Abstract
The ability to sense and respond to DNA damage is critical to maintenance of genomic stability and the prevention of cancer. In this study, we employed a genetic screen to identify a gene, NBA1 (new component of the BRCA1 A complex), that is required for resistance to ionizing radiation. The NBA1 protein localizes to sites of DNA damage and is required for G2/M checkpoint control. Proteomic analysis revealed that NBA1 is a component of the BRCA1 A complex, which also contains Brca1/Bard1, Abra1, RAP80, BRCC36, and BRE. NBA1 is required to maintain BRE and Abra1 abundance and for the recruitment of BRCA1 to sites of DNA damage. In depth bioinformatics analysis revealed that the BRCA1 A complex bears striking similarities to the 19S proteasome complex. Furthermore, we show that four members of the BRCA1-A complex possess a polyubiquitin chain-binding capability, thus forming a complex that might facilitate the deubiquitinating activity of the deubiquitination enzyme BRCC36 or the E3 ligase activity of the BRCA1/BARD1 ligase. These findings provide a new perspective from which to view the BRCA1 A complex.
Keywords: Abraxas, BRCA1 A complex, MPN domain, proteasome, VWA domain
Cells have evolved a complex sensory network called the DNA damage response (DDR) to respond to genotoxic stress. The DDR coordinates cell cycle regulation, DNA repair, chromatin reorganization, and transcription in response to damage. Once DNA damage is sensed, a phosphorylation signaling cascade is initiated by the central DNA damage kinases, ATM and ATR (Zhou and Elledge 2000; Harper and Elledge 2007). Recently, a DNA damage-induced ubiquitin signaling pathway has also emerged that works together with phosphorylation-dependent signaling to recruit essential repair factors to the sites of DNA damage (Harper and Elledge 2007). Damage-induced ATM/ATR phosphorylation on S139 of histone H2AX directly recruits MDC1 through MDC1’s BRCT domains. MDC1 itself is a substrate of ATM/ATR, and its phosphorylation leads to the recruitment of an ubiquitin ligase RNF8/UBC13 to damage sites. The subsequent ubiqutination events on the damaged chromatin create docking sites for BRCA1 complexes (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007).
The BRCA1 tumor suppressor is associated with hereditary breast and ovarian cancer and plays critical roles in DNA repair, cell cycle checkpoint control, and maintenance of genomic stability (Venkitaraman 2002; Narod and Foulkes 2004). BRCA1 contains two C-terminal BRCT repeats that bind pSPxF phospho-amino acid motifs and an N-terminal RING domain that binds BARD1, forming an E3 ubiqutin ligase (Wu et al. 1996; Hashizume et al. 2001; Ruffner et al. 2001; Baer and Ludwig 2002). We and others found that BRCA1 forms at least three distinct complexes—the BRCA1 A, BRCA1 B, and BRCA1 C complexes—through association with different adaptor proteins via their pSPxF motifs (Yu and Chen 2004; Greenberg et al. 2006; Wang et al. 2007). The adaptor proteins are Abraxas (Abra1) for the BRCA1 A complex, Bach1/Brip1 for the BRCA1 B complex, and CTIP for the BRCA1 C complex and each complex forms in a mutually exclusive manner consistent with the fact that they occupy the same physical location on Brca1. The BRCA1 A complex contains BRCA1/BARD1, Abraxas, Rap80, and BRCC36, a deubiquitination enzyme (Dong et al. 2003; Sobhian et al. 2007; Wang and Elledge 2007). In this complex, Abraxas contains a phosphorylated SPxF motif at its C terminus that associates with the BRCT domain of BRCA1 (Wang et al. 2007). Abraxas mediates the interaction of both Rap80 and BRCC36 with BRCA1 (Wang and Elledge 2007). Abraxas associates with Rap80 through the N-terminal domain (Wang and Elledge 2007). Rap80 contains two UIM (ubiquitin-interacting motif) domains that have been shown to bind K63-linked ubiquitin chains (Sobhian et al. 2007; Yan et al. 2007), and are required for the localization of the BRCA1 A complex to ionizing radiation (IR)-induced foci (IRIFs) (Kim et al. 2007; Sobhian et al. 2007; Wang et al. 2007). Abraxas binds BRCC36 through the coiled-coil domains on both proteins (Wang and Elledge 2007). BRCC36 is a deubiquitination enzyme that specifically catalyzes K63-linked polyubiquitin chains and contains a Mpr-1/Pad-1 N-terminal (MPN+)/JAMM domain (Dong et al. 2003; Sobhian et al. 2007).
In this study, using genetic and proteomic approaches we identified a new component of the BRCA1 A complex, NBA1. NBA1 plays a critical role in IR resistance, cell cycle regulation, and the recruitment of BRCA1 to sites of DNA damage. In depth bioinformatics analysis of the components of the A complex reveal that it has striking parallels to the lid complex of the 26S proteasome. Furthermore, we show that several members of the BRCA1-A complex possess a polyubiquitin chain-binding capability, thus forming a complex that might facilitate the deubiquitinating activity of the deubiquitination enzyme BRCC36 or the E3 ligase activity of the BRCA1/BARD1 ligase.
Results
Identification of a novel protein from an IR sensitivity screen
We recently developed bar-coded, microRNA-based shRNA libraries targeting the entire human genome that can be expressed from retroviral vectors for stable gene knockdown (Paddison et al. 2004; Silva et al. 2005; Schlabach et al. 2008; Silva et al. 2008). In this study, we performed a small scale loss-of-function screen to uncover novel players in the mammalian DDR pathway utilizing complex pools of shRNAs coupled with microarray deconvolution technology. Our screen is designed to identify genes whose inhibition leads to cellular sensitivity to IR (Fig. 1A). A pool of shRNAs was introduced into U2OS cells via retroviral transduction from an MSCV-driven vector at an approximate multiplicity of infection of 1 (Schlabach et al. 2008; Silva et al. 2008). Following selection of the cells with puromycin, the cells were treated with IR or left untreated. The cells were passaged in triplicate for ∼12 population doublings (14 d) and then harvested for genomic DNA isolation. The half-hairpin sequence was PCR recovered and the DNA from untreated cells and IR-treated cells were labeled with Cy5 and Cy3, respectively, and competitively hybridized to a custom microarray containing half-hairpin probes. Any shRNA that silences a gene that is necessary for the cell to respond to or repair DNA double-strand breaks will be deleterious and will drop out of the IR-treated population. In a preliminary screen, we identified two different shRNAs that target a previously uncharacterized ORF, c19orf62 (Fig. 1B), also noted as a hypothetical protein HSPC142. We will refer to c19orf62 as NBA1 for reasons described below.
