Abstract
ZO-1 is a multidomain protein involved in cell-cell junctions and contains three PDZ domains, which are necessary for its function in vivo. PDZ domains play a central role in assembling diverse protein complexes through their ability to recognize short peptide motifs on other proteins. We determined the structure of the second of the three PDZ domains of ZO-1, which is known to promote dimerization as well as bind to C-terminal sequences on connexins. The dimer is stabilized by extensive symmetrical domain swapping of β-strands, which is unlike any other known mechanism of PDZ dimerization. The canonical peptide-binding groove remains intact in both subunits of the PDZ2 dimer and is created by elements contributed from both monomers. This unique structure reveals an additional example of how PDZ domains dimerize and has multiple implications for both peptide binding and oligomerization in vivo.
PDZ (PSD-95/Discs Large/zonula occludens-1) domains are modular protein-binding motifs that are found in bacteria, plants, and animals. They play a key role in scaffolding protein complexes through their ability to recognize short polypeptide motifs on other proteins and in some cases their ability to crosslink through dimerization. Although found in proteins with functions as diverse as cell signaling, cytoskeletal structure, cell polarity, and trafficking, many PDZ proteins are associated with the plasma membrane, where they mediate the assembly of specific subcellular domains like synapses and cell-cell junctions (reviewed in Ref. 1). Thus, the mechanisms for peptide binding and dimerization of PDZ domains have important implications for several fields.
Although quite diverse at the primary sequence level, >150 known PDZ domain structures all share a conserved globular cluster of 5–6 β-strands and 1–2 α-helices (2). Peptide binding is mediated by a surface groove formed by an antiparallel β-strand (β2) and α-helix (α2) ((2) and reviewed in (3)). The majority of PDZ domain ligands are C-terminal peptides, which lie in this surface groove and form main chain interactions with α2 and β2. These interactions are highly specific and dependent on the terminal 3–4 amino acids of the peptide ligand (4). However, other residues within the ligand may also contribute to the specificity and affinity of the PDZ-ligand interaction (5). In addition, other binding modalities have also been characterized. For example, some PDZ domains can bind to internal peptide sequences or phosphoinositides (6–8). Finally, a limited number of PDZ domains can directly bind another PDZ domain to form homo- and hetero-oligomers (9–12). In contrast to the conserved mechanism of C-terminal peptide binding, there exist several distinct mechanisms for dimerization.
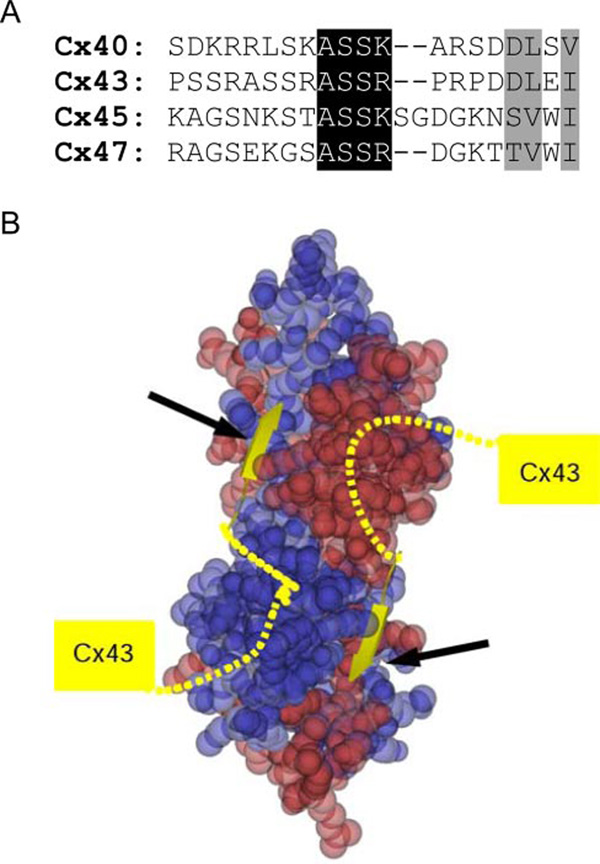
FIGURE 5. Binding of connexins to ZO PDZ2.
The connexin-PDZ2 interface involves both the canonical PDZ interaction plus an interaction supplied by more C-terminal residues in connexins. A, sequence alignment of approximately 20 C-terminal residues of human Cx40, Cx43, Cx45, and Cx47. Residues involved in the canonical PDZ interaction are shaded gray. Pronounced homology is observed N-terminal to this sequence, shaded black. B, a model of Cx43 binding to PDZ2. PDZ2 is represented as a space-filling model, with one monomer in blue and the other in red. The most six terminal residues of Cx43 (yellow coil and strand) were modeled based on the binding of peptide to the ZO-1 PDZ1 domain. Note how the Cx43 chain is predicted to extend from one PDZ molecule to the next, where the conserved ASSR/K sequence could be important for providing additional binding elements to the ZO-1 PDZ2 domain.
