Abstract
OBJECTIVE
The vascular ectonucleotidase ENTPD1 protects against renal injury and modulates glucose homeostasis in mouse models. We sought to determine whether human variation in ENTPD1 influences predisposition to diabetes or diabetic nephropathy.
RESEARCH DESIGN AND METHODS
We analyzed ENTPD1 single nucleotide polymorphisms (SNPs) in 363 African American control subjects, 380 subjects with type 2 diabetes and end-stage renal disease (DM-ESRD), and 326 subjects with ESRD unrelated to diabetes (non–DM-ESRD). Using human cell lines, we correlated disease-associated ENTPD1 haplotypes with ENTPD1 gene expression. Finally, we studied consequences of ENTPD1 deletion in a mouse model of type 2 diabetes (db/db).
RESULTS
A common ENTPD1 two-SNP haplotype was associated with increased risk for DM-ESRD (P = 0.0027), and an uncommon four-SNP haplotype was associated with protection against DM-ESRD (P = 0.004). These haplotypes correlated with ENTPD1 gene expression levels in human cell lines in vitro. Subjects with high ENTPD1-expressing haplotypes were enriched in the DM-ESRD group. By crossing ENTPD1-null mice with db mice, we show that ENTPD1 deletion has prominent effects on metabolic syndrome traits. Specifically, deletion of ENTPD1 lowered glucose levels in control (db/−) mice with one functional leptin receptor and dramatically lowered weights in db/db mice with no functional leptin receptors. Similar effects were seen in aged ENTPD1-null mice with normal leptin receptors.
CONCLUSIONS
ENTPD1 polymorphisms appear to influence susceptibility to type 2 diabetes and/or diabetic nephropathy in African Americans. Studies in human cell lines and in vivo mouse data support a potential role for ENTPD1 genetic variation in susceptibility to type 2 diabetes.
Diabetes and its complications are enormous sources of mortality, morbidity, and cost in the U.S. African Americans are especially prone to diabetes and diabetic kidney disease (1). Genetic variation plays a major role in susceptibility to both development of diabetes and onset of diabetic renal injury (2,3). Finding genes responsible for this predisposition to disease could help predict who may be at high risk and also identify important pathways involved in disease pathogenesis.
ENTPD1, also known as CD39, is an ectonucleotidase that hydrolyzes ATP and ADP to AMP, initiating an enzymatic cascade that leads to the generation of adenosine (4). By regulating nucleotide levels, ENTPD1 controls the activity of both purinergic receptors (P2X and P2Y) and downstream adenosine (P1) receptors. ENTPD1 is widely expressed, particularly in blood vessels and on cells of the immune system (5–7). ENTPD1 is present in glomeruli, afferent arterioles, and larger vessels of the kidney (8,9). Mice null for ENTPD1 sustained markedly more severe renal injury than wild-type mice in models of type 1 diabetes (9) and ischemia reperfusion injury (10).
Previously, a locus for susceptibility to diabetic and nondiabetic renal disease was located on human chromosome 10q24 in African Americans (11). Subsequently, others have also found loci in this chromosomal region associated with renal dysfunction in Caucasians with type 2 diabetes (12). It appears likely that one or more genes with effects on nephropathy are located in this region. ENTPD1 is located close to the linkage peak in both of these studies. More recently, we have reported that ENTPD1-null mice also exhibit abnormalities of glucose homeostasis (13). The close functional relationship between ENTPD1 and another ectonucleotidase that influences metabolic syndrome traits in humans, ENPP1, (14) suggests that ENTPD1 could affect glucose homeostasis in humans as well.
To determine whether variation in ENTPD1 might influence the development of diabetes or diabetic renal disease, we tested the frequency of single nucleotide polymorphisms (SNPs) in ENTPD1 in three groups of African American patients: 363 healthy control subjects, 326 patients with nondiabetic end-stage renal disease (non–DM-ESRD), and 380 patients with ESRD caused by type 2 diabetes (DM-ESRD). We support these genetic association results with functional studies in vitro using human cell lines and in vivo using mouse models with nonfunctional leptin receptors (db/db). We show that ENTPD1 gene expression in human cell lines correlates with the disease-associated genotypes, and mice null for ENTPD1 share metabolic phenotypes with humans who have low ENTPD1-expressing genotypes.
RESEARCH DESIGN AND METHODS
Collection of control and case subjects has been described previously (15,16). Briefly, all subjects were recruited in North Carolina, South Carolina, Georgia, Virginia, or Tennessee, and all were born in one of these five southeastern states. A total of 363 unrelated African American control subjects were recruited through community screening events, all without a history of type 2 diabetes or renal disease. A total of 380 unrelated African American patients with type 2 diabetes and ESRD were recruited (DM-ESRD group) from dialysis clinics. A total of 326 unrelated African American patients with ESRD but without a history of diabetes (non–DM-ESRD) were also recruited from dialysis clinics. A single investigator (B.I.F.) classified all ESRD patients into DM-ESRD or non–DM-ESRD groups. Non–DM-ESRD patients included those with hypertension-associated ESRD or primary glomerular diseases. The DM-ESRD groups included patients with either 1) diabetes and histologic evidence of diabetic nephropathy or 2) with diabetes ≥5 years before the initiation of renal replacement therapy and either diabetic retinopathy or proteinuria >500 mg per 24 h. DM-ESRD patients had no other known cause of renal disease. Patients with renal cystic diseases, hereditary nephritis, or ESRD related to urologic disease were all excluded from analysis.
Genotyping.
