Abstract
OBJECTIVE
Specific alleles of non-HLA genes INS, CTLA-4, and PTPN22 have been associated with type 1 diabetes. We examined whether some of these alleles influence development of islet autoimmunity or progression from persistent islet autoimmunity to type 1 diabetes in children with high-risk HLA-DR,DQ genotypes.
RESEARCH DESIGN AND METHODS
Since 1993, the Diabetes Autoimmunity Study in the Young (DAISY) has followed 2,449 young children carrying HLA-DR,DQ genotypes associated with type 1 diabetes. Of those, 112 have developed islet autoimmunity (persistent autoantibodies to insulin, GAD65, and/or IA-2), and 47 of these have progressed to type 1 diabetes. The influence of polymorphisms of INS(−23Hph1), CTLA-4(T17A), and PTPN22(R620W) on development of persistent islet autoimmunity and progression to type 1 diabetes was evaluated by parametric models and by survival analyses.
RESULTS
PTPN22(R620W) allele T was associated with development of persistent islet autoimmunity (hazard ratio 1.83 [95% CI 1.27–2.63]) controlling for ethnicity, presence of HLA-DR3/4,DQB1*0302, and having a first-degree relative with type 1 diabetes. Survival analyses showed a significantly (P = 0.002) higher risk of persistent islet autoimmunity by age 10 years for the TT genotype (27.3%) than for the CT or CC genotype (7.9 and 5.3%, respectively). Cumulative risk of persistent islet autoimmunity was slightly higher (P = 0.02) for the INS(−23Hph1) AA genotype (7.8%) than for the AT or TT genotype (4.2 and 6.4% risk by age 10 years, respectively).
CONCLUSIONS
Whereas the HLA-DR3/4,DQB1*0302 genotype had a dramatic influence on both development of islet autoimmunity and progression to type 1 diabetes, the PTPN22(R620W) T allele significantly influences progression to persistent islet autoimmunity in the DAISY cohort.
The INS (insulin), CTLA-4 (polymorphic cytotoxic T-lymphocyte–associated antigen), and PTPN22 (protein tyrosine phosphatase nonreceptor type 22) genes are some of the confirmed non-HLA genes associated with type 1 diabetes (1–4). The INS gene on chromosome 11p15 confers about 10% of the genetic susceptibility to type 1 diabetes; both a variable number of tandem repeats located ∼0.5 kb upstream of INS (1) and other polymorphisms in tight linkage disequilibrium (LD) such as −23HphI and +1140A/C (5) have been implicated as etiological factors. All of the polymorphisms lie outside coding sequences, suggesting that diabetes susceptibility derives from modulation of INS transcription (6).
The lymphoid-specific phosphatase (LYP), encoded by the PTPN22 gene on chromosome 1p13, is involved in preventing spontaneous T-cell activation. The C1858T single nucleotide polymorphism (SNP) results in a missense mutation that changes an arginine at position 620 to a tryptophan and, thereby, abrogates the ability of the molecule to bind to the signaling molecule Csk (c-Src tyrosine kinase) (3,7,8). The Trp620 variant is associated with autoimmune disorders including Graves' disease (9), rheumatoid arthritis (7), and systemic lupus erythematosus (10).
The CTLA-4 gene on chromosome 2q33 encodes a molecule that functions as a negative regulator of T-cell activation. The G allele of the first exon (Ala17Thr) has been most consistently associated with type 1 diabetes (11) and reduced control of T-cell proliferation (12). The (AT)n microsatellite marker in the 3′ untranslated region, in strong LD with Ala17Thr, is also associated with type 1 diabetes (2) and Graves' disease (13). In this study, we genotyped the children participating in the Diabetes Autoimmunity Study in the Young (DAISY) for polymorphisms in INS, CTLA-4, and PTPN22 to explore how these genes influence development of persistent islet autoimmunity and progression from persistent islet autoimmunity to type 1 diabetes.
