Abstract
Innate immune responses to bacterial or viral infection require rapid transition of large cohorts of inflammatory response genes from poised/repressed to actively transcribed states, but the underlying repression/derepression mechanisms remain poorly understood. Here, we report that, while the nuclear receptor corepressor (NCoR) and silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressors establish repression checkpoints on broad sets of inflammatory response genes in macrophages and are required for nearly all of the transrepression activities of liver X receptors (LXRs), they can be selectively recruited via c-Jun or the Ets repressor Tel, respectively, establishing NCoR-specific, SMRT-specific, and NCoR/SMRT-dependent promoters. Unexpectedly, the binding of NCoR and SMRT to NCoR/SMRT-dependent promoters is frequently mutually dependent, establishing a requirement for both proteins for LXR transrepression and enabling inflammatory signaling pathways that selectively target NCoR or SMRT to also derepress/activate NCoR/SMRT-dependent genes. These findings reveal a combinatorial, corepressor-based strategy for integration of inflammatory and anti-inflammatory signals that play essential roles in immunity and homeostasis.
Keywords: Inflammatory genes, NCoR, SMRT, TEL, cJun, p50
Inflammation is an adaptive host response that serves to direct the immune system and tissue repair processes to sites of infection and injury. Although inflammation normally resolves following elimination of infectious or traumatic stimuli, sustained inflammation can result in the induction or amplification of numerous chronic disease states that include atherosclerosis, insulin resistance, neurodegenerative disease, cancer, and autoimmune disorders. At a cellular level, genes that mediate inflammatory responses must be kept tightly repressed under normal conditions, but must also be rapidly and highly induced in the setting of infection or injury. Many of these genes are under the transcriptional control of signal-dependent transcription factors that include members of the nuclear factor κB (NFκB), activator protein 1 (AP1), and interferon regulatory factor (IRF) families of transcription factors (Karin and Greten 2005). The activities and/or expression of these factors can be induced by a large number of receptor systems, including pattern recognition receptors for pathogen-associated molecules, and receptors for cell-derived inducers of inflammation, such as IL1β, TNFα, and interferons (Akira and Takeda 2004). Upon activation, NFκB, AP1, and IRF proteins are capable of inducing the expression of hundreds of genes in diverse cell types that initiate and amplify inflammatory processes and promote the development of acquired immunity (Li and Verma 2002; Honda and Taniguchi 2006).
Recent studies have suggested that maintenance of at least some proinflammatory genes in a poised but repressed state in the absence of signal requires active repression by corepressor complexes that contain the nuclear receptor corepressor (NCoR) or the related silencing mediator of retinoic acid and thyroid hormone receptors (SMRT) (Hoberg et al. 2004; Ogawa et al. 2004; Perissi et al. 2004; Pascual et al. 2005). NCoR and SMRT were initially identified based on their ability to bind unliganded retinoic acid and thyroid hormone receptors and to mediate active repression of their respective target genes through the recruitment of additional corepressor molecules that include histone deacetylases (HDACs) (Chen and Evans 1995; Horlein et al. 1995; Guenther et al. 2000). In addition to nuclear receptors, NCoR and SMRT can exert repressive effects via interactions with numerous other transcription factors, including AP-1 proteins (Lee et al. 2000; Zhang et al. 2002), ETO-1/2 (Lutterbach et al. 1998; Wang et al. 1998), NF-κB (Lee et al. 2000; Baek et al. 2002; Perissi et al. 2004), and Ets proteins (Chakrabarti and Nucifora 1999; Guidez et al. 2000; Wang and Hiebert 2001). Consistent with this, analysis of NCoR-deficient macrophages revealed that NCoR acts to repress subsets of NF-κB and AP-1-dependent genes that regulate inflammation, cell migration, and collagen catabolism (Ogawa et al. 2004). Similarly, clearance of SMRT was found to be a requirement for NFκB-dependent activation of the cIAP-2 and IL-8 genes in DU145 cells in response to integrin attachment (Hoberg et al. 2004).
Recent studies indicate that NCoR complexes are not only required for active repression of inflammatory target genes in the absence of stimulus, but are also essential for ligand-dependent transrepression of at least some of these genes by the nuclear receptors PPARγ and liver X receptors (LXRs). Inhibition of lipopolysaccharide (LPS) activation of the iNOS gene by PPARγ and LXR agonists in macrophages and inhibition of IL1β activation of the CRP gene in hepatocytes by LXR agonists was found to result from their ability to prevent signal-dependent clearance of NCoR complexes from the promoters of these genes (Pascual et al. 2005; Blaschke et al. 2006; Ghisletti et al. 2007). LXRs and PPARγ have important biological roles as mediators of anti-inflammatory signaling pathways, but the global extent to which NCoR and/or SMRT contribute to these activities is unknown.
Although NCoR and SMRT share a common molecular architecture and form similar complexes, genetic deletion of either protein results in embryonic lethality, indicating that they cannot fully compensate for each other during development. Studies of NCoR−/− and SMRT−/− embyos indicate that NCoR is required for CNS, erythrocyte, and thymocyte development, while SMRT is required for cardiac development and maintenance of neural stem cells (Jepsen et al. 2000, 2007, 2008). The extent to which these biological requirements reflect distinct patterns of expression and/or are due to different interactions with transcriptional repressors is not known. SMRT and NCoR also differ in their response to kinase pathways that regulate their function. SMRT is regulated by MEKK1 and IKKα, leading to its redistribution from the nuclear to the cytoplasmic compartment (Hong and Privalsky 2000; Hoberg et al. 2004; Jonas and Privalsky 2004). In contrast, NCoR can be negatively regulated by the Akt and MEKK1/TAB2 pathways (Baek et al. 2002; Hermanson et al. 2002). The extent to which NCoR and SMRT exert overlapping or distinct roles in the regulation of inflammatory programs of gene expression remains largely undefined.
Here, we report combinatorial roles of NCoR and SMRT in the regulation of broad sets of inflammatory response genes in macrophages and demonstrate that they are required for nearly all of the transrepression activities of LXRs. Unexpectedly, both NCoR and SMRT are required to establish stable corepressor complexes on a large subset of these genes. As a consequence, both NCoR and SMRT are required for LXR transrepression of these genes. Conversely, this class of genes can also be derepressed by signals that selectively target NCoR or SMRT. Combinatorial interactions between NCoR and SMRT thus provide a widely used corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways.
