Abstract
In an earlier study, our group vaccinated rhesus macaques with vesicular stomatitis virus (VSV) vectors expressing Gag, Pol, and Env proteins from a hybrid simian/human immunodeficiency virus (SHIV). This was followed by a single boost with modified vaccinia virus Ankara (MVA) vectors expressing the same proteins. Following challenge with SHIV89.6P, vaccinated animals cleared challenge virus RNA from the blood by day 150 and maintained normal CD4 T cell counts for 8 months. Here we report on the long-term (>5 year post-challenge) status of these animals and the immunological correlates of long-term protection. Using real-time PCR, we found that viral DNA in peripheral blood mononuclear cells (PBMCs) of the vaccinees declined continuously and fell to below detection (< 5 copies/105 cells) by approximately 3 years post-challenge. SHIV DNA was also below the limit of detection in the lymph nodes of two of the four animals at 5 years post-challenge. We detected long-term persistence of multi-functional Gag-specific CD8+ T cells in both PBMCs and lymph nodes of the two protected animals with the Mamu A*01+ MHC I allele. All animals also maintained SHIV89.6P neutralizing antibody titers for 5 years. Our results show that this vaccine approach generates solid, long-term control of SHIV infection, and suggest that it is mediated by both cytotoxic T lymphocytes and neutralizing antibody.
1. Introduction
UNAIDS estimates that there are a total of 33.2 million people currently infected with HIV worldwide, of which 2.5 million were newly acquired infections in 2007 (www.unaids.org). Antiretroviral drug treatments have extended the lives of many people infected in the developed world, but are not as readily available for those infected in the developing world. The creation of an effective HIV vaccine is therefore extremely important. However, despite twenty-five years of intense research, an effective HIV vaccine is still not available. The recent failure of the phase IIb STEP trial, which tested recombinant adenovirus type 5 HIV vaccine vectors in a large number of human volunteers, illustrates the importance for further basic research in non-human primate models of AIDS [1].
Numerous vaccine approaches have been tested with varying degrees of success in simian immunodeficiency virus (SIV) and hybrid SIV/HIV (SHIV) non-human primate models of AIDS [2]. Several of these vaccine strategies have shown at least short-term control of challenge virus replication and protection from AIDS in the vaccine recipients, especially in the SHIV89.P model [3, 4]. Although the SHIV89.6P model has been considered a relatively easy model in which to obtain vaccine protection from AIDS [5], there has been very little long-term protection reported even in this model. Because the vaccinated animals become infected with SHIV89.6P after challenge, there can be continuous viral replication and eventual viral mutation to escape whatever immune responses initially control infection. One very well characterized virus escape mutation in a CTL epitope has been reported for a vaccinated macaque that initially controlled the SHIV89.6P challenge [6]. Also, depletion of CD8+ T cells in vaccinated animals controlling infection by SHIV89.6P for 2 years [7], or SHIV(KU-1) for 5 years [8] led to re-emergence of high viral loads. The possibility of SHIV escape from immune control in vaccinees compelled us to monitor the immunological and virological status of animals from a previous study where long-term protection from AIDS has now been achieved [9].
Our initial focus was on the development of recombinant vesicular stomatitis virus (VSV)-based vaccine vectors [9–11] for use in the SHIV89.6P rhesus macaque challenge model [4]. Our laboratory reported effective control of SHIV 89.6P replication in rhesus macaques that were first vaccinated with VSV vectors expressing SHIV Env and Gag proteins and boosted twice with VSV serotype exchange vectors expressing Env and Gag [10]. These animals remained AIDS free for over 3 years. However, two out of seven vaccinated animals developed high viral loads and AIDS between 3 and 4 years post-challenge (unpublished results), which is indicative of incomplete vaccine protection in those animals. When these animals developed AIDS the study was terminated.
In a subsequent study we reported that a single vaccination with recombinant VSV vectors expressing SHIV89.6P Env, Gag, and Pol, followed by a single boost with modified vaccinia virus Ankara (MVA) vectors expressing the same antigens, resulted in control of virus loads superior to those obtained in the earlier study [10], and superior to a single VSV prime-VSV vector boost [9]. In the VSV/MVA study, the vaccinated animals controlled SHIV challenge virus to levels below detection by day 150 post-challenge. They also maintained normal levels of CD4+ T cells for at least 8 months following challenge. We report here on the long-term protection from AIDS, gradual clearance of viral DNA, and immunological status of the VSV/MVA vaccinated animals more than 5 years after the initial SHIV 89.6P challenge.
2. Materials and Methods
2.1 Animals, tissue preparation, and viral RNA measurement
Animals were housed and cared for at the Tulane National Primate Research Center (TNPRC). All blood and tissue samples were isolated at the TNPRC, shipped in dry ice and stored in liquid nitrogen at Yale University. Plasma viral RNA levels were measured by branched DNA (bDNA) signal amplification by the Chiron Corporation (Emeryville, CA). The lower limit of detection for the bDNA assay was 125 copies/mL plasma. The two Mamu A*01 animals, V864* and AL86*, are denoted by the asterisks in all figures.
2.2 Real-time PCR
Genomic DNA (gDNA) was isolated from PBMCs or lymph node biopsied tissue using the Qiagen DNeasy blood and tissue kit (Valencia, CA). SIV gag copies in total cellular DNA were quantified using the TaqMan real-time PCR system. To eliminate background in the reactions, all reaction cocktails were prepared in a clean room away from the main laboratory where SIV plasmid DNAs were used and prepared. Cocktails contained the TaqMan universal PCR master mix (Applied Biosystems/Roche; Branchburg, NJ) and forward and reverse primers to a final concentration of 1 μM each. The FAM-TAMRA labeled gag-specific probe was added to a final concentration of 0.5 μM. Sequences for the primer/probe set were as follows: Forward, 5′-AGGCAGATTGGGACTTG-3′; Reverse, 5′-TGATCCTGACGGCTCCCTAA-3′; Probe, FAM-5′CCCACAACCAGCTCCACAACAAGGAA3′-TAMRA. Each reaction contained 3.5×104 cell equivalents of gDNA. Serial dilutions of a SIV gag containing plasmid served as a standard curve for all reactions, with a lower limit of detection of 5 copies per reaction. Further care was taken to prevent sample cross contamination by adding all genomic and plasmid templates in a chemical fume hood. Samples were run in an Applied Biosystem 7500 Real Time PCR machine and the data points were collected and analyzed using the 7500 system sequence detection software version 1.2.3. Each sample was run in triplicate a total of three times.