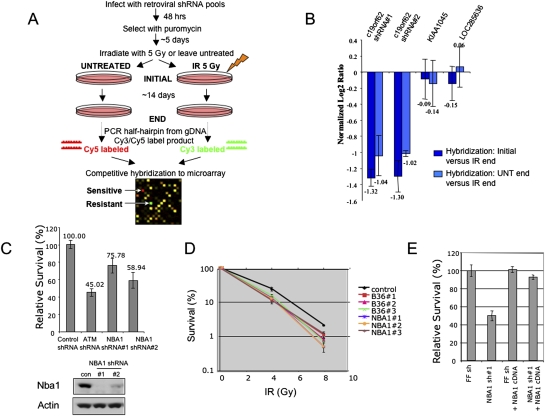
Figure 1.
NBA1 is required for radioprotection. (A) Overview of the pool-based shRNA screen for regulators of the DNA double-strand break response in mammalian cells. (B) Identification of NBA1 as a gene required for IR resistance. shRNAs are graphed based on their mean normalized log2 Cy5/Cy3 ratio, with a ratio less than −1 indicating a decrease in abundance greater than twofold. Two shRNAs targeting distinct regions of the NBA1 sequence scored in the screen. Two unrelated shRNAs that were considered unchanged are also shown for comparison. Error bars represent standard deviation across triplicates. (C) Validation of NBA1 shRNAs that lead to increased IR sensitivity. shRNAs were individually packaged into retroviruses and infected into cells in independent triplicates. Cells were either treated with 3 Gy IR or left untreated and cell viability was monitored ∼9 d later. An ATM shRNA that also scored in the screen and led to IR sensitivity is shown for comparison. (D) Validation of the NBA1 IR sensitivity phenotype using siRNAs. Three different pairs of siRNAs against NBA1 gene and BRCC36 gene were transfected into U2OS cells. Cell survival was measured by colony-forming assay. (E) NBA1 cDNA resistant to shRNA rescues IR sensitivity. U2OS cells expressing a control firefly shRNA or NBA1 shRNA#1 was infected with virus expressing a shRNA#1-resistant NBA1 cDNA or a control vector construct and cells were tested for sensitivity to IR.
To validate that NBA1 shRNAs lead to increased IR sensitivity, cells were infected with retroviruses expressing the individual shRNA and viability in response to IR was tested using a multicolored cell competition assay (Smogorzewska et al. 2007). Cells expressing the NBA1 shRNAs and dsRed were mixed equally with uncolored cells expressing a control shRNA that targets firefly luciferase. The mixed cells were treated with IR or left untreated and the viability of the dsRed-expressing cells was monitored by fluorescence-activated cell sorting (FACS) relative to untreated cells (Fig. 1C). Both NBA1 shRNAs, as well as an ATM shRNA, led to decreased cell viability in response to IR. Immunoblots of NBA1 protein levels show the efficiency of shRNA knockdown (Fig. 1C). To further address the specificity of the shRNA knockdown and to rule out that off target effects lead to the IR sensitivity phenotype, we also tested siRNAs that target distinct regions of the NBA1 sequence in an independent assay. Three siRNAs, which efficiently knocked down NBA1 expression, also led to increased IR sensitivity in a clonogenic survival assay in U2OS cells (Fig. 1D). In addition, when we expressed an NBA1 cDNA lacking the shRNA target site in cells treated with NBA1 shRNA#1, the shRNA-resistant cDNA reversed the IR sensitivity of the NBA1-shRNA (Fig. 1D). Furthermore, we found that depletion of NBA1 rendered cells more sensitive to other DNA-damaging agents, such as camptothecan (CPT), UV, and Mitomycin C (MMC) (Supplemental Fig. S1).
NBA1 localizes to sites of DNA damage
Proteins with roles in IR resistance can act directly or indirectly in the sensing and repair of DNA damage. Proteins acting in a direct manner in these processes often localize to chromatin at the sites of DNA damage. To investigate the localization of NBA1, we fused NBA1 in frame with a N-terminal GFP tag in MSCV. We found that in virally infected cells, GFP-NBA1 was localized in both the nucleus and cytoplasm, however, when extracted prior to fixation, the nuclear portion of GFP-NBA1 relocalized to foci in response to IR (Fig. 2A). To verify that these were indeed sites of DNA damage, we performed two experiments. We first examined colocalization with Brca1, which has been shown previously to localize to damage sites. We found that NBA1 colocalizes with Brca1 at sites of IR-induced damage (Fig. 2B). Second, we examined localization using a UV laser to microirradiate cells and found that NBA1 also localized to the laser-induced DNA damage stripes that overlaps with localization of histone γ-H2AX (Fig. 2C). These results indicate that NBA1 is likely to play a direct role in DNA damage sensing and/or repair.
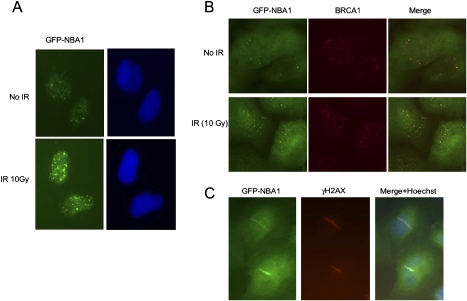
Figure 2.