RESEARCH DESIGN AND METHODS
Since 1993, DAISY has followed two cohorts of young children at increased risk of type 1 diabetes: the siblings and offspring cohort (SOC) of relatives of type 1 diabetes patients and the general population newborn cohort (NEC). The latter consists of children expressing type 1 diabetes susceptibility HLA-DR,DQ genotypes identified through screening of over 31,000 newborns at St. Josephs Hospital, Denver, Colorado. The details of screening and follow-up have been previously published (14). Children in this cohort have been followed from birth to an average age of 7.0 years (range 5 months to 19.7 years). Informed consent was obtained from the parents of each study subject. The Colorado Multiple Institutional Review Board approved all study protocols.
Islet autoantibodies.
Measurement of biochemical islet autoantibodies was performed in the laboratory of G.S.E. at the Barbara Davis Center. We used radioimmunoassays for autoantibodies to insulin, GAD65, and IA-2. The combined GAD65 autoantibody (GAA) and IA-2 autoantibody (ICA512AA) (or IA-2A) radioassay was done in duplicates on a 96-well filtration plate, and radioactivity was counted on a TopCount 96-well plate β-counter using methods previously described (15). The interassay coefficients of variation are 10 and 5% (n = 50) for GAA and ICA512AA, respectively. The upper limits of normal nondiabetic sera (0.032 for GAA, 0.049 for ICA512AA) were established as the 99th percentile of 198 healthy controls. In the 2005 Diabetes Autoantibody Standardization Program (DASP) workshop (16), the sensitivity and specificity were 76 and 99%, respectively, for GAA and 64 and 100%, respectively, for ICA512AA. Insulin autoantibodies were measured by a micro–insulin autoantibody (IAA) assay. An index was determined based on the difference in counts per minute between wells without and with cold insulin, with a positivity criterion of 0.010, which was the 99th percentile of 106 normal controls. The interassay coefficient of variation is 20% (n = 100) at low positive levels. In the 2005 DASP workshop, the sensitivity and specificity for micro-IAA were 58 and 99%, respectively.
Genotyping.
INS-23Hph1 (rs689), CTLA-4 T17A (rs231775), and PTPN22 R620W (rs2476601) polymorphisms were genotyped using a linear array (immobilized probe) method essentially as described in Mirel et al. (17). Briefly, ∼100 ng of genomic DNA was PCR amplified with biotinylated primers. The labeled amplicons were hybridized to an immobilized sequence-specific oligonucleotide probe (SSOP) array on a nylon membrane. The presence of bound amplicon to a specific probe is detected using streptavidin horseradish peroxidase and a soluble colorless substrate, tetramethylbenzidine, which can be converted in the presence of H2O2 to a blue precipitate.
Statistical analysis.
All analyses were performed in SAS version 9.1. (SAS Institute, Cary, NC). The SNPs were in Hardy-Weinberg equilibrium for both affected and unaffected subjects. Ongoing recruitment since 1994 and continuing follow-up have resulted in variable lengths of follow-up, producing right-censored data. Some of the affected children were positive for autoantibodies on their first blood draw, producing left-censored data. Multiple imputation was used to generate time of autoantibodies status change for the left-censored subjects in order to perform an analysis on progression to type 1 diabetes. Two outcomes were analyzed, i.e., time to persistent islet autoimmunity and time from persistent islet autoimmunity to type 1 diabetes. A parametric model, accounting for right and left censoring assuming a Weibull distribution, was used, adjusting for presence of HLA-DR3/4,DQB1*0302, ethnicity, sex, family history of type 1 diabetes, and age at detection of islet autoimmunity. Cumulative risk of development of persistent islet autoimmunity and progression from islet autoimmunity to type 1 diabetes by genotypes was estimated by survival analyses for each SNP. Follow-up time was defined as the age of the child at the first of the two consecutive positive visits for affected children and age of the child at the last visit for unaffected children. For the time to persistent islet autoimmunity, the survival curves were derived using Turnbull's algorithm, which is a nonparametric maximum likelihood estimator of survival function for arbitrarily truncated and censored data. For the time to type 1 diabetes from persistent islet autoimmunity, the Kaplan-Meier method was used to derive survival curves, which essentially approximates the interval-censored data. The Wilcoxon test was used to test differences in cumulative risk between groups. The analyses were based on genotypes using the additive model.