2.3 Cell culture stimulation and infection
Cryo-preserved PBMCs or cell suspensions from lymph node biopsies were thawed and pelleted by centrifugation. For functional assays, cells were washed and plated in RPMI containing 5% fetal calf serum at a concentration of 1×106 cells/mL in a 12 well dish. Expression of full length antigen using recombinant vaccinia virus (rVV) has been described previously[12, 13]. Briefly, cells were infected with rVV expressing either full-length SIV gag protein (rVV-Gag) or bacteriophage T7 RNA polymerase (rVV-T7), as a negative control, at an MOI of 2. In parallel, cells were either treated with PMA (50 ng/mL) and ionomycin (250 ng/mL) or mock infected to serve as positive and negative controls respectively. Cultures were stimulated for 16 hours (h) in a humidified incubator before being used in any assays.
2.4 Viral neutralization assays and titration of total envelope antibodies by ELISA
Neutralization was measured as a function of reductions in luciferase reporter gene expression after a single round of infection in TZM-bl cells as described [14]. TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program, as contributed by John Kappes and Xiaoyun Wu. Briefly, 200 TCID50 of virus was incubated with 3-fold serial dilutions of test sample in duplicate in a total volume of 150 μl for 1 h at 37°C in 96-well flat-bottom culture plates. Freshly trypsinized cells (1×105 cells in 100 μl of growth medium containing 75 μg/ml DEAE dextran) were added to each well. One set of control wells received cells and virus (virus control), and another set received cells only (background control). After 48 h incubation, 100 μl of cells were transferred to a 96-well black solid plate (Corning Costar, Lowell, MA) for measurements of luminescence using the Britelite Luminescence Reporter Gene Assay System (PerkinElmer Life Sciences, Waltham, MA). Neutralization titers are the dilution at which relative luminescence units (RLU) were reduced by 50% compared to virus control wells after subtraction of background RLUs. Assay stocks of molecularly cloned Env-pseudotyped viruses were prepared by transfection in 293T cells and were titrated in TZM-bl cells as described [14]. Heterologous clade B reference Env clones were described previously [14]. Total SHIV 89.6P envelope-specific antibody titer was measured by ELISA as described previously [9].
2.5 Flow cytometry
CD4 T cell counts were determined at the TNPRC as reported previously [9]. For cell surface marker and tetramer analysis, cryo-preserved cells were washed and stained immediately. For intracellular cytokine staining (ICS), cells were infected with rVVs as outlined above and then treated with GolgiStop (Becton Dickinson (BD), San Jose, CA) for 6 h to allow for the accumulation of cytokines. The cells were then washed in staining buffer (1x PBS containing 3% fetal calf serum and 0.1% sodium azide) and stained for surface marker expression. The directly conjugated monoclonal antibodies (BD, San Jose, CA), CD3-Pacific Blue, CD4-PerCP, CD8-APC-Cy7, and the Mamu-A*01 specific APC conjugated tetramer containing the SIV gag CM9 peptide (CTPYDINQM) (Beckman Coulter, Fullerton, CA) were used to discern T cell subsets. Following surface staining, cells were fixed and permeabilized using the BD Cytofix/Cytoperm solution and stained for intracellular cytokine expression. The directly conjugated anti-cytokine antibodies used were as follows: IFN-γ-FITC, IL2-PE, and TNF-α-Alexa 700 (BD). After washing, cells were resuspended in staining buffer and data acquired using a multicolor LSR II flow cytometer (BD). Acquired data was analyzed using FlowJo software version 6.4 (Treestar, Ashland, OR).
2.6 ELISPOT assay
The ELISPOT protocol has been described previously [9]. Briefly, MultiScreen HTS 96-well plates (Millipore, Bedford, MA) were coated with mouse anti-human IFN-γ antibody at a final concentration of 1μg/well. After overnight infection or stimulation as described above, 2×105 PBMCs were transferred to the antibody coated plates. After 5 h incubation at 37°C, the cells were washed off the plates using PBS-Tween (0.05% v/v) and then treated with an anti-mouse IgG biotinylated antibody (Invitrogen, Carlsbad, CA) for 1 h. Upon washing, plates were then treated with streptavidin-HRP (Southern BioTech, Birmingham, AL) and then exposed to one-step chromogenic substrate (Pierce, Rockford, IL). Plates were washed, allowed to air dry and finally counted for spot forming cells (SFC). Total gag-specific SFCs were determined by subtracting the total number of spots observed in PBMC cultures exposed to rVV-T7 (assayed in parallel) from those spots observed in PBMC cultures exposed to rVV-Gag. Data was analyzed and plotted using Prism software, version 4.0b (GraphPad Software, San Diego, CA).
