Abstract
The signal recognition particle (SRP) is a ribonucleoprotein composed of an Alu domain and an S domain. The S domain contains unique sequence SRP RNA and four SRP proteins: SRP19, SRP54, SRP68, and SRP72. SRP interacts with ribosomes to bring translating membrane and secreted proteins to the endoplasmic reticulum (ER) for proper processing. Additionally, SRP RNA is a member of a family of small nonribosomal RNAs found recently in the nucleolus, suggesting that the nucleolus is more plurifunctional than previously realized. It was therefore of interest to determine whether other SRP components localize to this intranuclear site. In transfected rat fibroblasts, green fluorescent protein fusions of SRP19, SRP68, and SRP72 localized to the nucleolus, as well as to the cytoplasm, as expected. SRP68 also accumulated in the ER, consistent with its affinity for the ER-bound SRP receptor. SRP54 was detected in the cytoplasm as a green fluorescent protein fusion and in immunofluorescence studies, but was not detected in the nucleolus. In situ hybridization experiments also revealed endogenous SRP RNA in the nucleolus. These results demonstrate that SRP RNA and three SRP proteins visit the nucleolus, suggesting that partial SRP assembly, or another unidentified activity of the SRP components, occurs at the nucleolus. SRP54 apparently interacts with nascent SRP beyond the nucleolus, consistent with in vitro reconstitution experiments showing that SRP19 must bind to SRP RNA before SRP54 binds. Our findings support the notion that the nucleolus is the site of assembly and/or interaction between the family of ribonucleoproteins involved in protein synthesis, in addition to ribosomes themselves.
The nucleolus long has been known as a dense subnuclear structure at which ribosomal genes are clustered and ribosomal transcription, rRNA processing, and ribosomal subunit assembly occurs (1). More recently, it has become clear that the RNA components of other ribonucleoproteins also may localize to the nucleolus (2, 3), including the signal recognition particle (SRP) (4). Protein components of both RNase P and RNase MRP also recently have been shown to be localized in the nucleolus (5). The hypothesis has been advanced that the nucleolus not only has evolved to carry out ribosome synthesis but, in fact, is the site of assembly of and/or interaction between multiple ribonucleoprotein machines, many of which are involved in protein synthesis (6). However, no information regarding the potential nucleolar localization of the other components of the SRP has been available.
The SRP is a ribonucleoprotein that arrests translation of secretory or membrane proteins and docks the nascent polypeptide–ribosome complex at receptors on the endoplasmic reticulum (ER), whereupon translation is resumed to direct the protein into the membrane assembly or secretory pathways (7–9). In mammalian cells, the SRP contains six proteins (7) and an ≈300-nt RNA (10) that previously had been known as 7S RNA or 7SL RNA (11–13). In an early study it was found that the SRP could be cleaved into two distinct ribonucleoprotein complexes by mild nuclease digestion (14). One of these complexes contained the portion of the folded SRP RNA known as the Alu domain (because of its homology with the Alu family of repeated DNA sequences in mammalian genomes) together with two of the SRP proteins, SRP9 and SRP14. The other subparticle produced by mild nuclease digestion contained the so-called S domain of SRP RNA and the SRP19, SRP54, SRP68, and SRP72 proteins. Subsequent studies demonstrated that the SRP RNA Alu domain and its associated SRP9 and SRP14 proteins constitute the translational arrest activity, whereas the S domain of SRP RNA and its associated proteins contain the nascent polypeptide signal recognition and ER docking activities, via SRP54, SRP68, and SRP72 (9, 15–20). In vitro experiments with purified SRP components revealed that (i) SRP9 and SRP14 form a heterodimer before binding to SRP RNA, (ii) SRP19 binding to SRP RNA is required for subsequent SRP54 binding, and (iii) SRP68 binding to SRP RNA enhances SRP72 binding (9, 10, 21–33).
We recently investigated the intracellular localization of fluorescent SRP RNA after its microinjection into the nucleus of mammalian cells. We observed a rapid localization in nucleoli, followed by a gradual dissipation of nucleolar signal and the progressive appearance of fluorescence in the cytoplasm (4). Nuclear microinjection of mutant SRP RNAs revealed that the nucleolar localization elements were associated with the Alu domain of SRP RNA as well as helix 8 in the S domain (4). In the current study we have investigated the intracellular distribution of endogenous SRP RNA in mammalian cells and found that it, too, is present in the nucleolus, in addition to its expected abundant presence in the cytoplasm. We also have investigated the intracellular localization of the four S-domain SRP proteins, SRP19, SRP54, SRP68, and SRP72, by expressing them as green fluorescent protein (GFP) fusions. We found that SRP19, SRP68, and SRP72, like SRP RNA, display a nucleolar localization whereas SRP54 does not. All four proteins also are found in the cytoplasm, as would be expected for SRP components. These results now expand the constellation of ribonucleoproteins known to be present in the nucleolus and make more attractive the hypothesis that many of the ribonucleoproteins involved in protein synthesis interact within and/or traffic through the nucleolus.