PDZ domains are particularly important in the organization of cellular tight junctions (TJ).3 Tight junctions form the paracellular barrier to the movement of ions, macromolecules, and cells across both endothelia and epithelia (13). Although several different classes of PDZ proteins are localized at the tight junction, the most directly relevant to TJ assembly and homeostasis are the zonula occludens (ZO) proteins ZO-1, -2, and -3. These proteins are members of the MAGUK (membrane-associated guanylate kinase) family, and all share a conserved set of protein- binding domains that includes three PDZ domains, an SH3 domain, and a region of homology to guanylate kinase (14) (Fig. 1A). The three PDZ domains are necessary for tight junction strand assembly (15), and ZO-1 polypeptides lacking PDZ1–3 cannot support TJ assembly in ZO-1 knockdown cells (16). The PDZ domains also appear to be an important component of signaling mechanisms at cell-cell contacts because overexpression of ZO-1 transgenes, encoding only the three PDZ domains, is sufficient to activate β catenin signaling and trigger an epithelial to mesenchymal transformation in cultured epithelial cells (17, 18). Thus, PDZ domains seem to serve dual role in cell signaling and structural properties of the ZO proteins.
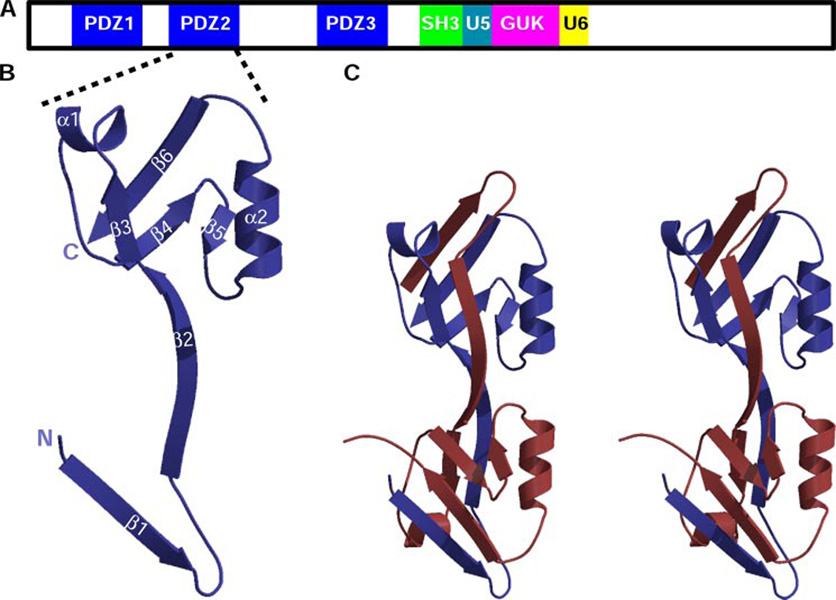
FIGURE 1. The second PDZ module of ZO-1 forms a domain-swapped dimer.
A, shown schematically is the modular arrangement of the MAGUK ZO-1. The unique regions 5 and 6 are labeled U5 and U6, respectively. B, a ribbon diagram of a ZO-1 PDZ2 monomer in blue with the secondary structure labeled. The N and C termini are marked with N and C, respectively. C, stereo view of a ZO-1 PDZ2 domain-swapped dimer. One monomer is depicted in blue and the other in red.
The functional role of the PDZ domains of the ZO proteins in these cellular processes is not clear, although it is most likely because of the fact that they bind to many of the known structural and signaling components of the tight junction. For example, the first PDZ domain (PDZ1) binds to both the claudin transmembrane proteins that form the intercellular barrier and to the signaling molecule ARVCF (19, 20), whereas PDZ3 binds to the TJ cell adhesion molecule JAM (junction adhesion molecule) and phospholipase Cβ(21, 22). Interestingly, the second PDZ domain appears to serve dual binding functions. It is necessary for homo- and hetero-oligomerization of the ZO proteins (12, 23), and it also binds to the C-terminal cytoplasmic domain of several gap junction-forming connexins (reviewed in (24)). Thus, we hypothesize that PDZ domains form the structural core that underlies the scaffolding and signaling properties of ZO proteins.
To better understand the structural basis for the role of PDZ2 in cell signaling and tight junction assembly we have solved the crystal structure of the ZO-1 PDZ2 homodimer (78 residues/monomer). The observed dimer is formed by a β-strand exchange mechanism that is unique among the known PDZ structures and which suggests that ZO protein dimers are extraordinarily stable. Although the predicted peptide-binding groove is not complete in a single PDZ monomer, it is reconstituted in the dimer by a β-strand from one molecule and a α-helix of the second. This unique structure reveals an additional example of how PDZ domains dimerize and has multiple implications for both peptide binding and oligomerization in vivo.
EXPERIMENTAL PROCEDURES
Cloning of ZO-1 PDZ2
The cDNA sequence encoding amino acids 186–262 of human ZO-1 was amplified using Taq polymerase (Promega, Madison, WI) and subcloned into the BamHI/EcoRI sites of pGEX-2T (GE Healthcare) with a stop codon following residue 162. Both strands of the resulting cDNA insert were sequenced to confirm accuracy of amplification.
Expression, Purification, and Crystallization of the ZO-1 PDZ2 Domains
The pGEX-2T vector carrying the gene for PDZ2 was transformed into Escherichia coli DH5α cells. Cells were grown at 25 °C in 2YT medium to an absorbance of 0.8, induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside, and allowed to continue growing for additional 16 h. Harvested cells were lysed by sonication and loaded onto a GST-Sepharose column (GE Healthcare). Following elution with glutathione, the fusion protein was cleaved by thrombin. The PDZ2 dimer was further purified on a S75 gel filtration column pre-equilibrated with 50 mm Tris/HCl, pH 7.5, 500 mm NaCl. The PDZ2 domain peak eluted at a volume consistent with the size of a dimer. The protein was concentrated to 4 mg/ml and directly used for crystallization screening using the NeXtal pre-formulated solution. A hit in the JCSG+ suite gave diffraction quality at the following condition: 0.1 m phosphate-citrate, pH 4.2, and 40% v/v polyethylene glycol 300. Crystals grew as cluster of very thin but large plates.