To determine whether inclusion of multiple siblings per family in this cohort affected our findings, we performed analyses treating siblings as correlated. As those results were the same, we are presenting the data treating siblings as independent. The level of significance was set at 0.05.
RESULTS
Of the 2,449 children included in this analysis, 112 have developed persistent islet autoimmunity, i.e., one or more islet autoantibody (IAA, GAA, or IA-2A), in samples collected on two consecutive visits 3–6 months apart and positive at the last visit; 47 of these children have progressed to type 1 diabetes, defined by a random blood glucose measurement >200 mg/dl and/or an A1C >6.3% in the presence of diabetes symptoms.
Descriptive characteristics of the DAISY cohort are shown in Table 1. Affected (n = 112) compared with unaffected (n = 2,337) children were more likely to be non-Hispanic white, be positive for the HLA-DR3/4,DQB1*0302 genotype, and have a first-degree relative (FDR) with type 1 diabetes. Allele frequencies of PTPN22, INS, and CTLA4 SNPs are shown in Table 2. Affected subjects with persistent islet autoimmunity more often carried the risk alleles for PTPN22 and INS with, respectively, unadjusted HR 1.81 (95% CI 1.26–2.59, P = 0.001) and 1.48 (1.06–2.06, P = 0.019).
TABLE 1.
Characteristics of the DAISY cohort
| Affected with IA (n = 112) | Unaffected (n = 2,337) | Unadjusted HR (95% CI) | P | |
|---|---|---|---|---|
| †HLA-DR3/4 DQB1*0302 | 47 (42.0) | 443 (19.0) | 3.10 (2.13–4.51) | <0.001 |
| First-degree relative with type 1 diabetes | 74 (66.1) | 1,004 (43.0) | 2.03 (1.37–3.00) | <0.001 |
| Sex (female) | 62 (55.4) | 1,106 (47.3) | 1.38 (0.95–2.00) | 0.093 |
| †Non-Hispanic white | 96 (85.7) | 1,667 (71.8) | 1.89 (1.11–3.21) | 0.018 |
| ‡Follow-up time (years) | 4.85 ± 3.6 | 7.11 ± 4.4 |
Data are n (%) or means ± SD unless otherwise indicated. Please note that DAISY cohort is highly enriched in HLA DR3/4 DQB1*0302 genotypes.
†HLA data missing for 4 children and ethnicity data missing for 16 children.
‡For affected, age of the child at the first of two consecutive positive visits; for unaffected, age of the child at the last visit. Boldface represents statistical significance.
TABLE 2.
Association of three SNPs with conversion to persistent islet autoimmunity and progression from persistent islet autoimmunity to type 1 diabetes
| Risk allele | Allele counts (freq.) |
Univariate analyses |
Conversion to persistent islet autoimmunity* (112 cases/2,449 subjects in total) |
Progression to type 1 diabetes† (47 cases/112 IA subjects) |
||||
|---|---|---|---|---|---|---|---|---|
| Affected (n = 112) | Unaffected (n = 2,337) | Unadjusted HR (95% CI) | P | Adjusted HR (95% CI) | P | Adjusted HR (95% CI) | P | |
| PTPN22 (T) | 37 (16.8) | 452 (9.7) | 1.81 (1.26–2.59) | 0.001 | 1.83 (1.27–2.63) | 0.001 | 0.98 (0.50–1.93) | 0.962 |
| INS (A) | 176 (78.6) | 3,323 (71.8) | 1.48 (1.06–2.06) | 0.020 | 1.39 (0.99–1.95) | 0.053 | 1.34 (0.72–2.52) | 0.353 |
| CTLA4 (G) | 95 (42.8) | 1,850 (39.8) | 1.11 (0.85–1.46) | 0.429 | 1.12 (0.86–1.46) | 0.415 | 0.54 (0.33–0.88) | 0.014 |
Data are n (%) unless otherwise indicated. *Parametric model controlled for HLA-DR3/4,DQB1*0302, ethnicity, sex, and first-degree relative with type 1 diabetes.
