Abstract
Post-translational processes are essential for the generation and dynamics of mammalian circadian rhythms. In particular, phosphorylation of the key circadian protein PER2 precisely controls the period and phase of circadian oscillations. However, the mechanisms underlying that control are poorly understood. Here, we identified in a high-throughput RNAi-based genetic screen casein kinase 2 (CK2) as a PER2-phosphorylating kinase and novel component of the mammalian circadian clock. When CK2 subunits are silenced by RNAi or when CK2 activity is inhibited pharmacologically, circadian rhythms are disrupted. CK2 binds to PER2 in vivo, phosphorylates PER2 specifically at N-terminal residues in vitro, and supports normal nuclear PER2 accumulation. Mutation of CK2 phosphorylation sites decreases PER2 stability and copies CK2 inhibition regarding oscillation dynamics. We propose a new concept of how PER2 phosphorylation and stabilization can set the clock speed in opposite directions, dependent on the phase of action.
Keywords: Circadian clock, casein kinase 2, period 2, RNAi screen, phosphorylation
Circadian rhythms in physiology, metabolism, and behavior are ubiquitous from cyanobacteria to humans (for review, see Young and Kay 2001). They are generated by endogenous clocks and entrained by environmental Zeitgebers (e.g., light, temperature, food), providing organisms with an internal representation of external time. Circadian clocks regulate a huge number of physiological processes, and interference with normal circadian rhythms by genetic or environmental perturbations likely has consequences for human health (for review, see Takahashi et al. 2008). The molecular basis of circadian clocks is cell-autonomous. In mammals, the machinery responsible for rhythm generation is present not only in neurons of the suprachiasmatic nucleus (SCN)—the master clock in the hypothalamus—but also in nearly every cell of the periphery, as well as in many standard tissue culture cell lines (Kowalska and Brown 2007).
In recent years, a number of components of the mammalian circadian clock have been identified. However, the intrinsic dynamics of their interaction generating molecular rhythms with a period close to 24 h are poorly understood. The current model is based on a gene regulatory network with a negative delayed feedback loop as the essential core module (for review, see Lowrey and Takahashi 2004): The transcription factor heterodimer CLOCK/BMAL1 activates the expression of the Period (PER1-3) and Cryptochrome (CRY1-2) genes, whose protein products form a complex, accumulate, and, after a delay of several hours, translocate in the nucleus to inhibit the expression of their own genes. Subsequent degradation of PER and CRY proteins leads to a disinhibition of CLOCK/BMAL1, and the cycle can restart. In a second positive feedback loop, Bmal1 transcription itself is oscillating through the rhythmic inhibition by the nuclear repressor REV-ERBα.
The dynamics of these networks are critically dependent on post-translational processes, with phosphorylation and dephosphorylation probably being most important (Harms et al. 2004; Gallego and Virshup 2007; Vanselow and Kramer 2007). Recently, it has been speculated that rhythmic phosphorylation and dephosphorylation may even comprise the heart of the circadian mechanism (Merrow et al. 2006). In mammals, most clock proteins are phosphorylated in vivo with a circadian variation in the degree of phosphorylation. Nevertheless, little is known about the dynamics of phosphorylation and dephosphorylation, about the number, location, and function of phosphorylation sites, as well as about the kinases and phosphatases involved. In fact, good genetic evidence exists only for casein kinase 1ɛ and 1δ (CK1ɛ/δ), which have been shown to phosphorylate and thereby destabilize PER proteins in vivo (Lowrey et al. 2000; Xu et al. 2005).
In a recent study by our laboratory (Vanselow et al. 2006), we found by extensive mass spectrometric screening and subsequent functional analyses that phosphorylation of the key circadian protein PER2 has at least two opposing effects: destabilization of PER2, but also its stabilization. Whereas destabilization of PER2 can be clearly attributed to CK1ɛ/δ function, it is completely unknown which kinase(s) are required for PER2 stabilization. It is important to note that interfering with PER2 stabilization—e.g., by mutation of the respective phosphorylation sites—substantially changes circadian dynamics. For example, in humans suffering from one form of familial advanced sleep phase syndrome (FASPS), a phosphorylation site of PER2 is mutated, which results in an inhibition of normal PER2 stabilization (Toh et al. 2001; Vanselow et al. 2006). Thus, elucidating the processes that affect PER2 stability would be a major step toward an understanding of the complex, however largely unknown, post-translational control of circadian timing in mammals.
Here, we report a systematic effort to elucidate unknown processes modulating PER2 stability. Initial attempts to chemically cross-link kinases with phosphorylation site-mutated versions of PER2 and a mechanism-based cross-linker (Maly et al. 2004) were not successful. Therefore, we decided to set up an unbiased large-scale RNAi screen with a functional readout, because perturbing the activity of these kinases is predicted to have a strong effect on circadian dynamics. We individually knocked down all known and predicted kinases (as well as phosphatases and F-box proteins as additional regulators of protein stability) and analyzed the effect on oscillation dynamics. We identified casein kinase 2 (CK2) as a new and essential component of the mammalian circadian clock that interacts with and phosphorylates PER2. When CK2 subunits are down-regulated by RNAi or when CK2 activity is pharmacologically inhibited, circadian rhythms are disrupted, probably due to an attenuated nuclear accumulation of PER2. Using a comparative mass spectrometric approach, we found that CK2 phosphorylates N-terminal residues of PER2. Thereby, PER2 is stabilized, which we show is required for normal circadian rhythms. We propose a new concept of how phosphorylation of the same clock protein (PER2) can set the clock speed in opposite directions, dependent on its phase of action.
Results
RNAi-mediated silencing of clock genes phenocopies “classical” mouse genetics
Prerequisites for the success of our large-scale RNAi screen are robustness of circadian oscillation in the cell culture model, efficient RNAi-mediated knockdown, as well as circadian phenotypes similar to those of “classical” loss-of-function mouse models. We chose human osteosarcoma U-2 OS cells because, firstly, circadian rhythms of these cells can be robustly monitored in 96-well plates via ∼7-d live-cell imaging of luciferase activity using a Bmal1-promoter fragment driving the expression of luciferase (Supplemental Fig. 1). Secondly, the RNAi library used in this study—a second-generation library of the Hannon-Elledge laboratories–was studied extensively and optimized in U-2 OS cells (Silva et al. 2005; Stegmeier et al. 2005, 2007; Draviam et al. 2007). MicroRNA-based RNAi constructs are lentivirally delivered and thus stably integrated into the genome, resulting in long-lasting gene knockdown.