Materials and Methods
In Situ Hybridization and Immunocytochemistry.
NRK cells were grown as described (4). In situ hybridization to SRP RNA was carried out by using as probes the following oligodeoxynucleotides, in which the underlined Ts denote 5-aminohexylthymidine:
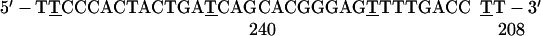 |
1 |
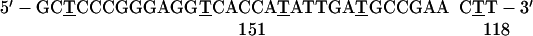 |
2 |
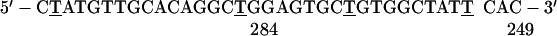 |
3 |
The numbers at the 5′ and 3′ ends denote the nucleotide positions at the ends of the SRP RNA region to which the oligo is complementary.
Control oligos used were:
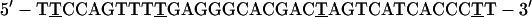 |
4 |
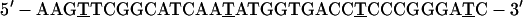 |
5 |
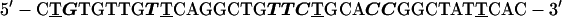 |
6 |
Oligo 4 is the reverse polarity sequence of oligo 1. Oligo 5 is the sense sequence of oligo 2. Oligo 6 is homologous to oligo 3 except for bold nucleotides, which have been changed to mimic the antisense sequence of Xenopus laevis SRP RNA. Oligos then were labeled with the cyanine dye, cy-3 (Amersham Pharmacia), as described (34).
Cells growing on glass coverslips were fixed in 4% formaldehyde and stored in 70% ethanol for up to 2 weeks. Before in situ hybridization, cells were permeabilized in 100% acetone at −20°C for 10 min. In situ hybridization was carried out in 40% formamide as described (35) except that after hybridization, cells were washed with 40% formamide, 2× SSC (0.15 M NaCl/0.015 M sodium citrate, pH 7.0), and then 20% formamide, 1× SSC, for 30 min each at 37°C, followed by three 15-min washes in 1× SSC at room temperature. For immunocytochemistry, cells were fixed and permeabilized as detailed (3) and incubated with IgG from a polymyositis patient autoimmune serum (36) provided by Frederick W. Miller (U.S. Food and Drug Administration, Bethesda, MD). This patient serum specifically immunoprecipitates the SRP from human cell extracts and also immunoprecipitates the SRP54 protein from 35S-methionine-labeled HeLa cell cytoplasmic supernatants (36). The IgG fraction from this patient serum was purified by protein A chromatography (37). A control IgG fraction was purified similarly from a polymyositis patient serum that lacked anti-SRP activity. Antibody binding was detected as detailed (3) except that the second antibody was cy3-labeled Fc-specific goat anti-human IgG (Sigma).
Construction of GFP Fusion Proteins and Transfection Protocol.
Enhanced green (F64 → L, S65 → T H231 → L) fluorescent protein (EGFP)-encoding plasmids pEGFP-N1 and pEGFP-C1 (CLONTECH) were used to construct SRP19, SRP54, SRP68, and SRP72 fusions with EGFP at the N or C terminus of each SRP protein. Human SRP19- and SRP54-encoding DNAs were amplified from the plasmids pET23d-19x and pEThSRP54, respectively (38, 39). The source of human SRP68- and SRP72-encoding sequences were plasmids pFBh68 and pFBh72, where the respective genes are inserted into the FastBac vector (GIBCO/BRL). The full-length coding regions were assembled from partial expressed sequence tag (EST) clones (Genome Systems) and clones identified in the screening of a λ-gt10 cDNA library of human HepG2 hepatoma cells obtained from the American Type Culture Collection (Manassas, VA; ATCC 77400). Four overlapping SRP68 clones were identified by using an approximately 300-bp probe derived from EST T48898. Similarly, a 191-bp HindIII fragment from EST H07969 was used to obtain the 5′ portion of the SRP72 gene. Screening of the cDNA library was carried out as described (39). Sequences of the two full-length clones were confirmed by using sequenase 2.0 (United States Biochemical).
For construction of plasmids encoding the fusion proteins, PCR primers were designed to introduce desired features (cloning sites, consensus translation initiation sites, or replacements for stop codons) into flanking regions of the SRP protein-encoding regions as follows.
H2N-SRP19-EGFP-COOH fusion.
The PCR primer 5′-GTTTAACTTTAAGATCTAGCGCCACCATGGCTTGCGC-3′ introduces a BglII site at position −15 to −10 from the start codon for SRP19, as well as a consensus translation initiation site at the proper position. The second primer, 5′-GCTGATACTAGGATCCTTCTTTTTCTTTCCTTTCCC-3′, replaces a stop codon for SRP19 with an aspartate codon and introduces a BamHI site. The amplified DNA was cloned into the BglII–BamHI sites of pEGFP-N1, resulting in a hexapeptide DPPVAT linker between SRP19 and EGFP.
H2N-EGFP-SRP19-COOH fusion.