Results
NCoR and SMRT are globally required for LXR transrepression in macrophages
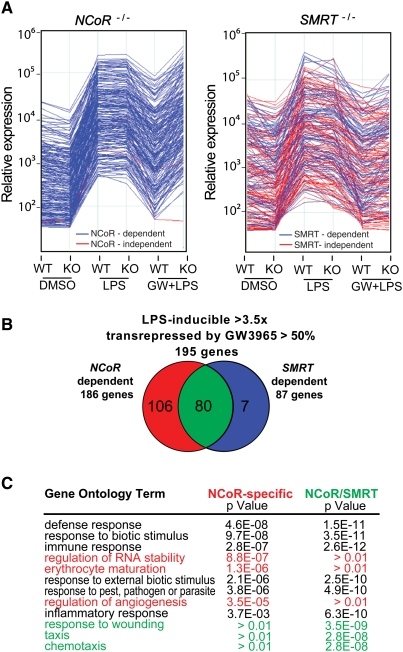
NCoR and SMRT are highly expressed in a variety of macrophage subtypes and cell lines, exemplified by elicited peritoneal macrophages, fetal liver-derived macrophages, and the RAW264.7 macrophage cell line (Supplemental Fig. S1A,B). LXRα and LXRβ are also expressed in each of these macrophages, and mediate repression of LPS responses to LXR-specific ligands such as GW3965 (Figs. 1A, 2C; Supplemental Fig. S1C). To evaluate the relative roles of NCoR and SMRT in LPS responses and LXR-dependent transrepression pathways, gene-expression profiling experiments were performed using fetal liver-derived macrophages from NCoR−/− or SMRT−/− embryos, comparing each knockout macrophage to control macrophages derived from wild-type embryos from the same litter. Although NCoR−/− and SMRT−/− macrophages exhibited quantitative differences in expression levels of a number of macrophage-specific genes (e.g., Supplemental Fig. S2), the response of each knockout macrophage to LPS treatment was similar to that of the corresponding wild-type control as assessed by gene expression profiling (Fig. 1A). Considering the 200 most highly LPS-inducible, GW3965-sensitive genes for each wild-type macrophage, NCoR was required for nearly all of the inhibitory function of GW3965, while inhibition of ∼40% of the LPS-inducible/GW3965-sensitive genes required SMRT (Fig. 1A).
Figure 1.
NCoR and SMRT are globally required for LXR transrepression. (A) Relative expression of the 200 most highly LPS-inducible, GW3965-sensitive genes in macrophages derived from fetal livers of wild-type (WT), NCoR−/− (KO), and SMRT−/− (KO) embryos under control conditions and after 6 h of LPS treatment in the absence or presence of LXR ligand (GW3965, GW 1 μM). Wild-type littermate embryos were used to derive control macrophages for each genotype. The NCoR-dependent or SMRT-dependent genes are color-coded in blue in each experiment. The NCoR-independent or SMRT-independent genes are color-coded in red in each experiment. (B) Venn diagram indicating relationships between LPS-inducible, GW3965-sensitive genes with respect to NCoR and SMRT dependence. The red sector respresents NCoR-specific genes, the blue sector SMRT-specific genes, and the green sector NCoR/SMRT-dependent genes. (C) GO analysis of the NCoR-specific and NCoR/SMRT-dependent sets of genes for selected biological process annotations.
The strain-specific, wild-type macrophages used as controls for NCoR−/− or SMRT−/− macrophages exhibited quantitative differences in the degree of induction of specific genes observed in response to LPS, as well as differences in the total number of genes induced. These differences are consistent with the distinct genetic backgrounds of the mouse strains that are required to maintain viability of knockout embryos to at least embryonic day 17. However, 195 genes were identified that were induced >3.5-fold by LPS in the wild-type cells corresponding to each knockout and that were also repressed by >50% by GW3965 (Fig. 1B; Supplemental Table T1). Nearly all of these genes (193 out of 195) required NCoR and/or SMRT for the repressive effects of GW3965. The majority of these genes (106 out of 195) exhibited loss of transrepression primarily in the NCoR KO, and are referred to as NCoR-specific. A small subset (seven out of 195) exhibited loss of transrepression primarily in SMRT−/− macrophages and are referred to as SMRT-specific. Surprisingly, a significant fraction of LXR-sensitive genes (80 out of 195) required both NCoR and SMRT for the inhibitory actions of GW3965 and are referred to as NCoR/SMRT-dependent.
Gene ontology (GO) analysis of the NCoR-specific and NCoR/SMRT-dependent sets of genes for biological process annotations revealed significant enrichment for general inflammation and immunity terms such as “defense response,” for both categories of genes, as expected (Fig. 1C). However, more specific annotations were differentially distributed among the three categories. For example, the GO terms “taxis” and chemotaxis were selectively enriched in the NCoR/SMRT-dependent group of genes and terms related to RNA metabolism and angiogenesis were selectively enriched in the NCoR-specific group of genes (Fig. 1C).
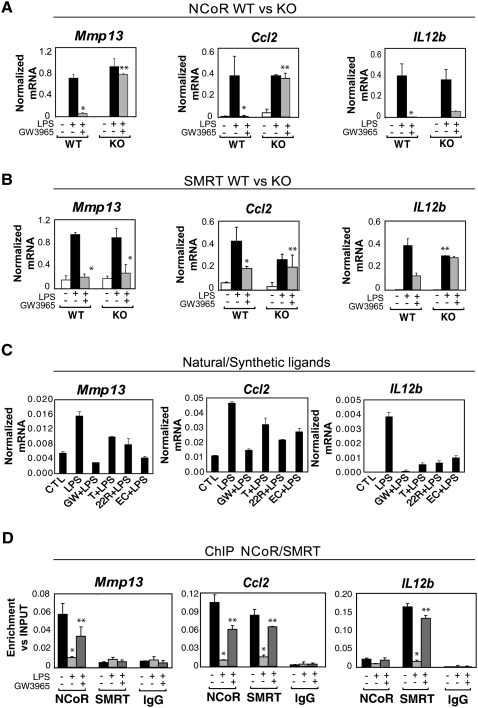
Validation of the gene-expression analysis by quantitative PCR confirmed the identification of three subsets of inflammatory response genes, exemplified by the NCoR-specific gene Mmp13, the SMRT-specific gene Il12b, and the NCoR/SMRT-dependent gene Ccl2 (Fig. 2A,B). This validation was extended by Q-PCR analysis of multiple additional genes belonging to these three categories (Supplemental Fig. S3; data not shown). To determine whether these gene-specific corepressor requirements for LXR transrepression identified in fetal liver macrophages were also required in other types of macrophages, siRNA knockdown experiments were performed in elicited peritoneal macrophages. LXR transrepression of Mmp13 was selectively impaired by the NCoR-specific siRNA, transrepression of Il12b was selectively impaired by knockdown of SMRT, and transrepression of Ccl2 was impaired by knockdown of either NCoR or SMRT, consistent with results obtained in SMRT−/− and NCoR−/− macrophages (Supplemental Fig. S4; knockdown efficiencies for all siRNA experiments are shown in Supplemental Fig. S13). In addition to being inhibited by GW3965, the LPS response of each of these genes was also repressed by a distinct synthetic LXR ligand (T1317), as well as by the natural LXR ligands 22R hydroxycholesterol and 24,25 epoxycholesterol (Fig. 2C).
