Abstract
The Saccharomyces cerevisiae Mod5 protein catalyzes isopentenylation of A to i6A on tRNAs in the nucleus, cytosol, and mitochondria. The substrate for Mod5p, dimethylallyl pyrophosphate, is also a substrate for Erg20p that catalyzes an essential step in sterol biosynthesis. Changing the distribution of Mod5p so that less Mod5p is present in the cytosol decreases i6A on cytosolic tRNAs and alters tRNA-mediated nonsense suppression. We devised a colony color/growth assay to assess tRNA-mediated nonsense suppression and used it to search for genes, which, when overexpressed, affect nonsense suppression. We identified SAL6, TEF4, and YDL219w, all of which likely affect nonsense suppression via alteration of the protein synthesis machinery. We also identified ARC1, whose product interacts with aminoacyl synthetases. Interestingly, we identified ERG20. Midwestern analysis showed that yeast cells overproducing Erg20p have reduced levels of i6A on tRNAs. Thus, Erg20p appears to affect nonsense suppression by competing with Mod5p for substrate. Identification of ERG20 reveals that yeast have a limited pool of dimethylallyl pyrophosphate. It also demonstrates that disrupting the balance between enzymes that use dimethylallyl pyrophosphate as substrate affects translation.
Keywords: N6-Δ2-isopentenyladenosine, sterol biosynthesis, assay for flux in sterol pathway
The Saccharomyces cerevisiae protein Mod5p catalyzes the addition of an isopentenyl group to adenosine (i6A) at position 37 of the anticodon loop of some tRNAs (1–5). There are two isoforms of Mod5p, Mod5p-I and Mod5p-II, that differ in the site of their translation initiation codon and in their distribution in the cell. Mod5p-I is translated starting at codon 1 of the MOD5 ORF and is localized to mitochondria and the cytoplasm. Mod5p-II, translated from codon 12 of the MOD5 ORF, is located in the nucleus and the cytoplasm (6, 7).
The i6A modification promotes the efficiency of SUP7 tRNA in cytosolic suppression of UAA nonsense mutations by the insertion of tyrosine (8). Cells possessing only Mod5p-I have limiting cytosolic amounts of isozyme, and changes in the subcellular distribution and/or the activity of this isozyme alter nonsense suppression. Hence, genetic screens/selections based on nonsense suppression can identify cells with altered cytosolic Mod5p-I activity (9).
Here we employed the genetic strategy of using overexpression to perturb a pathway (10) and developed a protocol for the selection of cells with lower than normal levels of cytosolic Mod5p-I activity. Using this strategy we were able to sample the entire yeast genome and identify genes that, when overexpressed, lead to lower than normal levels of cytosolic Mod5p-I activity.
As a result of that screen, we identified two categories of genes. The first category includes genes that affect nonsense suppression via alteration of the protein synthetic machinery. Our studies suggest that the yeast gene product encoded by YDL219w may function in protein synthesis and that the translation elongation factor EF1-γ may function in translational proofreading. SAL6 and ARC1, with previously established effects on protein synthesis, also were recovered.
The second category includes ERG20, involved in sterol biosynthesis. The mevalonate pathway generates sterols, prenylated proteins, heme A, dolichol, ubiquinone, and the substrate required for isopentenylation of tRNAs (Fig. 1). Both Mod5p and Erg20p use the same substrate, dimethylallyl pyrophosphate (DMAPP). Recovery of ERG20 coupled with subsequent biochemical assays demonstrating a reduction of i6A on tRNA is most easily explained by a model in which Mod5p and Erg20p compete for a limited pool of DMAPP. When more DMAPP is used to make sterols, less is available for modification of tRNA and a reduction in the efficiency of nonsense suppression results. Thus, we have demonstrated that the tRNA biosynthetic pathway and the sterol biosynthetic pathway are in apparent competition for substrate and that Erg20p and Mod5p must be balanced to optimally maintain the protein synthetic machinery. A practical consequence is that it should be possible to adapt the selection we developed to assess the effect of mutations and/or drugs that change the distribution of DMAPP between the sterol pathway and the tRNA biosynthetic pathway.
Figure 1.
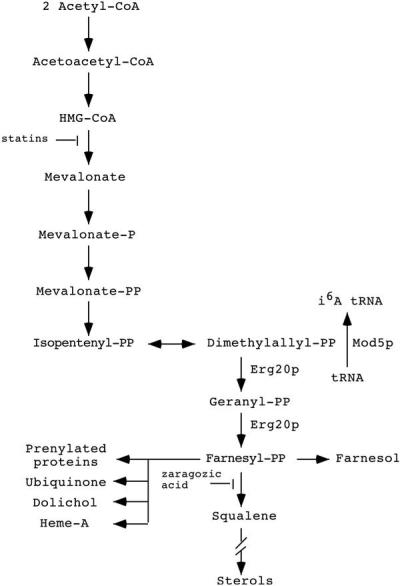
The mevalonate pathway (11).