NBA1 accumulates to DNA damage sites in response to IR. (A) NBA1 localizes to IRIF. U2OS cells that stably express a GFP-NBA1 fusion were treated with 10 Gy IR followed by a 4-h incubation at 37°C. Cells were then extracted with 0.5% Triton X-100, fixed with 3.7% formaldehyde, and stained with an anti-GFP antibody and Hoechst dye. (B) GFP-NBA1 forms IRIF that colocalize with BRCA1 foci. U2OS cells stably expressing GFP-NBA1 protein were treated with 10 Gy IR. After 2h incubation of cells post-IR, cells were fixed and stained with antibodies against GFP and BRCA1. (C) GFP-NBA1 accumulates to laser-induced DNA damage regions that colocalize with γ-H2AX. U2OS cells stably expressing GFP-NBA1 were treated with a laser for microirradiation.
NBA1 is a new component of the BRCA1 A complex
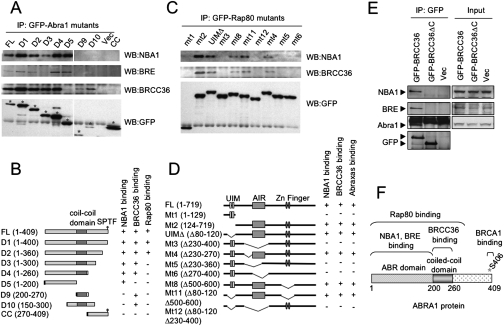
To explore the function of NBA1, we performed a tandem affinity purification (TAP) on 293T cells stably expressing Flag-HA-tagged NBA1. In the analysis of the associated proteins we identified a number of proteins including Abra1 (Fig. 3A; Supplemental Table S1). Abra1 is known to associate with Brca1. Consistent with an association with Abra1, we identified BRCA1 in the NBA1 pull-down by Western blotting (Fig. 3B). To further confirm this association we performed a purification of Abra1-associated proteins using TAP on 293T cells stably expressing endogenous levels of Flag-HA-tagged Abra1 using mass spectrometry analysis, all of the known components of the BRCA1 A complex were identified including BRCA1/BARD1, Rap80, BRE(BRCC45), and BRCC36 (Fig. 3C). Importantly, NBA1 was also identified as a prominent and potentially stoichiometric band in the Abra1 protein complex (Fig. 3C). Given its membership in the BRCA1 A complex we named this ORF NBA1. We then tested whether NBA1 associates with the known players in the A complex of BRCA1. We found that endogenous NBA1 associates with both Abra1 and Rap80 (Fig. 3D). We also found that NBA1 seems to be required to maintain the abundance of BRE and Abra1 as reduction of NBA1 results in a decrease in BRE and Abra1 levels (Fig. 3E). Like GFP-tagged BRE, immunoprecipitation of GFP-tagged NBA1 revealed that it associates with each member of the Brca1 A complex suggesting that all of the proteins are present in one complex as opposed to multiple different Abra1-containing complexes (Fig. 3F).
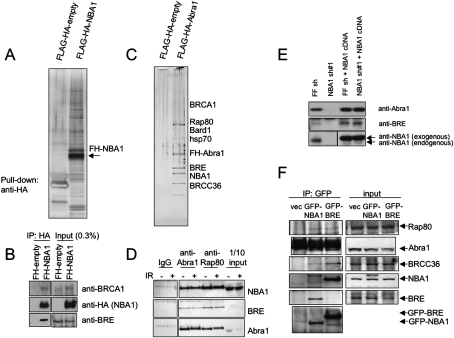
Figure 3.
NBA1 is a BRCA1 A complex component. (A) TAP purification of Flag-HA-NBA1. (B) NBA1 interacts with BRCA1. Immunoprecipitated Flag-HA-tagged NBA1 was fractionated in the gel and probed with antibodies against BRCA1. (C) TAP purification of Flag-HA-Abra1. (D) Endogenous NBA1 interacts with Rap80 and Abra1. Lysates from 293T cells treated with 10 Gy IR follwed with 2 h incubation or untreated were used for immunoprecipitation with antibodies against Abra1 or Rap80, franctionated with SDS-PAGE gel and Western blot was carried with antibodies against Abra1, Rap80, BRE, or NBA1. (E) NBA1 controls BRE and Abra1 abundance. U2OS cells were infected with control firefly (FF) hairpin or shRNA#1 targeting the 3′UTR region of NBA1 or 2 d later infected again with NBA1 cDNA lacking the 3′UTR region. Cell extracts were immunoblotted for Abra1, BRE, and NBA1 protein levels. (F) GFP-tagged NBA1 interacts with each member of the BRCA1-A complex. 293T cells stably expressing GFP-NBA1 or GFP-BRE were used to prepare lysates for the immunoprecipitation with antibodies against GFP. The immunoprecipitates were then separated in SDS-PAGE gel and tested for binding to Rap80, Abra1, and BRCC36 using Western blot and antibodies against Rap80, Abra1, BRCC36, BRE, and NBA1.
NBA1 is required for the DNA damage checkpoint
To explore the role of NBA1 in the DDR we examined the ability of cells depleted for NBA1 to arrest the cell cycle in G2 in response to IR, a function known to be dependent on Brca1. NBA1-depleted cells were unable to arrest efficiently in G2 in response to IR (Fig. 4A). This is similar to defects in cells depleted for another component of the Brca1 A complex, BRCC36 (Fig. 4A). This suggests that NBA1 plays an integral role in the function of the Brca1 A complex.
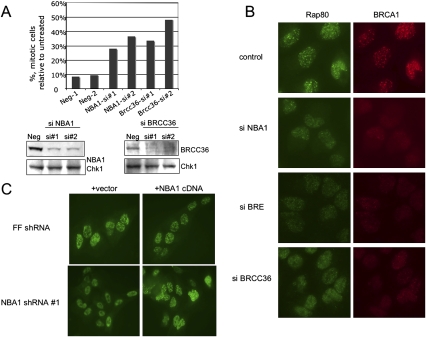
Figure 4.