†Additionally controlled for age at onset of islet autoimmunity.
‡PTPN22 (R620W) data missing for 5, CTLA4 (T17A) for 14, and INS-23HphI for 23 children. Boldface represents statistical significance. IA, islet autoimmunity.
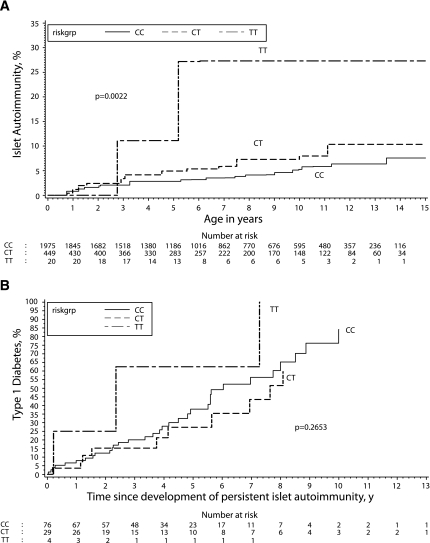
Cumulative incidence of the development of persistent islet autoimmunity and progression from islet autoimmunity to type 1 diabetes by genotypes for each SNP was estimated by survival analyses. The PTPN22 (R620W) TT genotype was associated with a significantly (P = 0.002) higher incidence of persistent islet autoimmunity (27.3% by age 10 years) than the CT (7.9%) or CC (5.3%) genotype (Fig. 1A). Cumulative incidence of progression to type 1 diabetes was also high in children with the PTPN22 TT genotype, with three of four children progressing to type 1 diabetes. However, these results are not statistically significant, likely due to the small sample size (Fig. 1B).
FIG. 1.
Cumulative risk of development of persistent islet autoimmunity (calendar age) by PTPN22 genotypes (A) and cumulative risk of progression from persistent islet autoimmunity to type 1 diabetes (follow-up time since first islet autoimmunity positivity) by PTPN22 genotypes (B) were estimated by survival analyses.
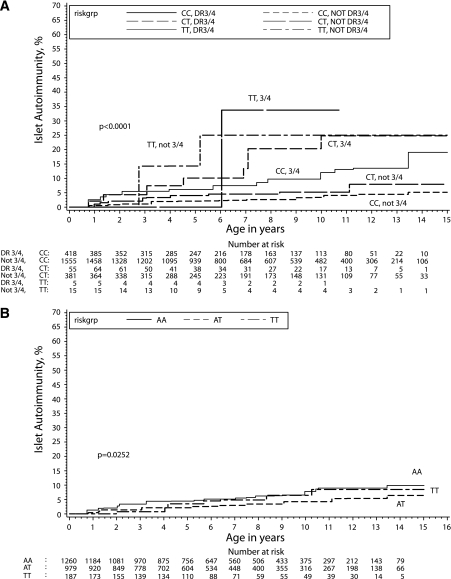
Analysis stratified by the presence of the HLA-DR3/4,DQB1*0302 genotype revealed the highest risk of islet autoimmunity in children carrying HLA-DR3/4,DQB1*0302 and PTPN22 TT genotypes (33.8% by the age of 10), although this is not statistically different from the risk in HLA-non-DR3/4,DQB1*0302, PTPN22 TT. The next highest risk genotypes were HLA-non-DR3/4,DQB1*0302, PTPN22 TT and HLA-DR3/4,DQB1*0302, PTPN22 CT, with the risks of islet autoimmunity by age 10 years being 25.0 and 24.8%, respectively (Fig. 2A).