Materials and Methods
Strains and Media.
Strain ALB1 (MATα mod5-M2 SUP7 ade2–1 can1–100 leu2–3, -112 lys1–1 lys2–1 trp1 ura3–1) was created from MD14A (6) by the replacement of the genomic mod5–1 through a pop-in/pop-out procedure (12). Plasmid YCfmod5-M2 (6) was used as the donor of the M2 mutation. The ARS1 and CEN4 sequences were removed from YCfmod5-M2 by a BglII/SmaI digestion. The BglII end of the remaining plasmid was filled in by using Klenow DNA Polymerase and then ligated to the SmaI blunt end. The resulting plasmid was linearized by SnaBI just upstream of the mod5-M2 ORF and then transformed into MD14A. Integration of the plasmid was selected for by growth on medium lacking uracil. Resulting cells contained two mod5 alleles: mod5–1 and mod5-M2. Cells in which intrachromosomal homologous recombination and loss of the mod5–1 mutation occurred were selected for by growth on medium containing 5-fluoro-orotic acid (5-FOA) and for the ability to grow on medium lacking adenine. The presence of the remaining mod5-M2 allele was confirmed by DNA blot hybridization.
Strain ALB8 (MATα SUP7 can1–100 ade2–1 leu2–3, -112 lys1–1 lys2–1 trp1 mod5∷TRP1 ura3–1∷MOD5) was created through integration of a plasmid containing MOD5 at the ura3–1 locus of strain MT-8 (6). Plasmid YCfMOD5 (6) was used as the donor of MOD5. The ARS1 and CEN4 sequences were removed as described above, and the resulting plasmid was linearized with NcoI to target it to ura3-1. MT-8 cells that acquired the plasmid were selected on medium lacking uracil. Correct integrants were assessed by DNA blot hybridization.
Strain SL680–9C (MATα asu9–1 sup45–2 ura3–52 can1–132 leu2–1 lys2–1 met8–1 trp1–1 or trp1–289) (13) was obtained from S. Liebman (University of Illinois, Chicago).
Yeast strains were propagated in YEPD (dextrose) medium or synthetic complete (SC) medium lacking certain nutrients to select for the presence of plasmid and to assay suppression. Canavanine sulfate salt (Sigma) was added to medium lacking arginine to a final concentration of 30 μg/ml. Yeast strains were transformed by following a modified TRAFO protocol (14).
Library DNAs and Plasmids.
The following DNA libraries were used: a genomic DNA library based in YEp13 (15), a genomic DNA library based in YEp24 (16), a cDNA library based in pMac561, a 2μ vector with the ADH1 promoter (17), and a cDNA library based in pRS316 (18).
YEpMOD5 contains the 1.8-kb DNA fragment encoding wild-type MOD5 gene and regulatory regions in the YEp24 vector (5). pRH127–3, a gift from R. Wright (University of Washington, Seattle), is a 2μ-based plasmid that contains a truncated HMG1 gene that causes increased HMG-CoA reductase activity and increased levels (≈10 times greater) of intermediates in the mevalonate pathway (19).
Sequencing of Inserts of Library Clones.
Library plasmids were isolated from yeast by the method of Ward (20). DNAs were sequenced by either the chain termination method (21) with Sequenase Version 2.0 DNA Sequencing Kit (United States Biochemical) or by automated cycle sequencing performed in the Pennsylvania State University College of Medicine Macromolecular Core Facility. Nucleotide sequences were identified by a blast (22) search at the Saccharomyces Genome Database blast server (http://genome-www2.stanford.edu/cgi-bin/SGD/nph-blast2sgd/).
Determination of the Levels of i6A in tRNA.
RNA was obtained (23) from overnight log-phase cultures of the following strains: ALB8, MD14A, and ALB1 with the pRS316 library plasmids containing SAL6, ARC1, TEF4, ERG20, YDL219w, or no cDNA insert. The RNA concentration was calculated from optical density measurement.