The first PCR primer, 5′-TTTTGTTAAGCTTTAGGAGGGGGATATACC-3′, introduces a HindIII site at −23 to −18 from the start codon for SRP19 and encodes LGGGYT at the NH2 terminus of SRP19. The second primer, 5′-CCACATACTGGATCCTGATACTAGG-3′, introduces a BamHI site 11–16 nt downstream from the stop codon of SRP19. This DNA was cloned into the HindIII–BamHI sites of pEGFP-C1, resulting in a pentadecapeptide SGLRSRAQALGGGYT linker between EGFP and SRP19.
H2N-SRP54-EGFP-COOH fusion.
The first primer, 5′-CTTTAAGCTGCAGCGCCACCATGGTTCTAGC-3′, introduces a PstI site at −13 to −8 from the start codon for SRP54, as well as a consensus translation initiation site at the proper position. The second primer, 5′-AGGACATACCGGTCCCATATTATTGAATCCC-3′, replaces a stop codon for SRP54 with a glycine codon and introduces an AgeI site immediately downstream. This DNA was cloned into the PstI–AgeI sites of pEGFP-N1, resulting in a pentapeptide GPVAT linker between SRP54 and EGFP.
H2N-EGFP-SRP54-COOH fusion.
The first primer, 5′-TAACTTCTCGAGGGGGAGCTACCATGGTTCTAGCAGACCTTGG-3′, introduces an XhoI site at position −17 to −12 from the start codon of SRP54 and the sequence encoding the pentapeptide RGGAT at the NH2 terminus of SRP54. The second primer, 5′-CAGTTTATCTGCAGGACATTATCTTTACATATTATTGAATCCC-3′, introduces a PstI site 11–16 nt downstream from the stop codon of SRP54. This DNA was cloned into the XhoI–PstI sites of pEGFP-C1, resulting in a decapeptide SGLRSRGGAT linker between EGFP and SRP54.
H2N-EGFP-SRP68-COOH fusion.
After digesting pFBh68 with BamHI and KpnI restriction enzymes sequentially, the fragment encoding SRP68 was purified from an agarose gel and cloned into the BglII–KpnI sites of pEGFP-C1. This resulted in an oligopeptide SGLRSRSEARGIQRPTSTSSLVAAA linker between EGFP and SRP68.
H2N-SRP72-EGFP-COOH fusion.
The plasmid pFBh72 was used as a template for PCR. The first PCR primer, 5′-CGAGCTCAGAATTCGCGGCCGCGATG-3′, introduced an EcoRI site at the position −15 to −10 from the start codon for SRP72. The second primer, 5′-CAAAAAGAGGTACCAGAAGAATATATGCATACCAG-3′, replaced two SRP72 stop codons with tyrosine and alanine codons and introduced a KpnI site 18–23 nt downstream from the last, tryptophan-encoding codon of SRP72. The PCR-derived fragment was digested with EcoRI and KpnI and cloned into the EcoRI–KpnI sites of pEGFP-N1, resulting in a heptadecapeptide YAYILIVPRARDPPVAT linker between SRP72 and EGFP.
Recombinant plasmids were amplified in Escherichia coli HB101 in LB containing kanamycin (30 μg/ml). Plasmid purification was performed by using the Qiagen Plasmid Mega kit (Chatsworth, CA). The GFP-SRP junction regions of these cloned DNAs were confirmed by DNA sequencing.
NRK cells plated at 1 × 105 cells per well (35-mm diameter, glass) in F12K medium (Sigma) containing 10% FBS were allowed to grow for 18–24 hr and then transfected by using Lipofectamine (2 μg DNA/5 μl Lipofectamine) or Lipofectamine 2000 (1 μg DNA/5 μl Lipofectamine 2000) following the manufacturer's instructions (GIBCO/BRL). After 3 hr of incubation (37°C), the transfection mix was replaced with fresh F12K medium containing 10% FBS, and the cells were examined 12–24 hr later.
Microscopy and Vital Dye Staining.
An inverted Leica microscope equipped with a ×40, 1.3-numerical aperture oil objective, both a regulated-intensity halogen lamp and a mercury arc lamp, and standard fluorescein and rhodamine filter sets (40) was used to visualize both live cells transfected with GFP plasmids and fixed cells after in situ hybridization and immunocytochemistry. Transfected cells were kept at 37°C, 5% CO2 while viewing (40). In some cases, the ER was stained during observation of transfected cells on the microscope stage by adding 3–5 μl of a 0.5-mg/ml solution of rhodamine B hexyl ester (Molecular Probes) directly to the 3 ml of medium overlaying the cells. Images were captured by using a Photometrics CH250 cooled charge-coupled device camera and processed by using metamorph software (Universal Imaging, Media, PA).