FIG. 2.
A: Cumulative risk of development of persistent islet autoimmunity (calendar age) by PTPN22 genotypes was estimated by survival analyses and stratified by high-risk HLA-DR3/4,DQB1*0302. B: Cumulative risk of development of persistent islet autoimmunity (calendar age) by INS genotypes was estimated by survival analyses.
The INS genotype appeared to modulate slightly (P = 0.02) the cumulative incidence of persistent islet autoimmunity (Fig. 2B). Within 10 years of detection of persistent islet autoimmunity, 7.8% of children with the INS AA genotype developed type 1 diabetes compared with 4.2 and 6.4% of those with AT and TT, respectively. There were no differences in the rate of progression from persistent islet autoimmunity to type 1 diabetes by the INS-23Hph1 genotypes (data not shown). Survival analyses of development of persistent islet autoimmunity and progression to type 1 diabetes showed no differences by CTLA-4 genotypes (data not shown).
We further analyzed the associations of these three SNPs with development of persistent islet autoimmunity and progression from persistent islet autoimmunity to type 1 diabetes in a multivariate parametric model controlling for the presence of the HLA-DR3/4,DQB1*0302 genotype, ethnicity, sex, family history of type 1 diabetes, and age at detection of islet autoimmunity (Table 2). The presence of the PTPN22 T allele was a significant independent predictor of the development of persistent islet autoimmunity (HR 1.83 [95% CI 1.27–2.63], P = 0.001). However, the PTPN22 T allele did not independently predict progression from islet autoimmunity to type 1 diabetes (0.98 [0.50–1.93], P = 0.96). The INS genotype did not independently predict islet autoimmunity or type 1 diabetes. The CTLA-4 G allele, normally associated with type 1 diabetes risk, did not independently predict islet autoimmunity but was negatively associated with progression from islet autoimmunity to type 1 diabetes (0.54 [0.33–0.88], P = 0.01). There was no interaction between the effect of the HLA-DR3/4,DQB1*0302 genotype and either PTPN22, INS, or CTLA-4 genotypes for the risk of islet autoimmunity or type 1 diabetes (data not shown).
We also performed analyses by cohort, i.e., including 1,371 children from the NEC and 1,078 children from the SOC (supplemental Tables 1 and 2, available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db08-1179/DC1). Affected NEC subjects with persistent islet autoimmunity more often carried the risk alleles for INS and CTLA4 with, respectively, unadjusted HR 1.91 (95% CI 1.08–3.36, P = 0.03) and 1.58 (1.02–2.45, P = 0.04), whereas affected SOC subjects with persistent islet autoimmunity more often carried the risk allele for PTPN22 with unadjusted HR 1.95 (1.27–3.00, P = 0.002). In a multivariate parametric model, the PTPN22 T allele was associated with progression to persistent islet autoimmunity in SOC subjects (HR 2.17 [95% CI 1.41–3.33], P < 0.001), whereas the CTLA4 G allele was an independent predictor of the development of persistent islet autoimmunity in NEC subjects (1.57 [1.01–2.44], P = 0.046). The CTLA-4 G allele, normally associated with type 1 diabetes risk, was negatively associated with progression from islet autoimmunity to type 1 diabetes in SOC subjects only (0.41 [0.22–0.78], P = 0.006). However, these results should be interpreted with caution due to the small sample size.