RNAs were resolved on a 12% polyacrylamide/7 M urea gel. The RNA was transferred to Hybond N+ Nylon Membrane (Amersham Pharmacia) in a Hoefer TE Series Transphor Apparatus containing TAE buffer (40 mM Tris-acetate, 1 mM EDTA). The RNA was fixed onto the membrane by UV crosslinking. The membrane was subjected to immunoblotting by following the protocol of the Renaissance Western Blot Chemiluminescence Kit (NEN) with slight variation. The primary antibody, anti-i6A isolated from rabbit (R. Clark and N.C.M., unpublished results) prepared by the method of Senapathy and Jacob (24), was diluted either 1:33.3 or 1:72.5 before use. Horseradish peroxidase-conjugated anti-rabbit IgG from donkey (Amersham Pharmacia) diluted 1:5,000 or 1:2,500 served as secondary antibody. The membrane also was utilized for RNA detection by using radiolabeled oligonucleotide complementary to a region of mature tRNATyr.
The intensities of the fluorescent and radioactive signals for each specimen on the immunoblots and RNA blots, respectively, were determined by densitometry using a Molecular Dynamics laser scanner with the Discovery Series Quantity One computer software (Bio-Rad). For each sample, the ratio of its signal with respect to that of the signal from the ALB1 with vector-only sample was calculated from the immunoblots (ratio 1) and from the RNA blots (ratio 2). The average values for ratio 1 to ratio 2 for samples from two membranes were calculated.
Results
Identification of Plasmids Causing Reduced Nonsense Suppression.
We employed the can1–100 and ade2–1 alleles in a mod5-M2 background (strain ALB1) to identify plasmids causing alterations in nonsense suppression. Mod5-M2 is an allele of MOD5 that has normal levels of mitochondrial but reduced levels of cytosolic Mod5p activity. In this strain, cytosolic tRNAs are only partially modified with i6A (60%; ref. 6) and further alterations in Mod5p levels are readily assessed by changes in nonsense suppression (9). The can1–100 and ade2–1 alleles contain UAA nonsense mutations. CAN1 encodes an arginine permease that allows the uptake of the arginine analog canavanine (25), and cells cannot grow in its presence. Therefore, cells with wild-type CAN1 are sensitive to canavanine (Cans), but cells with the mutant can1–100 are resistant to canavanine (Canr). Cells containing SUP7 tRNAs completely modified with i6A have functional arginine permease and, therefore, cannot grow in the presence of canavanine, whereas cells lacking i6A on their tRNAs are able to grow in the presence of this drug. Cells with intermediate levels of i6A show intermediate levels of growth in the presence of the drug.
ADE2 encodes an enzyme involved in the synthesis of adenine. Cells with ade2–1 turn red in color and fail to grow on defined medium lacking adenine (Ade−), whereas cells producing functional Ade2p can grow on such medium (Ade+) and are white (26). Cells with the ade2–1 allele and sufficient i6A-modified suppressor tRNA can grow in the absence of exogenous adenine and generate white colonies on rich medium, whereas cells with insufficient i6A-modified tRNA are unable to grow in the absence of exogenous adenine and generate red colonies on rich medium. Cells with intermediate levels of i6A modified tRNA have intermediate phenotypes in colony color and intermediate rates of growth in the absence of exogenous adenine.
Transformants able to grow on canavanine were selected from approximately 1.5 million independent ALB1 (relevant genotype: SUP7 ade2–1 can1–100 mod5-M2) transformed cells. One hundred and fifty-eight candidates that exhibited growth on medium containing canavanine were tested for lack of growth on medium lacking adenine to eliminate those gaining canavanine resistance because of additional nonsuppressible mutations in can1–100.
For each of the 71 canavanine-resistant candidates that also were unable to grow well on medium lacking adenine or were red-pink in color, an assay was performed to determine whether these phenotypes required the presence of the library plasmid. Thirty-three candidates showed dependence of suppression levels upon the presence of a plasmid. Analysis of the plasmids in these cells resulted in the identification of 11 with unique restriction patterns. These plasmids were retransformed into ALB1 and the transformants were reassayed for canavanine resistance and adenine prototrophy. One plasmid did not confer either of these phenotypes. Four pRS316 library clones were still able to prevent growth on medium lacking adenine, and six of the remaining clones still could produce both desired phenotypes in ALB1 as confirmed by a second set of co-loss experiments (Table 1). Sequencing of the inserts of these latter 10 clones led to the identification of five different genes: SAL6, ARC1, TEF4, YDL219W, and ERG20.
Table 1.
Growth of strains containing plasmids
| Strain | Growth on canavanine | Growth/colony color on SC-adenine
|
|
|---|---|---|---|
| Galactose | Glucose | Galactose | |
| MD14A | ++ | − r | −r |
| pRSvector/ALB1 | − | + w | + w |
| pRSSAL6/ALB1 | + | + w | +/− p |
| pRSERG20/ALB1 | + | + w | +/− p |
| pRSARC1/ALB1 | +/− | + w | +/− p |
| pRSYDL219w/ALB1 | +/− | + w | +/− p |
| pRSTEF4/ALB1 | +/− | +w | +/− p |
++, Very strong growth; +, strong growth; +/−, poor growth; −, no growth; r, red; p, pink; w, white.