Results
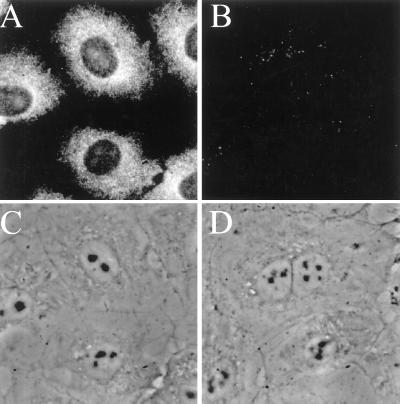
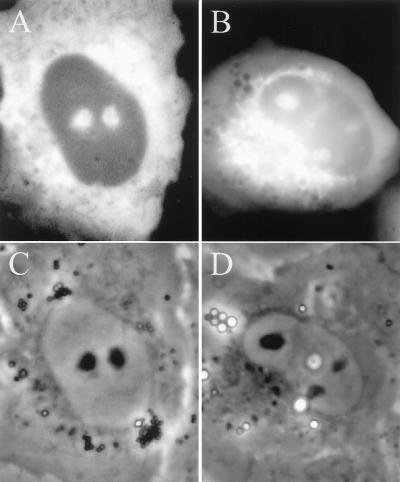
Fig. 1 shows typical in situ hybridization results for SRP RNA in NRK cells. In cells hybridized with oligos complementary to SRP RNA (Fig. 1A), the cytoplasm displayed a strong signal as would be expected from the known cytoplasmic location of the SRP. However, SRP RNA also was observed often in the nucleus, concentrated in the nucleoli (compare Fig. 1A with the image in Fig. 1C, in which the nuclei are apparent by their strong phase contrast). Approximately 80% of the cells in a given population showed signal in at least one nucleolus. When cells were hybridized with control oligonucleotides, only background levels of signal were observed (Fig. 1B).
Figure 1.
Detection of SRP RNA in rat NRK fibroblasts by in situ hybridization. (A) Cells hybridized with cy-3-labeled probes complementary to SRP RNA (see Materials and Methods). (B) Cells hybridized with control oligonucleotides (see Materials and Methods). (C) Phase-contrast image of the cells in A. (D) Phase-contrast image of the cells in B.
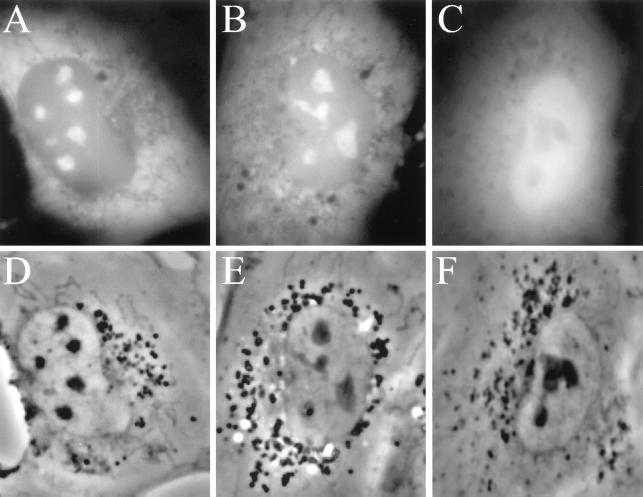
We next investigated the intracellular localization of the S domain SRP proteins, SRP19, SRP54, SRP68, and SRP72, by expressing them in NRK cells as GFP fusion proteins. Fig. 2 A and B shows that GFP-SRP19, with the GFP tag fused either to the NH2 or COOH terminus of the SRP19 protein, displayed prominent nucleolar localization as well as the cytoplasmic localization expected. This nucleolar localization did not appear to depend on expression level; 82% of the transfected cells observed in a given population displayed nucleolar signal. No nucleolar localization was seen in cells expressing GFP itself (Fig. 2C). It is of interest to note that both of the GFP-SRP19 fusion proteins also were present in the nucleoplasm (Fig. 2 A and B), although at about half the concentration of that in nucleoli (as judged by signal intensity after background subtraction).
Figure 2.
Nucleolar localization of SRP19-GFP protein. (A) Typical NRK cell expressing GFP-SRP19 protein with GFP fused to the N terminus of the SRP19 protein. (B) Same as A except GFP was fused to the C terminus of the SRP19 protein. (C) NRK cell expressing GFP itself. (D–F) Phase-contrast images of the cells shown in A, B, and C, respectively.
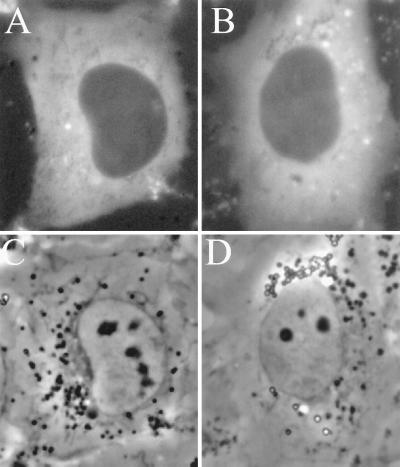
Different results were obtained with GFP-SRP54 fusion proteins. As can be seen in Fig. 3 A and B, neither of the two GFP-SRP54 protein fusions displayed nucleolar localization, though both were abundantly present in the cytoplasm. When average signal intensity was quantitated, the nucleoplasmic level of both GFP-SRP54 proteins was found to be about four times lower than that observed with the GFP-SRP19 proteins (Figs. 2 and 3). Similar results, i.e., nucleolar localization of GFP-SRP19 and not GFP-SRP54, also were observed in other cell lines, including HeLa and 293 (human kidney) cells.