A total of 18 SNPs tagging 80% of the known SNPs in the Yoruba (YRI) Hapmap population were selected using the program Tagger (www.broad.mit.edu/mpg/tagger). Tag SNPs all had minor allele frequency of ≥5%. rs3793744, the only common nonsynonymous coding variant in ENTPD1, was force inserted in SNP selection. Tag SNPs spanned the ENTPD1 gene on chromosome 10 from 97.504 to 97.613 Mb. We used a Sequenom MassArray platform (17) for genotyping.
Genetic analysis.
SNPs with call rates <96% were excluded from analysis. All SNPs were in conformation with Hardy-Weinburg equilibrium (P < 0.05) in control subjects. Determination of haploblock structures and associations were performed using Haploview (www.broad.mit.edu/mpg/haploview). Haplotypes with frequency >1% were included in the analysis. Permutation testing for haplotype associations was performed using Haploview software at 10,000 permutations.
In vitro ENTPD1 expression studies.
Genotyped, Epstein-Barr virus–transformed lymphoblasts from the YRI HapMap (www.hapmap.org) collection and Perlegen (www.perlegen.com) African American panel were used for analysis. Cells were grown in RPMI-1640 media with 2 mmol/l l-glutamine, 15% fetal bovine serum, penicillin-streptomycin mix, and 10 mmol/l Hepes buffer. Cells were seeded at 200,000 per ml and harvested 24–48 h later in log-phase growth. Then, mRNA was isolated using RNeasy (Qiagen) according to the manufacturer's instructions. A total of 0.5 μg of mRNA was reversed transcribed to cDNA using the Taqman Reverse Transcription Kit (Applied Biosystems). Probe-primer sets for ENTPD1 and the 18S ribosomal subunit (loading control) were obtained from Applied Biosystems. Real-time PCR was performed on an Applied Biosystems 7700 system. ENTPD1 mRNA expression was normalized using 18S expression.
For protein isolation, cells were washed twice with ice-cold PBS and then lysed in an NP-40–based buffer with protease inhibitors (Roche mini-tabs). A total of 40 μg of protein were separated on a gradient gel under nonreducing condtions and transferred to polyvinylidene fluoride paper (Millipore) using a semidry transfer apparatus. For ENTPD1, we used a mouse monoclonal antibody (BU-61; Ancell) and a goat anti-mouse secondary antibody (Pierce). We used anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (Ambion) as a loading control. Band density was quantified with a Kodak ImageStation 2000M, and protein level was expressed as a ratio of ENTPD1 to GAPDH.
In silico, expanded ENTPD1 expression studies.
For YRI HapMap subjects, ENTPD1 gene expression levels were downloaded from (http://www.sanger.ac.uk/humgen/genevar/). Methods for the generation of this data are described by Stranger et al. (18). Genotypes were obtained from www.hapmap.org. Statistics were performed using Student's t test on log2-transformed data.
Enzyme activity.
A total of 250,000 lymphocytes were harvested in log-phase growth. ATPase and ADPase activity were determined with a Malachite Green assay for phosphate production as described previously using 200 μmol/l nucleotide as the substrate during a 10-min incubation (19). Tetramisole was used to inhibit alkaline phosphatases (19).
Animal studies.
All mouse studies were done with institutional animal care and use committee approval at Beth Israel Deaconess Medical Center. Mice in all four db groups originated from crossing db/− heterozygote breeders on a C57/BL6 background from Jackson Labs with ENTPD1−/− breeders backcrossed more than six times on a C57/BL6 background. Diabetic mice were db/db offspring with or without ENTPD1, and db/− offspring were used as controls. In the aging study, male ENTPD1−/− mice backcrossed more than six times onto a C57/BL6 background were compared with age- and strain-matched C57/BL6 wild-type controls.
Animal phenotyping.
Albuminuria was quantified using an albumin-to-creatinine ratio. Albumin was measured using the Albuwell-M Kit from Exocell, and creatinine was measured using the Creatinine Companion from Exocell. Blood glucose was measured using a One Touch Ultra 2 glucometer (Lifescan). For the db-based studies, nonfasting glucose levels were measured at 3:00 p.m. For the non-db studies (aging), glucose levels were measured after an 8-h daytime fast.
For histopathology, mouse kidney tissue was fixed in 10% buffered formalin and embedded in paraffin then cut into 3-micron sections and stained with PAS stain. Glomerular area was calculated using Spot Imaging software. The glomerular sclerosis index has been described previously (9).
RESULTS
Using a set of markers chosen with the program Tagger, we genotyped 18 SNPs (minor allele frequency >5%) in ENTPD1 that were predicted to cover 80% of the known SNPs in the Yoruba (YRI) Hapmap population. Seventeen of 18 SNPs passed quality standards and conformed to Hardy-Weinburg equilibrium criteria as described in research design and methods above. These 17 SNPs tagged 79% of YRI HapMap SNPs.
Location of SNPs in the ENTPD1 gene, minor allele frequency of each SNP, and uncorrected P values in the populations in this study are shown in Table 1. When control subjects and subjects with non–DM-ESRD were compared, one SNP (rs6584026) showed nominal significance (P = 0.027). Comparing control subjects and subjects with DM-ESRD revealed two SNPS (rs12763743 and rs3897983) that were nominally significant (0.041 and 0.037, respectively).
TABLE 1.
