Abstract
TreX is an archaeal glycogen-debranching enzyme that exists in two oligomeric states in solution, as a dimer and tetramer. Unlike its homologs, TreX from Sulfolobus solfataricus shows dual activities for α-1,4-transferase and α-1,6-glucosidase. To understand this bifunctional mechanism, we determined the crystal structure of TreX in complex with an acarbose ligand. The acarbose intermediate was covalently bound to Asp363, occupying subsites -1 to -3. Although generally similar to the monomeric structure of isoamylase, TreX exhibits two different active-site configurations depending on its oligomeric state. The N terminus of one subunit is located at the active site of the other molecule, resulting in a reshaping of the active site in the tetramer. This is accompanied by a large shift in the “flexible loop” (amino acids 399-416), creating connected holes inside the tetramer. Mutations in the N-terminal region result in a sharp increase in α-1,4-transferase activity and a reduced level of α-1,6-glucosidase activity. On the basis of geometrical analysis of the active site and mutational study, we suggest that the structural lid (acids 99-97) at the active site generated by the tetramerization is closely associated with the bifunctionality and in particular with the α-1,4-transferase activity. These results provide a structural basis for the modulation of activities upon TreX oligomerization that may represent a common mode of action for other glycogen-debranching enzymes in higher organisms.
Debranching enzymes are divided into two groups. One includes isoamylase and pullulanase found in bacteria and plants, and the other includes glycogen-debranching enzymes (GDEs)4 found in mammals and yeast. Whereas the former has only a single α-1,6-glycosidic bond hydrolyzing activity for glycogen and amylopectin, the GDEs in eukaryotes are bifunctional, possessing both α-1,4-transferase (EC 2.4.1.25) and α-1,6-glucosidase (EC 3.2.1.33) activities within a single polypeptide chain (1, 2). The structure of GDE in eukaryotes has yet to be determined.
TreX is an archaeal enzyme in Sulfolobus that debranches the side chain of glycogen into maltodextrin, which is further converted to trehalose (3). It belongs to the GH13 (glycoside hydrolase 13) family and shows high sequence similarity to isoamylase and pullulanase. The amino acid sequence of TreX shows 74% homology to isoamylase from Sulfolobus acidocaldarium, 42% homology to the glycogen operon protein GlgX from Escherichia coli, and 30% homology to isoamylase from Pseudomonas.
However, despite its high sequence similarity to isoamylase, which has only a single α-1,6-glycosidic bond hydrolyzing activity, TreX exhibits α-1,4-transferase as well as α-1,6-glucosidase activity, similar to GDEs in mammals and yeast (4, 5). To date, it is the only GDE found in bacteria/archaea with α-1,4-transferase activity, transferring the α-1,4-glucan oligosaccharides from one molecule to another using various substrates, including glycogen and amylopectin (4).
In a previous study, we cloned the gene treX from the trehalose biosynthesis gene cluster of Sulfolobus solfataricus P2, a hyperthermophilic archaeon, and expressed the protein in E. coli (4). The treX gene is one of three trehalose operon genes, including treX, treY (maltooligosyltrehalose synthase), and treZ (maltooligosyltrehalose trehalohydrolase) (3, 6, 7).
TreX exists in two oligomeric states in solution, as a dimer and tetramer. The tetramer formed in the presence of dimethyl sulfoxide shows a 4-fold higher catalytic efficiency compared with the dimer (5). However, the mechanism underlying the increase in activity upon tetrameric rearrangement is not clearly understood. To elucidate the modulation of activity upon oligomerization and the mechanism of the dual functions of the enzyme, we carried out a structural study on the GDE TreX.
In this study, we describe the crystal structure of the archaeal GDE TreX from S. solfataricus at a resolution of 2.8 Å in complex with an acarbose ligand. Analysis of the structure revealed the unique tetrameric configuration of TreX reshaping the active site with an additional region from the N-terminal domain. This result provides a structural basis for the modulation of activity upon oligomerization and for the bifunctional mechanism of the archaeal GDE TreX.
EXPERIMENTAL PROCEDURES
Protein Purification—E. coli cells carrying pSGDE were cultured in LB medium plus ampicillin and harvested by centrifugation. The cells were resuspended in lysis buffer (50 mm Tris-HCl (pH 7.5), 300 mm NaCl, and 10 mm imidazole) and sonicated using a VC-600 sonicator (Sonics & Materials, Inc., Newtown, CT). The crude cell extract was centrifuged at 10,000 × g for 15 min at 4 °C. After sonication, the cell lysates were subjected to nickel-nitrilotriacetic acid affinity chromatography (Qiagen) as described (8).