(A) NBA1 is required for proper G2–M regulation in response to IR. U2OS cells were untreated or treated with 3 Gy IR, then incubated for 1 h at 37°C before fixation and p-H3 antibody staining. Two independent experiments were performed with two different pairs of siRNAs to NBA1 and BRCC36. (B) NBA1 is required for optitmal BRCA1 and Rap80 foci formation in response to IR. U2OS cells were transfected with control or siRNAs against NBA1, BRCC36, or BRE for 2 d, then irradiated with 10 Gy IR, incubated for 2 h, fixed and immunostained with the indicated antibodies. (C) The defect in BRCA1 IRIF formation in cells expressing shRNA to NBA1 can be rescued by expression of an shRNA-resistant NBA1 cDNA. U2OS cells expressing NBA1 shRNA#1 were infected with virus expressing NBA1 cDNA lacking the target sequence of shRNA#1, irradiated with 10 Gy IR, incubated for 4 h, fixed, and stained with Brca1 antibodies.
NBA1 is required for proper localization of BRCA1 protein in response to IR
Given NBA1’s central role in IR resistance and G2/M checkpoint control, we wished to determine what other functions of the Brca1 A complex were dependent on NBA1. Therefore, we examined whether NBA1 is required for Brca1 foci formation in response to IR. Similar to BRCC36 and BRE, we found that NBA1 is also required for optimal foci formation of BRCA1 in response to IR. In siRNA-treated cells lacking NBA1 expression, the percentage of cells forming IR-induced BRCA1 foci was decreased (Fig. 4B). This phenotype could be rescued by expression of a NBA1 cDNA that lacks the shRNA targets site present in the 3′ untranslated region (UTR) (Fig. 4C). Like Brca1, when NBA1 was depleted, Rap80 foci formation was also compromised, and this could also be rescued with the shRNA-resistant NBA1 cDNA (Supplemental Fig. S2). Therefore, NBA1 is a critical component of the BRCA1 A complex required for BRCA1/BARD1 E3 ligase recruitment to sites of DNA damage.
NBA1 is associated with the BRCA1 A complex through the ABR domain of Abra1
To explore how NBA1 is associated with the Brca1 A complex, we began a deletion analysis of the Abra1 protein, which appears to act as a scaffold for the A complex. Abra1 is known to mediate the interaction of Rap80 with BRCA1 (Wang et al. 2007). In addition BRCC36 also directly interacts with Abra1 through its coiled-coil domain (Wang and Elledge 2007). We found that, similar to BRE, NBA1 is incorporated into the Brca1 A complex via the N-terminal region of Abra1, through analysis of a series of deletion mutants of Abra1 (Fig. 5A,B; Wang and Elledge 2007). In addition, Abra1 mediates the interaction of NBA1 and BRE with Rap80 and BRCC36 (Fig. 5C–E; Wang and Elledge 2007). Thus, Abra1 binds multiple A complex components along its length; its N-terminal ABR domain binds NBA1 and BRE, the central coiled-coil domain binds BRCC36, the phosphorylated p-Ser406 binds Brca1 and the large N-terminal region spanning both ABR domain and the coiled-coil domain and beyond is responsible for binding to Rap80 (Fig. 5F). Therefore, Abra1 serves as a central organizing adaptor protein that bridges the interaction of each member of the BRCA1 A complex.
Figure 5.
NBA1 serves as an adaptor protein for the BRCA1 A complex. (A) NBA1 and BRE proteins interact with the N terminus of Abra1. Cell lysates from 293T cells stably expressing GFP-tagged mutants of Abra1 were immunoprecipitated with GFP antibodies and immunoblot analysis was carried out with antibodies against NBA1, BRE, or BRCC36. (B) A diagram for Abra1 deletion mutants. (C) NBA1–Rap80 interaction, similar to BRCC36–Rap80 interaction, depends on the AIR domain of Rap80 that is required for Rap80–Abra1 interaction. Cell lysates from 293T cells stably expressing GFP-tagged mutants of Rap80 were immunoprecipitated with GFP antibodies and Western blot was carried out with antibodies against NBA1 or BRCC36. (D) A diagram for Rap80 deletion mutants. (E) NBA1 and BRE interaction with BRCC36 is mediated through Abra1. Cell lysates from 293T cells stably expressing GFP-tagged NBA1 or BRE protein were immunoprecipitated with GFP antibodies and Western blot was carried out with antibodies against Abra1, BRE, or BRCC36. (F) A diagram mapping the regions of Abra1 that interacts with different members of the BRCA1-A complex.
Similarities between the 19S proteasome complex proteins and the Brca1 A complex components revealed through bioinformatic analysis
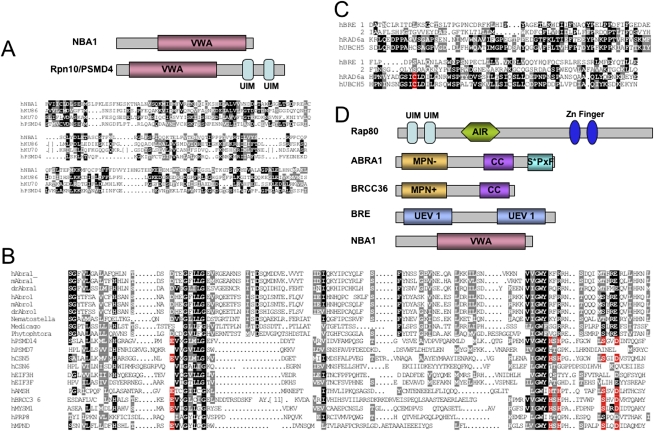
Bioinformatics analysis of the NBA1 protein revealed the presence of a VWA (von Willebrand factor A) domain in its N terminus (Fig. 6A) that is homologous to the VWA domain of the proteasome subunit Rpn10/PSMD4/S5a (Hofmann and Bucher 1998; Hofmann and Falquet 2001). The VWA domain of Rpn10/S5a/PSMD4 is required for maintaining the lid–base association of the 19S regulatory particle of the proteasome and certain aspects of proteasome function (Glickman et al. 1998; Fu et al. 2001; Verma et al. 2004; Hamazaki et al. 2007).
Figure 6.