ERG20, which encodes farnesyl diphosphate synthetase, was found once in each of the YEp13, pMac561, and pRS316 libraries. SAL6 cDNA, encoding a protein phosphatase, was discovered four times in the pRS316 library. ARC1, TEF4, and the putative ORF YDL219w were single candidates from the pRS316 library (Table 1). The pRS316 library is constructed such that the cloned genes are regulated by the GAL1 promoter. Thus, transcription is induced by galactose and repressed by glucose in the media. The effects of the identified genes upon nonsense suppression were dependent on having galactose in the medium (Table 1), showing that the phenotypes are a result of their expression.
SAL6, TEF4, and YDL219w Confer Antisuppressor Phenotypes.
SUP45 encodes a translation termination factor, and mutation of this gene can result in omnipotent suppression (27, 28). When SAL6 is overexpressed in a yeast strain containing sup45-2, an antisuppressor phenotype occurs (13). This suggested that the canavanine resistance conferred upon ALB1 by high levels of Sal6p also may be due to antisuppression. Therefore, all of the candidate genes were tested for effects upon translation by assessing whether they scored as antisuppressors of sup45–2.
SL680–9C (relevant genotype sup45–2 asu9–1 leu2–1 met8–1 ura3–52) is capable of growing on medium lacking methionine because the mutant sup45 permits translation through nonsense mutations such as met8–1 (13, 27, 28). The asu9 antisuppressor allele serves to weaken the effects of sup45 and permits better growth of cells with both mutations (13, 27).
SL680–9C cells transformed by each of the candidate genes in the pRS316 vector were analyzed for growth on galactose-containing or glucose-containing medium lacking uracil and methionine. The data (Table 2) indicate that overexpressed SAL6 and, to a lesser degree, TEF4 and YDL219w cause antisuppression, and, therefore, loss of suppression of the can1–100 and ade2–1 in ALB1 is likely Mod5p-I-independent. This phenotype was not observed consistently in the cells containing large amounts of Tef4p. Neither Arc1p nor Erg20p in high levels affected the suppression of met8–1 in SL680–9C, and, therefore, each probably affects can1–100 and ade2–1 suppression in ALB1 via another route, likely by alteration of Mod5p-I or tRNA activities or location.
Table 2.
Growth of SL680-9C cells overexpressing candidate genes
| Candidate gene/ORF | Growth on SC-uracil-methionine (galactose) | Growth on SC-uracil-methionine (glucose) |
|---|---|---|
| None (vector only) | + | + |
| SAL6 | − | + |
| ARC1 | + | + |
| TEF4 | +/−* | + |
| ERG20 | + | + |
| YDL219w | +/− | + |
Cells were grown at 28°C for 5–8 days; results are for three experiments. +, strong growth; +/−, poor growth; −, no growth.
*Galactose vs. glucose growth difference sometimes not apparent.
Overexpression of ERG20 Results in the Reduction of Isopentenyl Adenosine Present in tRNA.
The protein encoded by ERG20, farnesyl diphosphate synthetase, catalyzes the formation of geranyl pyrophosphate and farnesyl pyrophosphate by the condensation of units of DMAPP and its isomer isopentenyl pyrophosphate (IPP). DMAPP is the same molecule used by Mod5p as a substrate in the synthesis of i6A in tRNA (refs. 29 and 30; Fig. 1). It was possible that the large pool of Erg20p produced from the library clones used the DMAPP normally utilized for tRNA modification. This would cause reduced i6A-modified tRNA and, hence, reduced function of these tRNAs in nonsense suppression. It was also possible that others of the candidate genes, when overexpressed, inhibited the formation of i6A tRNA.