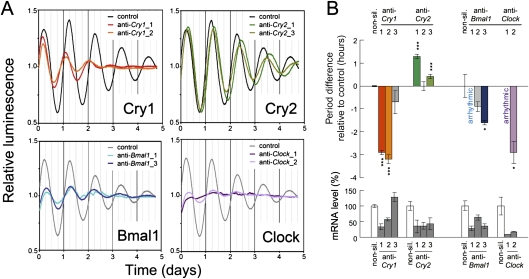
For proof of concept, we first tested whether silencing known clock components (Cry1, Cry2, Bmal1, and Clock) phenocopies the effects observed in mouse knockout studies (Fig. 1; for quantification and statistics of all time series, see Supplemental Table 1). Down-regulating Cry1 expression shortened the period and decreased the amplitude of the rhythms, whereas silencing Cry2 lengthened the period, very similar to the behavioral and cellular phenotypes of corresponding mouse mutants (Vitaterna et al. 1999; van der Horst et al. 1999; Liu et al. 2007). Knockdown of Bmal1 and Clock almost disrupted circadian rhythms completely, consistent with the arrhythmic behavioral and molecular phenotypes of Bmal1 −/− mice (Bunger et al. 2000; Liu et al. 2007) and the arrhythmic phenotype of Clock −/− peripheral cells (DeBruyne et al. 2007). Thus, RNAi-mediated gene knockdown is well-suited to detect circadian phenotypes.
Figure 1.
RNAi-based reverse genetics reproduces circadian phenotypes. (A) Human U-2 OS cells harboring a Bmal1-luciferase reporter were lentivirally transduced with RNAi constructs targeting the indicated transcripts, synchronized with dexamethasone, and luciferase activity was continuously monitored. Detrended average time series (n = 2–3) for two RNAi constructs and a nonsilencing control are given. (B) Quantification of circadian period and silencing efficiency. Error bars, mean ± standard deviation. (***) P < 0.001, (*) P < 0.01 (t-test). See also Supplemental Table 1.
RNAi screen reveals many new candidate clock components
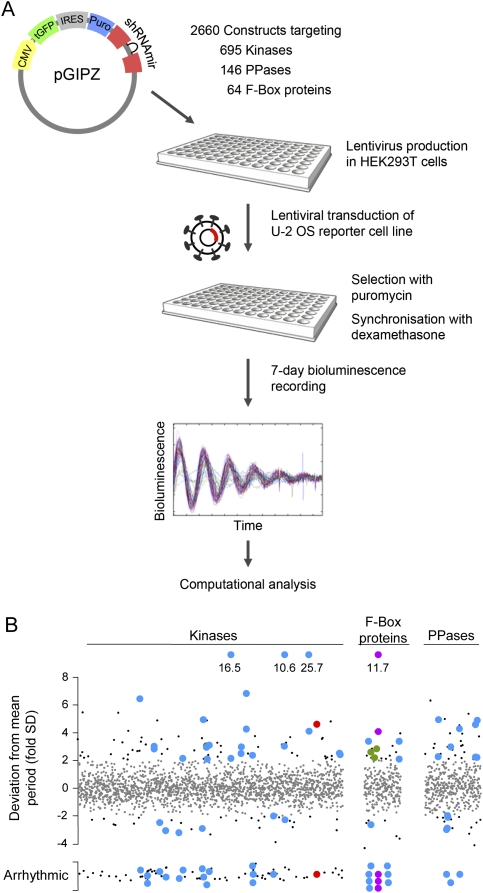
For identifying kinases and other components that regulate PER2 stability, 2660 RNAi constructs targeting 695 kinases or putative kinases, 146 phosphatases including regulatory subunits, as well as 64 F-box proteins were individually screened in high-throughput format for effects on circadian dynamics (Fig. 2A). 287 RNAi constructs led to period phenotypes or disrupted circadian rhythms. Due to potential off-target effects of RNAi constructs, we considered only those genes for further analysis for which at least two RNAi constructs resulted in a similar phenotype. Using these criteria, 22 kinases, seven phosphatases/regulatory subunits, and six F-box proteins were classified as candidate clock components (Fig. 2B).
Figure 2.
RNAi-based functional screen identifies new candidate clock components. (A) Schematic diagram of the large-scale RNAi-based screen for kinases, phosphatases, and F-box proteins required for normal circadian rhythms. For experimental details, see the Materials and Methods and the Supplemental Material. (B) Results of the screen described in A. Dots represent plate-normalized period values corresponding to an individual RNAi construct. Values within or outside a range of two standard deviations from the mean period are given as small gray or small black symbols, respectively. For hit evaluation details, see the text and the Supplemental Material. Among the 22 kinases, seven phosphatases, and six F-box proteins identified as potential clock genes (blue dots), we found FBXL3 (purple), β-TRCP2 (green), and CK2α (red) disrupted circadian rhythmicity in the reporter cell line.
Further support of the significance of our screen came from the fact that among the F-box candidates was FBXL3, recently reported to be involved in CRY protein degradation (Busino et al. 2007; Siepka et al. 2007). Its down-regulation lengthened the period up to 5 h (Supplemental Fig. 2), similar to phenotypes of hypomorphic mouse mutations (Godinho et al. 2007; Siepka et al. 2007). Silencing of another F-box gene, FBXW11 (β-TRCP2), also lengthened the circadian period and reduced oscillation amplitude. Simultaneous knockdown of both β-TRCP2 and its paralog β-TRCP1, previously reported to be involved in PER protein degradation (Eide et al. 2005; Shirogane et al. 2005; Reischl et al. 2007), led to a strongly reduced amplitude, indicating that both proteins have partly redundant functions (Supplemental Fig. 2).
CK2 is an essential component of the mammalian circadian clock
One result of our screen, CK2, especially attracted our attention as a potential candidate for being (one of) the unknown PER2 phosphorylating kinase(s). While a function for CK2 in the mammalian circadian clock has not yet been reported, CK2 plays important roles in the circadian systems of Arabidopsis (Sugano et al. 1999), Neurospora (Yang et al. 2002; Dunlap et al. 2007), and Drosophila (for a review, see Allada and Meissner 2005). In Drosophila, phosphorylation of PER by CK2 is required for normal nuclear entry dynamics (Lin et al. 2002; Akten et al. 2003; Smith et al. 2008). In mammals, CK2 is an ubiquitously expressed kinase involved in many cellular processes with a large number of substrates (Litchfield 2003). The holoenzyme is a tetramer consisting of two catalytic subunits (α and/or α′) and two regulatory β-subunits.