Members of the BRCA1 A complex contain ubiquitin-binding domains. (A) Alignment of the VWA domain of NBA1 and PSMD4/Rpn10. (B) Alignment of the MPN domains from various species of Abra1, Abro1, and other MPN domain-containing proteins. Residues outlined in red are required for enzymatic activity. (C) Alignment of the UEV domain from BRE with the UEV domain of human E2-conjugating enzyme Rad6a and UbcH5. (D) Protein domains present in the components of BRCA1 A complex. (UIM) Ubiquitin-interacting motif; (AIR) Abraxas-interacting region; (MPN−) Mpr1/Pad1 N-terminal region lacking catalytic activity; (MPN+) Mpr1/Pad1 N-terminal region with an intact protease catalytic site; (CC) coiled coil; (UEV) ubiquitin-conjugating enzyme variant domain; (VWA) von Willebrand factor type A domain.
The identification of the NBA1 VWA domain suggested a connection of the Brca1 A complex with the proteasome lid. This prompted us to perform a more thorough bioinformatics analyis of all of the components of the Brca1 A complex to search for more motifs that had escaped earlier detection. This analysis revealed that Abra1, and its paralog Abro1, contains a MPN domain in its N terminus. MPN domains are found in a group of proteins with a variety of functions: proteasome regulatory subunits, eukaryotic initiation factor 3 (eIF3) subunits, the signalosome, and regulators of transcription factors (Hofmann and Bucher 1998; Maytal-Kivity et al. 2002; Verma et al. 2002; Yao and Cohen 2002; Cope and Deshaies 2003). BRCC36 contains a MPN+/JAMM domain at its N terminus that is simailar to the Rpn11/PSDE of the poteasome lid complex and the Csn5 subunit of the COP9 signalosome, and displays a deubiqutination enzyme activity (Dong et al. 2003; Sobhian et al. 2007). Through protein sequence analysis of different species of Abra1, Abro1, and previously identified MPN domains including Rpn8/PSD7 subunit of the proteasome lid complex and Csn6 of the COP9 signalosome, we found that the MPN domain in Abra1 and Abro1 lacked conserved residues that coordinates Zn2+ binding required for enzymatic cleavage activity (Fig. 6B), and thus not likely to possess an enzymatic activity like the MPN+/JAMM domain (Ambroggio et al. 2004).
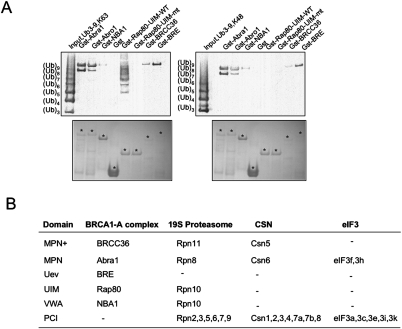
Bioinformatics analysis also revealed that BRE contains two UEV domains lacking the critical cysteine required for ubiquitination (Fig. 6C). Since both MPN and UEV domains are potential ubiquitin-binding domains (Hurley et al. 2006), we then tested whether Abra1, BRE, and BRCC36 possess the capacity to bind ubiquitin chains (Fig. 7A). In an in vitro ubiquitin-binding assay, the indicated GST-tagged proteins were purified from bacteria and incubated with either K63-linked polyubiquitin chains or K48-linked polyubiquitin chains. We found that purified Gst-tagged Abra1, Abro1, BRCC36, or BRE all bind to polyubiquitin chains with at least eight-ubiquitin subunits, both K63-linked and K48-linked (Fig. 7A). In comparison, wild-type UIM domains of Rap80 binds specifically only to K63-linked polyubiquitin chains of various lengths as shown previously (Fig. 7A; Sobhian et al. 2007; Yan et al. 2007). Therefore, four members of the BRCA1 A complex, Abra1, BRCC36, BRE, and Rap80 all possess polyubiquitin chain-binding activity, with Rap80 providing a binding specificity of K63-linked polyubiquitin chains for the complex.
Figure 7.
BRCA1 A complex components Rap80, Abra1, BRE, and BRCC36 bind to polyubiquitin chains. (A) In vitro assay to test the ability of each member of the BRCA1-A complex to bind ubiquitin chains. The indicated GST-tagged proteins were purified from bacteria and incubated with either K63-linked polyubiquitin chains or K48-linked polyubiquitin chains. After 1 h incubation, the pulled-down proteins were fractionated using SDS-PAGE gel, either stained with Coomassie blue for visulization of the expression of proteins, or immunoblotted with antibodies against ubiquitin. (B) A comparison of protein domains between the BRCA1 A, 19S proteasome, CSN, and eIF3 complexes. (PCI) Proteasome/CSN/IF3 domain.
Discussion
NBA1 is a critical component of the BRCA1 A complex
In this study we identified a gene, NBA1, that is required for resistance to IR. NBA1 localizes to sites of DNA damage and regulates the IR-induced G2–M checkpoint. Through mass spectrometry analysis we discovered NBA1 was a member of the BRCA1 A complex and, similar to all known A complex members, is required for optimal recruitment of the BRCA1/BARD1 E3 ligase to sites of DNA damage.
Abraxas appears to serve, in part, as a central scaffold protein that assembles the various components of the BRCA1 A complex. Abra1 has multiple protein interaction domains. NBA1 and BRE interact with Abraxas through the N-terminal ABR region of the Abra1 protein, which is shared with the NBA1-interacting protein Abro1, a Abra1 paralog. The BRCC36 protein associates with Abraxas through a central coiled-coil domain (Wang and Elledge 2007). RAP80 association with Abraxas requires N-terminal regions of Abraxas including ABR domain and the coiled-coil domain (Wang and Elledge 2007). The BRCA1/BARD1 ubiquitin ligase associates with Abraxas through a phospho-SPxF motif at is extreme C terminus that is recognized by the BRCT repeats of BRCA1 (Wang et al. 2007). Our data, together with others (Chen et al. 2006; Kim et al. 2007; Sobhian et al. 2007; Wang et al. 2007), suggests that each member of the BRCA1 complex is required for the recruitment of the BRCA1/BARD1 ligase to DNA damage sites and is responsible for IR resistance of the cells and proper cell cycle regulation.
