng, L., Kaveti, S., Kinter, M., Hazen, S. L., and Erzurum, S. C. (2006) J. Immunol. 176 5587-5597 [DOI] [PubMed] [Google Scholar]
- 57.Yamakura, F., Taka, H., Fujimura, T., and Murayama, K. (1998) J. Biol. Chem. 273 14085-14089 [DOI] [PubMed] [Google Scholar]
- 58.Rappopory, Z., (1984) CRC Handbook of Tables for Organic Compound Identification, 3rd Edition, p. 20, CRC, Cleveland, OH
- 59.Kurashima, K., Numata, M., Yachie, A., Sai, Y., Ishizaka, N., Fujimura, M., Matsuda, T., and Ohkuma, S. (1996) Lab. Investig. 75 689-698 [PubMed] [Google Scholar]
- 60.Lee, B., and Richards, F. M. (1971) J. Mol. Biol. 55 379-400 [DOI] [PubMed] [Google Scholar]