3. Results
3.1 Kinetics of SHIV viral RNA and DNA loss in PBMCs from vaccinated macaques
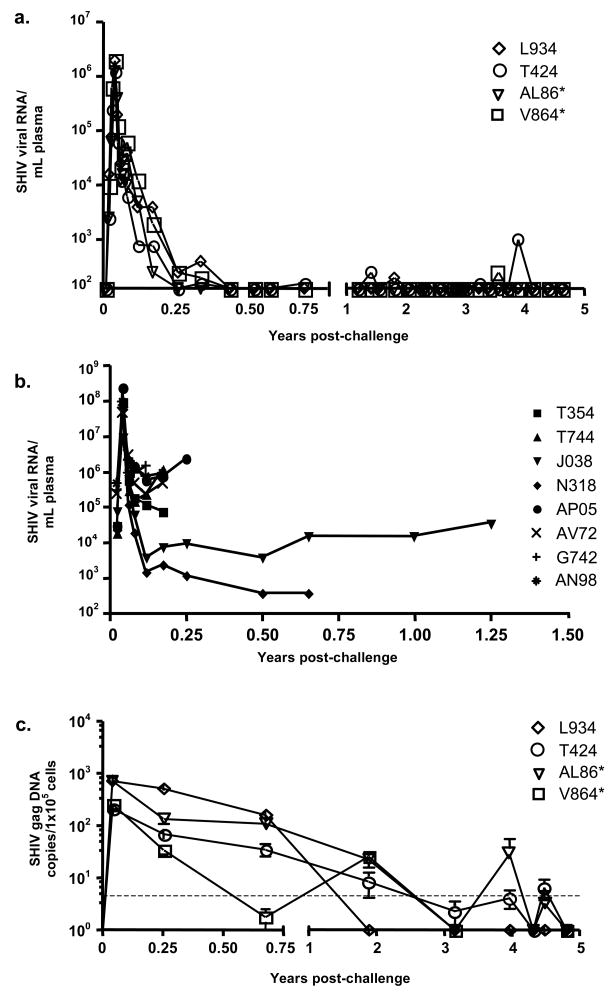
To measure SHIV viral load, we followed plasma viral RNA levels after SHIV 89.6P challenge for almost 5 years. Plasma viral RNA peaked approximately two weeks after SHIV 89.6P challenge and declined to undetectable levels by day 150 post-challenge (Fig. 1A and [9]). While plasma viral RNA remained undetectable for over 4 years in monkey AL86, occasional single RNA spikes near the detection threshold were seen at 1–3 time points in the other three animals (Fig. 1A). The viral load data are also shown for 7 control animals given the identical SHIV 89.6P challenge (Fig. 1B). These animals had much higher viral loads than the vacinees and developed AIDS with an average time of about six months.
Fig. 1. SHIV 89.6P RNA and DNA levels in the peripheral blood of vaccinated and control macaques after challenge.
(A) Serum SHIV 89.6P RNA levels were measured by the branched DNA (bDNA) assay for up to 4 years after viral challenge. The lower limit of detection for the assay was 125 copies/mL plasma. (B) Serum SHIV 89.6P RNA levels as measured by the bDNA assay for control animals [10], followed here until they developed AIDS and were euthanized. (C) SHIV 89.6P DNA levels were assayed in the PBMCs by quantitative real-time PCR using a probe specific for the viral gag gene. For real time PCR, each sample was run in triplicate a total of three times (n=9). The lower limit of detection for the real-time analysis was 5 copies per reaction and is represented by the dashed horizontal line. The Mamu A*01 positive animals are denoted by an asterisk following the designation.
We next examined the kinetics of viral DNA loss from genomic DNA in PBMCs of animals that showed long-term protection from AIDS. Gag-specific viral DNA sequences were quantified using real-time PCR. In all vaccinees, the highest levels of viral DNA were seen in the PBMCs at two weeks post-challenge (Fig. 1C), and declined thereafter. The decline of SHIV viral DNA in PBMCs was much slower than the viral RNA decline. In animals V864 and L934, viral DNA levels fell below the limit of detection 2–3 years after challenge and remained undetectable thereafter. In the remaining two animals, AL86 and T424, levels varied between undetectable and 5–50 copies/105 cells after 3 years, but were undetectable in both at 5 years.
3.2 SHIV DNA in lymph nodes and CD4 T cell counts
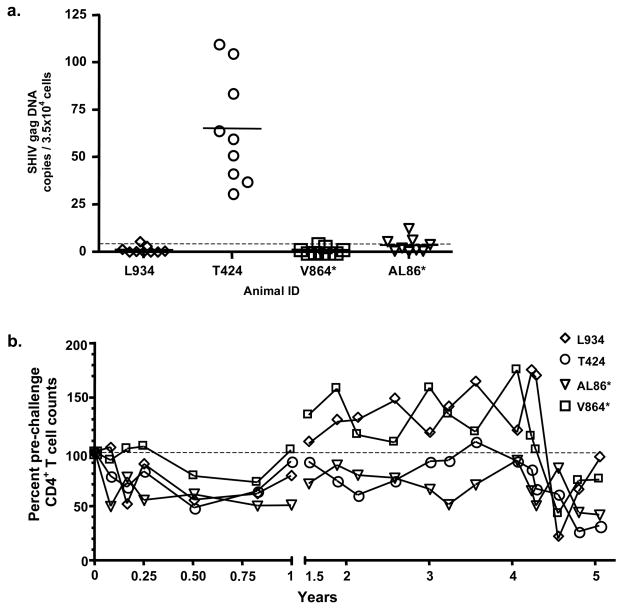
Because viral replication in known to occur within the lymph nodes in animals controlling SHIV infection [15], we next measured viral DNA levels in lymph nodes. We obtained lymph node biopsies from all four animals to determine if more viral DNA was sequestered in the lymph nodes than in the PBMCs at 5 years post-challenge. Two animals, L934 and V864, had an average SHIV gag DNA copy number below the limit of detection in their lymph nodes, while AL86 had an average copy number of SHIV gag DNA near the limit of detection (Fig. 2A). Consistently higher levels of SHIV gag DNA were detected in lymph node tissue of animal T424 as compared to the other three animals (Fig. 2A).
Fig. 2. Viral DNA levels in the lymph nodes and peripheral blood CD4+ T cell counts.