Data Collection, Structure Solution, and Refinement
Diffraction data to 1.7 Å resolution were collected at the Southeast Regional Collaborative Access Team ID beamline at the Advanced Photon Source (wavelength, 1.0 Å; temperature, 100 K). The mother liquor served as cryoprotectant so that crystals were picked directly from the drop and plunged into liquid nitrogen. Although the crystals diffracted to a very high resolution, the diffraction pattern revealed, at certain orientation, some very elongated spots and split spots. We interpret this to be because of the physical dimensions of the crystals (150 × 400 × 20 microns) and/or to a certain degree because of layering. Processing was accomplished with XDS (25).
The structure was solved using the molecular replacement method using Phaser (26). We tested numerous known PDZ structures as search models. In each case, we truncated the structure to where sequence alignment indicated similarity. Only one of the tested models (Protein Data Bank code 1WJL; converted to a poly-alanine model and truncated to 53 C-terminal residues) gave a single solution. The high resolution of our data allowed the automatic model tracing program Arp (27) to extend the model nearly to the entire molecule and dock the ZO-1 PDZ2 sequence. This initial model was subjected to several cycles of manual model adjustment usingO(28) and refinement with REFMAC (29).
The final model (Rwork = 20.9%/Rfree = 25.8%) has all residues in the allowed portion of the Ramachandran plot. The accuracy of our model was verified by simulated annealing omit maps calculated with Crystallography & NMR System; see supplemental Fig. S2 (30). For a plot of average B-factor versus residue number, see Fig. S3.
Data collection and refinement statistics are presented in Table 1. The coordinates have been deposited in the Protein Data Bank with accession code 2RCZ.
TABLE 1.
Data collection and refinement statistics
| ZO-1 PDZ2 | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 47.75, 33.61, 91.04 |
| α, β, γ (°) | 90.0, 103.64, 90.0 |
| Resolution (Å) | 20.0–1.70 |
| Rmergea (%) | 8.3 (55.8)b |
| I/σ I | 10.8 (4.0) |
| Completeness (%) | 99.2 (99.8) |
| Redundancy | 4.4 (3.7) |
| Refinement | |
| Resolution (Å) | 20.0–1.70 |
| No. of reflections (work/free) | 14032/1552 |
| Rwork/Rfree(%) | 20.9/25.8 |
| No. of atoms | |
| Protein | 1199 |
| Water | 103 |
| B-factors | |
| Protein main chain | 23.0 |
| Protein side chain | 26.2 |
| Water | 42 |
| r.m.s.c deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.492 |
| Ramachandran plot (%) | |
| Most allowed | 96.6 |
| Additionally allowed | 2.7 |
| Generously allowed | 0.7 |
| Disallowed | 0.0 |
The large size of the crystal enabled a low and high resolution sweep to be collected from a single crystal, exposing different surfaces.
Values in parentheses are for highest resolution shell.
r.m.s., root mean square.
RESULTS AND DISCUSSION
Structure Determination and Overall Fold of the ZO-1 PDZ2 Domain
The structure of the ZO-1 PDZ2 domain was solved to 1.7 Å resolution using the molecular replacement method. The challenge here was that a BLAST analysis revealed no known PDZ domain that possessed high sequence similarity to the ZO-1 PDZ2 sequence, except for the corresponding domains for ZO-2 and -3, which had not been solved. In addition, we speculated that the dimerization of this domain, a fact revealed by our previous studies (12, 23), might also make the molecular replacement process more complex. We decided on strategy that selects only the core of PDZ domains as search models. Numerous models were input into the molecular replacement program Phaser, and only one (Protein Data Bank code 1WJL) gave a unique solution. This partial model was sufficient as input into the automatic rebuilding program Arp (31) to allow nearly complete tracing of the dimer. After several rounds of manual model building and refinement, the R-factors converged to Rwork = 20.9% and Rfree = 25.8% (Table 1).
Homodimerization (10, 11) and heterodimerization (9) of PDZ domains have been previously observed, but are the exception although majority of PDZ domains are monomeric. In all of the previously observed dimers the overall fold of each PDZ monomer remained intact. In sharp contrast, in ZO-1 PDZ2 we observe a domainswapped dimer. A ribbon diagram of a monomer of PDZ2 is shown in Fig. 1B. The N-terminal portion, that includes strands β1 and β2, is not part of the core PDZ fold of this molecule. In fact, these segments complete the PDZ fold of the neighboring molecule and vice versa (Fig. 1C).