DISCUSSION
Whereas the associations between the INS, CTLA-4, and PTPN22 polymorphisms and type 1 diabetes are widely accepted, this study is the first comprehensive analysis of the effects of these genes on the age-specific incidence of persistent islet autoimmunity that precedes diagnosis of diabetes in all patients but may not necessarily lead to clinical diabetes. The distinction between the risk factors for islet autoimmunity versus type 1 diabetes is important because of potentially different mechanisms of gene-environment and gene-gene interaction in triggering islet autoimmunity versus epitope spreading and progressive loss of β-cell mass leading to overt diabetes. The results confirm the pivotal role of HLA-DR,DQ in both triggering islet autoimmunity and progression to type 1 diabetes. Whereas the PTPN22 1858 T allele may play a role at both stages leading to type 1 diabetes (18) and predict faster loss of the β-cell function afterward (19), in our population it appears to have an effect independent of the HLA-DR,DQ at the initial stage of islet autoimmunity development. PTPN22 (R620W) results in a missense mutation that changes an arginine at position 620 to a tryptophan and, thereby, abrogates the ability of the molecule to bind to the signaling molecule Csk (3,20). The LYP-Csk complex downregulates T-cell receptor signaling, and the diabetes-associated variant is reported to result in greater inhibition of T-cell receptor signaling (8,21,22). Consistent with an early and general effect on immune function is the finding that the minor tryptophan-encoding allele is associated with a series of autoimmune disorders including rheumatoid arthritis, systemic lupus erythematosus, Hashimoto thyroiditis, and Graves' disease (23,7,24). Hermann et al. (18) showed evidence that PTPN22 (R620W) regulates type 1 diabetes–specific autoimmunity and strongly affects the progression from preclinical to clinical diabetes in islet cell antibody–positive individuals.
Interestingly, when doing analyses separately by cohort, the PTPN22 T allele is a strong independent predictor of the development of persistent islet autoimmunity in the SOC cohort, whereas the CTLA-4 G allele is associated with progression to persistent islet autoimmunity in the NEC cohort. Different genetic loci influence the development of persistent islet autoimmunity and type 1 diabetes in individuals who have a family history of type 1 diabetes, and PTPN22 seems to be one of the genetic factors responsible for increased type 1 diabetes risk in relatives of type 1 diabetic subjects.
The number of subjects carrying the PTPN22 TT genotype is small, especially when analyzing progression from persistent islet autoimmunity to type 1 diabetes. Although three of four such children progress to type 1 diabetes, these results should be interpreted with caution due to the small sample size. This finding requires replication in independent populations.
Alternately, this study provides little support for a major independent effect of the INS(-23Hph1) or the CTLA-4 (T17A) polymorphisms on triggering islet autoimmunity or progression to type 1 diabetes. The insulin gene has been consistently associated with type 1 diabetes in almost all the populations that have been tested, with an odds ratio (OR) between 2 and 3 (1,5). We found a weak association of INS AA genotype with persistent islet autoimmunity, but not with progression to type 1 diabetes, after adjusting for the effect of the HLA-DR3/4,DQB1*0302 genotype and demographic factors. The adjustment or smaller sample size may account for weaker than expected effect (25,26). Although INS is a known factor associated with type 1 diabetes risk, its genetic effect seems to be rather weak overall and is therefore unlikely to help in identifying individuals at risk of type 1 diabetes in the general population. The OR for CTLA-4 association with type 1 diabetes is normally not greater than 1.5 (2,27). In this study, the decreased HR for progression from islet autoimmunity to type 1 diabetes with allele G (normally associated with risk) is likely a spurious finding. Overall, CTLA-4 appears to be a stronger determinant of Graves' disease than of type 1 diabetes (27).
In conclusion, the PTPN22 gene seems to have a large influence on the development of early islet autoimmunity associated with early progression to type 1 diabetes and may be useful in disease prediction using genetic markers. With more accurate prediction, intervention can be provided for individuals at greatest risk.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health Grants R37 DK32493 (DAISY) and P30 DK57516 (DERC Clinical Investigation Core). A.K.S. was supported by the O'Brien Fellowship Foundation.