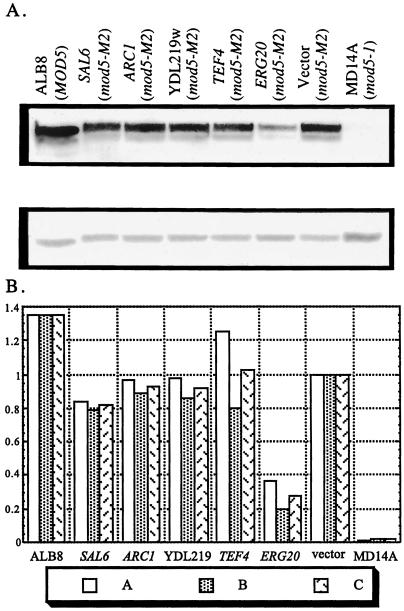
To determine the effect of overexpression of the candidate genes upon i6A modification of tRNA, we employed the Midwestern procedure (31) by using anti-i6A antibody. Low-molecular-weight RNA was prepared from cells with each plasmid and from control strains of ALB1 with vector only, ALB8, and MD14A. The RNAs were resolved on urea-polyacrylamide gels, transferred to membranes, and probed with anti-i6A antibody (Fig. 2A Upper). Hybridization with a radiolabeled oligonucleotide complementary to part of mature tRNATyr was used to assess the relative amount of tRNA in each lane (Fig. 2A Lower). Through densitometric analysis, the amount of i6A formed in tRNA of each strain as a fraction of the amount formed in the vector-only control strain was determined (Fig. 2B).
Figure 2.
The level of isopentenylated tRNA found in ALB1 overexpressing ERG20 is reduced substantially. (A) Low-molecular-weight RNA was prepared from ALB1 (mod5-M2) with each of the candidate genes or vector alone, ALB8 (MOD5), or MD14A (mod5–1). The RNAs were resolved on polyacrylamide gels, transferred to membranes, and probed with anti-isopentenyl adenosine antibody (Upper) or radiolabeled oligonucleotide complementary to mature tRNATyr (Lower). (B) The levels of isopentenyl adenosine tRNA found in ALB1 with each of the candidate genes or vector only or in the strain ALB8 or MD14A were assessed by densitometric analysis of two immunoblots and expressed as a fraction of the level found in the “vector” control. A, membrane 1 values; B, membrane 2 values; C, average values.
As expected (32), MD14A cells with the mod5–1 allele have barely detectable levels of modified tRNA. Also as anticipated, ALB8 cells with a wild-type MOD5 that encodes both Mod5p-I and Mod5p-II contained higher levels (≈35% more) of i6A-modified tRNA than ALB1 cells expressing only Mod5p-I (ref. 6; Fig. 2). The level of i6A tRNA in ALB1 overexpressing SAL6, ARC1, YDL219w, or TEF4 is similar to the level of i6A from ALB1 cells with vector only. However, the amount of i6A tRNA in ALB1 with excess Erg20p is significantly reduced to approximately 30% of that in ALB1 with vector alone. Overproduction of Erg20p results in reduced levels of i6A tRNA and causes a decrease in nonsense suppression.
If overproduction of Erg20p acts via competition with Mod5p-I for a common substrate (Fig. 3), then providing additional Mod5p should reverse the loss of suppression. To test this, we transformed ALB1 cells containing YEpERG20 with YEpMOD5 and assessed suppression of ade2–1 and can1–100. The results (Table 3) show that suppression indeed is restored by additional Mod5p. Likewise, if Erg20p and Mod5p are in competition for the same substrate, then alterations that increase the substrate should dampen the competition. To test this, we introduced a multicopy vector harboring a mutant HMG1 gene, pRH127–3, that causes elevated levels of intermediates of the mevalonate pathway (19) into ALB1 containing YEpERG20. We found that increased levels of the intermediates also restored nonsense suppression (Table 3). The data support the model that the mevalonate pathway and the i6A tRNA modification pathways are in competition.
Figure 3.
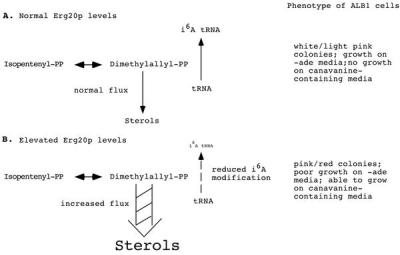
Model of competition between i6A modification of tRNA and sterol biosynthesis.
Table 3.
Growth of strains containing multicopy plasmids
| ALB1 with plasmids | Growth on canavanine | Growth/colony color on SC-adenine |
|---|---|---|
| YEp13 + YEpMOD5 | − | + w |
| YEpERG20 + YEp24 | +/− | +/− p |
| YEpERG20 + YEPMOD5 | − | + w |
| YEp13 + YEp24 | − | + w |
| YEp13 + pRH127-3 | − | + w |
| YEpERG20 + YEp24 | +/− | +/− p |
| YEpERG20 + pRH127-3 | +/− | + w |
+, strong growth; +/−, poor growth; −, no growth; p, pink; w, white.