Figure 2.
Expression profiles and promoter occupancy of NCoR- and SMRT-dependent target genes. (A,B) Expression profiles of NCoR-specific (Mmp13), SMRT-specific (Il12b), and NCoR/SMRT-dependent genes (Ccl2) analyzed by Q-PCR in wild-type, NCoR−/−, or SMRT−/− macrophages. Cells were treated with DMSO (white bars) or with LPS (1 μg/mL) for 6 h in the absence (black bars) or presence of LXR ligand (GW3965, 1 μM, gray bars) in wild-type versus NCoR−/− (A) or wild-type versus SMRT−/− macrophages (B). (*) P < 0.05 versus LPS; (**) P < 0.05 versus GW3965 + LPS wild type. (C) Effects of natural and synthetic LXR ligands (1 μM GW3965, 1 μM T1317, 5 μM 22R hydroxycholesterol, and 5 μM 24,25 epoxycholesterol) on LPS responses. (D) Promoter occupancy of the target genes described in A and B as evaluated by ChIP analysis. Primary macrophages were untreated (black bars) or treated with LPS (1 μg/mL) or GW3965 + LPS as indicated for 1 h. ChIP assays were performed with an antibody against NCoR or SMRT or control IgG. Immunoprecipitated DNA was analyzed by real-time PCR using primers specific for the indicated promoters. (*) P < 0.01 versus control; (**) P < 0.05 versus LPS. Results are expressed as the average of three independent experiments. Error bars represent standard deviations.
A common transrepression mechanism on NCoR-specific, SMRT-specific, and NCoR/SMRT-dependent genes
The observation that transrepression mediated by LXRs required either NCoR or SMRT and that about half of the LPS-inducible LXR-sensitive genes required both corepressors raised the questions whether NCoR and/or SMRT dependence correlated with NCoR and/or SMRT promoter occupancy. Chromatin immunoprecipitation (ChIP) experiments in primary macrophages confirmed this to be the case. NCoR but not SMRT was present under basal conditions on the NCoR-specific Mmp13 promoter, SMRT but not NCoR was recruited on the SMRT-dependent Il12b promoter, and both corepressors were found to be present on the NCoR and SMRT-dependent Ccl2 promoter (Fig. 2D). This pattern was confirmed for several other NCoR-specific, SMRT-specific, and NCoR/SMRT-dependent promoters (Supplemental Fig. S5).
The initial description of the LXR transrepression pathway based on studies of the inducible nitric oxide synthase (iNOS) gene identified several biochemical features in addition to the requirement for NCoR that included: (1) the requirement for SUMOylation of LXR, (2) the localization of LXR to target promoters, and (3) the prevention of NCoR clearance in response to LPS (Ghisletti et al. 2007). To determine whether SUMOylation is required for repression of each class of promoter, siRNAs were used to knockdown expression of Ubc9, the sole SUMO E3 ligase, in primary macrophages. This resulted in loss or reduction of GW3965-dependent inhibition of the responses of Mmp13, Ccl2, and Il12b to LPS treatment (Supplemental Fig. S6A). Similar results were observed for Il1a, Il1b, and Il18 (data not shown). ChIP experiments further documented recruitment of LXR to the Mmp13, Ccl2, and Il12b promoters in response to GW3965 treatment (Supplemental Fig. S6B). Finally, NCoR and/or SMRT were cleared upon LPS stimulation and treatment with LXR ligand prevented corepressor clearance (Fig. 2D). Thus, a common, SUMOylation-dependent mechanism appears to operate on each class of promoters and the gene-specific functional requirements for NCoR and SMRT for LXR transrepression are in accord with their gene-specific promoter occupancy under basal conditions.
c-Jun recruits NCoR to AP1 sites on NCoR-specific promoters
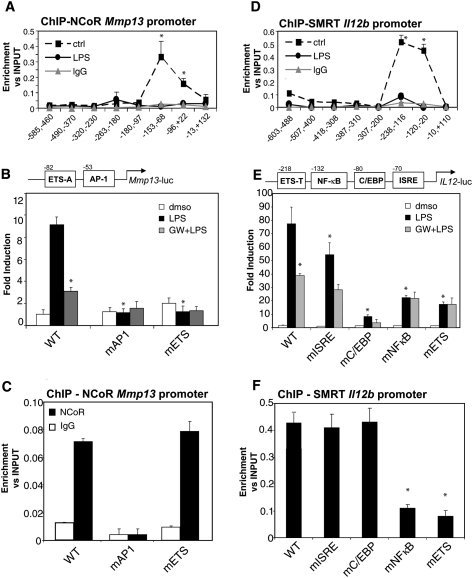
The findings that NCoR and SMRT bind to and regulate both distinct and common inflammatory response genes raised the question of how the two corepressors discriminate between different promoters. To address this question, Mmp13, Il12b, and Ccl2 were chosen for further study, based on the extensive knowledge regarding their cis-regulatory elements (diagrammed in Figs, 3B,E, 6A [below]) and their respective occupancy by NCoR only, SMRT only, or by both NCoR and SMRT. To investigate the mechanism by which NCoR was recruited to the Mmp13 gene, we first characterized the NCoR-binding region by ChIP–QPCR experiments using primers spanning the proximal region of the Mmp13 promoter. Figure 3A shows that NCoR binds to a region between the transcription start site and −100 base pairs (bp) that has been shown previously to contain a functionally important AP1 site and an ETS site (ETS-A) that binds ETS activators (Pendas et al. 1997; Tardif et al. 1997; Solis-Herruzo et al. 1999; de la Torre et al. 2005). Mutation of each of these sites abolished responsiveness of the Mmp13 promoter to LPS (Fig. 3B). In contrast, mutation of the AP-1 site, but not the ETS-A site, abolished binding of NCoR to the Mmp13 promoter under basal conditions (Fig. 3C). These results agree with previous studies indicating that c-Jun is an essential factor for the recruitment of NCoR to the Mmp9 promoter (Ogawa et al. 2004). We further confirmed that a member of the AP1 transcription factor family, c-Jun, and NCoR are both present on the Mmp13 promoter in primary macrophages and that, upon LPS stimulation, there is an exchange of c-Jun/NCoR corepressor complexes for c-Jun/c-Fos activator complexes (Supplemental Fig. S7A). Interestingly, AP1 consensus sequences are highly enriched in NCoR-dependent proinflammatory promoters identified by the microarray study, including Ccl4, Cxcl10, Cxcl2, Cxcl9, and iNOS (Supplemental Table S1). ChIP experiments confirmed the presence of c-Jun on five of five of NCoR-specific target genes tested (Supplemental Fig. S8A). Furthermore, knockdown c-Jun using specific siRNAs impaired NCoR recruitment on all these target genes (Supplemental Fig. S8B), suggesting a general role of c-Jun in mediating recruitment of NCoR to NCoR-specific promoters.