(A) Viral DNA was measured by real time PCR as described above. Nine determinations were made for each sample, and each value is represented as a data point. The arithmetic mean for the samples is represented by the thick, short bars. The lower limit of detection for the real time analysis was 5 copies per reaction and is represented by the dashed horizontal line. (B) CD4+ T cell counts were measured for each animal by FACS and represented as a percentage of the pre-challenge levels. The dashed line is set at 100% for reference.
After an initial decline in CD4+ T cell counts post-SHIV 89.6P challenge, all animals maintained counts near or above pre-challenge levels for 4 years (Fig. 2B). A decline in CD4+ T cell number was seen around 4.5 years for all animals; however, two animals (V864 and L934) rebounded to near their pre-challenge levels after 5 years (75.1 and 93.6 %, respectively; Fig. 2B). Interestingly, T424 and AL86, the two animals with detectable viral DNA in their lymph nodes, also had the largest reduction in CD4+T cell counts 5 years post-challenge (32.3% and 42% of pre-challenge values respectively).
3.3 Long-term persistence of SHIV 89.6P neutralizing antibody in all vaccinated animals
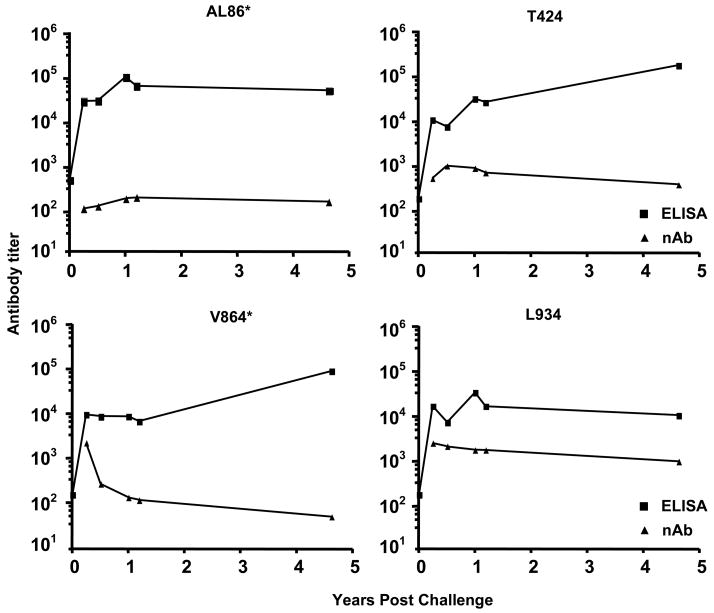
Because SHIV 89.6P neutralizing antibody (nAb) has the potential to control replication, we examined the levels of neutralizing and total anti-SHIV envelope antibodies in the serum of long-term protected macaques (Fig. 3). Neutralizing antibody titers persisted in all animals for 5 years post-challenge, but the levels were highly variable among animals. Serum samples from all animals, 5 years post-challenge, failed to neutralize a variety of heterologous tier 2 primary isolates of clade B HIV-1 (6535.3, QH0692.42, SC422661.8, PVO.4, TRO.11, AC10.0.29, RHPA4259.7 and THRO4156.18, data not shown). We also noted a lack of correlation between total anti-envelope antibody titers and nAb titers in two of the four animals, consistent with earlier results [9].
Fig. 3. Total envelope specific and neutralizing antibody titers.
Total serum antibody titers to SHIV89.6P envelope were measured by ELISA (squares). Serum neutralizing antibody titers (triangles) were determined for each animal by SHIV 89.6P neutralization assays. Pre-challenge neutralizing antibody titers undetectable for all animals and values are therefore not shown here. The animal identification number is indicated above each graph.
3.4 Long-term persistence of gag-specific CD8+ T cells in blood and lymph nodes
The importance of cytotoxic CD8+ T cells in control of SIV and SHIV infection is well established [16, 17]. Cytotoxic CD8+ T cells recognize specific viral peptides bound to MHC I molecules on the surface of infected cells. An immunodominant gag peptide (CM9) binds the Mamu A*01 MHC class I molecule [18]. Because two of the long-term protected animals in this study (AL86 and V864) were Mamu A*01 positive, we were able to monitor the number of CM9 gag-specific CD8+ T cells in these animals using MHC class I tetramer analysis.
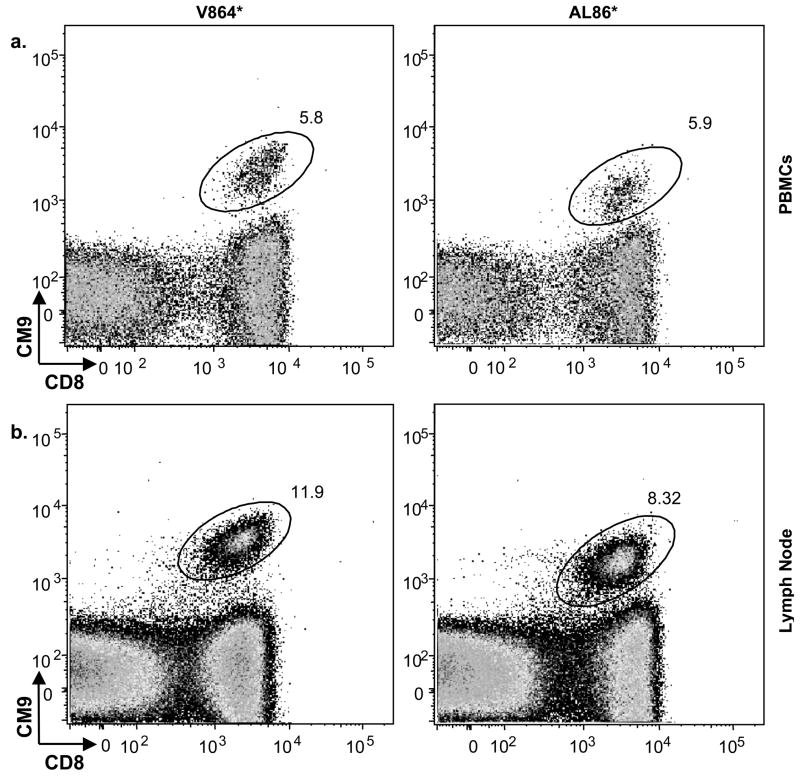
We initially examined T cells from PBMCs taken from animals 5 years post-challenge. The number of CD8+ gag-specific T cells was substantial, 5–6% of the total CD8+ cells (Fig. 4A). We also examined the frequency of tetramer positive cells in lymph node biopsies. There was approximately double the percentage of tetramer positive CD8+ T cells in the lymph node samples as in the peripheral blood (Fig. 4B). Background tetramer staining levels were less than 0.03% of the total CD8+ T cells in the two non-Mamu A*01 animals (data not shown).