Discussion
Our interest in the distribution of sorting isozymes led us to design a screen for gene products that function in the subcellular distribution of Mod5p or affect its ability to modify cytosolic tRNAs. We anticipated that we would uncover genes affecting transcription of MOD5 and/or tRNAs, nuclear export of tRNA, and the translation process, in addition to those involved in protein distribution. As anticipated, three gene products, Sal6p, Tef4p, and YDL219w, appear to affect nonsense suppression via function in protein translation and Arc1p could play a role in nuclear export of tRNA (see below). Surprisingly, no gene products affecting MOD5 or tRNA transcription or Mod5p-I subcellular distribution were uncovered. Rather, ERG20, important to sterol biosynthesis, was found.
Sal6p/Ppq1p is a serine–threonine protein phosphatase very similar to mammalian phosphatase PP1 (13, 33). Multiple copies of SAL6 cause antisuppression of nonsense mutations (13), and the sal6–1 allele acts as an allosuppressor (13, 27). Yeast cells containing a disrupted PPQ1 gene exhibit a slowed translation rate and hypersensitivity to inhibitors of protein synthesis (33). These findings suggest a role for Sal6p in the regulation of the fidelity of translation (13, 33).
Tef4p is the γ-subunit of elongation factor 1 (EF-1). Eukaryotic EF-1 functions in delivering incoming tRNAs to the ribosome and is composed of at least three subunits. EF-1α binds GTP and then the proper aminoacyl-tRNA and positions the aminoacyl tRNA in the ribosomal A site. Subsequently, GTP is hydrolyzed to GDP and EF-1β catalyzes the exchange of GDP for GTP to restore EF-1α to its initial state (34, 35). The exact function of EF-1γ has not been determined; however, in Artemia salina this subunit was observed to enhance the function of EF-1β catalysis. In the same study EF1-γ was found to interact with cell membranes and tubulin, suggesting that it might mediate the association of the translational machinery with the cell framework (36).
In bacteria, the EF-1 counterpart functions in translational proofreading. Incorrectly inserted aminoacyl tRNAs can be removed before GTP hydrolysis and before the departure of EF-1α from the ribosome (35). If the yeast EF-1 serves a similar function as the bacterial counterpart, it is unlikely that the α-subunit participates in this function as extra copies of the genes encoding EF-1α decrease fidelity (i.e., they enhance suppression of nonsense mutations; ref. 37). We show that overexpression of Tef4p reduces nonsense suppression and, therefore, it is possible that it is the EF1-γ subunit that functions in EF-1 proofreading.
ORF YDL219w is predicted to code for a 150-aa protein with no significant homology to any characterized protein. However, two lines of evidence indicate that this protein may function in the translation process. First, the gene possesses an intron (Saccharomyces Genome Database, http://genome-www.stanford.edu/cgi-bin/dbrun). Because introns are rare in yeast other than for approximately half of the genes encoding ribosomal proteins (38), the presence of the intron is suggestive of a role in translation. Second, we show that overexpression of YDL219w affects tRNA-mediated nonsense suppression.
Arc1p, or G4p1, originally was discovered as a quadruplex nucleic acid-binding protein (39). Subsequently, Arc1p was demonstrated to bind tRNA and to complex with methionyl and glutamyl tRNA synthetases (40, 41). Recent studies have shown that aminoacylation is important for tRNA nuclear export (42, 43). Moreover, Simos et al. (40) uncovered ARC1 by synthetic interactions with Los1p, the yeast tRNA exportin (41, 44–46). Thus, it is possible that overexpression of Arc1p interferes with the appropriate nucleus/cytosol distribution of tRNA. Alternatively, overexpressed Arc1p could interfere with the interaction of tRNA and ribosomes. Incubation of tRNAMet with large amounts of Arc1p in vitro resulted in the creation of supershifted complexes in a gel mobility assay, indicative of higher-order complexes (40). Perhaps large quantities of cellular Arc1p form complexes with tRNA, preventing the tRNA from functioning normally in translation.
Overexpression of Erg20p causes a decrease in i6A tRNA-mediated suppression in yeast and an approximately 70% decrease in i6A modification of tRNA. These effects are most likely due to the loss of Mod5p substrate to Erg20p. The data demonstrate the dependence of tRNA processing upon changes in components of the sterol pathway. The data also indicate that, at least in yeast, Erg20p may catalyze a rate-limiting step in sterol biosynthesis. Hence, the tRNA and the sterol biosynthetic pathways are in apparent competition, and a delicate balance in protein levels is required to maintain proper functioning of translation and mevalonate metabolism.