SNP analysis
| SNPs | Location on chromosome 10 (MBp) | Location within ENTPD1 | Major/minor allele (+) strand | Healthy control subjects (MAF) | Non–DM-ESRD case subjects (MAF) | Type 2 diabetic ESRD case subjects (MAF) | Control vs. non–DM-ESRD (P) | Odds ratio for non–DM-ESRD | Control vs. type 2 diabetic ESRD (P) | Odds ratio for DM-ESRD |
|---|---|---|---|---|---|---|---|---|---|---|
| rs3176894 | 97504449 | Promoter | A/G | * | * | * | * | * | * | * |
| rs12262943 | 97506434 | Intron | T/C | 0.106 | 0.134 | 0.121 | 0.112 | 1.30 (0.94–1.81) | 0.378 | 1.16 (0.84–1.60) |
| rs6584026 | 97530149 | Intron | C/T | 0.270 | 0.325 | 0.303 | 0.027† | 1.30 (1.03–1.65) | 0.157 | 1.18 (0.94–1.48) |
| rs10882665 | 97541718 | Intron | C/A | 0.360 | 0.398 | 0.382 | 0.154 | 1.17 (0.94–1.46) | 0.391 | 1.10 (0.89–1.36) |
| rs6584028 | 97548769 | Intron | T/C | 0.262 | 0.310 | 0.298 | 0.051 | 1.27 (0.99–1.61) | 0.124 | 1.20 (0.95–1.50) |
| rs7093453 | 97561565 | Intron | C/T | 0.096 | 0.085 | 0.088 | 0.484 | 0.88 (0.60–1.27) | 0.599 | 0.91 (0.64–1.30) |
| rs4918972 | 97569211 | Intron | G/A | 0.143 | 0.116 | 0.136 | 0.142 | 0.79 (0.57–1.08) | 0.694 | 0.94 (0.70–1.27) |
| rs10748648 | 97576857 | Intron | T/A | 0.457 | 0.471 | 0.450 | 0.613 | 1.06 (0.85–1.31) | 0.805 | 0.97 (0.79–1.20) |
| rs12763743 | 97582146 | Intron | A/T | 0.077 | 0.097 | 0.051 | 0.192 | 1.29 (0.88–1.89) | 0.041† | 0.64 (0.42–0.99) |
| rs1342790 | 97590529 | Intron | A/G | 0.474 | 0.484 | 0.478 | 0.703 | 0.84 (0.68–1.05) | 0.873 | 0.98 (0.80–1.21) |
| rs3176883 | 97594931 | Intron | T/C | 0.247 | 0.287 | 0.288 | 0.104 | 1.23 (0.96–1.57) | 0.087 | 1.23 (0.97–1.56) |
| rs12244606 | 97596321 | Intron | C/T | 0.068 | 0.092 | 0.084 | 0.094 | 1.40 (0.94–2.08) | 0.239 | 1.26 (0.86–1.87) |
| rs3181123 | 97597065 | Intron | C/T | 0.349 | 0.377 | 0.370 | 0.280 | 1.13 (0.91–1.41) | 0.395 | 1.10 (0.89–1.36) |
| rs3793744 | 97597256 | Exon 7 (V-I) | G/A | 0.043 | 0.050 | 0.029 | 0.600 | 1.15 (0.69–1.92) | 0.146 | 0.66 (0.38–1.16) |
| rs3897983 | 97608187 | Intron | G/A | 0.125 | 0.104 | 0.091 | 0.223 | 0.81 (0.58–1.14) | 0.037† | 0.70 (0.50–0.98) |
| rs3181121 | 97608570 | Intron | T/C | 0.096 | 0.088 | 0.086 | 0.612 | 0.90 (0.63–1.32) | 0.504 | 0.89 (0.62–1.27) |
| rs11598475 | 97611256 | Intron | G/A | 0.132 | 0.119 | 0.135 | 0.448 | 0.88 (0.64–1.22) | 0.897 | 1.02 (0.75–1.38) |
| rs3181118 | 97613366 | Intron | C/T | 0.032 | 0.029 | 0.027 | 0.725 | 0.89 (0.48–1.67) | 0.555 | 0.83 (0.45–1.53) |
Individual SNP results for the association study comparing African American control subjects with type 2 diabetic ESRD and nondiabetoc ESRD. *rs3176894 did not meet quality control criteria.
†P < 0.05 (boldface). When the odds ratio is >1, the minor allele is the risk allele; when the odds ratio is <1, the minor allele is the protective allele. MAF, minor allele frequency.
We considered the possibility that the two nominally significant SNPs for DM-ESRD might have additive effects. We used the two significant SNPs discovered in the DM-ESRD population to form a two-SNP haplotype for further analysis. Three common combinations accounted for all subjects tested (AG = 82.9%, AA = 10.8%, and TG = 6.3%). The most common haplotype (AG) appeared to confer increased risk of diabetes and/or diabetic nephropathy (P = 0.0027, P = 0.009 after permutation; odds ratio 1.51) compared with the other two haplotypes, which were both nominally associated with protection from diabetes and/or diabetic nephropathy (Table 2). We refer to the AG haplotype as the two-SNP risk haplotype, and the combined group of AA and TG haplotypes as the two-SNP protective haplotypes. No difference was seen with respect to these two-SNP risk or protective haplotypes when comparing control subjects with non–DM-ESRD subjects.
TABLE 2.