Both NBA1 and BRE associate with Abraxas through the ABR domain. NBA1 is required to maintian the abundance of BRE and Abra1 as reduction of NBA1 results in a decrease in BRE and Abra1 levels, most likely by controlling their stability. A direct association between NBA1 and BRE is supported by the fact that they were identified as an interacting pair in a genome scale yeast two-hybrid analysis (Rual et al. 2005). Together, these data suggest that NBA1 association with BRE or Abra1 directly regulates the stability of these proteins.
Multiple ubiquitin-binding motifs in the BRCA1 A complex
Bioinformatics analysis revealed multiple new motifs among BRCA1 A complex components. Abra1 possesses an MPN domain at its N terminus that lacks isopeptidase activity. Abra1 dimerization with BRCC36 through the coiled-coil domain on each protein positions a MPN+–MPN domain pair in the complex. In addition, another component of the A complex, BRE, contains two UEV domains that also lack a critical residue required for enzymatic activity. Abra1, Rap80, BRE, and BRCC36 all possess polyubiquitin chain-binding activity, which is likely due to their corresponding MPN−, UIMs, UEV, and MPN+ domains. The fact that only the UIM domains in this complex have the specificity to bind K63-linked polyubiquitin chains suggests that while other ubiquitin-binding proteins may help to hold the polyubiquitin chains in an appropriate position, the UIM domains of Rap80 may act as the key receptor for K63-linked polyubiqutinated substrates.
The BRCA1 A complex shares similarities with the 19S complex of the proteosome
The 26S proteasome contains two major subcomplexes, the 20S proteolytic core particle and the 19S regulatory particle. The 19S complex contains two subcomplexes, the base complex that binds and unfolds substrates (Glickman et al. 1998; Lam et al. 2002), and the lid complex that cleaves ubiquitin from substrates and may facilitate the entry of subtrates into the catalytic proteasome core (Maytal-Kivity et al. 2002; Verma et al. 2002; Yao and Cohen 2002). The proteasome lid contains nine subunits. All except one (Sem1) contain either of two structure motifs, the MPN domain or the PCI domain. Six subunits contain PCI domains that are thought to play a structural role allowing assembly of the lid complex, while two subunits, Rpn8 and Rpn11, contain a MPN domain. Rpn11 contains an active MPN+/JAMM motif that posess a Zn2+-dependent metalloprotease activity for substrate deubiquitination, while Rpn8 contains an enzymatically inactive MPN− domain. Two additional complexes, the Cop9 signalosome and the eIF3 complex share a similar archetecture with the proteasome.
Two components of the BRCA1 A complex contain a MPN domain, BRCC36 (MPN+) and Abra1 (MPN−). This MPN+–MPN− domain pair (BRCC36–Abra1) is also observed in both the proteasome lid complex (Rpn11–Rpn8) and the COP9 signalosome (Csn5–Csn6) (Scheel and Hofmann 2005). Unlike these, the eIF3 complex MPN–MPN pair (eIF3f–eIF3h) does not possess the isopeptidase activity. We speculate that the MPN+–MPN− pair may provide both affinity and help orient the ubiquitin chains to allow efficient cleavage by the MPN+ metalloprotease activity.
Rap80 possesses two UIM domains at its N terminus, while NBA1 contains a VWA domain at its N terminus. Both UIM domains and a VWA domain are present in a single protein of the 19S regulatory complex, Rpn10/S5a. Rpn10 contains a VWA domain at its N terminus and two UIM domains at its C terminus and has been suggested to act as a ubiquitin receptor that, together with other ubiquitin receptor proteins, Rad23 and Dsk2, deliver the ubiquitinated protein to the proteasome (Elsasser and Finley 2005). The UIM domains of Rpn10 specifically bind to K48-linked polyubiquitin chains and determine the substrate specificity of the proteosome (Elsasser and Finley 2005). Similarly, the UIM domains of Rap80 provide the specificity allowing the BRCA1 A complex to bind K63-linked polyubiquitin chains.
Unlike the proteasome lid complex, the COP9 signalosome, and the eIF3 complex, the BRCA1 A complex does not contain PCI domain proteins to serve as structural organizers. Instead, Abra1 may play an analogous role, acting as a adaptor to organize the structure of the complex to recognize both K48- and K63-linked polyubiquitinated proteins.
What is the significance of the similarity to the 19S proteasome (Fig. 7B) and why does the BRCA1 A complex contain so many ubiquitin-binding domains? One possibility is that the different UIMs might allow integration of signals from different E3 ubiquitin ligases to control, for example, the proper localization of the BRCA1 A complex within an IRIF. Furthermore, it is unlikely that the Brca1 A complex would be recruited to every K63 chain in the cell through the RAP80 UIM motifs. Perhaps these UIM domains of Rap80 do not bind K63 chains with sufficient affinity to localize the A complex to a particular K63 chain and the complex then employs additional interactions to provide specificity. The ability to interact with multiple different Ub chains might provide the additional binding energy to allow the Brca1 A complex to be recruited only to regions where multiple distinct Ub chains are located, such as sites of DNA damage. A third possibility is that once the complex is localized, these domains may be required to recognize certain ubiquitin chains in the vicinity to promote the deubiquitination of specific substrates as a further signaling event, which might be important for retaining DNA repair proteins such as BRCA1 on the damaged chromatin.
What is the role of the BRCA1 A complex in DNA repair? While the complex localizes to sites of DNA damage, its precise biochemical role there is unclear. As it has both ubiquitinating and deubiquitinating activities, it may function to amplify or remodel the existing ubiquitin chains already in place to promote DNA repair. A key to unraveling the role of this complex may reside in understanding the role of the VWA domain in NBA1. A similar VWA domain located in the DNA repair protein Ku80 has been speculated to act as a protein–protein interaction domain that recruits the WRN helicase to sites of damage (Doherty and Jackson 2001). Perhaps, the NBA1 VWA domain plays a similar role to recruit repair factors to sites of damage to influence DNA repair, like the BRCA1 B and BRCA1 C complexes, which contain the helicase Brip1 and the nuclease CTIP, respectively. Alternatively, a second related VWA domain in the 19S proteosome lid appears to link the lid to the 20S core particle of the proteasome responsible for proteolysis. It is possible that at some stage in the repair process, the BRCA1 A complex recruits the 26S proteosome to sites of damage to promote repair or terminate structures through proteolysis. It is clear there is much to be learned about the BRCA1 A complex that is likely to shed light on the role of Brca1 in DNA repair and cancer.