Figure 3.
NCoR is recruited to an AP-1 site in the Mmp13 promoter, and SMRT is recruited to an ETS site in the Il12b promoter. (A). NCoR occupancy of the Mmp13 proximal promoter region evaluated by ChIP assay. Primary macrophages were untreated (solid square) or treated with LPS (1 μg/mL, solid circles) for 1 h. ChIP assays were performed with an antibody against NCoR and control IgG. Immunoprecipitated DNA was analyzed by real-time PCR using primers that amplified the indicated regions of the Mmp13 promoter. (B) AP1 and ETS mutations abolished LPS induction of the Mmp13 promoter. RAW264.7 cells were transfected with luciferase reporter genes driven by the wild-type (WT) Mmp13 promoter or Mmp13 promoters with mutant AP1 or ETS sites. Twenty-four hours after transfection, cells were treated with DMSO (white bars) or with LPS (1 μg/mL) for 12 h in the absence (black bars) or presence of LXR ligand (GW3965, 1 μM, gray bars). (*) P < 0.05 versus LPS wild type. (C) The AP1 site is necessary for NCoR recruitment on the Mmp13 promoter. RAW cells were transfected with the wild-type or indicated mutant Mmp13-luciferase vectors. ChIP assays were performed with an antibody against NCoR and control IgG. Immunoprecipitated DNA was analyzed by real-time PCR using primers designed to amplify a region across the junction of the luciferase reporter coding region and the Mmp13 promoter. (*) P < 0.01 versus wild type. (D) SMRT occupancy of the Il12b proximal promoter region evaluated by ChIP assay. Primary macrophages were untreated (solid square) or treated with LPS (1 μg/mL, solid circles). ChIP assay was performed with an antibody against SMRT and control IgG. Immunoprecipitated DNA was analyzed by real-time PCR using primers that amplified the indicated regions of the Il12b proximal region. (*) P < 0.05 versus LPS. (E) LXR transrepression is impaired by ETS and NFκB mutations of the Il12b promoter. RAW264.7 cells were transfected with luciferase reporter genes driven by the wild-type (WT) Il12b promoter or Il12b promoters in which the ETS, NFκB, C/EBP, or ISRE sites were mutated. Twenty-four hours after transfection, cells were treated with DMSO (white bars) or with LPS (1 μg/mL) for 12 h in the absence (black bars) or presence of LXR ligand (GW3965, 1 μM, gray bars). (*) P < 0.05 versus LPS wild type. (F) The κB and ETS sites are necessary for SMRT recruitment to the Il12b promoter. RAW cells were transfected with the indicated wild-type or mutant Il12b-promoter/luciferase vectors, and ChIP assays were performed with an antibody against SMRT and control IgG. Immunoprecipitated DNA was analyzed by real-time PCR using primers designed to amplify a region between the luciferase and the proximal promoter region. (*) P < 0.05 versus wild type. Results are expressed as average of three independent experiments. Error bars represent standard deviations.
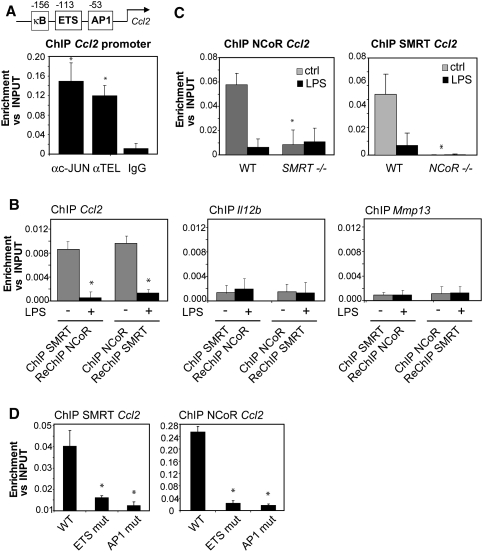
Figure 6.
NCoR and SMRT bind interdependently to the Ccl2 promoter. (A) c-Jun and TEL occupy the Ccl2 promoter as shown by ChIP in primary macrophages. (*) P < 0.05 versus IgG. (B) NCoR and SMRT are present in the same complex as shown by ReChIP assays in unstimulated primary macrophages (gray bars). Immunoprecipitated DNA was analyzed by real-time PCR primers specific for the Ccl2 promoter. The Il12b and Mmp13 promoters were used as negative controls (*) P < 0.01 versus control. (C) SMRT and NCoR are mutually required for binding to the Ccl2 promoter, as shown by ChIP assay using liver-derived macrophages from wild-type, NCoR−/−, and SMRT−/− embryos. Cells were untreated (gray bars) or treated with LPS (black bars) for 1 h. (*) P < 0.05 versus wild-type control. (D) Mutations in the ETS and AP-1 sites of the Ccl2 promoter each lead to loss of NCoR and SMRT occupancy. RAW cells were transfected with luciferase reporter genes under the control of the wild-type Ccl2 promoter or Ccl2 promoters mutated at the AP1 or ETS sites. ChIP assays were performed using antibodies against NCoR, SMRT, and control IgG. Immunoprecipitated DNA was analyzed by real-time PCR using primers amplifying a region corresponding to the junction between the Ccl2 promoter and the luciferase reporter coding region. (*) P < 0.05 versus wild type. Results are expressed as the average of three independent experiments. Error bars represent standard deviations.