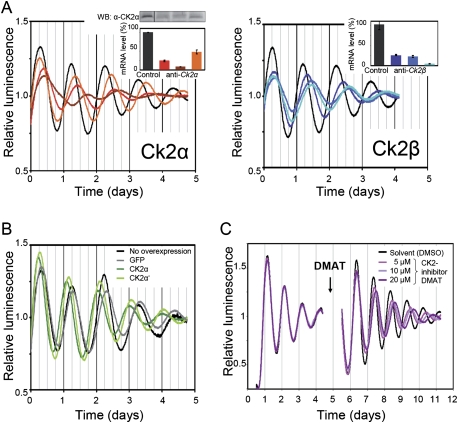
Down-regulation of both catalytic subunits CK2α and CK2α′ as well as the regulatory subunit CK2β lengthened circadian periods by a degree that is correlated with the silencing efficiency of the respective RNAi construct (Fig. 3A; Supplemental Fig. 3). Low-amplitude residual rhythmicity was observed when both CK2 catalytic subunits were targeted (Supplemental Fig. 3).
Figure 3.
CK2 levels and activity are crucial for normal circadian rhythmicity. (A) Circadian phenotypes of RNAi-mediated knockdown of CK2α and CK2β in U-2 OS reporter cells. The experiment was essentially performed as described in Fig. 1A. The insets show the degree of protein or mRNA down-regulation. Error bars, mean ± SEM (n = 2–3). (B) Circadian phenotypes of U-2 OS cells ectopically expressing CK2α and CK2α′ or GFP as control. Shown are detrended average time series (n = 2–3). (C) Lung tissue explanted from Per2∷Luc knock-in mice (Yoo et al. 2004) was cultured first without and after 5 d in the presence of the specific CK2 inhibitor DMAT.
Overexpression of CK2α and CK2α′ had the opposite period effect, leading to a period shortening of ∼2 h (Fig. 3B). This phenotype was dependent on the kinase activity, since overexpression of kinase-inactive dominant-negative versions of the catalytic subunits (Vilk et al. 1999) resulted in a period lengthening of ∼1 h (Supplemental Fig. 3).
In addition to its abundance, the activity of CK2 is crucial for normal circadian rhythms, since pharmacological inhibition of CK2 with the specific inhibitor 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT) (Pagano et al. 2004) dose-dependently lengthened circadian periods in lung explants of Per2∷Luc knock-in mice (Fig. 3C; Yoo et al. 2004) in U-2 OS cells (Supplemental Fig. 4) as well as in murine NIH3T3 cells (data not shown).
CK2 binds to the clock protein PER2
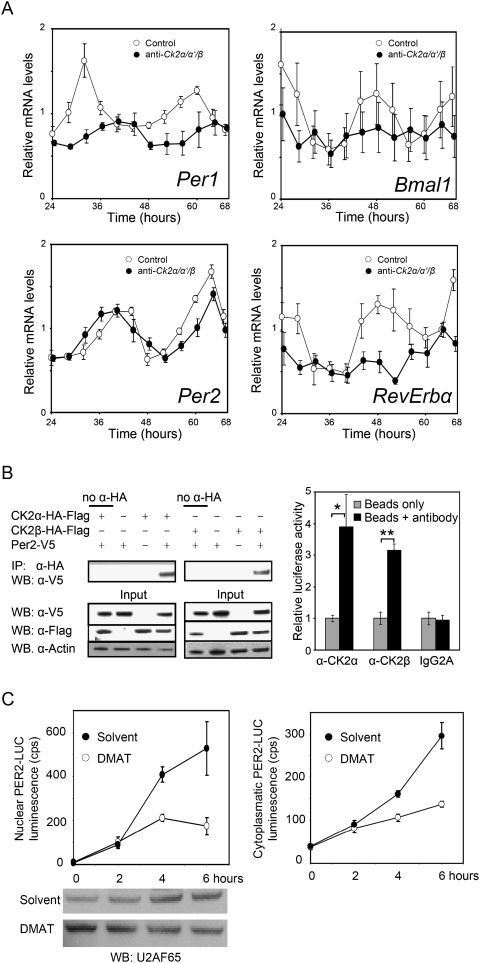
The rhythmicity and expression levels of clock gene transcripts are differentially affected in CK2α/CK2α′/CK2β triple-knockdown U-2 OS cells, as revealed by quantitative RT–PCR analysis (Fig. 4A). Bmal1 and Rev-Erbα transcript levels are arrhythmic and low in these cells, while damped rhythms persist for Per1 and (to a lesser extent) also for Per2 with a very low expression level and substantially altered phase for Per1. These molecular phenotypes suggest that the positive feedback loop is severely damaged upon CK2 knockdown, and resemble those found in Per2 mutant mice (Shearman et al. 2000; Oster et al. 2002).
Figure 4.
PER2 is a target of CK2. (A) Expression levels of clock genes were analyzed in CK2α/α′/β triple knockdown cells (or control cells transduced with a nonsilencing RNAi construct) by quantitative RT–PCR. Expression levels of clock genes are given relative to Gapdh and normalized to the average expression level of the control cells. Error bars, mean ± SEM (n = 3). Whereas Per1 (P < 0.01) and Per2 (P < 0.001) transcript levels are still significantly rhythmic, Bmal1 (P > 0.05) and RevErbα (P > 0.05) levels cease to oscillate (CircWave analysis; Oster et al. 2006). (B) (Left) Lysates from HEK293 cells transfected with the indicated constructs were subjected to IP with or without anti-HA antibody and analyzed by Western blotting (WB). (Right) Liver lysate from Per2∷Luc mice was subjected to IP with or without anti-CK2α, anti-CK2β, as well as an idiotypic control antibody (IgG2A). Binding to PER2 was analyzed using luciferase activity of PER2-LUC. Error bars, mean ± SEM (n = 3); (*) P < 0.05; (**) P < 0.01 (t-test). (C) Primary fibroblasts from Per2∷Luc mice were treated with CHX for 15 h and subsequently released in medium with or without DMAT. Cell nuclei and cytoplasmic fractions were analyzed for PER2-LUC abundance. Western blots for the nuclear marker protein U2AF65 serve as loading control. Error bars, mean ± SEM (n = 3). Two-way ANOVA revealed significant differences between DMAT treatment and solvent control (P < 0.01).