To gain insight into the ramifications of domain swapping of strands β1 and β2 on the PDZ-fold we compared the dimeric ZO-1 PDZ2 to the previously published monomeric structure of ZO-1 PDZ1 (Protein Data Bank code 2H2B) (32). The overlay of PDZ1 onto PDZ2 reveals a very good superposition of secondary structure elements. Strand β1 of PDZ1 is basically at the same position as strand β1 of PDZ2 (Fig. 2A, arrow 1). Likewise, strand β2 of PDZ1 is aligned with strand β2 of PDZ2 (Fig. 2A, arrow 2). In the canonical PDZ-fold as observed in the ZO-1 PDZ1 domain, strand β2 is followed by a loop that turns ~ 180°, which enables the following sequence to form the antiparallel strand β3 (Fig. 2A, green elements). In contrast, in ZO-1 PDZ2 strand β2 continues in the same direction, away from the core of the first PDZ molecule. The longer β2 directly connects to a short β3, which starts the core of the second PDZ molecule. Topology diagrams in Fig. 2, B (PDZ2) and C (PDZ1) illustrate the change in folding pattern that occurs upon domain swapping of strands β2 and β3.
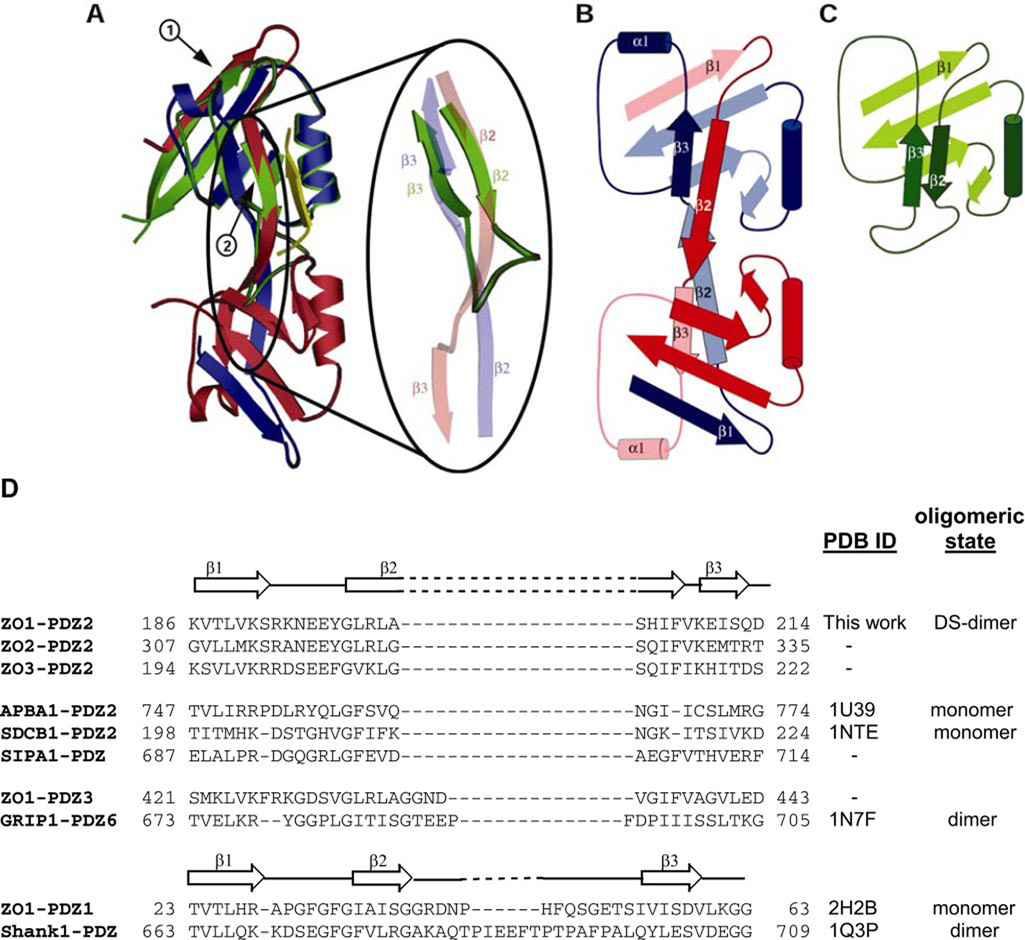
FIGURE 2. The ZO-1 PDZ2 domain maintains the canonical PDZ-fold, but with elements from both monomers.
A, overlay of ZO-1 PDZ2 and ZO-1 PDZ1 (Protein Data Bank code 2H2B). One monomer of the PDZ2 dimer is depicted in blue and the other in red. The ZO-1 PDZ1 domain is in green. The ligand bound to the ZO-1 PDZ1 domain is depicted as a yellow strand. The overlay shows a good fit between the PDZ1 and PDZ2 domains, albeit in the case of PDZ2, the structural elements are contributed by both molecules. Arrow 1 points at strand β1, and arrow 2 at strand β2. Importantly, despite the domain swapping that occurs in ZO-1 PDZ2, the peptide-binding grove of this module is not affected. The region where domain swapping commences is magnified. B and C, topology diagrams of ZO-1 PDZ2 and ZO-1 PDZ1, respectively. Color scheme as in A. D, amino acid sequence alignment of the N-terminal region of PDZ domains from ZO-1, -2, and -3 PDZ2; APBA1-PDZ2 (myeloid β A4 precursor protein-binding family A member 1); SDCB1- PDZ2 (syntenin-1); SIPA1-PDZ (signal-induced proliferation-associated protein 1); ZO-1-PDZ3; GRIP1-PDZ6; ZO-1-PDZ1; and Shank1-PDZ (SH3 and multiple ankyrin repeat domains protein 1). All sequences are of the human proteins and were aligned using ClustalW. The secondary structural elements for ZO-1 PDZ2 and PDZ1 are shown above their respective sequences. The secondary structure of APBA1, SDCB1, GRIP1, and Shank1 PDZ domains resemble that of ZO-1 PDZ1. DS-dimer, domain-swapped dimer.