The mevalonate pathway generates sterols, prenylated proteins, heme A, dolichol, ubiquinone, and isopentenyl tRNAs. Inhibitors of HMG-CoA reductase, catalyzing a rate-limiting step of the pathway, have been developed for administration to patients with hypercholesterolemia (47, 48). The “statin” class (e.g., compactin and lovastatin) of reductase inhibitors is safe and effective in lowering total cholesterol levels and preventing the progression and reducing the occurrence of coronary disease events (48–51). Zaragozic acid inhibits squalene synthetase, catalyzing the first step of the pathway committed solely to sterol biosynthesis, but appears not to be used currently in the clinical setting. A novel type of zaragozic acid, ZAD3, also inhibits farnesyl protein transferase and, therefore, protein prenylation as well (52). There appears to be no drug in current clinical use that affects solely cholesterol biosynthesis without affecting other farnesyl-PP-derived products.
We have demonstrated that increased levels of an enzyme of the sterol biosynthetic pathway can be assessed indirectly simply by colony color and/or growth on particular medium because changes in nonsense suppression occur. Thus, additional mutations and/or drugs that affect the pathway could be identified by a modification of the screening protocol we describe. The effect of prospective new regulators of pathway enzymes on the suppressor ability of a strain with the ALB1 genotype (mod5-M2 SUP7 can1–100 ade2–1) could be assessed by adding them to canavanine-containing or adenine-lacking medium and monitoring cell growth. If the presence of a substance enhances colony growth on canavanine or deters colony formation on medium lacking adenine, in comparison with the growth observed on medium without the test substance, then the substance may be increasing the flow of DMAPP through the mevalonate pathway and decreasing tRNA modification. Likewise, by using the mod5-M2,KR6 allele, we previously have shown (9) that increased cytosolic Mod5p activity results in acquisition of the ability to grow on medium lacking lysine. Therefore, compounds that decrease the use of DMAPP by the sterol pathway also may be assessed easily. In this way, potential enhancers and inhibitors of the mevalonate pathway could be screened initially for their efficacy quickly and inexpensively.
Acknowledgments
We thank F. A. Benko for his excellent technical assistance. This work was supported by grants from the National Science Foundation to A.K.H. and N.C.M.
Abbreviations
- i6A
N6-(Δ2-isopentenyl)adenosine
- DMAPP
dimethylallyl pyrophosphate
- EF-1
elongation factor 1
- SC medium
synthetic complete medium
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kline L K, Fittler F, Hall R H. Biochemistry. 1969;8:4361–4371. doi: 10.1021/bi00839a021. [DOI] [PubMed] [Google Scholar]
- 2.Bartz J K, Kline L K, Söll D. Biochem Biophys Res Commun. 1970;40:1481–1487. doi: 10.1016/0006-291x(70)90035-5. [DOI] [PubMed] [Google Scholar]
- 3.Rosenbaum N, Gefter M L. J Biol Chem. 1972;247:5675–5680. [PubMed] [Google Scholar]
- 4.McCloskey J A, Nishimura S. Acc Chem Res. 1977;10:403–410. [Google Scholar]
- 5.Dihanich M E, Najarian D, Clark R, Gillman E C, Martin N C, Hopper A K. Mol Cell Biol. 1987;7:177–184. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillman E C, Slusher L B, Martin N C, Hopper A K. Mol Cell Biol. 1991;11:2382–2390. doi: 10.1128/mcb.11.5.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boguta M, Hunter L A, Shen W-C, Gillman E C, Martin N C, Hopper A K. Mol Cell Biol. 1994;14:2298–2306. doi: 10.1128/mcb.14.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laten H, Gorman J, Bock R M. Nucleic Acids Res. 1978;5:4329–4342. doi: 10.1093/nar/5.11.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoladek T, Vaduva G, Hunter L A, Boguta M, Go B D, Martin N C, Hopper A K. Mol Cell Biol. 1995;15:6884–6894. doi: 10.1128/mcb.15.12.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rine J. Methods Enzymol. 1991;194:239–251. doi: 10.1016/0076-6879(91)94019-9. [DOI] [PubMed] [Google Scholar]