Thus, we hypothesized that CK2 exerts its function in the mammalian clock on PER2. If PER2 is a substrate of CK2, a direct interaction of the two components is predicted. Indeed, coimmunoprecipitation (co-IP) assays with overexpressed and with endogenous proteins revealed that both CK2α and CK2β are in a complex with PER2 in tissue culture cells and in mouse liver (Fig. 4B; Supplemental Fig. 5), suggesting that PER2 may be a substrate for CK2 in vivo.
CK2 activity is necessary for PER2 accumulation
According to the current model (Lowrey and Takahashi 2004), PER and CRY proteins accumulate in the nucleus several hours after their synthesis to repress their own transcription. If CK2-mediated phosphorylation of PER2 is required for protein accumulation, as has been shown in the Drosophila circadian system (Lin et al. 2002; Akten et al. 2003; Smith et al. 2008), interference with CK2 activity should alter nuclear accumulation. As an accurate readout for PER2 abundance, we exploited the fact that in (behaviorally normal) Per2∷Luc knock-in mice (Yoo et al. 2004) endogenous PER2 is represented as a PER2-LUC fusion protein, and thus luciferase activity can be directly correlated with endogenous PER2 levels. We treated primary fibroblasts from Per2∷Luc mice with the translation inhibitor cycloheximide (CHX) for 15 h to deplete the cells of endogenous PER2. After removal of CHX, nuclear accumulation of newly synthesized PER2 was followed by analyzing PER2-LUC activity in isolated cell nuclei as well as in the cytoplasm. Indeed, when CK2 activity is inhibited, both cytoplasmic and nuclear accumulation of PER2 is significantly attenuated (Fig. 4C). These results support the hypothesis that CK2 phosphorylation of PER2 early in the PER2 life cycle stabilizes PER2 and thus allows for timely cytoplasmic as well as eventually nuclear accumulation.
CK2 phosphorylates PER2 at N-terminal residues
Which residues of PER2 are phosphorylated by CK2? To map those, we used a comparative mass-spectrometric approach, which should (at least to a certain extent) circumvent the intrinsically limited specificity of in vitro phosphorylation assays. We compared the phosphorylation profiles of PER2 phosphorylated either by endogenous kinases, as described in our previous study (Vanselow et al. 2006), with those of CK2 holoenzyme, CK1δ, or GSK-3β phosphorylation in vitro (Table 1). Interestingly, most in vivo PER2 phosphorylation sites can be phosphorylated by at least two of the tested kinases in vitro. Only the very N-terminal region of PER2 (Ser10 to Thr15) is phosphorylated both in vivo and in vitro by CK2 but not by other kinases tested (Table 1; Fig. 5A).
Table 1.
Phosphorylated residues of mPER2
aPhosphorylation by endogenous kinases in HEK293 cells (Vanselow et al. 2006).
bPhosphorylation in vitro by CK1δ (Schlosser et al. 2005).
cTwo of the sites in brackets are phosphorylated.
dOne of the sites in brackets is phosphorylated.
Figure 5.
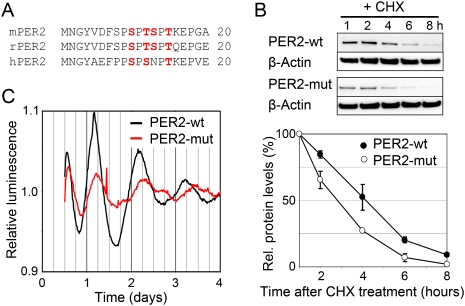
Mutation of N-terminal phosphorylation sites alters PER2 stability as well as circadian dynamics. (A) Two serine and/or threonine residues (red) of the highly conserved N-terminal region of PER2 are phosphorylated in living cells (Vanselow et al. 2006). The mass spectra do not allow a more exact location of the phosphorylated residues in this region. When phosphorylated in vitro, this region is phosphorylated only by CK2, but not by CK1δ or GSK-3β (see Supplemental Table 1). (B) NIH3T3 circadian reporter cells expressing PER2-wt or a mutant version of PER2 (PER2-mut: Ser10, Thr12, Ser13, and Thr15 were exchanged by Ala) from the same chromosomal location (Vanselow et al. 2006) were treated with the protein translation inhibitor CHX. Protein abundance was analyzed by immunoblotting and normalized to β-Actin-levels. Error bars, mean ± SEM (n = 3). (C) NIH3T3 reporter cells expressing either PER2-wt or PER2-mut were synchronized with dexamethasone and circadian oscillations were recorded by luminometry. Shown are detrended average time series (n = 3).
Given these data, we hypothesize that this N-terminal region of PER2 may be a target site for CK2 activity. If this N-terminal region is stabilizing PER2 upon phosphorylation, mutation of phosphoacceptor sites by alanine is predicted to destabilize the protein. Indeed, when the four serine and threonine residues of this region (Ser10, Thr12, Ser13, and Thr15) are exchanged by alanine (PER2-mut), the stability of the mutant PER2 is substantially decreased (Fig. 5B), consistent with the hypothesis that CK2-mediated phosphorylation at the PER2 N terminus contributes to the stabilization of the protein.
Mutation of in vitro defined CK2 phosphorylation sites phenocopies CK2 down-regulation
Next, we tested whether a phosphorylation defect of PER2 at the N terminus might cause the disrupted rhythms and long periods, which we observe in cells with reduced CK2 level or activity. To ensure that potential differences in circadian dynamics are not artifacts due to chromosomal position effects or variation of integration events, we used our previously described NIH3T3 reporter cells harboring a Flp Recombinase Target (FRT) site, in which either wild-type Per2 or Per2-mut was integrated (Vanselow et al. 2006). Indeed, cells expressing PER2-mut have reduced amplitude and longer periods compared with those expressing PER2 wild type (Fig. 5C). Based on these results, it seems very likely that phosphorylation by CK2 is stabilizing PER2 in vivo, which is required for normal nuclear accumulation and thereby a normal circadian period and amplitude.