Haplotypes
| Haplotype | Frequency | Control subjects | Case subjects | χ2 | P | Odds ratio (95% CI) |
|---|---|---|---|---|---|---|
| Two-SNP haplotypes | ||||||
| AG | 0.829 | 0.799 | 0.857 | 9.0 | 0.0027 | 1.51 (1.15–1.99) |
| AA | 0.108 | 0.124 | 0.091 | 4.2 | 0.0404 | 0.70 (0.51–0.98) |
| TG | 0.063 | 0.077 | 0.051 | 4.1 | 0.0431 | 0.64 (0.42–0.98) |
| Four-SNP haplotypes | ||||||
| GAGG | 0.432 | 0.428 | 0.436 | 0.10 | 0.7495 | 1.03 (0.84–1.27) |
| GTAG | 0.393 | 0.386 | 0.398 | 0.23 | 0.6323 | 1.05 (0.85–1.30) |
| ATAG | 0.139 | 0.141 | 0.138 | 0.04 | 0.8336 | 0.97 (0.72–1.30) |
| GAGA | 0.023 | 0.035 | 0.012 | 8.26 | 0.0041 | 0.34 (0.16–0.73) |
| GTGA | 0.013 | 0.010 | 0.016 | 0.09 | 0.3259 | 1.67 (0.65–4.26) |
Association of DM-ESRD with two-SNP haplotypes consisting of the two nominally significant SNPs rs12763743 and rs3897983. The AG haplotype is referred to as the two-SNP risk haplotype, and the AA and TG haplotypes are referred to as the two-SNP protective haplotypes. Association of DM-ESRD with four-SNP haplotypes consisting of rs4918972, rs10748648, rs1342790, and rs3793744. The GAGA haplotype is referred to as the four-SNP protective haplotype.
A broader, unbiased haplotype analysis using all successfully genotyped SNPs showed that 16 of the SNPs fell into a single haplotype block defining 10 haplotypes with frequency >1%. One haplotype was associated with protection against diabetes and/or diabetic nephropathy with P = 0.004 (odds ratio 0.34). This statistical significance of this protective haplotype remained unchanged when reduced to four SNPs (rs4918972, rs10748648, rs1342790, and rs3793744; GAGA in Table 2). After permutation analysis, the P value remained significant at 0.017. The GAGA haplotype is referred to as the four-SNP protective haplotype. This protective haplotype included the uncommon variant rs3793744, a coding SNP in exon 7 causing a valine to isoleucine substitution. No association was seen for this haplotype when comparing control subjects to non–DM-ESRD subjects.
We made several additional comparisons to help define whether our haplotypes were associated with diabetes or diabetic nephropathy. First, we compared healthy control subjects with all ESRD subjects (both DM-ESRD and non–DM-ESRD) and found much weaker associations for both the two-SNP and four-SNP haplotypes (supplementary Table 1 [available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db08-1214/DC1]). Then we compared non–DM-ESRD with DM-ESRD subjects: the two-SNP (AG) risk haplotype remained robust (P = 0.0038, odds ratio 1.53), while the four-SNP (GAGA) protective haplotype lost some statistical power (P = 0.024), though the odds ratio was unchanged (odds ratio 0.37) when non-ESRD subjects were substituted for control subjects. We also pooled healthy control subjects and non–DM-ESRD subjects into a nondiabetic group and compared them with DM-ESRD subjects. Here, the P value of the two-SNP (AG) risk haplotype improved to 0.0008 (odds ratio 1.51), while the four-SNP (GAGA) protective haplotype was similar to the control versus DM-ESRD comparison (P = 0.0056, odds ratio 0.37). Taken together, these analyses tend to support a genetic association with diabetes rather than diabetic nephropathy.
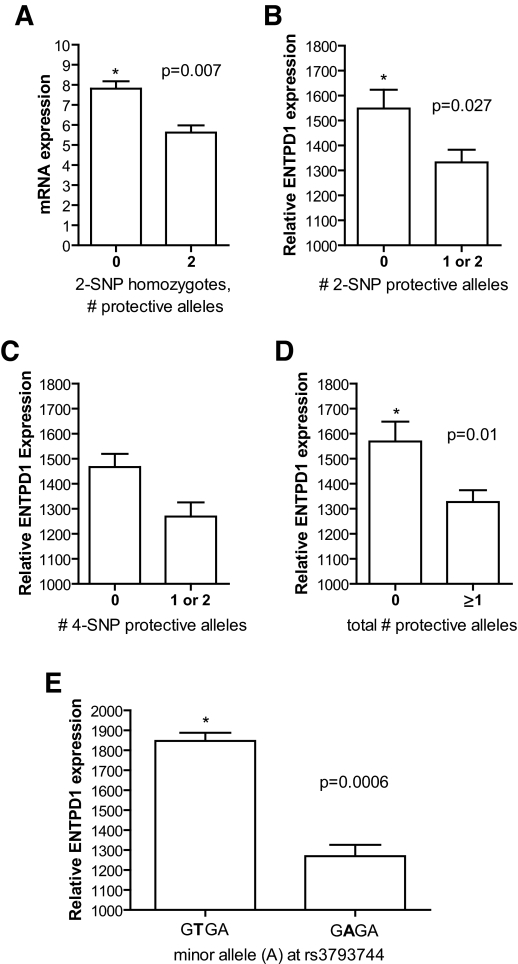
To determine whether either of these disease-associated haplotypes correlated with general expression levels of ENTPD1, we took advantage of the fact that ENTPD1 is highly expressed on B-lymphocytes (6). For the two-SNP haplotype, we examined YRI Hapmap lymphoblasts for subjects who were homozygous for the three different haplotype groups. While many subjects in the YRI population were homozygous for the AG risk haplotype, only two were homozygous for the AA protective haplotype and only one was homozygous for the TG protective haplotype. Since the AA and TG haplotypes were almost identical in terms of risk (P = 0.04, nominally protective) and odds ratios (0.70 and 0.64, respectively), we pooled these into a single group and compared ENTPD1 expression from their lymphocytes with those from the risk (AG) haplotype group using real-time PCR (Fig. 1A). Lymphocytes homozygous for the AG risk haplotype had 39% higher ENTPD1 expression levels than homozyogotes for the protective AA or TG haplotypes (protective 5.62 ± 0.36 vs. risk 7.81 ± 0.36; P = 0.007), with no overlap between groups (range 5.09–6.30 for protective and 6.91–9.53 for risk). Not enough cell lines homozygous for the four-SNP protective haplotype (one cell line) were available for comparison.
FIG. 1.