In the case of the PDZ2 domain of ZO-1, strand β3 is followed by a short helix (α1) (Fig. 2B), which is missing in the ZO-1 PDZ1 domain (C). This difference is not related to the domain-swapping event, and the presence or absence of this helix is a common variation between many different PDZ domains.
Inspection of the primary sequence of ZO proteins suggests an explanation for the unique topology observed in PDZ2. Most PDZ sequences include several residues that form a loop that connects strand β2 to strand β3 (Fig. 2D and supplemental Fig. S1A). In some instances, as in the case of ZO-1 PDZ1, this region can be rather large (Fig. 2D, bottom) and forms a long loop that connects the two strands (32) (this segment is even longer in the Shank1 (10) PDZ domain). Some PDZ domains have a shorter sequence to connect the two strands, for example, PDZ6 of GRIP1 (11). In contrast, this region is completely missing from PDZ2 sequences in ZO-1, -2, and -3 proteins. We propose that it is the lack of these residues that induces the continuation of β2 in the same direction.
We analyzed the sequences of PDZ domains in this region so that we could predict additional PDZ domains that may undergo domain swapping. We discovered that in fact the sequence of ZO PDZ2 domains had the largest deletion between β2 and β3 of any of the recorded sequences. Interestingly, the PDZ2 domain of APBA1 (also known as X11/MINT) contains a deletion of the same magnitude as the ZO-1 PDZ2 sequence. However, the NMR structure of this domain (Protein Data Bank code 1U39) reveals a monomeric PDZ domain (33), albeit with a rather short β2 and β3 strands, which are connected by a tight turn. Likewise, the second PDZ domain of SDCB1 (syntenin-1) is monomeric (34), despite having acomparable deletion in this region in comparison to ZO-1 PDZ2. Notably, both in the case of APBA1 and SDCB1, a glycine residue, which is a common feature of hairpin loops, is present in the sequence connecting β2 to β3 (Fig. 2D). In contrast, the corresponding residue in PDZ2 domains is either a histidine or a glutamine. This suggests that although deletion of residues at the region that typically connects strands 2 and 3 is required for domain swapping to occur, it is not sufficient, and other factors such as lack of a glycine residue in this region are important.
The Domain-swapped Dimer Is Stabilized by Extensive Interactions
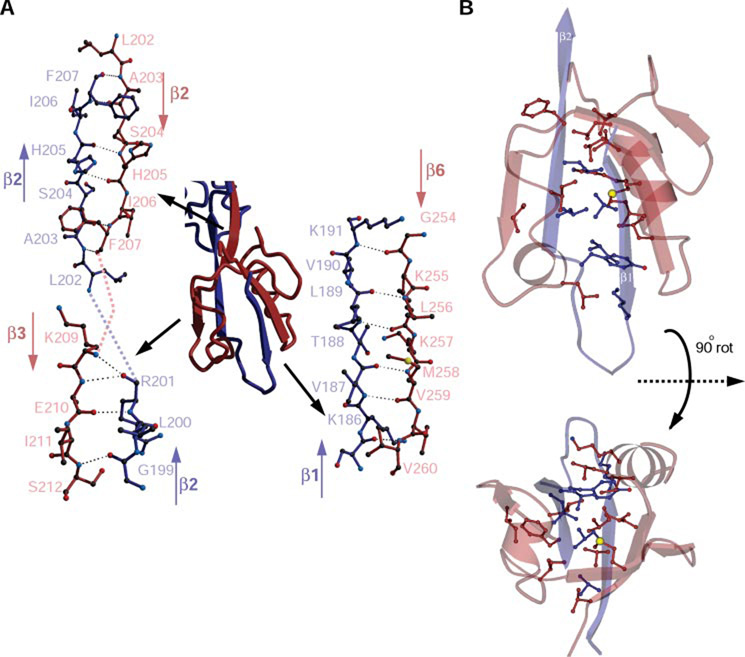
Our previous work demonstrated a very high affinity interaction between ZO-1 PDZ2 domains (12). Analysis of the monomer-monomer interactions indicates that dimerization is maintained by numerous interactions between antiparallel strands contributed by the two monomers. Strand β1 of one monomer interacts with strand β6 of the other monomer (for a total of six interactions; Fig. 3A, bottom right), and a segment of strand β2 interacts with strand β3 (four interactions; Fig. 3A, bottom left) and the remainder of strand β2 with its symmetrical partner from the second monomer (2 × 3 interactions; Fig. 3A, top left). The total number of hydrogen bonds built by the dimmer-forming antiparallel strands is 26.
FIGURE 3. The intermonomer interactions of ZO-1 PDZ2 are comprehensive and are mainly mediated by antiparallel strands.
A, a detailed representation of the three regions of interaction. On the bottom right, the interactions between β1 of one monomer and β6 of the other are shown. Likewise, interactions are shown on the bottom left, between a section of β2 and β3 and top left, between β2 of one monomer and its symmetrical segment from the second monomer. Only half of the total interactions per dimer are shown; a corresponding half is formed in the other part of the dimer. B, hydrophobic side chains from strands β1 and β2 of one molecule blue) are buried by residues from the complementary molecule (red). For clarity only the relevant parts of each molecule are displayed. Two views related by a 90° rotation are shown.