α-1,4-Transferase Activity—The disproportionation activity of TreX and its mutants was determined by a glucose oxidase/peroxidase method (9) with slight modification. The reaction mixture contained 800 μl of glucose oxidase solution from a glucose kit (Youngdong Pharmaceutical Co., Seoul, Korea) and 1 μm enzyme in 50 mm sodium acetate buffer (pH 5.5). This reaction mixture was incubated at 40 °C for 10 min. The reaction was started by adding 40 μl of substrate (10 and 20 mm maltotriose). The absorbance of the reaction mixture was measured for 30 min at 500 nm. The equivalent of the hydrolyzed glycosidic bonds was based on glucose equivalents using a glucose equilibration curve. Enzyme activity was defined as the amount of glucose that 1 μm enzyme liberated per min.
α-1,6-Glucosidase Activity—The α-1,6-hydrolysis activity of TreX and its mutants was assayed at 40 and 70 °C in 50 mm sodium acetate buffer (pH 5.5) plus glycogen by determining the reducing power of the hydrolysis products using the dinitrosalicylic acid method (10). One unit of enzyme was defined as the micromoles of maltose equivalents produced by 1 ml of enzyme/1 min. The specific activity (units/mg of enzyme) of each enzyme was calculated using the enzyme units. The enzyme reaction mixture (500 μl) was composed of 0.5% (w/v) glycogen (final concentration) as the substrate and 1 μm enzyme solution (final concentration) in 50 mm sodium acetate buffer (pH 5.5). The enzyme solution was added to the reaction and incubated for 10 min before the reaction was stopped by the addition of 500 μl of dinitrosalicylic acid solution. A reaction without addition of enzyme was used as a control. The reaction mixture was boiled for 5 min and cooled by placing the tube under running water. Absorbance was measured at 575 nm. The micromole equivalent of split bonds was based on the micromole of maltose equivalents using a maltose calibration curve.
Crystallization and Data Collection—Crystallization trials of TreX were set up using the microbatch method under oil at 18 °C. The native protein was crystallized in the two different crystal forms, the dimeric and tetrameric forms, in space groups P3121 and P321, respectively. In both crystal forms, the asymmetric unit consisted of one dimer. The dimeric form of the TreX crystal was grown from 16% polyethylene glycol 8000, 0.2 m NaCl, and 0.1 m CHES buffer (pH 9.5). The tetrameric form of the crystal was grown from 2.2 m ammonium phosphate and 0.1 m Tris-HCl buffer (pH 8.5). For the acarbose intermediate complex crystal, 0.1% acarbose (kindly provided by Professor Seiya Chiba, Hokkaido University) was added to the protein, followed by incubation for 1 h prior to the setup of the crystal in 8% polyethylene glycol 3000, 0.2 m lithium sulfate, and 0.1 m imidazole buffer (pH 8.0). Before data collection, the crystals were cryocooled to 95 K using a cryoprotectant consisting of mother liquor supplemented with 20% glycerol. For the acarbose crystal, 0.1% acarbose was added to the cryoprotectant. The native and ligand-bound data sets were collected at the MX4II and MX6C beamlines of the Pohang Accelerator Laboratory, respectively.
Structure Determination and Refinement—The diffraction data were processed and scaled with the program HKL2000 (11). The native structure was determined by the molecular replacement method using the Phaser CCP4 suite (12) and the structure of Pseudomonas isoamylase (Protein Data Bank code 1BF2) as a search model. The resulting model was refined in conjunction with model rebuilding using the program CNS (13). The structure of the complex was solved by direct refinement of the structure of native TreX against the diffraction data. The program Coot (14) was used for manual model building, and the stereochemical quality of the model was monitored using the program PROCHECK (15). The current atomic model of a native TreX dimer contains residues 9-718 with disordered N-terminal tails (residues 1-8) and has an R factor of 20.6% (Rfree = 23.1%). The data from the native crystal in the P321 space group were initially collected to 2.5-Å resolution. However, because of the presence of pseudo-merohedral twinning in the crystal, it was refined to only 3 Å, and the final R factors were relatively high in comparison with the resolution of the data. Molecular images were created using the program PyMOL (16), and electron density maps were generated in Coot. The statistics for the diffraction data and the final refined structures are given in Table 1.
TABLE 1.