- 11.Dimster-Denk D, Thorsness M K, Rine J. Mol Biol Cell. 1994;5:655–665. doi: 10.1091/mbc.5.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothstein R. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 13.Vincent A, Newnam G, Liebman S W. Genetics. 1994;138:597–607. doi: 10.1093/genetics/138.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gietz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasmyth K A, Tatchell K. Cell. 1980;19:753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- 16.Carlson M, Botstein D. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 17.McKnight G L, McConaughy B L. Proc Natl Acad Sci USA. 1983;80:4412–4416. doi: 10.1073/pnas.80.14.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Krizek J, Bretscher A. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donald K A, Hampton R Y, Fritz I B. Appl Environ Microbiol. 1997;63:3341–3344. doi: 10.1128/aem.63.9.3341-3344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward A C. Nucleic Acids Res. 1990;18:5319. doi: 10.1093/nar/18.17.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Hopper A K, Schultz L D, Shapiro R A. Cell. 1980;19:741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- 24.Senapathy P, Jacob M T. J Biol Chem. 1981;256:11580–11584. [PubMed] [Google Scholar]
- 25.Whelan W L, Gocke E, Manney T R. Genetics. 1979;91:35–51. doi: 10.1093/genetics/91.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasse-Messenguy F, Fink G R. Genetics. 1973;75:459–464. doi: 10.1093/genetics/75.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stansfield I, Tuite M F. Curr Genet. 1994;25:385–395. doi: 10.1007/BF00351776. [DOI] [PubMed] [Google Scholar]
- 28.Stansfield I, Jones K M, Kushnirov V V, Dagkesamanskaya A R, Poznyakovski A I, Paushkin S V, Nierras C R, Cox B S, Ter-Avanesyan M D, Tuite M F. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown M S, Goldstein J L. J Lipid Res. 1980;21:505–517. [PubMed] [Google Scholar]
- 30.Voet D, Voet J G. Biochemistry. New York: Wiley; 1990. [Google Scholar]
- 31.Rasmussen T P, Culbertson M R. Gene. 1996;182:89–96. doi: 10.1016/s0378-1119(96)00519-7. [DOI] [PubMed] [Google Scholar]
- 32.Martin N C, Hopper A K. J Biol Chem. 1982;257:10562–10565. [PubMed] [Google Scholar]
- 33.Chen M X, Chen Y H, Cohen P T W. Eur J Biochem. 1993;218:689–699. doi: 10.1111/j.1432-1033.1993.tb18423.x. [DOI] [PubMed] [Google Scholar]
- 34.Kinzy T G, Ripmaster T L, Woolford J L., Jr Nucleic Acids Res. 1994;22:2703–2707. doi: 10.1093/nar/22.13.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinnebusch A G, Liebman S W. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics. Broach J R, Pringle J R, Jones E W, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. pp. 627–735. [Google Scholar]
- 36.Janssen G M C, Möller W. Eur J Biochem. 1988;171:119–129. doi: 10.1111/j.1432-1033.1988.tb13766.x. [DOI] [PubMed] [Google Scholar]
- 37.Song J M, Picologlou S, Grant C M, Firoozan M, Tuite M F, Liebman S. Mol Cell Biol. 1989;9:4571–4575. doi: 10.1128/mcb.9.10.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woolford J L, Jr, Warner J R. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics. Broach J R, Pringle J R, Jones E W, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. pp. 587–626. [Google Scholar]
- 39.Frantz J D, Gilbert W. J Biol Chem. 1995;270:20692–20697. doi: 10.1074/jbc.270.35.20692. [DOI] [PubMed] [Google Scholar]
- 40.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt E C. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 41.Simos G, Sauer A, Fasiolo F, Hurt E C. Mol Cell. 1998;1:235–242. doi: 10.1016/s1097-2765(00)80024-6. [DOI] [PubMed] [Google Scholar]
- 42.Lund E, Dahlberg J E. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- 43.Sarkar S, Azad A, Hopper A K. Proc Natl Acad Sci USA. 1999;96:14366–14371. doi: 10.1073/pnas.96.25.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arts G, Fornerod M, Mattaj I W. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 45.Kutay U, Lipowsky G, Izaurralde E, Bischoff F R, Schwarzmaier P, Hartmann E, Görlich D. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 46.Sarkar S, Hopper A K. Mol Biol Cell. 1998;9:3041–3055. doi: 10.1091/mbc.9.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldstein J L, Brown M S. Nature (London) 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 48.Endo A. J Lipid Res. 1992;33:1569–1582. [PubMed] [Google Scholar]
- 49.Brown G, Albers J J, Fisher L D, Schaffer S M, Lin J T, Kaplan C, Zhao X Q, Bisson B D, Fitzpatrick V F, Dodge H T. N Eng J Med. 1990;323:1289–1298. doi: 10.1056/NEJM199011083231901. [DOI] [PubMed] [Google Scholar]
- 50.Nash D T. Am J Cardiol. 1996;78, Suppl. 6A:26–31. doi: 10.1016/s0002-9149(96)00659-5. [DOI] [PubMed] [Google Scholar]
- 51.Olsson A G. Eur Heart J. 1998;19, Suppl. M:M29–M35. [PubMed] [Google Scholar]
- 52.Tanimoto T, Ohya S, Tsujita Y. J Antibiot. 1998;51:428–431. doi: 10.7164/antibiotics.51.428. [DOI] [PubMed] [Google Scholar]