A: ENTPD1 mRNA expression by real-time PCR comparing YRI Hapmap cell lines homozygous for the two-SNP risk haplotype AG (n = 7) versus the protective haplotypes AA and TG (n = 3), normalized for 18S expression. ENTPD1 mRNA levels are 39% higher in cell lines with zero protective haplotypes than in cell lines with two protective haplotypes. B: In silico ENTPD1 expression for YRI cell lines with zero (n = 31) versus one or two (n = 29) copies of the two-SNP protective haplotypes AA and TG. Zero copies of the protective haplotype conferred 16% higher levels of ENTPD1 expression. C: ENTPD1 expression for YRI cell lines with zero (n = 53) versus one or two (n = 7) copies of the four-SNP protective (GAGA) haplotype. Zero copies of the protective haplotype were associated with 16% higher ENTPD1 expression. Though nonsignificant, cell lines with one or two protective copies had lower expression than all other groups tested. D: Protective index incorporating both the two- and four-SNP haplotypes. The total number of protective two- and four-SNP protective alleles was summed for each cell line. Cell lines with zero protective alleles (n = 29) had 18% more ENTPD1 expression than cell lines with at least one protective allele (n = 31). E: Comparison of ENTPD1 mRNA expression of protective four-SNP haplotype (GAGA, n = 7) and uncommon four-SNP haplotype (GTGA, n = 3) that share the minor allele (A) at rs3793744 that codes for a valine to isoleucine substitution at amino acid 293. The difference indicates that rs3793744 is not itself the causal SNP.
To provide additonal support for a correlation between haplotype and expression level, we used gene expression data from Stranger et al. (18) generated from YRI HapMap cell lines, which allowed us to incorporate haplotype heterozygotes into the analysis. For consistency, we analyze the two- and four-SNP haplotypes in terms of number of protective alleles. We pooled subjects with one or two protective alleles for the two-SNP haplotype and compared them with subjects with no protective alleles (two risk alleles). Subjects with zero protective haplotypes (n = 31) expressed 16% more ENTPD1 than subjects with one or two protective haplotypes (n = 29; P = 0.027) (Fig. 1B). We then pooled subjects with one or two copies of the protective four-SNP allele (n = 7) and compared them with those with no copies (n = 53). While the difference was not significant (P = 0.09), those with no protective alleles expressed 16% more ENTPD1 than those with one or two protective alleles (Fig. 1C). These cell lines with at least one protective four-SNP allele had the lowest mRNA expression levels of any haplotype group, including the protective two-SNP haplotype. To account for the fact that we were comparing subjects with protective four-SNP alleles versus a pool that contained many more subjects with one or more protective two-SNP allele, we then compared subjects based on their total number of protective alleles (two-SNP protective alleles plus four-SNP protective alleles) to integrate the effects of two-SNP and four-SNP haplotypes on ENTPD1 expression. Those with no protective alleles (n = 29) expressed 18% more ENTPD1 than subjects with at least one protective two-SNP allele or one protective four-SNP allele (n = 31) (P = 0.01). Both haplotypes appear to correlate with ENTPD1 expression and were especially predictive of mRNA expression levels when used in tandem. The small number of subjects with the protective four-SNP allele prevented us from determining whether the effects of the two-SNP and four-SNP haplotypes were independent.
The protective four-SNP haplotype (GAGA) included a SNP causing an amino acid change (rs3793744: V to I at position 293). A second, uncommon haplotype (GTGA) also included the minor allele (A) at rs3793744 but was not associated with DM-ESRD when compared with control subjects. We compared expression levels between cell lines with one or more copies of each of these haplotypes. Remarkably, the nonprotective (GTGA) four-SNP haplotype including the minor allele at rs3793744 expressed 45% more ENTPD1 mRNA than the protective (GAGA) four-SNP allele with the same SNP at rs3793744 (n = 3 vs. n = 7, P = 0.0006). This finding suggests that differences in mRNA expression underly the effect of the four-SNP haplotype rather than the amino acid substitution. We did note, though, that protein expression in the homozygotes for the protective four-SNP haplotype were surprisingly low compared with other cell lines (supplementary Fig. 1A and B). This finding was demonstrated in single samples from two separate collections (HapMap and Perlegen), and activity assays showed that the effect was not due to differential antibody binding at the amino acid change at position 293 (rs3793744) (supplementary Fig. 1C and D).
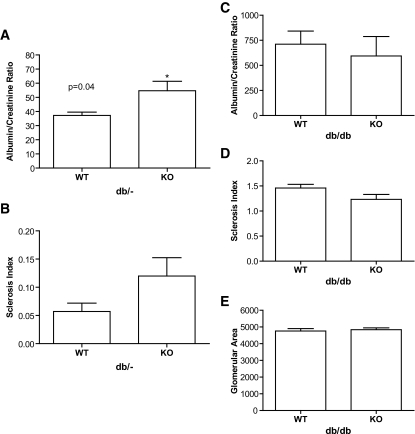
To further investigate the effects of ENTPD1 on either diabetes or diabetic nephropathy, we crossed ENTPD1-null mice with db/db mice to create a type 2 model of diabetes and type 2 diabetic nephropathy. Assessment for renal injury at 8 months of age demonstrated that in the nondiabetic mice (db/−), ENTPD1 deletion caused mildly increased albuminuria (wild type = 37.2 ± 2.3 μg/mg vs. knockout = 54.7 ± 6.7 μg/mg; P = 0.028) and a trend toward increased glomerular sclerosis (Fig. 2A and B). In the diabetic groups, there were no differences in albuminuria, sclerosis index, or glomerular area between db/db mice with and without ENTPD1 (Fig. 2C–E).
FIG. 2.