Data collection and structure solution parameters
| Crystal type | Native (dimer) | Native (tetramer) | Acarbose complex |
|---|---|---|---|
| Unit cell parameters (Å) | a = 203.633, b = 203.633, c = 89.428 | a = 136.077, b = 136.077, c = 173.545 | a = 204.814, b = 204.814, c = 89.558 |
| Resolution (Å) | 50.0-3.0 | 30.0-3.0 | 30.0-2.8 |
| Space group | P321 | P3121 | P321 |
| Completeness (%) | 98.4 (95.8)a | 99.8 (99.5)a | 96.6 (93.0)a |
| Rsym (%)b | 8.0 (24.0) | 7.3 (25.3) | 8.8 (27.0) |
| I/σ(I) | 12.8 | 10.3 | 15.4 |
| No. of refined atoms: protein/water | 11,634/0 | 11,154/0 | 11,685/401 |
| Rfactor/Rfree (%)c | 20.3/23.1 | 21.3/28.7 | 21.4/26.6 |
| r.m.s.d. bond length (Å) | 0.006 | 0.009 | 0.006 |
| r.m.s.d. bond angle | 1.2° | 0.97° | 0.6° |
| Ramachandran plot (%) | |||
| Most favored region | 91.5 | 86.6 | 86.8 |
| Additionally allowed region | 8.5 | 13.4 | 13.2 |
The numbers in parentheses are statistics from the highest resolution shell.
Rsym = ∑|Iobs - Iavg|/Iobs, where Iobs is the observed intensity of individual reflection and Iavg is the average over symmetry equivalents.
R factor = ∑||Fo| - |Fc||/∑|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated using 5% of the data.
RESULTS
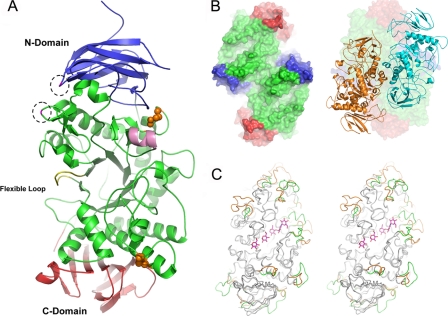
Overall Structural Characteristics of TreX—The overall structure of the TreX monomer shows similarity to its homolog debranching enzyme, Pseudomonas isoamylase, with a root mean square deviation of 1.13 Å over 562 C-α atoms. It consists of three major regions: the N-terminal, central, and C-terminal domains (Fig. 1). The N-terminal domain (amino acids (aa) 1-153) comprises six β-strands forming a β-sandwich. The central domain contains the characteristic (β/α)8-barrel motif found in many members of the α-amylase family, consisting of eight parallel β-strands surrounded by eight parallel α-helices. The superposition of TreX on the isoamylase shows similarity in most regions, whereas some parts of the substrate-binding groove, including the helix region of aa 231-237, an inserted region of aa 601-612, and deleted regions of aa 392-395 and 371-374, show variations (Fig. 1C). The conserved calcium ion observed in isoamylase and other α-amylases is not found in the TreX structure, whereas it has two clear cysteine disulfide bridges between Cys505 and Cys519 and between Cys254 and Cys261.
FIGURE 1.
Overall structure of TreX. A, schematic overview of TreX with the active site in the center: The N- and C-terminal domains are shown in blue and red, respectively. The flexible loop in the active site is shown in yellow. Disulfide bonds between cysteines are drawn as spheres. The mutated residues in this study are shown in dashed circles. Helix α4 is shown in pink. For clarity, loops were smoothed in PyMOL (16). B, surface representation of the dimeric structure in the P321 space group with its substrate-binding groove facing up (left) and the tetrameric arrangement as the dimer of the dimer in the P3121 space group (right). C, stereo view of the superposition of TreX with ligand and isoamylase from Pseudomonas (Protein Data Bank code 1BF2). The different regions are highlighted in TreX (green) and in isoamylase (orange).
Previously, we reported that TreX exists in two oligomeric forms in solution (5). We obtained both types of crystals and determined the structures of the dimer and tetramer. In the dimeric structure, each monomer is oriented side by side with the active site of each monomer facing the same side (Fig. 1B). The dimeric arrangement of TreX produces a buried interface of 1523 Å2, corresponding to 5.6% of the solvent-accessible surface per monomer, with numerous polar interactions and van der Waals contacts. In the tetrameric structure, two dimers face each other with a slight offset as a dimer of dimers, orienting the substrate-binding grooves of each subunit such that they are connected to the inside of the tetramer. The tetrameric arrangement of TreX produces buried interfaces of 2435 Å2 in which most of the interacting residues differ from those involved in the dimerization process (Fig. 2).
FIGURE 2.