In addition to these polar interactions, hydrophobic forces play a major role in dimer stabilization. Hydrophobic residues donated by one molecule bury hydrophobic side chains present in strands β1 and β2 of the other molecule (Fig. 3B). We conclude that dimerization is of very high affinity with essentially no monomer present in the cell. This is consistent with our previous equilibrium centrifugation studies of pure ZO-1, which failed to detect monomers (12).
Based on sequence homology (see supplemental Fig. S1B) we predict that both ZO-2 and -3 will likewise form domain-swapped PDZ dimers. Gel filtration experiments performed with the PDZ2 domain of ZO-2 showed an elution volume consistent with a dimeric protein (data not shown).
Homodimerization Versus Heterodimerization of ZO proteins
Co-immunoprecipitation studies suggest that in addition to homodimers, ZO-1 can also form heteromultimers with ZO-2 and -3 (35), and that the PDZ2 domain of each of these proteins is required for this interaction (23, 36). These observations suggest that the strand-exchange mechanism may also regulate the formation of heteromultimers in vivo. Interestingly, the same studies have also demonstrated that ZO-2 does not form multimers with ZO-3 (35). The structural basis for this observation is not obvious and cannot be rationalized from the sequence and structural information detailed here; the overall homology between the domains in ZO-1, -2, and -3 is very high (supplemental Fig. S1B), and there are no obvious differences in critical residues involved in strand exchange. Thus, we propose that other regions within the ZO polypeptides may also promote heteromultimerization in vivo.
Comparing Mechanisms of PDZ Dimerization
As mentioned previously, PDZ domains have been observed to form both homodimers and heterodimers. An example of the latter is the interaction between the PDZ domains of neuronal nitricoxide synthase (nNOS) with syntrophin. In this case, heterodimerization is mediated by residues in nNOS that follow the canonical PDZ sequence and form what has been termed a “β-finger” (9). The β-finger of nNOS inserts into the peptide-binding groove of the syntrophin PDZ domain. The primary sequence of residues that follow the ZO-1 PDZ2 domain are not similar to those that build the β-finger of nNOS; we predicted at the onset of our work that dimerization of the ZO-1 PDZ2 domains follows a different mechanism.
Homodimerization of PDZ domains was first detected in the PDZ domain of the glutamate receptor-interacting protein GRIP) (11). The dimerization interface stabilizing the GRIP1 dimer is much less extensive than that present in ZO-1 PDZ2 (Fig. 4A). Key to GRIP1 dimerization is an antiparallel orientation between strands β1 of the molecules forming the dimer (Fig. 4B). Shortly thereafter, dimerization of the PDZ domain of Shank1 is reported (10). Here too dimerization was predominantly mediated by strands β1 (Fig. 4C). Interestingly, in the three known cases of PDZ homodimerization (i.e. ZO-1 PDZ2, GRIP1-PDZ6, and Shank1 PDZ), the relative orientation of the monomers is different. Fig. 4D presents an overlay of the three dimers that is prepared by calculating the superpositioning matrix using only residues in the boxed molecule. These molecules overlay well between the structures. In contrast, the molecule completing the dimer is oriented differently in each case. Even for the PDZ domains that homodimerize largely because β1-β1 interactions, the orientation is very different. Because these domains are parts of multidomain proteins involved in recruiting other protein partners, it is likely that each orientation is optimized to accommodate the other parts of the molecule and those of its particular binding protein.
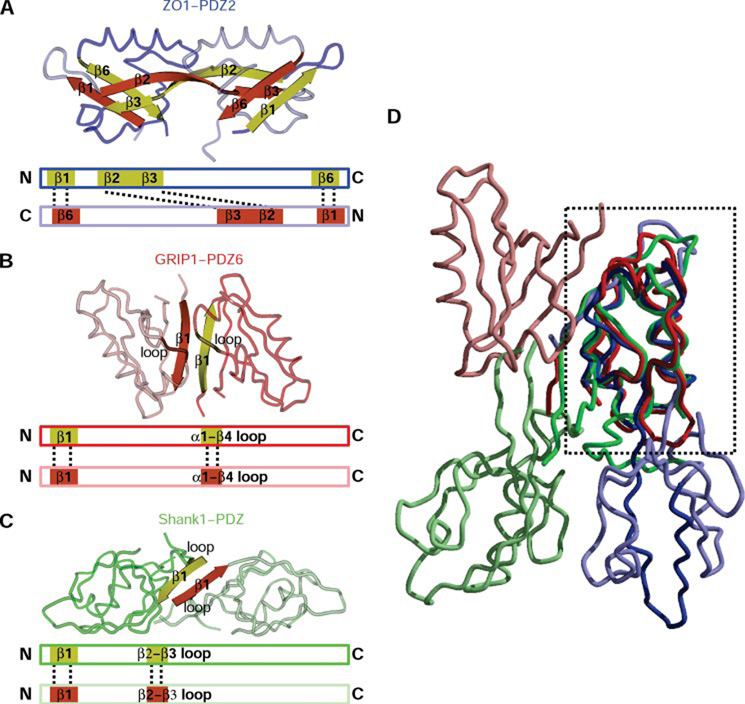
FIGURE 4. Homodimerization in PDZ domains.