A: Albumin-to-creatinine ratio for db/− nondiabetic mice. n = 7 for ENTPD1 wild type and n = 12 for ENTPD1 knockout. B: Sclerosis index for db/− nondiabetic mice. n = 7 for ENTPD1 wild type and n = 12 for ENTPD1 knockout. C: Albumin-to-creatinine ratio for db/db mice. n = 13 for ENTPD1 wild type and n = 9 for ENTPD1 knockout. D: Sclerosis index for db/db mice. n = 13 for ENTPD1 wild type and n = 10 for ENTPD1 knockout. E: Glomerular area for db/db mice. n = 10 for ENTPD1 wild type and n = 8 for ENTPD1 knockout.
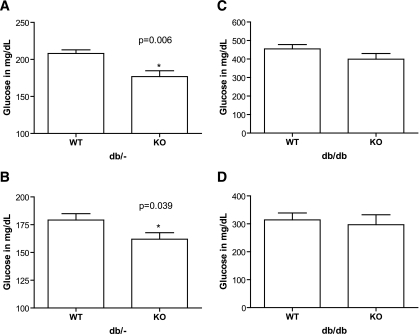
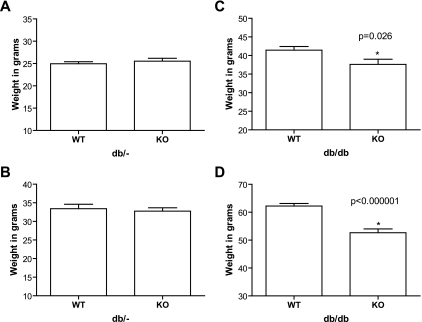
Metabolic syndrome parameters revealed more dramatic differences. In nondiabetic mice, nonfasting (3:00 p.m.) glucose was lower in ENTPD1-null mice at both 8 weeks (wild type = 208.1 ± 4.8 mg/dl vs. knockout = 176.8 ± 7.8 mg/dl; P = 0.006) and 6 months (wild type = 179.2 ± 5.7 mg/dl vs. knockout = 161.9 ± 5.8 mg/dl; P = 0.039) (Fig. 3A and B). No differences in glucose levels were seen in the db/db groups at these time points with or without ENTPD1 (Fig. 3C and D). While weights of nondiabetic mice were no different (Fig. 4A and B), large differences were seen in db/db mice, with deletion of ENTPD1 protecting against obesity in the db/db mice at both 8 weeks (wild type = 41.4 ± 1.0 g vs. knockout = 37.6 ± 1.4 g; P = 0.026) and 6 months (wild type = 62.4 ± 0.9 g vs. knockout = 52.0 ± 1.3 g; P < 0.000001) (Fig. 4C and D).
FIG. 3.
A and B: Blood glucose levels for db/− nondiabetic mice at 8 weeks (A) and 6 months (B). n = 9 for ENTPD1 wild type and n = 12 for ENTPD1 knockout. C and D: Blood glucose levels for db/db mice at 8 weeks (C) and 6 months (D). n = 19–20 for ENTPD1 wild type and n = 12–13 for ENTPD1 knockout.
FIG. 4.
A and B: Weights for db/− nondiabetic mice at 8 weeks (A) and 6 months (B). n = 9 for ENTPD1 wild type and n = 12 for ENTPD1 knockout. C and D: Weights for db/db mice at 8 weeks (C) and 6 months (D). n = 19–20 for ENTPD1 wild type and n = 13–15 for ENTPD1 knockout.
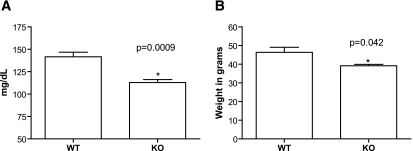
We also aged wild-type and ENTPD1-null mice (leptin receptor normal) to see if aging would cause similar metabolic differences as we saw in the db/db model. At 2 years of age, ENTPD1-null mice had markedly lower fasting glucoses (wild type = 141.6 ± 5.1 mg/dl vs. knockout = 112.8 ± 3.3 mg/dl; P = 0.0009) and lower weights (wild type = 46.4 ± 2.7 g vs. knockout = 39.2 ± 0.7; P = 0.042) than wild type controls (Fig. 5A and B). Consistent with the db experiments, we did also note increased albuminuria in aged ENTPD1-null mice despite lower weights and blood glucoses (D.J.F., unpublished data).
FIG. 5.
A: Fasting glucose levels in mg/dl for 2-year-old wild-type (n = 8) and ENTPD1-null (n = 6) mice. B: Weight at 2 years in wild-type (n = 7) and ENTPD1-null (n = 11) mice.
DISCUSSION
Our previous studies in ENTPD1-null mice suggested a role for this ectonucleotidase in both resistance to renal injury and in glucose homeostasis. We tested polymorphic markers in the ENTPD1 gene in humans for association with diabetic ESRD and nondiabetic ESRD in African Americans, a group of Americans with high rates of both diseases. We found two common polymorphisms (rs12763743 and rs3897983) that are nominally associated with diabetic but not nondiabetic ESRD. When used together, these two polymorphisms showed even stronger association (P = 0.0027) with subjects in the diabetic nephropathy group. These two SNPs may represent independent, additive effects or they may together tag a causal SNP better than either of the SNPs alone. More general haplotype analysis also identified an uncommon four-SNP haplotype that appears to protect against diabetes or diabetic nephropathy (P = 0.004). The effect size of this uncommon haplotype is strong, conferring a relative risk of 0.34. Additional groupwise comparisons using pooled groups further suggested that the association was due to diabetes rather than nephropathy in the DM-ESRD groups.