Sequence alignment between TreX, GlgX, and isoamylase. A sequence of six debranching enzymes was aligned using ClustalW. The black circles above the sequence indicate the essential active-site residues conserved in the debranching enzymes. The two mutated residues in this study are indicated by asterisks above the sequence. Red and dark green boxes indicate the residues involved in dimerization and tetramerization, respectively, for TreX. Helix α4 observed in the TreX active site is shown in pink. The flexible loop observed in the tetramer is shown in orange. Green, cyan, and yellow highlighting indicates the residues conserved in 100, 80, and 55% of the proteins, respectively. The secondary structure shown here corresponds to that of TreX (S. solfataricus). The accession numbers for these sequences are NP_343483.1 (TreX from S. solfataricus (S. Solf)), YP_256058.1 (TreX from Sulfolobus acidocaldarius (S. Acid)), AAS00512.1 (TreX from Metallosphaera hakonensis (M. Hako)), A1AGW4.1 (GlgX from E. coli), CAA00929.1 (isoamylase (Isoa) from Pseudomonas (Pseduo) sp.), and AAB63356.1 (isoamylase from Flavobacterium (Flavob)).
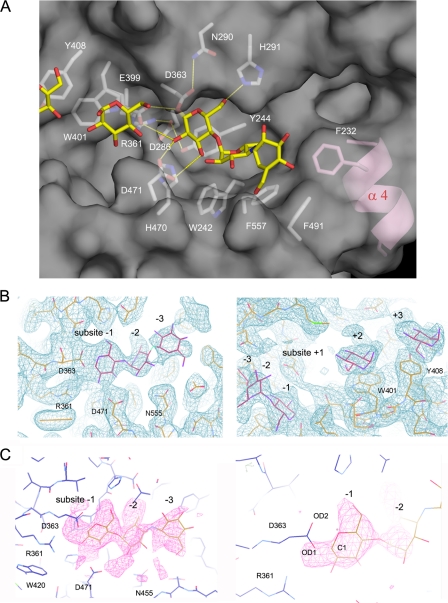
Structure of the Active Site with the Acarbose Intermediate—Three catalytic residues (Asp363, Glu399, and Asp471), invariant essential residues in the GH13 family, are located at the bottom of the active-site cleft. A crystal soaked in acarbose shows an electron density at -1 to -3 in the active site, whereas the density at -1 clearly appears to be connected to Asp363 with no density at +1 (Fig. 3). The nomenclature for the subsites follows that of Davies et al. (17). The glucose moiety at the reducing end of the acarbose ligand is thought to be cleaved, resulting in the covalent intermediate bound to Asp363. The inhibitory acarviosine moiety binds to subsites -2 to -3, showing a similar binding mode to the acarbose intermediate observed in the amylomaltase structure (18, 19). The plane of the ester moiety connecting Asp363 and the C-1 atom of the -1 sugar is approximately parallel to the plane of the -1 sugar ring. The covalent intermediate was possibly trapped by crystallization at pH 8.0, a pH value at which acarbose is rarely cleaved by TreX.
FIGURE 3.
Active site with the acarbose covalent intermediate. A, the acarbose intermediate covalently bound to Asp363 shows tight interactions with the residues at subsite -1 in the dimer. The inhibitory acarviosine moiety is bound to subsites -2 and -3. The catalytic residues (Asp363, Glu399, and Asp471) and other conserved residues in the active site are indicated by sticks. The ligand molecules are shown in yellow. Helix α4 unique to TreX is in pink. B, the electron density map of 2Fo - Fc (1.0 σ) shows the acarbose intermediate molecule at subsites -1, -2, and -3 (left) and the glucose molecules at subsites +2 and +3 (right). C, the omit map of Fo - Fc drawn without the ligand shows the covalent binding of the acarbose intermediate to Asp326, contoured at 3.0 σ (left) and 5.0 σ (right).
Tyr244, His291, Arg361, and His470 form the deeply buried -1 subsite. Tyr244, part of the motif NYWDYDP that is essential for the interaction with substrate glucose rings, forms a hydrogen bond with Asp286. The side chains of the three carboxylate residues Asp363, Glu399, and Asp471 are located in close proximity to subsite -1, in which the O-ε2 atom of the catalytic acid/base Glu399 is at a distance of 3.8 Å to the O-1 atom of the -1 glucose, possibly bridging the -1 and +1 rings. Glu399 and Asp363 are located in the appropriate range for the retaining enzyme at distances of 3.5 and 2.9 Å in the acarbose-free and acarbose complex structures, respectively.