A, the ZO-1 PDZ2 dimer is shown with one monomer in dark blue and the other in light blue. The strands mediating dimerization are in yellow and red. Below the structure figure is a schematic of the interactions (displayed by dotted lines) between the monomer in the dimer. B, GRIP1-PDZ6 (Protein Data Bank code 1N7F) also forms a homodimer (dark and light red). However, here the interactions are predominately provided by residues from β1, with only additional few from the loop connecting α1 with β4. A schematic as in A is below the structure. C, dimerization of Shank1 PDZ (Protein Data Bank code 1Q3P) also depends on β1, with minor contribution from the loop connecting β2 and β3. A schematic as in A is below the structure. D, overlay of the three dimers done by superpositioning one of the monomers in each dimer (region used for superposition is boxed). Although the overall fold of the PDZ domains is nearly identical, due to different dimerization motifs, the orientation of the molecule completing the dimer is very different among the three homodimers.
Implications of Dimerization for Ligand Binding
One highly conserved function of PDZ domains is to bind to short peptide sequences at the C terminus of transmembrane proteins. Notably, inspection of the ZO-1 PDZ2 dimer structure suggests that the peptide-binding groove of the domain-swapped PDZ2 is not obstructed. This has important consequences for ZO-1 function, since the PDZ2 domain of ZO-1 has been reported to bind to several of the gap junction-forming connexin (Cx) proteins (e.g. Cx43 (37); Cx45 (38); Cx47 (39)). ZO-1 binding to at least one connexin, Cx43, is required for phospholipase C-mediated regulation of gap junction signaling (22) and has been further implicated in the regulation of connexin 43 gap junction organization (40). The PDZ2 domain is therefore predicted to elicit a clustering of connexin 43 molecules by having two adjacent binding sites on the domain-swapped PDZ2.
Interestingly, for both ZO-1 (41) and ZO-2 (37) PDZ2 domain binding to Cx43, it was observed that this protein-protein interaction is not only dependent on the most C-terminal Cx 43 residues (as is the case for the canonical binding of proteins to PDZ domains), but that a region encompassing approximately 20 C-terminal residues is also involved in this interaction. As expected, sequence alignment of the C-terminal portion of several connexins shows sequence conservation at the terminal residues that would form the canonical PDZ interaction (Fig. 5A, gray shaded residues). However, a patch of four residues N-terminal to this sequence that is highly conserved is striking (Fig. 5A, black shaded residues). To understand how these residues could play a role in Cx43-PDZ2 interaction, we modeled the canonical PDZ-binding residues of Cx43 into our ZO-1 PDZ2 structure (arrows, Fig. 5B). As mentioned before, two Cx43 C-terminal regions (Fig. 5B, yellow strands) would bind, one to each of the PDZ2 monomers. This binding would be in an antiparallel fashion. Our modeling suggests that residues N-terminal to the canonical PDZ interaction sequence (i.e. the last four amino acids) would extend to the neighboring PDZ2 molecule. In other words, both PDZ2 domains are predicted to participate in the binding of each Cx43 C-terminal peptide and by sequence analogy (Fig. 5A) to other connexins such as Cx40, Cx45, and Cx47. Moreover, this model suggests that surface residues on the ZO proteins can play a role in the selectivity of binding of connexins to PDZ2 domains. Experiments to test this model are underway.
In conclusion, the structure of the PDZ2 dimer of ZO-1 reveals a form of domain swapping unique among PDZ domains. Dimerization is based on extensive exchange of β-strand interactions. The dimeric nature of PDZ2 rationalizes the unique binding determinants of several connexin proteins to this domain, implicating both molecules in the dimer as forming the connexin-PDZ2 interaction surface.
Supplementary Material
Supplemental Material can be found at: http://www.jbc.org/cgi/content/full/M707255200/DC1
Acknowledgments
We thank the staff at the Southeast Regional Collaborative Access Team and acknowledge discussions over refinement with Peter Zwart.
Footnotes
This work was supported in part by National Institutes of Health Grant DK61397 and by the Universities of North Carolina and Illinois.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Supported in part by NIDDK Institutional Training Grant T32DK07739, “Training Program in Signal Transduction and Cellular Endocrinology,” from the National Institutes of Health.
The atomic coordinates and structure factors (code 2RCZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The abbreviations used are: TJ, tight junctions; ZO, zonula occludens; SH3, Src homology 3; nNOS, nitric– oxide synthase; GRIP, glutamate receptor-interacting protein; Cx, connexin.