Discussion
In this study, we searched for as-yet-unknown factors that ensure that a relatively short-lived clock protein such as PER2 can create rhythms that last an entire 24-h day. We knew from our previous study (Vanselow et al. 2006) that there are likely yet undiscovered PER2-stabilizing kinases, and we also knew (from many studies in various clock models) that the precise fine-tuning of phosphorylation events is of critical importance for circadian dynamics. Therefore, we predicted that systematic down-regulation of all kinases should lead us to the discovery of unknown kinase(s) important for PER2 protein stability and dynamics.
The unbiased RNAi screen in cell culture, which we set up here, has several features that makes it complementary to classic forward genetic screens in mice: (1) high throughput at relatively low costs—we can screen ∼2000 RNAi constructs per week. (2) The circadian clock can be studied in a human model system. Our results provide direct evidence that the molecular mechanism of the human circadian clock is likely to be very similar to that of mice (Fig. 1). (3) The sensitivity for detecting circadian phenotypes is predicted to be higher, since loss-of-function genetics results in more severe circadian phenotypes in uncoupled cells compared with behavioral outputs (Liu et al. 2007). (4) Clock components, whose ablation is lethal during development (as is probably the case for CK2 subunits, given the many roles CK2 plays in cellular physiology) (Litchfield 2003), or whose loss-of-function phenotypes are masked by developmental compensation effects, may be uncovered. Therefore, we are currently extending our screen to a genome-wide scale to identify more clock components that are predicted by genetic studies, suggesting as-yet-unknown quantitative trait loci regulating circadian rhythms (Lowrey and Takahashi 2004; Takahashi 2004).
Our RNAi screen led to the discovery of many potential new clock components, most of them kinases. One of them, CK2, especially caught our eyes because of its role for PER regulation in Drosophila as well as its importance for the circadian clocks of Neurospora (Yang et al. 2002; Dunlap et al. 2007) and Arabidopsis (Sugano et al. 1999). In Drosophila, PER protein level, phosphorylation, as well as nuclear entry dynamics are substantially altered in hypomorphic mutations of Drosophila CK2 subunits displaying long circadian periods (Lin et al. 2002; Akten et al. 2003; Smith et al. 2008). Therefore, we focused our subsequent work on CK2. For most of the other new candidates, we have not yet accumulated enough functional data to unambiguously prove their role within the circadian clockwork. Nevertheless, since RNAi-based perturbations copied “classical” phenotypes from mouse genetics (Fig. 1), it seems reasonable to assume that at least some of the newly discovered clock candidates will turn out to be true clock components. While future studies will nail down the function of these components within the mammalian clockwork, the sheer number of candidates suggests that the post-translational control of mammalian circadian rhythms is very complex.
Our results clearly demonstrate that normal CK2 levels and activity are essential within the mammalian circadian clockwork. Down-regulation of CK2 catalytic and regulatory subunits either individually or simultaneously leads to long circadian periods or to arrhythmicity, respectively. In contrast, overexpression of CK2 catalytic subunits but not kinase-inactive forms results in shortening of the circadian period. Given the fact that CK2 is involved in many cellular processes with a large number of substrates (Litchfield 2003), we cannot exclude other functions of CK2 within the mammalian clock in addition to those we uncovered in this study. The data presented here indicate that one function of CK2 is to bind PER2 and to promote its phosphorylation at N-terminal positions. This seems to stabilize PER2 early in its life cycle, which leads to normal nuclear accumulation. Thus, CK2 is most likely a PER2-stabilizing kinase.
How does phosphorylation of PER2 regulate the circadian period, and how is PER2 phosphorylation correlated with stability? Previous studies as well as the present study suggest that the answer to that question is not simple and one-dimensional. In contrast, interference with PER2 phosphorylation can lead to either short or long periods, dependent on which phosphorylation sites are affected. For example, when phosphorylation of Ser662 is inhibited by mutation (as occurs in FASPS patients), the circadian period gets shorter (Jones et al. 1999; Vanselow et al. 2006; Xu et al. 2007), while pharmacologic inhibition of CK1ɛ/δ activity (Eide et al. 2005; Vanselow et al. 2006; Reischl et al. 2007) lengthens the period. In both cases, PER2 is less phosphorylated, but with opposite effects on circadian period. Since PER2 in FASPS is unstable and CK1ɛ/δ inhibition stabilizes PER2, one might assume that the circadian period is directly correlated with PER2 stability. This, however, is not the case. As in FASPS, mutation of the N-terminal phospho-acceptor sites also leads to an unstable PER2 (Fig. 5B), but the resulting circadian periods are either short (in FASPS) or long (upon interference with CK2 phosphorylation).
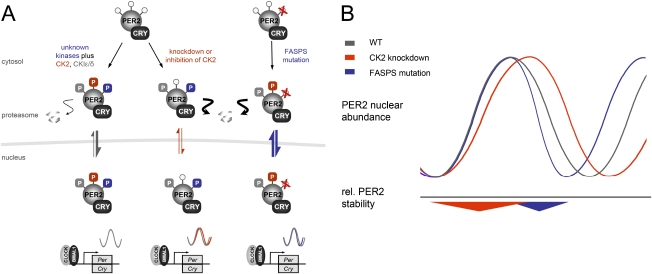
We propose that not PER2 stability, but nuclear localization of the PER/CRY repressor complex, is a major determinant of circadian period (Fig. 6). Our data on the delayed nuclear accumulation upon CK2 inhibition (Fig. 4C) and the premature nuclear clearance of PER2-FASPS (Vanselow et al. 2006) indicate that the circadian period is critically determined by the beginning and the end of nuclear repression. When the beginning of nuclear repression is delayed, because, for example, a signal for nuclear accumulation is missing, the circadian period gets longer. On the other hand, if the end of nuclear repression happens earlier because PER2 disappears earlier from the nucleus, the period gets shorter. In both cases, PER2 stays longer in the cytoplasm, either at a phase when it should already be in the nucleus or at a phase when it should still be in the nucleus. Since proteasomal degradation of PER2 happens primarily in the cytoplasm (Vanselow et al. 2006; Meng et al. 2008), this phase-dependent shift to a more cytoplasmic localization of PER2 protein would explain its reduced stability in both situations.
Figure 6.