No potential conflicts of interest relevant to this article were reported.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Bell GI, Horita S, Karam JH: A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes 33: 176– 183, 1984 [DOI] [PubMed] [Google Scholar]
- 2.Marron MP, Raffel LJ, Garchon HJ, Jacob CO, Serrano-Rios M, Martinez Larrad MT, Teng WP, Park Y, Zhang ZX, Goldstein DR, Tao YW, Beaurain G, Bach JF, Huang HS, Luo DF, Zeidler A, Rotter JI, Yang MC, Modilevsky T, Maclaren NK, She JX: Insulin-dependent diabetes mellitus (IDDM) is associated with CTLA4 polymorphisms in multiple ethnic groups. Hum Mol Genet 6: 1275– 1282, 1997 [DOI] [PubMed] [Google Scholar]
- 3.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T: A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 36: 337– 338, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661– 678, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barratt BJ, Payne F, Lowe CE, Hermann R, Healy BC, Harold D, Concannon P, Gharani N, McCarthy MI, Olavesen MG, McCormack R, Guja C, Ionescu-Tirgoviste C, Undlien DE, Ronningen KS, Gillespie KM, Tuomilehto-Wolf E, Tuomilehto J, Bennett ST, Clayton DG, Cordell HJ, Todd JA: Remapping the insulin gene/IDDM2 locus in type 1 diabetes. Diabetes 53: 1884– 1889, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Pugliese A, Zeller M, Fernandez A, Jr, Zalcberg LJ, Bartlett RJ, Ricordi C, Pietropaolo M, Eisenbarth GS, Bennett ST, Patel DD: The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet 15: 293– 297, 1997 [DOI] [PubMed] [Google Scholar]
- 7.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, Conn MT, Chang M, Chang SY, Saiki RK, Catanese JJ, Leong DU, Garcia VE, McAllister LB, Jeffery DA, Lee AT, Batliwalla F, Remmers E, Criswell LA, Seldin MF, Kastner DL, Amos CI, Sninsky JJ, Gregersen PK: A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 75: 330– 337, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, Nika K, Tautz L, Tasken K, Cucca F, Mustelin T, Bottini N: Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet 37: 1317– 1319, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, Howson JM, Vella A, Nutland S, Rance HE, Maier L, Barratt BJ, Guja C, Ionescu-Tirgoviste C, Savage DA, Dunger DB, Widmer B, Strachan DP, Ring SM, Walker N, Clayton DG, Twells RC, Gough SC, Todd JA: Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes 53: 3020– 3023, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, Chang M, Ramos P, Baechler EC, Batliwalla FM, Novitzke J, Williams AH, Gillett C, Rodine P, Graham RR, Ardlie KG, Gaffney PM, Moser KL, Petri M, Begovich AB, Gregersen PK, Behrens TW: Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet 75: 504– 507, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kavvoura FK, Ioannidis JP: CTLA-4 gene polymorphisms and susceptibility to type 1 diabetes mellitus: a HuGE Review and meta-analysis. Am J Epidemiol 162: 3– 16, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Kouki T, Sawai Y, Gardine CA, Fisfalen ME, Alegre ML, DeGroot LJ: CTLA-4 gene polymorphism at position 49 in exon 1 reduces the inhibitory function of CTLA-4 and contributes to the pathogenesis of Graves' disease. J Immunol 165: 6606– 6611, 2000 [DOI] [PubMed] [Google Scholar]
- 13.Howson JM, Dunger DB, Nutland S, Stevens H, Wicker LS, Todd JA: A type 1 diabetes subgroup with a female bias is characterised by failure in tolerance to thyroid peroxidase at an early age and a strong association with the cytotoxic T-lymphocyte-associated antigen-4 gene. Diabetologia 50: 741– 746, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rewers M, Bugawan TL, Norris JM, Blair A, Beaty B, Hoffman M, McDuffie RS, Hamman RF, Klingensmith G, Eisenbarth GS, Erlich HA: Newborn screening for HLA markers associated with IDDM: Diabetes Autoimmunity Study in the Young (DAISY). Diabetologia 39: 807– 812, 1996 [DOI] [PubMed] [Google Scholar]