Fig. 4. High levels of MHC I tetramer positive cells in PBMCs and lymph nodes.
PBMCs (A) and lymph node biopsies (B) from Mamu A*01 positive animals (AL86 and V864) at 5 years post-challenge were stained with the gag-specific tetramer and analyzed by FACS. Scatter plots represent gated CD3+CD8+ lymphocyte populations and plot CD8 expression versus tetramer binding. Values next to elliptical gates represent the percentage of the total CD8+ cells that are tetramer positive.
3.5 Functional analysis of T cells in the peripheral blood
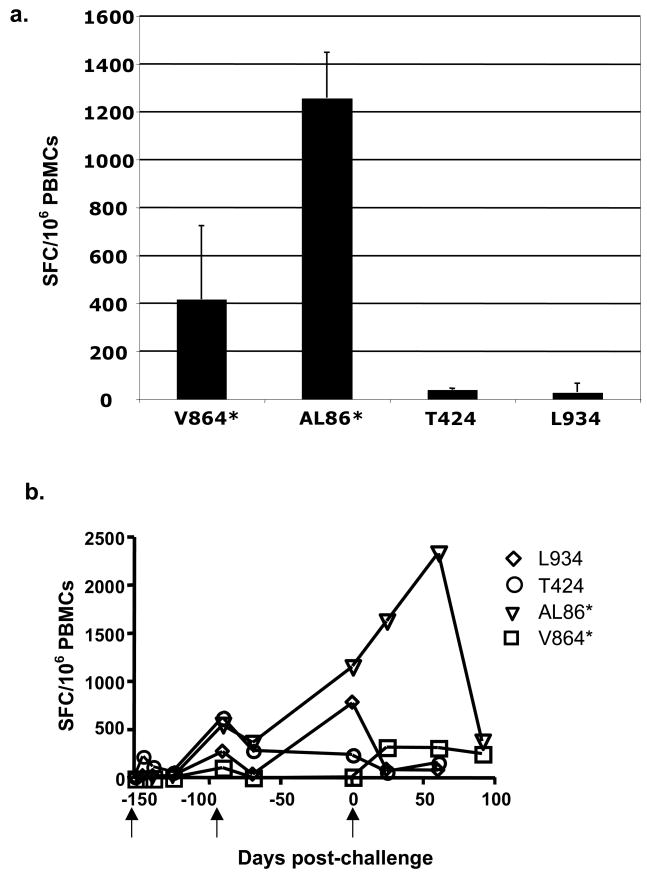
To determine if there were functional gag-specific T cells in the vaccinees, we performed an ELISPOT assay for IFN-γ on bulk PBMCs in vitro, after infection with recombinant vaccinia expressing full-length Gag protein or T7 polymerase, as a negative control. The results (Fig. 5A) showed strong functional responses from the two Mamu A*01 animals (V864 and AL86) but only low responses in the other two animals (T424 and L934). Very low backgrounds from control cells infected with vaccinia-T7 run in parallel were subtracted from those generated by the vaccinia-Gag infected cultures to give the values shown (Fig. 5A). The previously reported ELISPOT data [9] from these animals pre-challenge and post challenge are included for completeness (Fig. 5B) The first two arrows indicate the days of prime and boost, and the third arrow indicates the day of challenge.
Fig. 5. ELISPOT analysis reveals functional T cells.
(A) 5 years post-challenge, PBMCs from each animal were used to quantify the ability of the animals to mount a response to gag antigen expressed through rVV-gag infection for 16 h. An IFN-γ specific ELISPOT assay was used to quantify numbers of responding cells and is given as represented as spot forming cells (SFC)/1×106 cells (n=3). Values shown have background SFCs generated by parallel control rVV-T7 cultures subtracted. (B) Previously published [9] pre- and early post-challenge IFN-γ ELISPOT levels are represented here for reference. Animals were primed on day -153, boosted on day -97, and challenged on day 0 (arrows).
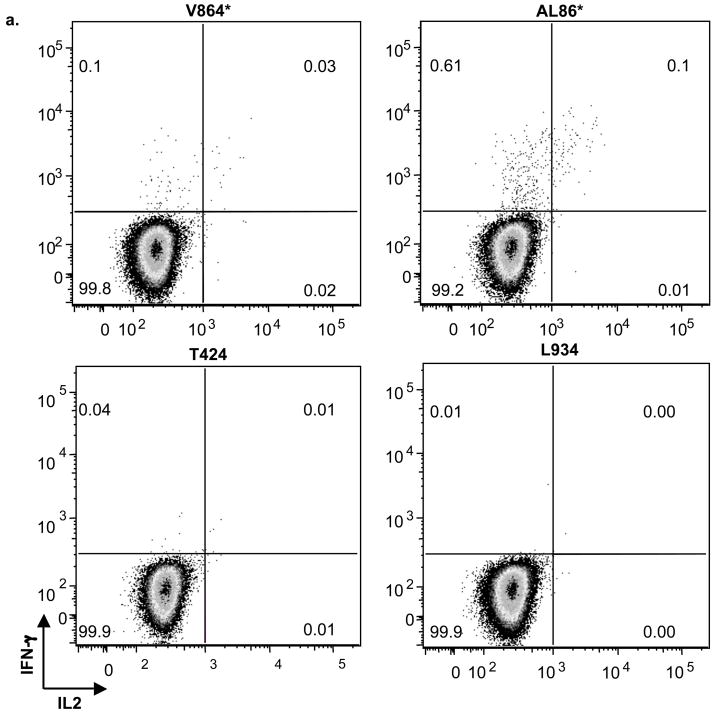
Next we examined the CD8+ T cell populations by intracellular cytokine staining (ICS) to determine if the Mamu A*01 animals made polyfunctional cytokine responses. When CD3+CD8+ cells were examined for their expression of the cytokines IFN-γ and IL-2, the Mamu A*01 positive animals (V864 and AL86) had cells expressing IFN-γ alone, and cells that expressed both IFN-γ and IL-2 (Fig. 6B). The non-Mamu A*01 (T424 and L934) animals had only background levels of cytokine staining (data not shown), consistent with the ELISPOT data (Fig. 5).