TEL recruits SMRT to ETS sites of SMRT-specific promoters
We next evaluated the molecular mechanism by which SMRT is recruited to the Il12b promoter. By ChIP analysis using primers tiling the proximal region of the Il12b promoter, SMRT was found to be recruited to a region between −10 bp and −250 bp from the transcription start site (Fig. 3D). This region has previously been demonstrated to contain ISRE, NFκB, C/EBP, and ETS elements (Murphy et al. 1995; Plevy et al. 1997; Gri et al. 1998; Sanjabi et al. 2000, 2005; Wang et al. 2000; Bradley et al. 2003; Maruyama et al. 2003; Zhu et al. 2003). Mutation of each of these sites individually reduced, but did not abolish, promoter responsiveness to LPS (Fig. 3E), consistent with previous studies (Murphy et al. 1995; Plevy et al. 1997; Wang et al. 2000; Maruyama et al. 2003; Zhu et al. 2003). Intriguingly, mutation of the NF-κB and ETS sites not only resulted in reduced responsiveness to LPS, but also abolished LXR transrepression (Fig. 3E). ChIP experiments evaluating the binding of SMRT to the wild-type and mutant Il12b promoters revealed that mutation of the ETS and κB sites completely abolished SMRT recruitment (Fig. 3F). These results provide an explanation for the loss of LXR transrepression of the residual promoter activity of Il12b promoters containing mutations in the ETS and NFκB motifs (Fig. 3E), and indicate that SMRT recruitment to the Il12b promoter is dependent on these sites.
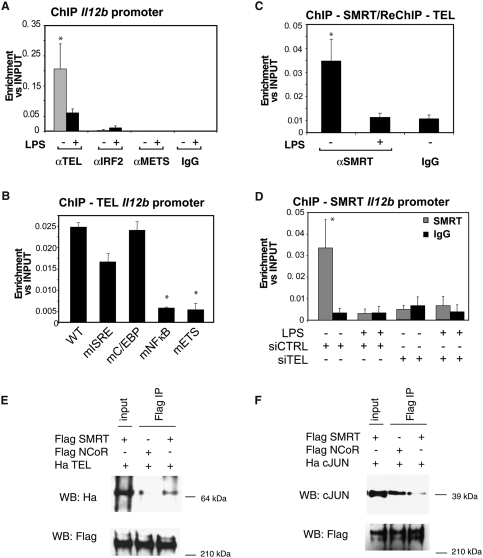
Previous studies demonstrated that IRF1 and IRF8 are recruited to the ETS site on the Il12b promoter upon IFNγ and LPS stimulation in macrophages (Wang et al. 2000), but factors present on this site under basal conditions that could serve to recruit SMRT are not known. ETS proteins form one of the largest families of signal-dependent transcription regulators, mediating cell proliferation, differentiation, and tumorigenesis. Most of the known ETS proteins have been shown to activate transcription, but a subset can act as transcriptional repressors (Mavrothalassitis and Ghysdael 2000). In addition, members of the IRF family have been shown to be able to bind to ETS sites and in particular IRF2 has been proposed to act as a repressor (Masumi and Ozato 2001). IRF2 and the ETS repressors METS and TEL are expressed in macrophages, and were therefore evaluated for their potential roles in recruiting SMRT to the Il12b promoter.
ChIP experiments demonstrated that TEL, but not METS or IRF2, specifically bound to the Il12b promoter in unstimulated macrophages, and that this recruitment was dependent on the ETS and NFκB sites (Fig. 4A,B). In contrast, ChIP experiments documented recruitment of METS and IRF2 to known target genes (Supplemental Fig. S7B; Hester et al. 2007; Yang et al. 2007). By ReChIP analysis of the Il12b promoter, TEL was found to be present in the same complex with SMRT and both SMRT and TEL were cleared in response to LPS treatment (Fig. 4C). To test whether SMRT binds to the ETS consensus sequence through TEL or vice versa, specific siRNAs were used to knockdown TEL and SMRT in primary macrophages (Supplemental Fig. S13B,E). ChIP experiments demonstrated that SMRT was not present on the Il12b promoter when TEL expression was reduced by the use of TEL-specific siRNA (Fig. 4D). In contrast, TEL recruitment on Il12b promoter was unaffected by SMRT-specific siRNA (data not shown). Experiments evaluating the expression of Il12b by QPCR in primary macrophages indicated that LXR transrepression is partially impaired by knocking down TEL, consistent with the lack of SMRT on the promoter (Supplemental Fig. S9A). Although TEL has previously been demonstrated to interact with NCoR, coimmunoprecipitation (co-IP) assays using Flag-tagged proteins (thereby eliminating differences that could reflect different antibodies), indicated that full-length SMRT interacted much more robustly with TEL than NCoR (Fig. 4E). Conversely, c-Jun interacted preferentially with NCoR (Fig. 4F). These results suggest that different affinities of c-Jun and TEL for NCoR and SMRT account, at least in part, for the preferential binding of NCoR to the Mmp13 promoter and SMRT to the Il12b promoter.
Figure 4.
SMRT is recruited to the Il12b promoter through TEL (A) TEL binds specifically to the Il12b promoter. ChIP analysis for binding of TEL, IRF2, and METS to the Il12b promoter was performed with untreated (gray bars) and LPS-treated (black bars) primary macrophages using TEL, IRF2, and METS antibodies or control IgG (*) P < 0.05 versus IgG. (B) The κB and Ets sites are necessary for TEL binding to the Il12b promoter. RAW cells were transfected with the indicated wild-type and mutant Il12b-luciferase reporter plasmids. ChIP assay was performed with antibody against TEL and control IgG. Immunoprecipitated DNA was analyzed by real-time PCR using primers designed to amplify a region between the luciferase and the proximal promoter region. (*) P < 0.01 versus wild-type. (C) TEL and SMRT are present in the same complex on Il12b promoter as shown by ReChIP assay done in primary macrophages. (*) P < 0.05 versus IgG. (D) TEL is the beacon for the recruitment of SMRT on Il12b promoter. Primary macrophages were transfected with siRNA control and siRNA against TEL. ChIP assays were performed using antibodies against SMRT (top panel, gray bars) or control IgG (black bars, top and bottom panels). (*) P < 0.05 versus IgG. Results are expressed as average of three independent experiments. Error bars represent standard deviations. (E) Preferential co-IP of HA-TEL by Flag-SMRT. (F) Preferential co-IP of c-Jun by Flag-NCoR.