Figure 3.
SRP54-GFP protein does not display nucleolar localization. (A) NRK cell expressing EGFP-SRP54 protein with GFP fused to the N terminus of the SRP54 protein. (B) Same as A except the GFP was fused to the C terminus of the SRP54 protein. (C and D) Phase-contrast images of the cells shown in A and B, respectively.
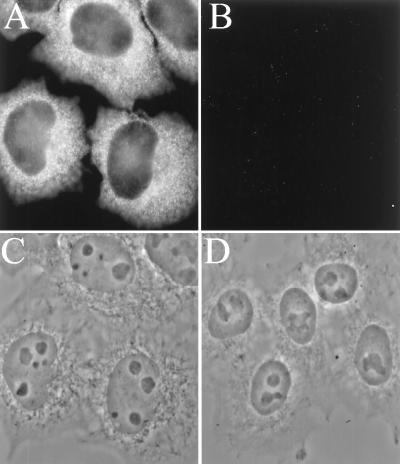
To confirm the absence of SRP54 from nucleoli by an independent method, we used an antibody specific for the SRP54 protein (36) to immunocytochemically detect the endogenous SRP54 protein in these cells. As shown in Fig. 4A, this antibody displayed strong cytoplasmic staining, as expected. However, the nuclear pattern consisted of faint nucleoplasmic staining and no detectable nucleolar signal (compare Fig. 4A with phase-contrast image in Fig. 4C). Cells stained with a control antibody that is known not to react with the SRP displayed no signal (Fig. 4B). These results provide further evidence that SRP54 does not traffic through the nucleolus to an extent detectable by these methods.
Figure 4.
Immunostaining of NRK cells with SRP54 protein-specific antibody. (A) Cells incubated with a human IgG specific for SRP54 (see Materials and Methods) and then a fluorescent goat anti-human IgG second antibody. (B) Cells incubated with IgG from a human patient serum that lacks anti-SRP activity and then fluorescent second antibody as in A. (C and D) Phase-contrast images of the cells in A and B, respectively.
The localization of SRP68 and SRP72 GFP fusion proteins was explored next. Fig. 5 shows that both proteins localized to the nucleolus as well as the cytoplasm. Approximately 80% of cells transfected with SRP72 showed nucleolar signal (Fig. 5A), and the cytoplasmic SRP72 was distributed similarly to cytoplasmic SRP19. Cells transfected with SRP68 also showed nucleolar signal, although the percentage was lower (40%, Fig. 5B). In addition, a distinct perinuclear accumulation of SRP68 was present in ≈50% of the transfected cells (which often also contained nucleolar signal). The remaining cells showed a more diffuse distribution of SRP68, similar to the other three SRP proteins tested (not shown). To test the nature of this accumulation, the ER (and also other organelles) was stained by using rhodamine B hexyl ester (41) in cells expressing SRP68-EGFP (Fig. 6). Under the staining conditions used (see Materials and Methods), the dye first brightly stained mitochondria and then, within 2–3 min, also stained the ER and other vesicular bodies (see ref. 41). The bright perinuclear SRP68 signal overlapped with perinuclear structures stained with the dye after the 2- to 3-min incubation (Fig. 6D), but did not overlap with stained mitochondria, indicating that the transfected SRP68 was associated with the ER.
Figure 5.
SRP68 and SRP72 localize to the nucleolus as well as the cytoplasm. (A) NRK cell expressing EGFP-SRP72 protein. (B) NRK cell expressing EGFP-SRP68 protein. (C and D) Phase-contrast images of the cells in A and B, respectively.
Figure 6.