To support association the association of ENTPD1 with diabetes and/or diabetic nephropathy, we tested HapMap cell lines derived from African subjects for ENTPD1 expression. Cell lines homozygous for the two-SNP (AG) risk haplotype expressed 39% more ENTPD1 mRNA than those cell lines with two protective haplotypes (AA and TG). We performed additional bioinformatic analysis that showed similar results: zero alleles of the two-SNP protective haplotypes led to 16% greater ENTPD1 expression than one or two protective alleles. The protective four-SNP haplotype also showed a trend toward lower ENTPD1 expression levels, though this analysis was limited by fewer representative cell lines and was confounded by comparison with a group including many protective two-SNP haplotypes. In fact, the protective four-SNP haplotype showed the lowest ENTPD1 expression of any group. By using an index that took into account the total number of both the two- and four-SNP protective haplotypes, we identified a strong correlation between genotype and ENTPD1 mRNA expression level using both haplotypes derived from our genetic association study. Additional experiments measuring ENTPD1 protein levels confirmed that the protective four-SNP haplotype was associated with very low ENTPD1 levels and may suggest an effect beyond mRNA expression, though we did not have sufficient samples available to us to prove this hypothesis definitively.
We also used genetic studies in mice to assess the role of ENTPD1 on diabetes and diabetic nephropathy. While ENTPD1-null control mice did have increased albuminuria compared with wild-type mice, perhaps the more striking findings related to metabolic syndrome traits. In particular, ENTPD1-null mice had lower glucose levels in the absence of the homozygous db/db mutation and dramatically lower weights in the presence of the db/db mutation. Notably, previous work demonstrated that ENTPD1-null mice have higher leptin levels than wild-type mice (13), potentially a compensatory response that is disabled in the absence of one or both copies of the normal leptin receptor. The exact reason for this differential effect in db/db and db/− mice on blood glucose and weight still remains to be elucidated. However, even in the presence of normal leptin signaling, using wild-type mice rather than db/− heterozygotes as controls, aging accentuated both phenotypes (i.e., difference in glucose homeostasis and weight gain). Our mouse findings again suggest that the effect we discovered in humans may relate to low ENTPD1 protecting against diabetes in control subjects, though increased albuminuria in the nondiabetic CD39-null mice may point toward an effect on renal disease as well.
Further support for association between ENTPD1 and diabetes and diabetic nephropathy is the finding that polymorphisms in another ectonucleotidase with a remarkably similar enzymatic mechanism, ENPP1, have also been associated with both diabetes and diabetic nephropathy. Nephropathy has been associated with ENPP1 polymorphisms, first in Caucasian populations (20–22), and subsequently in an African American population that included our subjects (23). Polymorphisms in the same gene also have an important influence on metabolic syndrome traits (14), and there appears to be an important interaction between weight and glucose homeostasis (24,25), just as we see with ENTPD1 in mice. While ENPP1 hydrolyzes extracellular ATP to AMP and pyrophosphate, ENTPD1 hydrolyzes extracellular ATP to AMP and two phosphate molecules. Low functional expression of either of these ectoenzymes could increase extracellular levels of ATP and decrease levels of the nucleoside adenosine.
The strength of our study rests on mutually supportive data built from genetic association of human disease with genetic variants, gene expression levels that correlate with the disease-associated haplotypes, and a mouse model that shows phenotypic differences based on expression level of the gene. Ultimately, replication in larger cohorts will be needed to validate our findings. Our data, along with previous studies with ENPP1, suggest that extracellular nucleotide metabolism may be a critical determinant of diabetes and/or diabetic nephropathy.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants K08-DK-076868 (to D.J.F.) and P01 HL-076540 (to S.C.R). We acknowledge Coriell Institute, Perlegen, and the National Human Genome Research Institute (NHGRI) for their role in providing human cell lines to the research community.
No potential conflicts of interest relevant to this article were reported.
We thank Barbara Stranger, Emmanouil Dermitzakis, and their colleagues for making their gene expression data publicly available.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Satko SG, Freedman BI, Moossavi S: Genetic factors in end-stage renal disease. Kidney Int Suppl S46– S49, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Freedman BI, Bostrom M, Daeihagh P, Bowden DW: Genetic factors in diabetic nephropathy. Clin J Am Soc Nephrol 2: 1306– 1316, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Florez JC, Hirschhorn J, Altshuler D: The inherited basis of diabetes mellitus: implications for the genetic analysis of complex traits. Annu Rev Genomics Hum Genet 4: 257– 291, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann HBN, Heine P, Kohring K, Marxen M, Sevigny J, Robson SC: Ecto-ATPases and Related Nucleotidases Maastrict, Netherlands, Shaker Publishing, 2000 [Google Scholar]