The mechanism by which the NFκB site in the Il12b promoter affects SMRT recruitment is complex, because mutation of this site leads to loss of TEL binding (Fig. 4B). We confirmed previous ChIP studies (Sanjabi et al. 2000; Sanjabi et al. 2005) demonstrating that p50 is present in the basal state on the Il12b promoter and c-Rel is recruited after LPS treatment (Supplemental Fig. S9B). p50 could thus directly participate in SMRT recruitment on the Il12b promoter as well as indirectly participate by influencing the binding of TEL. In either case, these data collectively support a model in which TEL functions to recruit SMRT to the ETS site of the Il12b promoter, with the neighboring NFκB site playing a contributing role. Consistent with these data, ETS and NFκB sites similar to those in the Il12b promoter are found in the promoters of other SMRT-specific genes, such as Il10, Il19, EphA4, Ptx3, and LCN2 (Supplemental Table S1). ChIP experiments confirmed that TEL is present on the proximal regions of these SMRT-dependent genes (Supplemental Fig. S10A). Reduction of TEL expression in primary macrophages using specific siRNA impaired SMRT recruitment to Il19, Ptx3, and LCN2 (Supplemental Fig. S10B), suggesting a frequent, but not exclusive, role of TEL in recruitment of SMRT to SMRT-specific promoters.
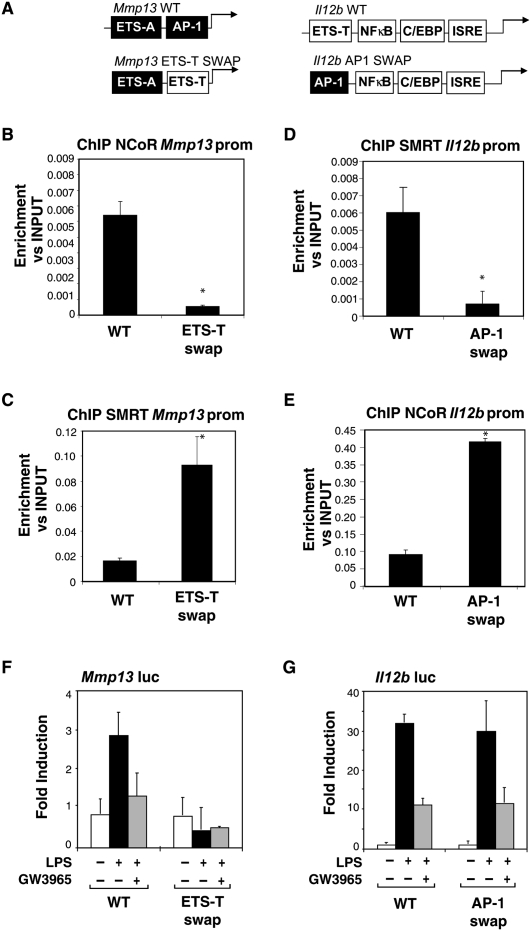
The observation that NCoR is specifically recruited to an AP1 consensus sequence on the Mmp13 promoter while SMRT binds to an ETS site on the Il12b promoter raised the question as to whether swapping the two sites would result in a corresponding change in corepressor recruitment. To test this hypothesis, Mmp13 and Il12b promoter constructs were generated in which the AP-1 site in the Mmp13 promoter responsible for NCoR recruitment was mutated to the sequence of the ETS TEL-binding site (ETS-T) in the Il12b promoter that is required for SMRT recruitment. Conversely, the ETS-T site in the Il12b promoter was mutated to the sequence of the AP1 site in the Mmp13 promoter required for NCoR recruitment (Fig. 5A). The wild-type and mutant promoter constructs were transfected into RAW macrophages, and subjected to ChIP analysis for SMRT and NCoR. In parallel, luciferase assays were performed to assess the LPS responses and transrepression of these chimeric promoters. As expected, SMRT recruitment was observed on the wild-type Il12b promoter and NCoR recruitment on wild-type Mmp13 promoter (Fig. 5D,B, respectively). Remarkably, when the ETS site of Il12b was mutated to the AP1 site of the Mmp13 promoter, SMRT recruitment was impaired while NCoR recruitment was observed (Fig. 5D,E). This chimeric promoter continued to exhibit low basal levels of activity, was still inducible by LPS and was sensitive to repression by GW3965 (Fig. 5G). Conversely, when the AP1 site of the Mmp13 promoter was mutated to the ETS-T site of the Il12b promoter, NCoR recruitment was significantly decreased while SMRT was recruited (Fig. 5B,C). In contrast to the chimeric Il12b promoter, however, the chimeric Mmp13 promoter was no longer inducible by LPS (Fig. 5F). These results are consistent with the observation that mutating the AP-1 site in the Mmp13 promoter (Fig. 3B) completely abolished inducibility of Mmp13, indicating that the AP-1 site in the Mmp13 gene is necessary for both NCoR binding and signal-dependent activation. While the corepressor recruitment function of this site can be replaced by the TEL site from the Il12b promoter, the TEL site does not replace the activation function of the Mmp13 AP1 site, implying a crucial role of c-Jun/c-Fos heterodimers in this promoter context. In concert, these results demonstrate that AP1 and ETS sites can dictate the specificity of NCoR and SMRT recruitment to NCoR-specific or SMRT-specific promoters, respectively, and that the repression checkpoint functions of these elements are distinct from their potential activation functions.