Fig. 6. Polyfunctional T cells were present in MamuA*01 animals.
(A) Examples of dot plots showing intracellular cytokine staining for IFN-γ and IL-2 in CD3+CD8+ T cells in response to gag antigen expressed through rVV-gag exposure for 16 h. Animal numbers are indicated above each plot.
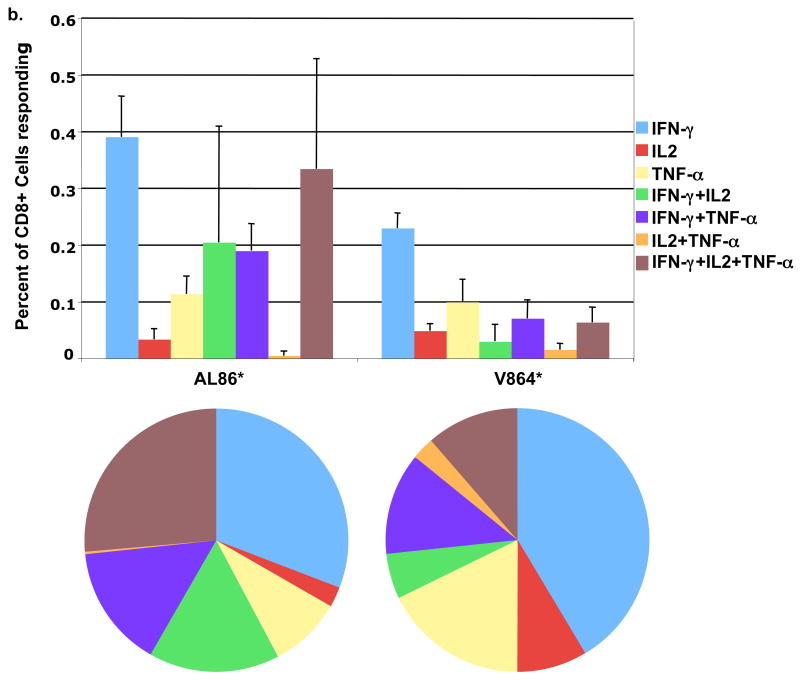
(B) Quantitation by ICS of rVV-gag stimulated (16 h), polyfunctional expression of IFN-γ, IL-2, and TNF-α in CD3+CD8+ T cells from Mamu A*01 animals (AL86 and V864), the standard deviation is shown for a total of three experiments (n=3). Pie charts represent the average values of three experiments for each animal.
Further analysis of the CD8+ T cells from the Mamu A*01 positive animals showed a distinct subset of polyfunctional cells with respect to cytokine production in response to Gag protein (Fig. 6). While both animals had detectable dual and polyfunctional T cell responses, animal AL86 had the highest number of CD8+ T cells that co-expressed the three cytokines tested (TNF-α, IFN-γ, and IL-2). These results are consistent with the ELISPOT assay, where animal AL86 had higher numbers of IFN-γ secreting cells than animal V864. Greater than 50% or 25% of the responding CD8+ T cells, from animals AL86 or V864 respectively, were either dual or polyfunctional.
4. Discussion
We have previously shown that rhesus macaques vaccinated with recombinant VSVs expressing the gag, pol, and env genes of SHIV 89.6P and singly boosted with either VSV serotype exchange vectors or MVA expressing the same genes were able to control a SHIV 89.6P challenge infection for at least 8 months [9]. We have now extended those findings and show that all animals in the VSV/MVA vaccine group have remained healthy, have normal CD4+ T cell counts, and have controlled their infection with SHIV 89.6P for more than 5 years. Two of the four animals in a separate VSV vector prime/VSV vector boost group [9] failed to control viral loads and eventually developed AIDS by 2 years post-challenge. Study of this group was then terminated because of complications unrelated to AIDS.
In the VSV/MVA group, all animals showed peak levels of SHIV89.6P RNA and DNA in their PBMCs by two weeks (Fig. 1 and [9]). Viral RNA levels fell below the limit of detection by 5 months (150 days). In contrast, the levels of viral DNA in PBMCs declined much more slowly (Fig. 1B) presumably because of retention of DNA in long-lived memory T cells. Infected, resting, memory T cells, which do not produce viral antigens, are known to be a major long-term reservoir of integrated viral DNA and these cells have a very long half-life [19, 20].
SHIV 89.6P nAbs were maintained in the serum of all four macaques out to 5 years post-challenge, and presumably contributed to control of challenge virus. There was no correlation, however, between the level of nAbs and the level of total Ab measured by ELISA. The lack of breadth in cross-neutralizing titers found after 5 years suggests low antigen levels and little viral evolution over time in all four animals.