Speculative model of a phase-dependent effect of PER2 stability on circadian period. (A, left) In the wild-type situation, PER2 is phosphorylated by multiple kinases including CK2 and CKIɛ/δ, allowing normal nuclear accumulation kinetics. (Middle) Depletion of CK2 destabilizes PER2 early in its life cycle, resulting in a later nuclear enrichment and thus a prolonged period. (Right) The FASPS mutation of PER2 precludes phosphorylation at Ser662, which leads to premature nuclear clearance (Vanselow et al. 2006) and thereby to a shortened circadian cycle. (B) Reduction in PER2 stability at the beginning of nuclear accumulation by CK2 knockdown (red) has a period lengthening effect due to delayed nuclear repression of CLOCK/BMAL1 transactivation. Accelerated nuclear clearance as it occurs in FASPS patients (blue) is also correlated with destabilization of PER2, but has the opposite effect on circadian period.
Based on our findings, we speculate that CK2-mediated phosphorylation of PER2 provides a signal that allows the timely nuclear accumulation of the repressor complex. While the exact mechanism of the nuclear accumulation dynamics of the PER/CRY repressor complex is elusive, we know that complex formation of PER and CRY proteins leads to nuclear localization and probably thereby to stabilization of both proteins (Kume et al. 1999). However, several questions remain to be investigated: (1) Is a signal required to allow nuclear accumulation of a cytoplasmic complex that is otherwise constantly degraded by CK1ɛ/δ in the cytoplasm? (2) Does the complex need to reach a concentration threshold or a certain stoichiometry in the cytoplasm to enter the nucleus, while CK1ɛ/δ-mediated degradation of PER proteins is slowing down this process? (3) Do PER/CRY complexes shuttle between the cytoplasm and nucleus and is a signal needed to allow nuclear retention? (4) Do PER and CRY proteins dissociate before entering the nucleus as has been reported for PER and TIM in Drosophila (Meyer et al. 2006)? In principle, CK2-mediated phosphorylation of PER2 could play a role in each of these—not mutually exclusive—scenarios. Future studies investigating the spatio-temporal pattern of phosphorylation, complex formation, stoichiometry, and subcellular localization are required to elucidate the mechanism of how and when nuclear repression is initiated—a critical determinant for setting the clock speed.
Materials and methods
RNAi screen
RNAi constructs were purchased from Open Biosystems. Lentiviruses were produced in HEK293T cells in a 96-well plate format essentially as described (Brown et al. 2008). Virus-containing supernatants were filtered and U-2 OS (human, American Type Culture Collection [ATCC] # HTB-96) reporter cells were transduced with 100 μL of virus filtrate plus 8 ng/μL protamine sulfate. After 1 d, medium was exchanged to puromycin-containing (10 μg/mL) medium. 3 d later, cells were synchronized with dexamethasone (1 μM) for 30 min. Bioluminescence was recorded for 5–7 d in a TopCount luminometer with a stacker unit (sampling rate: ∼30 min). Raw data were detrended by dividing the 24-h-running average. Periods and amplitudes were estimated by fitting the cosine wave function y = a * exp(b * t) * cos(2 *pi * t * 24/c + d), which includes an exponential term for damping (a = amplitude, b = damping, c = period, d = phase). For a detailed description of hit evaluation, see the Supplemental Material.
Generation of reporter cells/tissue
U-2 OS cells (human, ATCC # HTB-96) stably expressing firefly luciferase from a 0.9-kb Bmal1 promoter fragment (kind gift from Steve Brown, University of Zurich, Switzerland) were generated by lentiviral transduction of the respective expression plasmid. A single subclone was used for further experiments. Primary fibroblasts and lung tissues were derived from Per2∷Luc knock-in animals (Yoo et al. 2004). Care of animals was in accordance with the guidelines of our institute. To generate primary fibroblasts, lung and ear tissue was digested with liberase blendzyme 3 (Roche) in medium containing 20% fetal calf serum for 16 h. Subsequently, cells were washed and plated in standard tissue culture flasks. Adherent cells were passaged and considered as primary fibroblasts.
Bioluminescence recording
Reporter cells or tissues were cultured essentially as described (Vanselow et al. 2006) in phenol-red-free DMEM medium containing 10% fetal calf serum, antibiotics, and 250 μM D-luciferin (Biothema). Bioluminescence recordings were performed at 35°C–37°C either in 96-well plate luminometers (TopCount, PerkinElmer), Orion II (Berthold Detection Systems), a LumiCycle (Actimetrics), or light-tight boxes using a single photomultiplier tube (Hamamatsu Photonics).
RNAi studies
Lentiviruses containing RNAi constructs (Open Biosystems) were generated essentially as described (Brown et al. 2008). RNAi constructs targeting hCK2β were designed using the rules given in the RNAi codex database (http://codex.cshl.edu/scripts/newmain.pl). Oligos were cloned into pGIPZ lentiviral shRNAmir empty vector (Open Biosytems). Reporter cells were transduced either with single virus or mixes containing several viruses in 35-mm dishes. After 3 d, medium was exchanged to puromycin-containing (10 μg/mL) medium. The next day, cells were synchronized with dexamethasone (1 μM) for 30 min. Bioluminescence was recorded for 5–7 d.
Pharmacological perturbation
Lung tissue from one Per2∷Luc knock-in mouse (Yoo et al. 2004) was sliced with a tissue chopper. Eight slices each were cultured in 35-mm dishes, and bioluminescence was recorded in light-tight boxes with a photomultiplier tube (Hamamatsu). After 5 d, medium was replaced by medium with or without various concentrations of 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT) in 0.2% dimethyl sulfoxide (DMSO) and bioluminescence recording was continued. The concentration of the solvent DMSO was kept constant in all samples. U-2 OS reporter cells were seeded in complete medium into a white 96-well plate. The next day, cells were synchronized by adding 1 μM dexamethasone for 1 h. After synchronization, the medium was exchanged to complete medium with luciferin without phenol red. DMAT (1 μM, 5 μM, or 10 μM) or DMSO was added. The concentration of the solvent DMSO was kept constant in all samples. Luminescence was monitored in a 96-well plate luminometer Orion II for 7 d.