Figure 5.
cJun and TEL sites are sufficient to confer NCoR and SMRT binding to the Il12b and Mmp13 promoters, respectively. (A) Schematic diagram of the wild-type and mutant Mmp13 and Il12b promoters indicating the swapped AP1 and ETS sites. ETS-T is the TEL-binding site in the Il12b promoter. ETS-A is a binding site for ETS activators in the Mmp13 promoter. AP-1 is the cJun-binding site in the Mmp13 promoter. (B,C) Replacing the ETS-A site in the Mmp13 promoter with the ETS-T site from Il12b inhibits the recruitment of NCoR and promotes SMRT binding to the Mmp13 promoter as shown by ChIP assays performed in RAW cells transfected with the plasmids shown in A. (D,E) NCoR is recruited to the Il12b promoter upon a swap of the AP1 Mmp13 site into the ETS-T on the ETS site as shown by ChIP assays. (*) P < 0.05 versus wild type. Results are expressed as the average of three independent experiments. Error bars represent standard deviations. (F,G) Functional analysis of the indicated wild-type and mutant Mmp13 and Il12b promoters in RAW264.7 cells.
c-Jun and TEL work in a combinatorial manner to recruit NCoR/SMRT complexes on NCoR/SMRT-dependent promoters
Based on the studies of the Mmp13 and Il12b promoters, we hypothesized that an NCoR and SMRT-dependent promoter such as Ccl2 should have both ETS and AP1 consensus sequences. Consistent with this, the Ccl2 promoter has proximal AP1 and ETS sites (indicated in Fig. 6A) and ChIP experiments documented that both TEL and c-Jun occupy this promoter under basal conditions (Fig. 6A). To determine whether NCoR and SMRT bind to the Ccl2 promoter simultaneously, ChIP–ReChIP experiments were performed, using the Il12b and Mmp13 promoters as negative controls. These exeperiments demonstrated concurrent occupancy of the Ccl2 promoter by NCoR and SMRT (Fig. 6B). Surprisingly, ChIP experiments in NCoR−/− and SMRT−/− macrophages demonstrated that NCoR recruitment on the Ccl2 promoter was impaired in SMRT−/− macrophages, while SMRT was no longer able to bind to the Ccl2 promoter in NCoR−/− macrophages (Fig. 6C), suggesting a model in which the two corepessors bind in a highly cooperative manner. To explore this relationship further, ChIP experiments were performed to evaluate NCoR and SMRT binding to Ccl2 promoters mutated at either the AP1 or ETS sites. As expected, mutation of the ETS site decreased SMRT recruitment and mutation of the AP1 site abolished NCoR recruitment (Fig. 6D). Furthermore, mutation of the AP1 site also impaired SMRT recruitment and mutation of the ETS site decreased NCoR binding. These findings are consistent with the consequences of genetic deletion of NCoR or SMRT and support the hypothesis that NCoR and SMRT bind to the Ccl2 promoter in an interdependent manner. These results were further confirmed by a functional assay (data not shown), where each mutation impaired LXR transrepression, consistent with the absence of both corepressors. Interestingly, the combination of ETS and AP1 sites is frequently observed in NCoR/SMRT-dependent promoters analyzed so far, including the Il1a, Il1b, Ccl7, Ccl12, and Il6 promoters (Supplemental Table S1). ChIP studies of these representative NCoR/SMRT-dependent target genes revealed that c-Jun is present on each of these promoters, and TEL is present on all but the Il6 promoter (Supplemental Fig. S11; data not shown). Knockdown of c-Jun reduced both NCoR and SMRT binding in all cases, while knockdown of TEL reduced SMRT binding on Il1a, Ccl7, and Ccl12 and to a lesser extent reduced NCoR binding on Il1a, Il1b, and Ccl7 (Supplemental Fig. S11). These results are consistent with the relative roles of c-Jun and TEL in establishing NCoR-specific and SMRT-specific promoters, and suggest that this combination of binding sites also provides a common, but not exclusive, mechanism for establishing NCoR/SMRT-dependent promoters.
Signal-specific clearance of corepressors on proinflammatory promoters
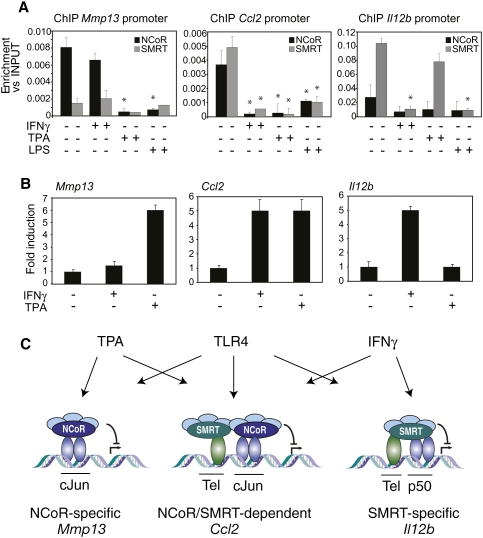
The observation that NCoR and SMRT regulate broad sets of both common and distinct proinflammatory target genes by being recruited to promoters through distinct transcriptional repressors raised the question about the biological roles of these corepressors in integrating cellular responses to distinct inflammatory stimuli. The early steps of TLR4 stimulation by LPS involve the activation of a broad number of kinases that ultimately induce NCoR and SMRT clearance by mechanisms that remain to be elucidated (Glass and Ogawa 2006; O'Neill and Bowie 2007). However, previous studies in CV1 cells demonstrated that activation of MEKK1 leads to phosphorylation of SMRT and its redistribution from the nucleus to a cytoplasmic compartment (Hong and Privalsky 2000). Interestingly, NCoR does not detectably change its subcellular distribution in response to MEKK1 signaling (Jonas and Privalsky 2004). We therefore hypothesized that an inflammatory stimulus specifically activating MEKK1 should induce SMRT clearance from SMRT-specific and SMRT/NCoR-dependent target genes. We therefore evaluated IFNγ as an inflammatory stimulus, because previous studies have demonstrated that it induces MEKK1–MEK1–ERK1/2 kinases in macrophages (Roy et al. 2002). ChIP experiments demonstrated that NCoR occupancy of the Mmp13 promoter was not affected by IFNγ treatment, while SMRT was dismissed from the Il12b and Ccl2 promoters in response to IFNγ (Fig. 7A). In parallel, IFNγ treatment promoted SMRT translocation to the cytosolic compartment in macrophages, as expected (Supplemental Fig. S12). Corepressor clearance was correlated with gene activation, as IFNγ induced Il12b and Ccl2 gene expression, but not Mmp13 expression (Fig. 7B). In contrast, TPA, a stimulus that regulates the activity of AP-1 (Angel et al. 1987), induced Mmp13 and Ccl2 gene expression, but not Il12b expression (Fig. 7B). This activity was correlated with the clearance of NCoR from the Mmp13 and Ccl2 promoters. NCoR and SMRT were also required for transrepressive functions of LXR and PPARγ ligands on the Mmp13, Ccl2, and Il12b genes in the context of TNFa or TLR3 activation with poly I:C (Supplemental Figure S14).