REFERENCES
- 1.Fanning AS, Anderson JM. J. Clin. Investig. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 3.Jemth P, Gianni S. Biochemistry. 2007;46:8701–8708. doi: 10.1021/bi7008618. [DOI] [PubMed] [Google Scholar]
- 4.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 5.Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. Science. 2007;317:364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuppen E, Gerrits H, Pepers B, Wieringa B, Hendriks W. Mol. Biol. Cell. 1998;9:671–683. doi: 10.1091/mbc.9.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shieh BH, Zhu MY. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann P, Zhang Z, Degeest G, Mortier E, Leenaerts I, Coomans C, Schulz J, N'Kuli F, Courtoy PJ, David G. Dev. Cell. 2005;9:377–388. doi: 10.1016/j.devcel.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 10.Im YJ, Lee JH, Park SH, Park SJ, Rho SH, Kang GB, Kim E, Eom SH. J. Biol. Chem. 2003;278:48099–48104. doi: 10.1074/jbc.M306919200. [DOI] [PubMed] [Google Scholar]
- 11.Im YJ, Park SH, Rho SH, Lee JH, Kang GB, Sheng M, Kim E, Eom SH. J. Biol. Chem. 2003;278:8501–8507. doi: 10.1074/jbc.M212263200. [DOI] [PubMed] [Google Scholar]
- 12.Utepbergenov DI, Fanning AS, Anderson JM. J. Biol. Chem. 2006;281:24671–24677. doi: 10.1074/jbc.M512820200. [DOI] [PubMed] [Google Scholar]
- 13.Anderson JM, Van Itallie CM, Fanning AS. Curr. Opin. Cell Biol. 2004;16:140–145. doi: 10.1016/j.ceb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Funke L, Dakoji S, Bredt DS. Annu. Rev. Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 15.Fanning AS, Little BP, Rahner C, Utepbergenov D, Walther Z, Anderson JM. Mol. Biol. Cell. 2007;18:721–731. doi: 10.1091/mbc.E06-08-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M. Cell. 2006;126:741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 17.Reichert M, Muller T, Hunziker W. J. Biol. Chem. 2000;275:9492–9500. doi: 10.1074/jbc.275.13.9492. [DOI] [PubMed] [Google Scholar]
- 18.Polette M, Gilles C, Nawrocki-Raby B, Lohi J, Hunziker W, Foidart JM, Birembaut P. Cancer Res. 2005;65:7691–7698. doi: 10.1158/0008-5472.CAN-04-4230. [DOI] [PubMed] [Google Scholar]
- 19.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. J. Cell Biol. 1999;147:1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kausalya PJ, Phua DC, Hunziker W. Mol. Biol. Cell. 2004;15:5503–5515. doi: 10.1091/mbc.E04-04-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bazzoni G, Martinez-Estrada OM, Orsenigo F, Cordenonsi M, Citi S, Dejana E. J. Biol. Chem. 2000;275:20520–20526. doi: 10.1074/jbc.M905251199. [DOI] [PubMed] [Google Scholar]
- 22.van Zeijl L, Ponsioen B, Giepmans BN, Ariaens A, Postma FR, Varnai P, Balla T, Divecha N, Jalink K, Moolenaar WH. J. Cell Biol. 2007;177:881–891. doi: 10.1083/jcb.200610144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fanning AS, Anderson JM. Curr. Top. Microbiol. Immunol. 1998;228:209–233. doi: 10.1007/978-3-642-80481-6_9. [DOI] [PubMed] [Google Scholar]
- 24.Giepmans BN. Cardiovasc. Res. 2004;62:233–245. doi: 10.1016/j.cardiores.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. J. Appl. Crystallogr. 1993;24:795–800. [Google Scholar]
- 26.Read RJ. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 27.Lamzin VS, Perrakis A, Wilson KS. In: International Tables for Crystallography. Vol. F: Crystallography of Biological Macromolecules. Rossmann MG, Arnold E, editors. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2001. pp. 720–722. [Google Scholar]
- 28.Jones TA, Zhou J-Y, Cowan SW, Kjeldgaard M. Acta Crystallogr. Sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewskli J, Nilges N, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. Sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Perrakis A, Morris R, Lamzin VS. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 32.Appleton BA, Zhang Y, Wu P, Yin JP, Hunziker W, Skelton NJ, Sidhu SS, Wiesmann C. J. Biol. Chem. 2006;281:22312–22320. doi: 10.1074/jbc.M602901200. [DOI] [PubMed] [Google Scholar]
- 33.Long JF, Feng W, Wang R, Chan LN, Ip FC, Xia J, Ip NY, Zhang M. Nat. Struct. Mol. Biol. 2005;12:722–728. doi: 10.1038/nsmb958. [DOI] [PubMed] [Google Scholar]
- 34.Kang BS, Cooper DR, Devedjiev Y, Derewenda U, Derewenda ZS. Structure. 2003;11:845–853. doi: 10.1016/s0969-2126(03)00125-4. [DOI] [PubMed] [Google Scholar]
- 35.Wittchen ES, Haskins J, Stevenson BR. J. Biol. Chem. 1999;274:35179–35185. doi: 10.1074/jbc.274.49.35179. [DOI] [PubMed] [Google Scholar]
- 36.Itoh M, Morita K, Tsukita S. J. Biol. Chem. 1999;274:5981–5986. doi: 10.1074/jbc.274.9.5981. [DOI] [PubMed] [Google Scholar]
- 37.Singh D, Solan JL, Taffet SM, Javier R, Lampe PD. J. Biol. Chem. 2005;280:30416–30421. doi: 10.1074/jbc.M506799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kausalya PJ, Reichert M, Hunziker W. FEBS Lett. 2001;505:92–96. doi: 10.1016/s0014-5793(01)02786-7. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Ionescu AV, Lynn BD, Lu S, Kamasawa N, Morita M, Davidson KG, Yasumura T, Rash JE, Nagy JI. Neuroscience. 2004;126:611–630. doi: 10.1016/j.neuroscience.2004.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunter AW, Barker RJ, Zhu C, Gourdie RG. Mol. Biol. Cell. 2005;16:5686–5698. doi: 10.1091/mbc.E05-08-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorgen PL, Duffy HS, Sahoo P, Coombs W, Delmar M, Spray DC. J. Biol. Chem. 2004;279:54695–54701. doi: 10.1074/jbc.M409552200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material can be found at: http://www.jbc.org/cgi/content/full/M707255200/DC1