Co-IP and immunoblotting
Overexpressed epitope-tagged proteins
HEK293 cells were grown to 60% confluence, transfected with equal amounts of DNA (mPer2-V5: pcDNA-DEST40 [Invitrogen]; rCK2α/β-HA-Flag: VP5 [Clontech]) with FuGene 6 (Roche) according to the manufacturer's protocol. 48 h after transfection, cells were harvested in co-IP buffer (20 mM Tris-HCl at pH 8.0; 140 mM NaCl; 1.5 mM MgCl2; 1 mM TCEP; 1% Triton-X-100; 10% glycerin) containing protease and phosphatase inhibitor cocktails (Sigma). For each condition, 500 μg of total protein were incubated with 2 μg of anti-HA antibody (sc-7392, Santa Cruz Biotechnologies) and equivalent amounts of Protein A-agarose or G PLUS-agarose beads (Santa Cruz Biotechnologies) overnight at 4°C under constant agitation. Beads were washed three times in washing buffer (20 mM Tris-HCl at pH 8.0; 150 mM NaCl; 0.5% Igepal CA-630). Proteins were denatured in SDS-loading buffer (Invitrogen) by boiling for 5 min, separated by SDS-PAGE using 4%–12% Bis-Tris gels (Invitrogen), transferred to nitrocellulose membrane, and incubated with the following primary antibodies according to the manufacturer's instructions: anti-V5 (R960-25, Invitrogen), anti-Flag (F7425, Sigma), and anti-β-Actin (A3853, Sigma). The next day, membranes were probed with HRP-conjugated secondary antibodies (Santa Cruz Biotechnologies), and a chemiluminescence assay was performed using Super Signal West Pico substrate (Pierce) followed by protein detection with a Lumi-Imager (Roche).
Endogenous components
Liver tissue from Per2∷Luc mice was processed under nondenaturing conditions according to the protocol available at http://www.abcam.com/technical. Co-IP was performed as described above using the following antibodies: anti-CK2α (sc-6479, Santa Cruz Biotechnologies), anti-CK2β (sc-12793, Santa Cruz Biotechnologies), IgG2A (sc-3878, Santa Cruz Biotechnologies). Bioluminescence recordings were performed by incubation of the bead pellets for 10 sec with 20 μL of LARI (Promega) using the Beta Scout (PerkinElmer) detection system. Binding of CK2 subunits to PER2 was confirmed independently by co-IP followed by Western blot analysis (see Supplemental Fig. 5). Detection of coimmunoprecipitated PER2-LUC was performed with the anti-LUC antibody (sc-32896, Santa Cruz Biotechnologies). Nonspecific binding of recombinant luciferase generated in HEK293 cells to Protein A-agarose or G PLUS-agarose beads (Santa Cruz Biotechnologies) was excluded previously (data not shown).
Nuclear accumulation assay
Primary fibroblasts from Per2∷Luc mice were treated with cycloheximide (final concentration 355 μM) for 15 h. Cell viability was confirmed by visual inspection of the cells as well as detection of robust circadian rhythmicity after CHX removal (data not shown). Then medium was removed, and cells were washed and treated with either DMAT (20 μM) or DMSO. At indicated time points, nuclei were isolated by adding 200 μL of Hypotonic Lysis Buffer. After centrifugation, the supernatant was used as cytoplasmic fraction. The nuclear pellet was washed and resuspended. Nuclei and cytoplasmic fractions were analyzed for PER2-luciferase activity using standard luminometry.
For additional methods, see the Supplemental Material.
Acknowledgments
We thank D. Lohse, A. Grudziecki, M. Mette, and N. Thierfelder for excellent technical support. We are grateful to J.S. Takahashi, S.A. Brown, M. Flajolet, H. Rebholz, and E.E. Wanker for materials, and S.A. Brown and H. Herzel for critical discussions. This study was supported by the Deutsche Forschungsgemeinschaft (SFBs 618 and 740), as well as by the 6th EU framework program EUCLOCK.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.512209.
Supplemental material is available at http://www.genesdev.org.
References
- Akten B., Jauch E., Genova G.K., Kim E.Y., Edery I., Raabe T., Jackson F.R. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- Allada R., Meissner R.A. Casein kinase 2, circadian clocks, and the flight from mutagenic light. Mol. Cell. Biochem. 2005;274:141–149. doi: 10.1007/s11010-005-2943-1. [DOI] [PubMed] [Google Scholar]
- Brown S.A., Kunz D., Dumas A., Westermark P.O., Vanselow K., Tilmann-Wahnschaffe A., Herzel H., Kramer A. Molecular insights into human daily behavior. Proc. Natl. Acad. Sci. 2008;105:1602–1607. doi: 10.1073/pnas.0707772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger M.K., Wilsbacher L.D., Moran S.M., Clendenin C., Radcliffe L.A., Hogenesch J.B., Simon M.C., Takahashi J.S., Bradfield C.A. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L., Bassermann F., Maiolica A., Lee C., Nolan P.M., Godinho S.I., Draetta G.F., Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- DeBruyne J.P., Weaver D.R., Reppert S.M. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam V.M., Stegmeier F., Nalepa G., Sowa M.E., Chen J., Liang A., Hannon G.J., Sorger P.K., Harper J.W., Elledge S.J. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat. Cell Biol. 2007;9:556–564. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C., Loros J.J., Colot H.V., Mehra A., Belden W.J., Shi M., Hong C.I., Larrondo L.F., Baker C.L., Chen C.H., et al. A circadian clock in Neurospora: How genes and proteins cooperate to produce a sustained, entrainable, and compensated biological oscillator with a period of about a day. Cold Spring Harb. Symp. Quant. Biol. 2007;72:57–68. doi: 10.1101/sqb.2007.72.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide E.J., Woolf M.F., Kang H., Woolf P., Hurst W., Camacho F., Vielhaber E.L., Giovanni A., Virshup D.M. Control of mammalian circadian rhythm by CKIɛ-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M., Virshup D.M. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Godinho S.I., Maywood E.S., Shaw L., Tucci V., Barnard A.R., Busino L., Pagano M., Kendall R., Quwailid M.M., Romero M.R., et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- Harms E., Kivimae S., Young M.W., Saez L. Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms. 2004;19:361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- Jones C.R., Campbell S.S., Zone S.E., Cooper F., DeSano A., Murphy P.J., Jones B., Czajkowski L., Ptacek L.J. Familial advanced sleep-phase syndrome: A short-period circadian rhythm variant in humans. Nat. Med. 1999;5:1062–1065. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- Kowalska E., Brown S.A. Peripheral clocks: Keeping up with the master clock. Cold Spring Harb. Symp. Quant. Biol. 2007;72:301–305. doi: 10.1101/sqb.2007.72.014. [DOI] [PubMed] [Google Scholar]