Two additional glucose molecules are observed at subsites +2 and +3 with stacking interactions with Trp401 and Tyr408, respectively (Fig. 3B). Residues 401 and 408 are located in a row next to Glu399. Given the stacking interaction of the sugar ring and the binding mode of the acarbose intermediate molecule at the active site, the branched substrate is likely to bind in a curved manner, making a sharp bend at the bond between the -1 and +1 rings.
Analysis of the substrate-binding region showed an interesting feature in that the region of residues (aa 228-238) forming helix α4 protrudes at the bottom of the substrate groove. Helix α4 is observed only in TreX and is not found in either the isoamylase or pullulanase structures. Phe232, adjacent to Phe557 and Phe491, is positioned in subsite -4 and is involved in a stacking interaction with the substrate. Given the typical left-handed helical configuration of the substrate, helix α4 may provide a platform for the stable binding of the longer substrate, explaining the substrate specificity of TreX. In comparison with isoamylase and pullulanase, TreX is unique in exhibiting higher activity on branched substrates with longer maltooligosaccharides (5).
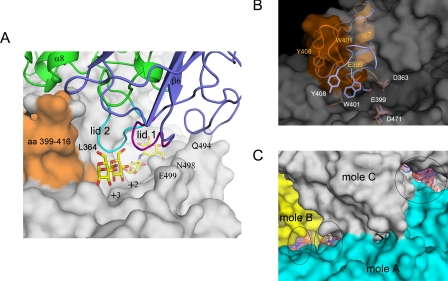
Reshaping the Active Site by Tetrameric Arrangement—The tetrameric structure of TreX reveals a novel configuration that appears to differ from any other amylase structure previously reported. In the tetramer, two dimers face each other such that the active sites are oriented to each other at a slight offset. The region comprising lid 1 (aa 92-97) and lid 2 (aa 315-322) of one dimer is located at the active site of the other dimer molecule, reshaping the geometry of the substrate-binding groove. Although the substrate-binding cleft is wide and open in the dimer, the association of lids and lid 2 with the active site in the tetramer results in a channel-like cavity with structural lids on the active site. Lid 1 of the N-terminal domain is placed right above the position corresponding to the glucose molecule at subsites +2 and +3 in the complex structure, whereas lid 2 is placed close to Leu364 and Glu399 near helix α7 (Fig. 4, A and B).
FIGURE 4.
Reshaped active site in the tetrameric structure. A, the substrate cleft is covered with lid 1 (aa 92-97; purple) and lid 2 (aa 315-322; cyan) generated by the tetrameric arrangement. The N-terminal domain is shown in blue. The glucose and acarbose intermediate ligands in the complex structure are superposed and indicated by sticks. The flexible loop (aa 399-416) is orange. B, the conformational change in the flexible loop results in a shift of Glu399 and Tyr408 away from the other catalytic residues Asp363 and Asp471. The region in the comparison is blue for the dimeric structure and orange for the tetramer. C, the active sites (black circles) highlighted by the superposed ligands are seen along the connected substrate-binding grooves inside the tetramer. mole, molecule.
The formation of the channel-like cavity generated by the structural lids is accompanied by a conformational change of the loop (aa 399-416) in the active site. This loop shifts outward from the active center and moves away from helix α7 (aa 364-370) in the tetrameric form, causing the substrate-binding groove to be extended. The corresponding loop is, however, adjacent to helix α7 in the dimeric form, closing the edge of the cleft. Because of the shift of the loop, the substrate-binding groove is open on one side and extends to the nearby groove of the adjacent molecule to form connected channels with an approximate width of 10-15 Å. Given the strong electrostatic potential distribution of the negative charge along this groove and the geometrical length of the holes, the connected channel appears to be a suitable architecture for the binding of a branched substrate (Fig. 4C).
The loop region (aa 399-416) was previously found to adopt a flexible conformation with an induced fit motion to accommodate the binding of different substrates in many enzymes of this family (18-21). However, the conformational change observed in the TreX tetramer takes a significantly different conformation with a large deviation of C-α. No conformational change has been reported upon oligomerization. This loop contains essential residues such as the catalytic nucleophile Glu399 and substrate-interacting residues Trp401 and Tyr408. In the dimeric form, the side chains of Trp401 and Tyr408 rotate by ∼90° to allow stacking interactions at subsites +3 and +2. In the tetramer, the side chain of Trp401 folds back, and Glu399 and Tyr408 separate at 4.5 and 8.3 Å, respectively, resulting in Glu399 being too far from the other catalytic residues to function as a nucleophile (Fig. 4B). It is assumed that TreX adopts this conformation with an extended groove to allow binding of branched substrates such as glycogen, and subsequent conformational change should occur for catalysis.