Materials and methods
A loss-of-function screen for radioprotective genes using shRNA libraries
The screens were performed as described previously (Schlabach et al. 2008) except cells were treated with IR or left untreated and were passaged in triplicate for ∼12 population doublings (14 d) and then harvested for genomic DNA isolation. Any shRNA that silences a gene necessary for repair of DNA double-strand breaks will be deleterious and will drop out of the IR-treated population and will be identified by microarray analysis (Schlabach et al. 2008). shRNAs with log2 ratios greater than 1 or less than −1 were considered enriched or depleted from the population, respectively. The details of the screens will be reported elsewhere (K. Hurov and S. Elledge, in prep.).
Plasmids, siRNAs, and shRNAs
Retroviral constructs expressing GFP-NBA1, HA-Flag-NBA1, and HA-Flag-Abra1 were made using a MSCV vector. Retroviral constructs expressing GFP-Rap80 and its various mutants, GFP-Abra1 and its various mutants were described previously (Wang and Elledge 2007). Gst-Abra1, Gst-Abro1, Gst-BRCC36, GST-BRE, GST-NBA1, GST-Rap80-UIM-WT (Rap80 1–129 amino acids), and Gst-Rap80-UIM-mt (Rap80 1–129 amino acids A88S, S92A, A113S, S117A) were generated with pDEST15 vector (Invitrogen). The following siRNA used in experiments were purchased from Invitrogen: Stealth RNAi Negative Control Med GC (12935-300), Stealth RNAi for NBA1(hspc142) (HSS120850, HSS120851, HSS150852) (si1-UCAUCUUCUGGGAGACAUUGAGGGC, si2-AAUCGUCUGCACGUUCUCUGUGACC, and si3-AACAACGUCAAAGAAGAAAUAUGGG), BRCC36 (HSS128338, HSS128339, HSS128340) (si1-AAUGAAUGUGAACAUUCUGACGGC and si2-UAUGGAUUGGGAUUUCGAUUCUCUC, si3-UAAGGCUGUGGAUCCUCCUAUACGC), BRE (HSS114298, HSS114299, HSS114300) (si1-AUUUGAAGGAUUCCAGGAGGCAAGA, si2-UUUCCAUGUUCUCUCCAUACUGUGG, and si3-AAAGCUGGGAUAUGAAGAGCUGAGG). shRNA sequences used were ATM (TTAAATGACTGTATAGTCACCA), NBA1 sh#1 (ATTATCCATGACAAACTAGGAG), and NBA1 sh#2 (AAGTCCTTCCAGATTGAAGGTG)
Antibodies
NBA1 antibody was purchased from Genway (hspc142 antibody, catalog no. 15-288-22374A). Antibodies against Brca1 used in this study are Brca1 D9 (Santa Cruz Biotechnologies) and Brca1 BL319 (Bethyl). Anti-GFP antibody was purchased from Invitrogen and anti-p-H3 antibody was from Upstate Biotechnologies. Other antibodies used were Rap80 antibodies (Bethyl, BL2839) BRCC36 antibodies (Invitrogen #40-1300), BRE antibodies (Santa Cruz Biotechnologies, sc-48847; or ProteinTech Group, Inc., #11702-1-AP), and ubiquitin antibodies (Santa Cruz Biotechnologies, sc-9133). Polyubiquitin chains were obtained from Boston Biochem [poly-Ub(3-7)-K48-linked, UC-220; poly-Ub(3-7)-K63-linked, UC-320)].
Cell culture
U2OS cells were grown in MyCoy5’s medium supplemented with 10% fetal bovine serum (FBS). 293T and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS. All cells were cultivated at 37°C in a humidified incubator with 5% CO2.
TAP purification
NBA1 was cloned into a N-terminal Flag-HA-tagged doxycycline-inducible vector and transduced via retrovirus into 293T-Rex cells. Stable cells were induced with 2 μg/mL final concentration of doxycycline for 24 h followed by treatment with 10 Gy IR for 4 h before harvesting and lysates were prepped as previously described and immunoprecipitated with HA antibodies and protein was eluted with HA peptide (500 μg/mL final; Sigma). The elution was TCA precipitated and analyzed by mass spectrometry. Flag-HA-tagged Abra1 was purified similarly, except samples were separated in a SDS-PAGE gel and gel slices were excised for mass spectrometry analysis.
Cell lysis and immunoprecipitation
Cells were lysed in NETN buffer (50 mM Tris-HCl at pH 8.0, 0.15 M NaCl, 1 mM EDTA, 0.5% NP-40) with protease inhibitors (Roche) and protein phosphatase inhibitors, 10 mM NaF and 50 mM β-glycerophosphate. Immunoprecipitations were carried out in the same buffer with appropriate antibodies for 3 h to overnight at 4°C.
Colony-forming assay
U2OS cells transfected with siRNAs were grown for 2 d, then seeded at low density and irradiated with various doses of ionizing or UV radiation. Cells were left for 14 d at 37°C to allow colonies to form. Colonies were stained with 2% methylene blue/50% ethanol and counted. Colonies were defined as containing 50 or more cells.
G2/M checkpoint assay
U2OS cells transfected with control oligos or siRNAs against NBA1 or BRCC36 were grown for 2 d, irradiated with 3 Gy IR, and incubated for 1 h. Assays were performed as described (Wang et al. 2007).
Laser-induced DNA DSBs
Cells were treated with 10 μM BrdU (BD) for 24 h prior to laser irradiation on a Zeiss Observer.Z1 inverted microscope integrated with Palm microbeam laser microdissection workstation. Following laser treatment, the cells were incubated for 30 min at 37°C, fixed, permeabilized, and stained with GFP (Invitrogen) and γ H2AX (Upstate Biotechnologies) antibodies and Alexa 488 and Alexa 594 secondary antibodies, respectively.