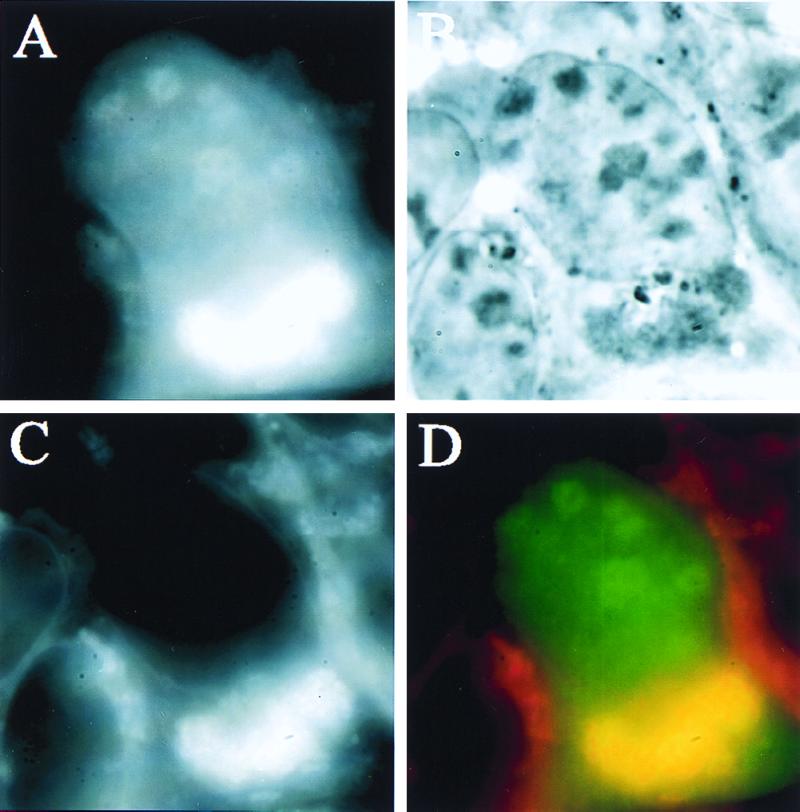
SRP68 perinuclear signal overlaps with ER. (A) Human kidney 293 cell transfected with EGFP-SRP68 before dye was added. (B) Phase image of same cell. (C) Image of same cell showing distribution of rhodamine B hexyl ester ER dye. (D) Merged images of A and B; overlapping regions are yellow.
Discussion
We find that three of the four S domain SRP proteins, SRP19, SRP68, and SRP72, localize to the nucleolus (as well as the cytoplasm) as GFP fusion proteins. In addition, this investigation confirms and considerably extends our initial discovery that nucleus-microinjected SRP RNA localizes in the nucleolus (4) and also confirms recent biochemical results showing the presence of SRP RNA in HeLa cell nucleolar fractions (42, 43). The present in situ hybridization results show that endogenous SRP RNA also visits the nucleolus in NRK cells. Although SRP RNA undergoes a limited amount of 3′ end processing (42), neither the SRP itself nor any of its protein components have a known nucleolar function. It therefore is more likely that the nucleolar localization of these SRP components represents a step in SRP assembly and/or transport to the cytoplasm.
In vitro reconstitution studies characterizing SRP assembly are compatible with the idea that partial SRP assembly may be taking place in the nucleolus. SRP19 protein is known to bind SRP RNA independently of other SRP proteins or factors (9, 30). The SRP68/SRP72 complex from the SRP also binds SRP RNA independently from the other SRP proteins, but the interaction is stabilized by SRP19 and/or SRP54 binding (27). When in vitro expressed proteins are studied, SRP68 binds SRP RNA before SRP72, but its binding is stabilized by SRP72 binding (27, 28). Our results here show that all three of these proteins visit the nucleolus and, therefore, can interact with SRP RNA. One plausible interpretation of our results, then, is that SRP19, SRP68, and SRP72 bind to nucleolar SRP RNA to form a partially assembled SRP.
The contrasting pattern of intracellular localization observed with SRP54 indicates that this component of the final SRP does not have a nucleolar phase that is sufficiently abundant or long-lived to be detected. The lack of nucleolar SRP54 was observed both in the GFP-SRP54 expression studies as well as by immunocytochemistry with an SRP54-specific antibody, showing that neither the expressed GFP-SRP54 nor the endogenous SRP54 protein traffic through the nucleolus to a detectable extent. The nucleoplasmic level of SRP54 also was very low in both the GFP-SRP54 experiments and in the endogenous SRP54 immunocytochemistry, suggesting that SRP54 traffics through the nucleus very rapidly or perhaps not at all. Interestingly, SRP54 binding to SRP RNA has been shown to require prior SRP19 binding in vitro. SRP19 probably effects a conformational change in the SRP RNA that promotes SRP54 binding (32). Taken together with our results, then, it seems very possible that only after SRP RNA-SRP19 assembly has occurred in the nucleolus does SRP54 subsequently bind elsewhere in the cell, either in the nucleoplasm or in the cytoplasm. SRP54 is unique among the SRP proteins in other ways: it can bind to signal sequences even without being complexed into the SRP (44–46) and it is the homologue to the only SRP protein known in E. coli, Ffh (45, 47).
Although it seems unlikely that these SRP proteins and the SRP RNA all would colocalize to the nucleolus but not actually interact with one another, we do not have direct evidence for such interaction. It is possible that the observed colocalization may represent unidentified activities of the SRP RNA and the individual SRP proteins. For example, SRP19 binds to bacterial 5S RNA in vitro (30), so there exists the formal possibility that SRP19 is present in the nucleolus because it is bound to eukaryotic 5S RNA.