The two Mamu A*01 positive animals, V864 and AL86, maintained high levels of gag-specific CD8+ T cells in both blood and lymph nodes for more than 5 years (Fig. 4). Both animals retained very high levels of tetramer positive gag-specific CD8+ T cells. These levels were between 5 and 6% of the total CD8+ T cells in the PBMCs (Fig. 4A) and between 8 and 11% of the total CD8+ T cells in the lymph node (Fig. 4B). In another long-term follow up of DNA-MVA vaccinated/SHIV 89.6P challenged animals, the percentage of CD8+ tetramer+ cells varied between 0.3 and 5.4% in PBMCs [21]. The preservation of high levels of gag-specific CD8+ T cells suggests that gag-specific CTLs recognizing the CM9 epitope are important for the continued elimination of SHIV infected cells in the animals. Other studies have also shown maintenance of gag-specific CTLs in macaques during control of SHIV 89.6P infection [22, 23].
While animal AL86 maintained levels of CM9 specific T cells similar to animal V864, tetramer positivity does not always correlate with virus-specific CTL function [6, 24, 25]. We therefore examined the ability of CD8+ T cells in these animals to produce IFN-γ in response to gag antigen without prior expansion of the cells in vitro. These cells were readily detected using an ELISPOT assay. T424 and L934, the MamuA*01 negative animals that do not present the dominant CM9 epitope, had only low level Gag responses. These animals are likely still responding to Env or other SHIV protein epitopes [9], but we did not have sufficient samples to screen for other responses.
Polyfunctional T cells are thought to be the most effective cells controlling SIV and SHIV replication in macaques [26–28], and HIV-1 replication and disease progression in humans [29, 30]. Of the responding CD8+ T cells in the Mamu A*01 animals, a substantial percentage was either dual or polyfunctional with respect to their cytokine production (Fig. 6B). The total number of CD8+ T cells producing cytokines in the ICS assay was only a small percentage of the overall tetramer positive population. These results are similar to those obtained in another long-term vaccine study using SHIV 89.6P challenge [21].
Our results show that vaccination with recombinant VSV followed by a single MVA boost generates solid control of SHIV 89.6P challenge in macaques for at least 5 years. The longest control of SHIV 89.6P challenge reported previously was 3.8 years using two DNA vaccinations, followed by an MVA vector boost [21]. Recently, viral escape from CTL control and progression to AIDS for the two vaccinees remaining from that study was reported at approximately 5 years post-challenge [6].
While the percentage of CD4+ cells in two of our animals (AL86 and T424) was below their individual pre-challenge levels 5 years after SHIV 89.6P challenge (Fig. 2B), both animals remain healthy. However, the detection of SHIV viral DNA in the lymph nodes of these same two animals (AL86 and T424; Fig. 2A) suggests greater potential for viral replication and generation of mutants able to escape immune control. Therefore, additional follow-up of these animals as well as of the two animals with SHIV DNA at the limit of detection is needed.
Our results and those of others show the importance of long-term follow-up of vaccinated macaques after challenge. Vaccines that provide short-term protection (less than 4 years) from challenge, clearly may not be sufficient for the longer-term control of viral replication that will likely be required in an effective AIDS vaccine. We will continue to follow the virological and immunological status of the animals in this study to determine the durability of protection.
Acknowledgments
This work was supported by NIH grants R37-AI40357, R01-AI45510, R01-AI30034, and the TNPRC base grant and by the Intramural Research Program of the NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat Med. 2008 Jun;14(6):617–21. doi: 10.1038/nm.f.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letvin NL. Progress toward an HIV vaccine. Annu Rev Med. 2005;56:213–23. doi: 10.1146/annurev.med.54.101601.152349. [DOI] [PubMed] [Google Scholar]
- 3.Robinson HL. New hope for an AIDS vaccine. Nat Rev Immunol. 2002 Apr;2(4):239–50. doi: 10.1038/nri776. [DOI] [PubMed] [Google Scholar]
- 4.Reimann KA, Li JT, Veazey R, Halloran M, Park IW, Karlsson GB, et al. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996 Oct;70(10):6922–8. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feinberg MB, Moore JP. AIDS vaccine models: challenging challenge viruses. Nat Med. 2002 Mar;8(3):207–10. doi: 10.1038/nm0302-207. [DOI] [PubMed] [Google Scholar]
- 6.Sadagopal S, Amara RR, Kannanganat S, Sharma S, Chennareddi L, Robinson HL. Expansion and exhaustion of T-cell responses during mutational escape from long-term viral control in two DNA/modified vaccinia virus Ankara-vaccinated and simian-human immunodeficiency virus SHIV-89.6P-challenged macaques. J Virol. 2008 Apr;82(8):4149–53. doi: 10.1128/JVI.02242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amara RR, Ibegbu C, Villinger F, Montefiori DC, Sharma S, Nigam P, et al. Studies using a viral challenge and CD8 T cell depletions on the roles of cellular and humoral immunity in the control of an SHIV-89.6P challenge in DNA/MVA-vaccinated macaques. Virology. 2005 Dec 20;343(2):246–55. doi: 10.1016/j.virol.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 8.Mackay GA, Liu Z, Singh DK, Smith MS, Mukherjee S, Sheffer D, et al. Protection against late-onset AIDS in macaques prophylactically immunized with a live simian HIV vaccine was dependent on persistence of the vaccine virus. J Immunol. 2004 Sep 15;173(6):4100–7. doi: 10.4049/jimmunol.173.6.4100. [DOI] [PubMed] [Google Scholar]