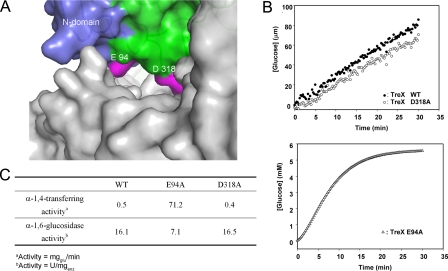
Involvement of Lid 1 in Bifunctional Activities—In structural comparisons with other enzymes with α-1,4-transferase activity, we noticed an interesting feature: enzymes with α-1,4-transferase activity share a structural lid above the substrate-binding cleft as a common motif similar to that observed in the TreX tetramer (see “Discussion”). To investigate whether the structural lid over the active site is related to the α-1,4-transferase activity of TreX, Glu94 and Asp318, which are positioned in the center of lid 1 (aa 92-97) and lid 2 (aa 315-322), respectively (Fig. 5A), were mutated to alanines. Enzyme assay of the mutants revealed a surprising result: the mutant enzyme E94A showed sharply increased α-1,4-transferase activity compared with the wild type or the mutant enzyme D318A, indicating the critical involvement of this region in the α-1,4-transferase activity (Fig. 5B). In comparison, the mutant E94A showed a relatively minor change in the α-1,6-glucosidase activity, and the mutant D318A showed an activity level identical to that of the wild type (Fig. 5C).
FIGURE 5.
Involvement of the structural lid in the activity of TreX. A, surface representation of the active site. The mutated residues Glu94 and Asp318 in lid 1 (aa 92-97) and lid 2 (aa 315-322) are shown in pink. B, enzyme assay of the wild type (WT) and mutants E94A and D318A for α-1,4-transferase activity. C, comparison of the α-1,4-transferase and α-1,6-glucosidase activities of each mutant.
Any possibility of the involvement of Glu94 in the catalytic mechanism of the active site of the same molecule has been excluded because it is located at a considerable distance from the active site (45 Å). Moreover, the size of the substrate used in the assay is not long enough to reach the region from the active center. Unless there is another unknown activity associated with this N-terminal domain region of lid 1, the change in the α-1,4-transferase activity in the TreX mutant E94A is most likely caused by the change in the interactions of this region with the substrate in the active site. This provides evidence for modulation of the bifunctional activities by tetramerization of its subunits.
DISCUSSION
TreX is a member of the GH13 family of glycoside hydrolases, to which isoamylase also belongs (22, 23). TreX is of special interest because, unlike its homologs, it exhibits two activities, the α-1,4-transferase activity and the α-1,6-glucosidase activity. In an attempt to understand the bifunctional activities of TreX, we carried out an extensive analysis of the active sites of those enzymes in the GH13 family containing α-1,4-transferase or α-1,6-glucosidase activity for which structures are available. For α-1,4-transferase activity, cyclodextrin glucanotransferase, 4-α-glucanotransferase, and neopullulanase were investigated because they represent enzymes with high transglycosylation activity for α-1,4-linkages, as does TreX. In the structural analysis, an interesting feature was noticed among the enzymes with α-1,4-transferase activity (48 Protein Data Bank files for cyclodextrin glucanotransferases, 14 for 4-α-glucanotransferase, and 29 for neopullulanases): they all contain a structural lid region located close to the active site. For example, neopullulanase has a narrow cleft at the active site with a structural lid that is formed through dimerization. This oligomerization is thought be essential for the enzyme to exhibit unique substrate specificity (21, 24, 25). In the structure of Thermotoga maritima 4-α-glucanotransferase, a loop in the N-terminal domain from the same molecule forms a clamp over the active site that captures the sugar rings of the substrate at the acceptor-binding site (26). In the amylomaltase structure, the corresponding loop (aa 246-258) is involved in acceptor binding (18), whereas the lid (aa 627-630) protruding from the helix bundle deeply penetrates the active site upon binding of the substrate in the 4-α-glucanotransferase structure (27). In cyclodextrin glucanotransferase, tyrosine or phenylalanine corresponding to the same region also protrudes, whereas various mutants of that region affect cyclization, demonstrating its involvement in transfer activity (28).
The structure of the TreX tetramer shows the same structural lid on the active site generated by the oligomerization, whereas no similar structural component exists in the dimer, suggesting that the α-1,4-transferase activity may be conferred by the oligomeric arrangement. Mutation in this region showed a remarkable change in α-1,4-transferase activity without significant alteration of the α-1,6-glucosidase activity of TreX. Based on the structural comparison, the putative role of the lid region is to interact with and stabilize the acceptor molecule during catalysis (21, 29, 30). Through the effective combination of these two activities, TreX may produce long maltooligosaccharides from a glycogen substrate in the process of trehalose biosynthesis.