- 5.Sevigny J, Sundberg C, Braun N, Guckelberger O, Csizmadia E, Qawi I, Imai M, Zimmermann H, Robson SC: Differential catalytic properties and vascular topography of murine nucleoside triphosphate diphosphohydrolase 1 (NTPDase1) and NTPDase2 have implications for thromboregulation. Blood 99: 2801– 2809, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Maliszewski CR, Delespesse GJ, Schoenborn MA, Armitage RJ, Fanslow WC, Nakajima T, Baker E, Sutherland GR, Poindexter K, Birks C, et al. : The CD39 lymphoid cell activation antigen. Molecular cloning and structural characterization. J Immunol 153: 3574– 3583, 1994 [PubMed] [Google Scholar]
- 7.Mizumoto N, Kumamoto T, Robson SC, Sevigny J, Matsue H, Enjyoji K, Takashima A: CD39 is the dominant Langerhans cell-associated ecto-NTPDase: modulatory roles in inflammation and immune responsiveness. Nat Med 8: 358– 365, 2002 [DOI] [PubMed] [Google Scholar]
- 8.Kishore BK, Isaac J, Fausther M, Tripp SR, Shi H, Gill PS, Braun N, Zimmermann H, Sevigny J, Robson SC: Expression of NTPDase1 and NTPDase2 in murine kidney: relevance to regulation of P2 receptor signaling. Am J Physiol Renal Physiol 288: F1032– F1043, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Friedman DJ, Rennke HG, Csizmadia E, Enjyoji K, Robson SC: The vascular ectonucleotidase ENTPD1 is a novel renoprotective factor in diabetic nephropathy. Diabetes 56: 2371– 2379, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Grenz A, Zhang H, Hermes M, Eckle T, Klingel K, Huang DY, Muller CE, Robson SC, Osswald H, Eltzschig HK: Contribution of E-NTPDase1 (CD39) to renal protection from ischemia-reperfusion injury. FASEB J 21: 2863– 2873, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Freedman BI, Rich SS, Yu H, Roh BH, Bowden DW: Linkage heterogeneity of end-stage renal disease on human chromosome 10. Kidney Int 62: 770– 774, 2002 [DOI] [PubMed] [Google Scholar]
- 12.Placha G, Poznik GD, Dunn J, Smiles A, Krolewski B, Glew T, Puppala S, Schneider J, Rogus JJ, Rich SS, Duggirala R, Warram JH, Krolewski AS: A genome-wide linkage scan for genes controlling variation in renal function estimated by serum cystatin C levels in extended families with type 2 diabetes. Diabetes 55: 3358– 3365, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Enjyoji K, Ko K, Thukral C, Blumel B, Sun X, Wu Y, Imai M, Friedman D, Csizmadia E, Bleibel W, Kahn BB, Robson SC: Deletion of Cd39/Entpd1 results in hepatic insulin resistance. Diabetes 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyre D, Bouatia-Naji N, Tounian A, Samson C, Lecoeur C, Vatin V, Ghoussaini M, Wachter C, Hercberg S, Charpentier G, Patsch W, Pattou F, Charles MA, Tounian P, Clement K, Jouret B, Weill J, Maddux BA, Goldfine ID, Walley A, Boutin P, Dina C, Froguel P: Variants of ENPP1 are associated with childhood and adult obesity and increase the risk of glucose intolerance and type 2 diabetes. Nat Genet 37: 863– 867, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sale MM, Smith SG, Mychaleckyj JC, Keene KL, Langefeld CD, Leak TS, Hicks PJ, Bowden DW, Rich SS, Freedman BI: Variants of the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in an African-American population enriched for nephropathy. Diabetes 56: 2638– 2642, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Freedman BI, Bowden DW, Rich SS, Valis CJ, Sale MM, Hicks PJ, Langefeld CD: A genome scan for all-cause end-stage renal disease in African Americans. Nephrol Dial Transplant 20: 712– 718, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Gabriel S, Ziaugra L: SNP genotyping using Sequenom MassARRAY 7K platform. Curr Protoc Hum Genet Chapter 2:Unit 2 12, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, Montgomery S, Tavare S, Deloukas P, Dermitzakis ET: Population genomics of human gene expression. Nat Genet 39: 1217– 1224, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, Chen JF, Enjyoji K, Linden J, Oukka M, Kuchroo VK, Strom TB, Robson SC: Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med 204: 1257– 1265, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Cosmo S, Trevisan R, Dalla Vestra M, Vedovato M, Argiolas A, Solini A, Saller A, Damone F, Tiengo A, Trischitta V, Fioretto P: PC-1 amino acid variant Q121 is associated with a lower glomerular filtration rate in type 2 diabetic patients with abnormal albumin excretion rates. Diabetes Care 26: 2898– 2902, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Canani LH, Ng DP, Smiles A, Rogus JJ, Warram JH, Krolewski AS: Polymorphism in ecto-nucleotide pyrophosphatase/phosphodiesterase 1 gene (ENPP1/PC-1) and early development of advanced diabetic nephropathy in type 1 diabetes. Diabetes 51: 1188– 1193, 2002 [DOI] [PubMed] [Google Scholar]
- 22.De Cosmo S, Argiolas A, Miscio G, Thomas S, Piras GP, Trevisan R, Perin PC, Bacci S, Zucaro L, Margaglione M, Frittitta L, Pizzuti A, Tassi V, Viberti GC, Trischitta V: A PC-1 amino acid variant (K121Q) is associated with faster progression of renal disease in patients with type 1 diabetes and albuminuria. Diabetes 49: 521– 524, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Keene KL, Mychaleckyj JC, Smith SG, Leak TS, Perlegas PS, Langefeld CD, Freedman BI, Rich SS, Bowden DW, Sale MM: Association of the distal region of the Ectonucleotide Pyrophosphatase/Phosphodiesterase 1 (ENPP1) gene with type 2 diabetes in an African American population enriched for nephropathy. Diabetes 123: 333– 41, 2008 [DOI] [PubMed] [Google Scholar]
- 24.McAteer JB, Prudente S, Bacci S, Lyon HN, Hirschhorn JN, Trischitta V, Florez JC: The ENPP1 K121Q polymorphism is associated with type 2 diabetes in European populations: evidence from an updated meta-analysis in 42,042 subjects. Diabetes 57: 1125– 1130, 2008 [DOI] [PubMed] [Google Scholar]
- 25.Stolerman ES, Manning AK, McAteer JB, Dupuis J, Fox CS, Cupples LA, Meigs JB, Florez JC: Haplotype structure of the ENPP1 Gene and Nominal Association of the K121Q missense single nucleotide polymorphism with glycemic traits in the Framingham Heart Study. Diabetes 57: 1971– 1977, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.