- 9.Ramsburg E, Rose NF, Marx PA, Mefford M, Nixon DF, Moretto WJ, et al. Highly effective control of an AIDS virus challenge in macaques by using vesicular stomatitis virus and modified vaccinia virus Ankara vaccine vectors in a single-boost protocol. J Virol. 2004 Apr;78(8):3930–40. doi: 10.1128/JVI.78.8.3930-3940.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rose NF, Marx PA, Luckay A, Nixon DF, Moretto WJ, Donahoe SM, et al. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell. 2001 Sep 7;106(5):539–49. doi: 10.1016/s0092-8674(01)00482-2. [DOI] [PubMed] [Google Scholar]
- 11.Rose NF, Roberts A, Buonocore L, Rose JK. Glycoprotein exchange vectors based on vesicular stomatitis virus allow effective boosting and generation of neutralizing antibodies to a primary isolate of human immunodeficiency virus type 1. J Virol. 2000 Dec;74(23):10903–10. doi: 10.1128/jvi.74.23.10903-10910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donahoe SM, Moretto WJ, Samuel RV, Metzner KJ, Marx PA, Hanke T, et al. Direct measurement of CD8+ T cell responses in macaques infected with simian immunodeficiency virus. Virology. 2000 Jul 5;272(2):347–56. doi: 10.1006/viro.2000.0404. [DOI] [PubMed] [Google Scholar]
- 13.Moretto WJ, Drohan LA, Nixon DF. Rapid quantification of SIV-specific CD8 T cell responses with recombinant vaccinia virus ELISPOT or cytokine flow cytometry. Aids. 2000 Nov 10;14(16):2625–7. doi: 10.1097/00002030-200011100-00034. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005 Aug;79(16):10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A, Mukherjee S, Shen J, Buch S, Li Z, Adany I, et al. Immunization of macaques with live simian human immunodeficiency virus (SHIV) vaccines conferred protection against AIDS induced by homologous and heterologous SHIVs and simian immunodeficiency virus. Virology. 2002 Sep 30;301(2):189–205. doi: 10.1006/viro.2002.1544. [DOI] [PubMed] [Google Scholar]
- 16.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999 Feb 5;283(5403):857–60. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 17.Matano T, Shibata R, Siemon C, Connors M, Lane HC, Martin MA. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J Virol. 1998 Jan;72(1):164–9. doi: 10.1128/jvi.72.1.164-169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen TM, Sidney J, del Guercio MF, Glickman RL, Lensmeyer GL, Wiebe DA, et al. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998 Jun 15;160(12):6062–71. [PubMed] [Google Scholar]
- 19.Shen A, Zink MC, Mankowski JL, Chadwick K, Margolick JB, Carruth LM, et al. Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J Virol. 2003 Apr;77(8):4938–49. doi: 10.1128/JVI.77.8.4938-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sedaghat AR, Siliciano RF, Wilke CO. Low-level HIV-1 replication and the dynamics of the resting CD4+ T cell reservoir for HIV-1 in the setting of HAART. BMC Infect Dis. 2008;8:2. doi: 10.1186/1471-2334-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadagopal S, Amara RR, Montefiori DC, Wyatt LS, Staprans SI, Kozyr NL, et al. Signature for long-term vaccine-mediated control of a Simian and human immunodeficiency virus 89.6P challenge: stable low-breadth and low-frequency T-cell response capable of coproducing gamma interferon and interleukin-2. J Virol. 2005 Mar;79(6):3243–53. doi: 10.1128/JVI.79.6.3243-3253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevceva L, Moniuszko M, Alvarez X, Lackner AA, Franchini G. Functional simian immunodeficiency virus Gag-specific CD8+ intraepithelial lymphocytes in the mucosae of SIVmac251- or simian-human immunodeficiency virus KU2-infected macaques. Virology. 2004 Feb 20;319(2):190–200. doi: 10.1016/j.virol.2003.08.046. [DOI] [PubMed] [Google Scholar]
- 23.Bertley FM, Kozlowski PA, Wang SW, Chappelle J, Patel J, Sonuyi O, et al. Control of simian/human immunodeficiency virus viremia and disease progression after IL-2-augmented DNA-modified vaccinia virus Ankara nasal vaccination in nonhuman primates. J Immunol. 2004 Mar 15;172(6):3745–57. doi: 10.4049/jimmunol.172.6.3745. [DOI] [PubMed] [Google Scholar]
- 24.Kostense S, Vandenberghe K, Joling J, Van Baarle D, Nanlohy N, Manting E, et al. Persistent numbers of tetramer+ CD8(+) T cells, but loss of interferon-gamma+ HIV-specific T cells during progression to AIDS. Blood. 2002 Apr 1;99(7):2505–11. doi: 10.1182/blood.v99.7.2505. [DOI] [PubMed] [Google Scholar]
- 25.Shankar P, Russo M, Harnisch B, Patterson M, Skolnik P, Lieberman J. Impaired function of circulating HIV-specific CD8(+) T cells in chronic human immunodeficiency virus infection. Blood. 2000 Nov 1;96(9):3094–101. [PubMed] [Google Scholar]
- 26.Genesca M, Rourke T, Li J, Bost K, Chohan B, McChesney MB, et al. Live attenuated lentivirus infection elicits polyfunctional simian immunodeficiency virus Gag-specific CD8+ T cells with reduced apoptotic susceptibility in rhesus macaques that control virus replication after challenge with pathogenic SIVmac239. J Immunol. 2007 Oct 1;179(7):4732–40. doi: 10.4049/jimmunol.179.7.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tasca S, Tsai L, Trunova N, Gettie A, Saifuddin M, Bohm R, et al. Induction of potent local cellular immunity with low dose X4 SHIV(SF33A) vaginal exposure. Virology. 2007 Oct 10;367(1):196–211. doi: 10.1016/j.virol.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwissa M, Amara RR, Robinson HL, Moss B, Alkan S, Jabbar A, et al. Adjuvanting a DNA vaccine with a TLR9 ligand plus Flt3 ligand results in enhanced cellular immunity against the simian immunodeficiency virus. J Exp Med. 2007 Oct 29;204(11):2733–46. doi: 10.1084/jem.20071211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potter SJ, Lacabaratz C, Lambotte O, Perez-Patrigeon S, Vingert B, Sinet M, et al. Preserved central memory and activated effector memory CD4+ T-cell subsets in human immunodeficiency virus controllers: an ANRS EP36 study. J Virol. 2007 Dec;81(24):13904–15. doi: 10.1128/JVI.01401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007 Oct 1;204(10):2473–85. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]