Reshaping the active site by oligomeric arrangement as observed in TreX is not a newly discovered phenomenon in maltogenic amylases. In fact, the N-terminal domain of the pullulanase molecule is located near the active site of the other subunit in the dimeric structure, with several residues of the N-terminal domain interacting with the sugar moieties bound to the other monomer (20). The formation of this dimer is thought to occur only in the presence of longer sugar chains in solution. Also, the role of the N-terminal domain in oligomer formation and substrate specificity has been reported in maltogenic α-amylase (31), α-amylase from Thermo-actinomyces vulgaris (32), and neopullulanase (33). Thus, by adopting an oligomeric arrangement, TreX can reshape the active site for modulation of the dual activities and also generate a connected hollow cavity inside the tetramer molecule, possibly providing a suitable conformation for the binding of a bulky branched substrate.
Based on a previous mutational study, two activities of yeast GDE were suggested to be independent and located at different sites on a single polypeptide chain, with the N- and C-terminal regions involved in the transferase and glucosidase activities, respectively (2, 34). Here, we observed that α-1,4-transferase and α-1,6-glucosidase activities take place at the same catalytic site in TreX. In this respect, it would be interesting to determine whether GDEs in higher organisms have a similar configuration of the active site.
In conclusion, we were able to dissect the dual activities of the archaeal GDE TreX by mutational study and provided evidence that the α-1,4-transferase activity is closely associated with the structural lid generated by tetramerization. This study presents the structural basis for the modulation of the dual activities of TreX upon oligomerization.
Acknowledgments
We thank the staff at beamlines HFMX4A and HFMX6C, Pohang Accelerator Laboratory, for data collection and technical assistance.
The atomic coordinates and structure factors (codes 2VUY, 2VR5, and 2VNC) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by a grant from the Korea Research Institute of Bioscience and Biotechnology Research Initiative, by the Marine and Extreme Genome Research Center Program, Ministry of Marine Affairs and Fisheries, Republic of Korea, and in part by Korea Research Foundation Grant KRF-2006-005-J04703. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GDE, glycogen-debranching enzyme; aa, amino acids; CHES, 2-(cyclohexylamino)ethanesulfonic acid.
References
- 1.Bates, E. J., Heaton, G. M., Taylor, C., Kernohan, J. C., and Cohen, P. (1975) FEBS Lett. 58 181-185 [DOI] [PubMed] [Google Scholar]
- 2.Nakayama, A., Yamamoto, K., and Tabata, S. (2001) J. Biol. Chem. 276 28824-28828 [DOI] [PubMed] [Google Scholar]
- 3.Maruta, K., Hattori, K., Nakada, T., Kubota, M., Sugimoto, T., and Kurimoto, M. (1996) Biochim. Biophys. Acta 1289 10-13 [DOI] [PubMed] [Google Scholar]
- 4.Park, H. S., Park, J.-T., Kang, H. K., Cha, H., Kim, D. S., Kim, J. W., and Park, K.-H. (2007) Biosci. Biotechnol. Biochem. 71 1348-1352 [DOI] [PubMed] [Google Scholar]
- 5.Park, J.-T., Park, H. S., Kang, H. K., Hong, J. S., Cha, H., Woo, E.-J., Kim, J. W., Kim, M. J., Boos, W., Lee, S., and Park, K.-H. (2008) Biocatal. Biotransform. 26 76-85 [Google Scholar]
- 6.Maruta, K., Kubota, M., Fukuda, S., and Kurimoto, M. (2000) Biochim. Biophys. Acta 1476 377-381 [DOI] [PubMed] [Google Scholar]
- 7.Maruta, K., Mitsuzumi, H., Nakada, T., Kubota, M., Chaen, H., Fukuda, S., Sugimoto, T., and Kurimoto, M. (1996) Biochim. Biophys. Acta 1291 177-181 [DOI] [PubMed] [Google Scholar]