We do not think the localization patterns we see are artifacts from overexpression or alteration of the SRP proteins because of the presence of GFP. All of our observations were recorded from transiently transfected cells, and the patterns of signal distribution observed were routinely evident in cells expressing the fusion proteins at low levels as well as more intermediate levels. In this regard, it is interesting to note that about half of the cells transferred with SRP68 exhibited signal colocalized with the ER. This colocalization suggests that the SRP68 fusion protein retains its native binding affinities, because the SRP68/SRP72 complex has been shown to interact with the SRP receptor complex located in the ER membrane (19). We did not see prominent signal accumulation in the ER of cells transfected with SRP72, perhaps suggesting that SRP68 has a higher affinity for the SRP receptor. That SRP54 is not found in the nucleolus under our transfection conditions also argues that the conditions used did not cause the nonspecific localization of proteins to the nucleolus.
In vivo studies defining the intracellular localization sites of the remaining two SRP proteins, SRP9 and SRP14, may prove more difficult because the Alu domain to which they bind is a highly abundant RNA sequence in human cell lines. However, it is known that the SRP RNA Alu domain is necessary for nuclear export of the SRP and that a deletion that abolishes SRP9/SRP14 binding also abolishes export (42, 48). It therefore has been proposed that SRP9/SRP14 binds to SRP RNA in the nucleus early in the assembly process (28), perhaps before final SRP RNA 3′ end processing (42).
The intranuclear location of the SRP RNA gene(s) is not known; it may lie close to the nucleolus or at a distant chromosomal site. If the transcription site is remote from the nucleolus, it is possible that SRP RNA transcripts and the SRP proteins do not encounter one another in the nucleoplasm, perhaps because SRP RNA transcripts move rapidly from their transcription site to the nucleolus and/or because the SRP proteins imported from the cytoplasm immediately traffic to the nucleolus, thus leaving the steady-state nucleoplasmic concentrations of SRP RNA and SRP proteins well below their equilibrium association-binding constants. Alternatively, it is possible that newly synthesized SRP RNA assembles with certain SRP proteins at or near the SRP RNA transcription site and the resulting complex moves rapidly to the nucleolus. In either case, it is the nucleolar localization altogether that is the most intriguing issue. It is tempting to speculate that the nucleolar localization of SRP RNA and SRP proteins we have discovered is related to the nuclear export pathway of nascent SRP. For example, partially assembled SRPs may piggyback with ribosomal subunits for export from the nucleus. In this connection, it is worth noting that the SRP has a demonstrable affinity for ribosomes independent of its interaction with the nascent polypeptide signal sequence (9, 45, 49).
Acknowledgments
We thank Frederick W. Miller (U.S. Food and Drug Administration, Bethesda, MD) for providing the autoimmune patient sera. We thank Marty R. Jacobson for helpful participation in early immunocytochemistry experiments and Christina Alavian for dedicated technical assistance. This work is supported by National Institutes of Health National Research Service Award postdoctoral fellowship AR-08361 (J.C.P.) and National Institutes of Health Grant GM-21595-23 (T.P.). Cloning of the original SRP protein cDNAs by K.G. and C.Z. was supported by National Institutes of Health Grant GM-49034.
Abbreviations
- SRP
signal recognition particle
- GFP
green fluorescent protein
- EGFP
enhanced GFP
- ER
endoplasmic reticulum
Footnotes
References
- 1.Chambliss G, Craven G R, Davies J, Davis K, Kahan L, Nomura M. Ribosomes: Structure, Function and Genetics. Baltimore: Univ. Park Press; 1980. [Google Scholar]
- 2.Jacobson M R, Cao L G, Taneja K, Singer R H, Wang Y L, Pederson T. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson M R, Cao L G, Wang Y L, Pederson T. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobson M R, Pederson T. Proc Natl Acad Sci USA. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarrous N, Wolenski J S, Wesolowski D, Lee C, Altman S. J Cell Biol. 1999;146:559–571. doi: 10.1083/jcb.146.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pederson T. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter P, Blobel G. Proc Natl Acad Sci USA. 1980;77:7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter P, Blobel G. Nature (London) 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 9.Walter P, Johnson A E. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 10.Walter P, Blobel G. Cell. 1983;34:525–533. doi: 10.1016/0092-8674(83)90385-9. [DOI] [PubMed] [Google Scholar]
- 11.Reddy R, Li W-Y, Henning D, Choi Y C, Nohga K, Busch H. J Biol Chem. 1981;256:8452–8457. [PubMed] [Google Scholar]