Color competition viability assay
U2OS cells were retrovirally transduced with MSCV-dsRed (no selection marker) and FACs sorted to obtain a more homogeneous population. The dsRed cells were then infected with retrovirus expressing individual ATM, NBA1, or control shRNAs, selected with puromycin, and then mixed equally with U2OS cells (uncolored) expressing a control shRNA targeting FF luciferase. The mixed population was left untreated or treated with 3 Gy γ irradiation and following approximately six population doublings, harvested for FACs analysis. For graphical representation, the percent of dsRed-expressing cells in IR-treated samples was normalized to the behavior of untreated cells as well as to cells carrying a control shRNA (both dsRed and uncolored cells carrying the control shRNA were equally mixed). For the rescue experiment, a control FF shRNA or a NBA1 shRNA that targets the 3′UTR were introduced into dsRed-expressing U2OS cells via retrovirus and the cells were stably selected with puromycin. The cells were then infected with retrovirus that express the NBA1 cDNA and equally mixed with U2OS (uncolored) expressing a control FF shRNA and further processed as above.
Immunofluoresence
Cells were fixed with 3% paraformaldehyde/2% sucrose for 10 min, permeablized with 0.5% Triton X-100 solution, and then immunostained with primary antibodies against various proteins and the appropriate Alexa 488-conjugated (green; Molecular Probes) and Cy3-conjugated (red; Amersham Biosciences) secondary antibodies. Images were taken with a Zeiss microscope. For the rescue experiment, a control FF shRNA or a NBA1 shRNA that targets the 3′UTR were introduced into U2OS cells via retrovirus and the cells were stably selected with puromycin. The cells were subsequently infected with retrovirus expressing a NBA1 cDNA or an empty cassette and processed as above.
Acknowledgments
We thank Dr. Jianping Jin for the MSCV-N-GFP gateway expression construct, Dr. Mark Vidal for the ORFeome clones for NBA1 and BRE, C. Cotta-Ramusino for contributing to the IR screening, and M. Schlabach for help with microarray hybridizations. B.W. is a recipient of an NCI Howard Temin Award (1KO1, CA116275-01). K.H. is a Special Fellow of the Leukemia and Lymphoma Society. This work was supported by a CMCR grant (1U19A1067751-01), a DOD grant, and a NIH grant to S.J.E. S.J.E. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1770309.
Supplemental material is available at http://www.genesdev.org.
References
- Ambroggio X.I., Rees D.C., Deshaies R.J. JAMM: A metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 2002;12:86–91. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
- Chen X., Arciero C.A., Wang C., Broccoli D., Godwin A.K. BRCC36 is essential for ionizing radiation-induced BRCA1 phosphorylation and nuclear foci formation. Cancer Res. 2006;66:5039–5046. doi: 10.1158/0008-5472.CAN-05-4194. [DOI] [PubMed] [Google Scholar]
- Cope G.A., Deshaies R.J. COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- Doherty A.J., Jackson S.P. DNA repair: How Ku makes ends meet. Curr. Biol. 2001;11:R920–R924. doi: 10.1016/S0960-9822(01)00555-3. [DOI] [PubMed] [Google Scholar]
- Dong Y., Hakimi M.A., Chen X., Kumaraswamy E., Cooch N.S., Godwin A.K., Shiekhattar R. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell. 2003;12:1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- Elsasser S., Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Fu H., Reis N., Lee Y., Glickman M.H., Vierstra R.D. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M.H., Rubin D.M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V.A., Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Greenberg R.A., Sobhian B., Pathania S., Cantor S.B., Nakatani Y., Livingston D.M. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes & Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki J., Sasaki K., Kawahara H., Hisanaga S., Tanaka K., Murata S. Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol. Cell. Biol. 2007;27:6629–6638. doi: 10.1128/MCB.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Elledge S.J. The DNA damage response: Ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., Yabuki Y., Ogata H., Ohta T. The RING heterodimer BRCA1–BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- Hofmann K., Bucher P. The PCI domain: A common theme in three multiprotein complexes. Trends Biochem. Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- Hofmann K., Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H., Lee S., Prag G. Ubiquitin-binding domains. Biochem. J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Chen J., Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Y.A., Lawson T.G., Velayutham M., Zweier J.L., Pickart C.M. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V., Reis N., Hofmann K., Glickman M.H. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narod S.A., Foulkes W.D. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- Paddison P.J., Silva J.M., Conklin D.S., Schlabach M., Li M., Aruleba S., Balija V., O'Shaughnessy A., Gnoj L., Scobie K., et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Ruffner H., Joazeiro C.A., Hemmati D., Hunter T., Verma I.M. Cancer-predisposing mutations within the RING domain of BRCA1: Loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. 2001;98:5134–5139. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel H., Hofmann K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics. 2005;6:71. doi: 10.1186/1471-2105-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlabach M.R., Luo J., Solimini N.L., Hu G., Xu Q., Li M.Z., Zhao Z., Smogorzewska A., Sowa M.E., Ang X.L., et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.M., Li M.Z., Chang K., Ge W., Golding M.C., Rickles R.J., Siolas D., Hu G., Paddison P.J., Schlabach M.R., et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Silva J.M., Marran K., Parker J.S., Silva J., Golding M., Schlabach M.R., Elledge S.J., Hannon G.J., Chang K. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R., 3rd, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D'Andrea A.D., et al. Identification of the FANCI protein, a monoubiqutinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B., Shao G., Lilli D.R., Culhane A.C., Moreau L.A., Xia B., Livingston D.M., Greenberg R.A. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman A.R. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W.H., Yates J.R., III, Koonin E.V., Deshaies R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Verma R., Oania R., Graumann J., Deshaies R.J. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Wang B., Elledge S.J. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., Elledge S.J. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.C., Wang Z.W., Tsan J.T., Spillman M.A., Phung A., Xu X.L., Yang M.C., Hwang L.Y., Bowcock A.M., Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- Yan J., Kim Y.S., Yang X.P., Albers M., Koegl M., Jetten A.M. Ubiquitin-interaction motifs of RAP80 are critical in its regulation of estrogen receptor α. Nucleic Acids Res. 2007;35:1673–1686. doi: 10.1093/nar/gkl1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T., Cohen R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Yu X., Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.B., Elledge S.J. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]