- 8.Kim, T. J., Kim, M. J., Kim, B. C., Kim, J. C., Cheong, T. K., Kim, J. W., and Park, K.-H. (1999) Appl. Environ Microbiol. 65 1644-1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, J. D., and Robyt, J. F. (1991) Anal. Biochem. 195 93-96 [DOI] [PubMed] [Google Scholar]
- 10.Miller, G. L. (1959) Anal. Chem. 31 426-428 [Google Scholar]
- 11.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 12.(1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-763 [DOI] [PubMed] [Google Scholar]
- 13.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 14.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 15.Laskowski, R. A., Moss, D. S., and Thornton, J. M. (1993) J. Mol. Biol. 231 1049-1067 [DOI] [PubMed] [Google Scholar]
- 16.DeLano, W. L. (2002) The PyMOL Molecular Graphic System, DeLano Scientific, San Carlos, CA
- 17.Davies, G. J., Wilson, K. S., and Henrissat, B. (1997) Biochem. J. 321 557-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barends, T. R., Bultema, J. B., Kaper, T., van der Maarel, M. J., Dijkhuizen, L., and Dijkstra, B. W. (2007) J. Biol. Chem. 282 17242-17249 [DOI] [PubMed] [Google Scholar]
- 19.Przylas, I., Terada, Y., Fujii, K., Takaha, T., Saenger, W., and Strater, N. (2000) Eur. J. Biochem. 267 6903-6913 [DOI] [PubMed] [Google Scholar]
- 20.Mikami, B., Iwamoto, H., Malle, D., Yoon, H. J., Demirkan-Sarikaya, E., Mezaki, Y., and Katsuya, Y. (2006) J. Mol. Biol. 359 690-707 [DOI] [PubMed] [Google Scholar]
- 21.Hondoh, H., Kuriki, T., and Matsuura, Y. (2003) J. Mol. Biol. 326 177-188 [DOI] [PubMed] [Google Scholar]
- 22.Stam, M. R., Danchin, E. G., Rancurel, C., Coutinho, P. M., and Henrissat, B. (2006) Protein Eng. Des. Sel. 19 555-562 [DOI] [PubMed] [Google Scholar]
- 23.MacGregor, E. A., Janecek, S., and Svensson, B. (2001) Biochim. Biophys. Acta 1546 1-20 [DOI] [PubMed] [Google Scholar]
- 24.Kim, J. S., Cha, S. S., Kim, H. J., Kim, T. J., Ha, N. C., Oh, S. T., Cho, H. S., Cho, M. J., Kim, M. J., Lee, H. S., Kim, J. W., Choi, K. Y., Park, K.-H., and Oh, B. H. (1999) J. Biol. Chem. 274 26279-26286 [DOI] [PubMed] [Google Scholar]
- 25.Lee, H. S., Kim, M. S., Cho, H. S., Kim, J. I., Kim, T. J., Choi, J. H., Park, C., Lee, H. S., Oh, B. H., and Park, K.-H. (2002) J. Biol. Chem. 277 21891-21897 [DOI] [PubMed] [Google Scholar]
- 26.Roujeinikova, A., Raasch, C., Sedelnikova, S., Liebl, W., and Rice, D. W. (2002) J. Mol. Biol. 321 149-162 [DOI] [PubMed] [Google Scholar]
- 27.Imamura, H., Fushinobu, S., Yamamoto, M., Kumasaka, T., Jeon, B. S., Wakagi, T., and Matsuzawa, H. (2003) J. Biol. Chem. 278 19378-19386 [DOI] [PubMed] [Google Scholar]
- 28.Schmidt, A. K., Cottaz, S., Driguez, H., and Schulz, G. E. (1998) Biochemistry 37 5909-5915 [DOI] [PubMed] [Google Scholar]
- 29.Watanabe, Y., Makino, Y., and Omichi, K. (2006) J. Biochem (Tokyo) 140 135-140 [DOI] [PubMed] [Google Scholar]
- 30.Strokopytov, B., Knegtel, R. M., Penninga, D., Rozeboom, H. J., Kalk, K. H., Dijkhuizen, L., and Dijkstra, B. W. (1996) Biochemistry 35 4241-4249 [DOI] [PubMed] [Google Scholar]
- 31.Park, K.-H., Kim, T. J., Cheong, T. K., Kim, J. W., Oh, B. H., and Svensson, B. (2000) Biochim. Biophys. Acta 1478 165-185 [DOI] [PubMed] [Google Scholar]
- 32.Yokota, T., Tonozuka, T., Kamitori, S., and Sakano, Y. (2001) Biosci. Biotechnol. Biochem. 65 401-408 [DOI] [PubMed] [Google Scholar]
- 33.Cheong, K. A., Kim, T. J., Yoon, J. W., Park, C. S., Lee, T. S., Kim, Y. B., Park, K.-H., and Kim, J. W. (2002) Biotechnol. Appl. Biochem. 35 27-34 [DOI] [PubMed] [Google Scholar]
- 34.Liu, W., Madsen, N. B., Braun, C., and Withers, S. G. (1991) Biochemistry 30 1419-1424 [DOI] [PubMed] [Google Scholar]