- 12.Li W-Y, Reddy R, Henning D, Epstein P, Busch H. J Biol Chem. 1982;257:5136–5142. [PubMed] [Google Scholar]
- 13.Ullu E, Murphy S, Melli M. Cell. 1982;29:195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- 14.Gundelfinger E D, Krause E, Melli M, Dobberstein B. Nucleic Acids Res. 1983;11:7363–7364. doi: 10.1093/nar/11.21.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegel V, Walter P. Nature (London) 1986;320:81–84. doi: 10.1038/320081a0. [DOI] [PubMed] [Google Scholar]
- 16.Krieg U C, Walter P, Johnson A E. Proc Natl Acad Sci USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurzchalia T V, Wiedmann M, Girshovich A S, Bochkareva E S, Bielka H, Rapoport T A. Nature (London) 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- 18.Wiedmann M, Kurzchalia T V, Bielka H, Rapoport T A. J Cell Biol. 1987;104:201–208. doi: 10.1083/jcb.104.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel V, Walter P. Cell. 1988;52:39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- 20.Siegel V, Walter P. Proc Natl Acad Sci USA. 1988;85:1801–1805. doi: 10.1073/pnas.85.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwieb C. Nucleic Acids Res. 1985;13:6105–6124. doi: 10.1093/nar/13.17.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwieb C, Ullu E. Nucleic Acids Res. 1986;14:4639–4657. doi: 10.1093/nar/14.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Römisch K, Webb J, Lingelbach K, Gausepohl H, Dobberstein B. J Cell Biol. 1990;111:1793–1802. doi: 10.1083/jcb.111.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strub K, Walter P. Mol Cell Biol. 1990;10:777–784. doi: 10.1128/mcb.10.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zwieb C. Nucleic Acids Res. 1991;19:2955–2960. doi: 10.1093/nar/19.11.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strub K, Moss J, Walter P. Mol Cell Biol. 1991;11:3949–3959. doi: 10.1128/mcb.11.8.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lütcke H, Prehn S, Ashford A J, Remus M, Frank R, Dobberstein B. J Cell Biol. 1993;121:977–985. doi: 10.1083/jcb.121.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janiak F, Walter P, Johnson A E. Biochemistry. 1992;31:5830–5840. doi: 10.1021/bi00140a019. [DOI] [PubMed] [Google Scholar]
- 29.Hsu K, Chang D-Y, Maraia R J. J Biol Chem. 1995;270:10179–10186. doi: 10.1074/jbc.270.17.10179. [DOI] [PubMed] [Google Scholar]
- 30.Walker K P I, Black S D, Zwieb C. Biochemistry. 1995;34:11989–11997. doi: 10.1021/bi00037a041. [DOI] [PubMed] [Google Scholar]
- 31.Gowda K, Chittenden K, Zwieb C. Nucleic Acids Res. 1997;25:388–394. doi: 10.1093/nar/25.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gowda K, Zwieb C. Nucleic Acids Res. 1997;25:2835–2840. doi: 10.1093/nar/25.14.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang D-Y, Newitt J A, Hsu K, Bernstein H, Maraia R J. Nucleic Acids Res. 1997;25:1117–1122. doi: 10.1093/nar/25.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Politz J C, Singer R H. Methods. 1999;18:281–285. doi: 10.1006/meth.1999.0785. [DOI] [PubMed] [Google Scholar]
- 35.Taneja K L, Lifshitz L M, Fay F S, Singer R H. J Cell Biol. 1992;119:1245–1260. doi: 10.1083/jcb.119.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Targoff I N, Johnson A E, Miller F W. Arthritis Rheum. 1990;33:1361–1370. doi: 10.1002/art.1780330908. [DOI] [PubMed] [Google Scholar]
- 37.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. p. 311. [Google Scholar]
- 38.Chittenden K, Black S D, Zwieb C. J Biol Chem. 1994;269:20497–20502. [PubMed] [Google Scholar]
- 39.Gowda K, Black S D, Moeller I, Sakakibara Y, Liu M-C, Zwieb C. Gene. 1998;207:197–207. doi: 10.1016/s0378-1119(97)00627-6. [DOI] [PubMed] [Google Scholar]
- 40.Jacobson M R, Pederson T. In: Analysis of mRNA Formation and Function. Richter J D, editor. New York: Academic; 1997. pp. 341–359. [Google Scholar]
- 41.Terasaki M, Reese T S. J Cell Sci. 1992;3:315–322. doi: 10.1242/jcs.101.2.315. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Sinha K, Perumal K, Gu J, Reddy R. J Biol Chem. 1998;273:35023–35031. doi: 10.1074/jbc.273.52.35023. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell J R, Cheng J, Collins K. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hauser S, Bacher G, Dobberstein B, Lutcke H. EMBO J. 1995;14:5485–5493. doi: 10.1002/j.1460-2075.1995.tb00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bui N, Strub K. Biol Chem. 1999;380:135–145. doi: 10.1515/BC.1999.021. [DOI] [PubMed] [Google Scholar]
- 46.Clemons W J, Gowda K, Black S D, Zwieb C, Ramakrishnan V. J Mol Biol. 1999;292:697–705. doi: 10.1006/jmbi.1999.3090. [DOI] [PubMed] [Google Scholar]
- 47.Keenan R J, Freymann D M, Walter P, Stroud R M. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 48.He X-P, Bataille N, Fried H M. J Cell Sci. 1994;107:903–912. doi: 10.1242/jcs.107.4.903. [DOI] [PubMed] [Google Scholar]
- 49.Ogg S C, Walter P. Cell. 1995;81:1075–1084. doi: 10.1016/s0092-8674(05)80012-1. [DOI] [PubMed] [Google Scholar]