- Kume K., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert S.M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lin J.M., Kilman V.L., Keegan K., Paddock B., Emery-Le M., Rosbash M., Allada R. A role for casein kinase 2α in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- Litchfield D.W. Protein kinase CK2: Structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003;369:1–15. doi: 10.1042/BJ20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.C., Welsh D.K., Ko C.H., Tran H.G., Zhang E.E., Priest A.A., Buhr E.D., Singer O., Meeker K., Verma I.M., et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey P.L., Takahashi J.S. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey P.L., Shimomura K., Antoch M.P., Yamazaki S., Zemenides P.D., Ralph M.R., Menaker M., Takahashi J.S. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly D.J., Allen J.A., Shokat K.M. A mechanism-based cross-linker for the identification of kinase-substrate pairs. J. Am. Chem. Soc. 2004;126:9160–9161. doi: 10.1021/ja048659i. [DOI] [PubMed] [Google Scholar]
- Meng Q.J., Logunova L., Maywood E.S., Gallego M., Lebiecki J., Brown T.M., Sladek M., Semikhodskii A.S., Glossop N.R., Piggins H.D., et al. Setting clock speed in mammals: The CK1 ɛτ mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M., Mazzotta G., Chen Z., Roenneberg T. The right place at the right time: Regulation of daily timing by phosphorylation. Genes & Dev. 2006;20:2629–2633. doi: 10.1101/gad.1479706. [DOI] [PubMed] [Google Scholar]
- Meyer P., Saez L., Young M.W. PER–TIM interactions in living Drosophila cells: An interval timer for the circadian clock. Science. 2006;311:226–229. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- Oster H., Yasui A., van der Horst G.T., Albrecht U. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes & Dev. 2002;16:2633–2638. doi: 10.1101/gad.233702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H., Damerow S., Hut R.A., Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J. Biol. Rhythms. 2006;21:350–361. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- Pagano M.A., Meggio F., Ruzzene M., Andrzejewska M., Kazimierczuk Z., Pinna L.A. 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole: A novel powerful and selective inhibitor of protein kinase CK2. Biochem. Biophys. Res. Commun. 2004;321:1040–1044. doi: 10.1016/j.bbrc.2004.07.067. [DOI] [PubMed] [Google Scholar]
- Reischl S., Vanselow K., Westermark P.O., Thierfelder N., Maier B., Herzel H., Kramer A. β-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J. Biol. Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- Schlosser A., Vanselow J.T., Kramer A. Mapping of phosphorylation sites by a multi-protease approach with specific phosphopeptide enrichment and NanoLC-MS/MS analysis. Anal. Chem. 2005;77:5243–5250. doi: 10.1021/ac050232m. [DOI] [PubMed] [Google Scholar]
- Shearman L.P., Sriram S., Weaver D.R., Maywood E.S., Chaves I., Zheng B., Kume K., Lee C.C., van der Horst G.T., Hastings M.H., et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shirogane T., Jin J., Ang X.L., Harper J.W. SCFβ-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- Siepka S.M., Yoo S.H., Park J., Song W., Kumar V., Hu Y., Lee C., Takahashi J.S. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.M., Li M.Z., Chang K., Ge W., Golding M.C., Rickles R.J., Siolas D., Hu G., Paddison P.J., Schlabach M.R., et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Smith E.M., Lin J.M., Meissner R.A., Allada R. Dominant-negative CK2α induces potent effects on circadian rhythmicity. PLoS Genet. 2008;4:e12. doi: 10.1371/journal.pgen.0040012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Hu G., Rickles R.J., Hannon G.J., Elledge S.J. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl. Acad. Sci. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Rape M., Draviam V.M., Nalepa G., Sowa M.E., Ang X.L., McDonald E.R., III, Li M.Z., Hannon G.J., Sorger P.K., et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Sugano S., Andronis C., Ong M.S., Green R.M., Tobin E.M. The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc. Natl. Acad. Sci. 1999;96:12362–12366. doi: 10.1073/pnas.96.22.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi J.S. Finding new clock components: Past and future. J. Biol. Rhythms. 2004;19:339–347. doi: 10.1177/0748730404269151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi J.S., Hong H.K., Ko C.H., McDearmon E.L. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat. Rev. Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh K.L., Jones C.R., He Y., Eide E.J., Hinz W.A., Virshup D.M., Ptacek L.J., Fu Y.H. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- van der Horst G.T., Muijtjens M., Kobayashi K., Takano R., Kanno S., Takao M., de Wit J., Verkerk A., Eker A.P., Van Leenen D., et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vanselow K., Kramer A. Role of phosphorylation in the mammalian circadian clock. Cold Spring Harb. Symp. Quant. Biol. 2007;72:167–176. doi: 10.1101/sqb.2007.72.036. [DOI] [PubMed] [Google Scholar]
- Vanselow K., Vanselow J.T., Westermark P.O., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A. Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes & Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilk G., Saulnier R.B., St Pierre R., Litchfield D.W. Inducible expression of protein kinase CK2 in mammalian cells. Evidence for functional specialization of CK2 isoforms. J. Biol. Chem. 1999;274:14406–14414. doi: 10.1074/jbc.274.20.14406. [DOI] [PubMed] [Google Scholar]
- Vitaterna M.H., Selby C.P., Todo T., Niwa H., Thompson C., Fruechte E.M., Hitomi K., Thresher R.J., Ishikawa T., Miyazaki J., et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Padiath Q.S., Shapiro R.E., Jones C.R., Wu S.C., Saigoh N., Saigoh K., Ptacek L.J., Fu Y.H. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- Xu Y., Toh K.L., Jones C.R., Shin J.Y., Fu Y.H., Ptacek L.J. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Cheng P., Liu Y. Regulation of the Neurospora circadian clock by casein kinase II. Genes & Dev. 2002;16:994–1006. doi: 10.1101/gad.965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.H., Yamazaki S., Lowrey P.L., Shimomura K., Ko C.H., Buhr E.D., Siepka S.M., Hong H.K., Oh W.J., Yoo O.J., et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.W., Kay S.A. Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]