- 15.Yu L, Rewers M, Gianani R, Kawasaki E, Zhang Y, Verge C, Chase P, Klingensmith G, Erlich H, Norris J, Eisenbarth GS: Antiislet autoantibodies usually develop sequentially rather than simultaneously. J Clin Endocrinol Metab 81: 4264– 4267, 1996 [DOI] [PubMed] [Google Scholar]
- 16.Torn C, Mueller PW, Schlosser M, Bonifacio E, Bingley PJ: Diabetes Antibody Standardization Program: evaluation of assays for autoantibodies to glutamic acid decarboxylase and islet antigen-2. Diabetologia 51: 846– 852, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Mirel DB, Valdes AM, Lazzeroni LC, Reynolds RL, Erlich HA, Noble JA: Association of IL4R haplotypes with type 1 diabetes. Diabetes 51: 3336– 3341, 2002 [DOI] [PubMed] [Google Scholar]
- 18.Hermann R, Lipponen K, Kiviniemi M, Kakko T, Veijola R, Simell O, Knip M, Ilonen J: Lymphoid tyrosine phosphatase (LYP/PTPN22) Arg620Trp variant regulates insulin autoimmunity and progression to type 1 diabetes. Diabetologia 49: 1198– 1208, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Petrone A, Spoletini M, Zampetti S, Capizzi M, Zavarella S, Osborn J, Pozzilli P, Buzzetti R: The PTPN22 1858T gene variant in type 1 diabetes is associated with reduced residual β-cell function and worse metabolic control. Diabetes Care 31: 1214– 1218, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Cloutier JF, Veillette A: Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med 189: 111– 121, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH: Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol 179: 4704– 4710, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Aarnisalo J, Treszl A, Svec P, Marttila J, Oling V, Simell O, Knip M, Korner A, Madacsy L, Vasarhelyi B, Ilonen J, Hermann R: Reduced CD4(+)T cell activation in children with type 1 diabetes carrying the PTPN22/Lyp 620Trp variant. J Autoimmun 31: 13– 21, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Criswell LA, Pfeiffer KA, Lum RF, Gonzales B, Novitzke J, Kern M, Moser KL, Begovich AB, Carlton VE, Li W, Lee AT, Ortmann W, Behrens TW, Gregersen PK: Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet 76: 561– 571, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee AT, Li W, Liew A, Bombardier C, Weisman M, Massarotti EM, Kent J, Wolfe F, Begovich AB, Gregersen PK: The PTPN22 R620W polymorphism associates with RF positive rheumatoid arthritis in a dose-dependent manner but not with HLA-SE status. Genes Immun 6: 129– 133, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Hermann R, Laine AP, Veijola R, Vahlberg T, Simell S, Lahde J, Simell O, Knip M, Ilonen J: The effect of HLA class II, insulin and CTLA4 gene regions on the development of humoral beta cell autoimmunity. Diabetologia 48: 1766– 1775, 2005 [DOI] [PubMed] [Google Scholar]
- 26.Walter M, Albert E, Conrad M, Keller E, Hummel M, Ferber K, Barratt BJ, Todd JA, Ziegler AG, Bonifacio E: IDDM2/insulin VNTR modifies risk conferred by IDDM1/HLA for development of Type 1 diabetes and associated autoimmunity. Diabetologia 46: 712– 720, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, Herr MH, Dahlman I, Payne F, Smyth D, Lowe C, Twells RC, Howlett S, Healy B, Nutland S, Rance HE, Everett V, Smink LJ, Lam AC, Cordell HJ, Walker NM, Bordin C, Hulme J, Motzo C, Cucca F, Hess JF, Metzker ML, Rogers J, Gregory S, Allahabadia A, Nithiyananthan R, Tuomilehto-Wolf E, Tuomilehto J, Bingley P, Gillespie KM, Undlien DE, Ronningen KS, Guja C, Ionescu-Tirgoviste C, Savage DA, Maxwell AP, Carson DJ, Patterson CC, Franklyn JA, Clayton DG, Peterson LB, Wicker LS, Todd JA, Gough SC: Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423: 506– 511, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.