Figure 7.
NCoR and SMRT function as integrators of multiple signals. (A) IFNγ and TPA induce SMRT and NCoR clearance, respectively, on Il12b, Ccl2, and Mmp13 promoters. Primary macrophages were treated with IFNγ (10 U/mL), TPA (100 ng/mL), or LPS (1 μg/mL) for 1 h, and ChIP assay was performed using antibodies against NCoR, SMRT, and control IgG. Immunoprecipitated DNA was analyzed by real-time PCR using primers for Mmp13, Ccl2, and Il12b promoters. (*) P < 0.05 versus control. (B) Mmp13, Ccl2, and Il12b expression analysis by Q-PCR of primary macrophages treated for 6 h with IFNγ (10 U/mL), TPA (100 ng/mL), or LPS (1 μg/mL). (C) Model depicting differential recruitment of NCoR and SMRT on target promoters by c-Jun, Tel, and p50, and differential clearance by TPA, LPS, and IFNγ. See the text for discussion.
Discussion
Roles of NCoR and SMRT in the regulation of inflammatory gene expression
While mechanisms responsible for activation of sequence-specific transcription factors that drive inflammatory responses have been intensively studied, corresponding mechanisms that maintain inflammatory response genes in a poised but actively repressed state under basal conditions remain poorly understood. Here, we demonstrate that NCoR and SMRT are required for nearly all of the transrepression activities of LXR agonists in activated macrophages, thereby extending the concept of corepressor clearance as an essential prerequisite for transcriptional activation to broad sets of inflammatory response genes. Although the absence of PPARγ expression in fetal liver-derived macrophages precludes a similar analysis of transrepression activities of PPARγ ligands, analysis of individual genes in elicited macrophages following NCoR or SMRT knockdown suggests that NCoR and SMRT will also be broadly required. These observations are in contrast to the apparent lack of a requirement of NCoR for glucocorticoid receptor-mediated transrepression (Ogawa et al. 2005).
The well-established basal repression functions of NCoR and SMRT were initially identified based on their discovery as nuclear receptor corepressors, which function in a combinatorial manner with other sequence-specific activators. Deletion of NCoR or SMRT is in many cases sufficient to lead to derepression of genes under the control of nuclear receptors even in the absence of ligand because of constitutive activities of cooperating transcription factors that are no longer under the influence of NCoR/SMRT-mediated repression. In contrast, transcriptional activation of inflammatory response genes generally requires signal-dependent recruitment of factors that include NFkB and IRFs. Because of this, deletion of NCoR or SMRT is in most cases not sufficient in itself to allow effective gene activation, consistent with the results of gene expression profiling experiments (Fig. 1A). The repression functions of NCoR (and now SMRT) on these genes are thus only revealed by perturbations, such as LXR activation, that prevent their signal-dependent turnover. In this context, LXR-dependent transrepression can be viewed as the maintenance of basal repression in the presence of an activating signal. Because loss of NCoR/SMRT does not in itself lead to constitutive activation of most LPS target genes, but does result in the profound loss of transrepression, we refer to this function as a transcriptional “checkpoint.” We propose that this “checkpoint” function plays an important role in enabling appropriate integration of proinflammatory and anti-inflammatory signaling pathways, which we have now extended to a large cohort of inflammatory response genes in macrophages.
As highly related corepressors, NCoR and SMRT share the same molecular architecture, interact with many of the same transcription factors and assemble into similar corepressor complexes, raising the question of the extent to which they serve redundant functions. The present studies demonstrate that NCoR and SMRT can function independently to regulate specific subsets of inflammatory response genes in macrophages as a consequence of being targeted to the promoters of these genes through distinct sequence-specific transcription factors. Furthermore, a large cohort of inflammatory response genes was found to bind NCoR and SMRT in a mutually dependent manner, establishing an unexpected requirement for both corepressors for appropriate regulation by inflammatory and anti-inflammatory signaling inputs. The full extent of NCoR/SMRT occupancy on LPS target genes remains to be established because there appear to be at least some genes that are occupied by NCoR and/or SMRT that are not LXR-sensitive (C.K. Glass, unpubl.). These potentially represent genes on which NCoR and SMRT do function in a redundant manner.
Molecular determinants for NCoR and SMRT recruitment on inflammatory target genes
Based on the gene-specific and interdependent functions of NCoR and SMRT, a major goal is to understand the mechanisms by which they are recruited to their respective promoters either selectively or in combination. Prior biochemical studies have demonstrated that NCoR and SMRT can interact with many of the same transcription factors in vitro, including AP-1, NFκB, and ETS factors (Lee et al. 2000; Wang and Hiebert 2001; Zhang et al. 2002). In contrast, using a combination of ChIP and promoter mutational analysis, we found remarkable specificity in the cis-active elements and corresponding transcription factors that function to recruit NCoR and SMRT to specific target genes in macrophages. Here, we find that c-Jun functions to recruit NCoR to AP1 sites on all NCoR-specific target promoters examined, consistent with previous studies of a limited set of phorbol-ester-responsive genes (Ogawa et al. 2004). Conversely, we found that the ETS repressor TEL recruits SMRT, but not NCoR, to the ETS site of the Il12b promoter, as well as to most of the other SMRT-dependent genes analyzed. The specificity of the SMRT/TEL interaction appears to be mediated, at least in part, by a relatively higher affinity of TEL for SMRT than NCoR. Overall, our findings indicate that in addition to its established roles as a tumor suppressor and regulator of hematopoiesis, TEL serves as a negative regulator of inflammatory response genes. Sequence analysis of NCoR- and SMRT-dependent promoters indicates the presence of AP-1 sites in nearly all NCoR-dependent promoters and Ets sites in nearly all SMRT-dependent promoters, suggesting widely used roles of c-Jun and TEL in NCoR and SMRT recruitment, respectively. Deletion or loss of function of c-Jun and TEL, coupled to genome-wide location analysis for NCoR and SMRT, will be required to establish this point. Because NCoR and SMRT do not appear to interact in solution, we speculate that their codependence for binding to NCoR/SMRT-dependent promoters is established by cooperative interactions of recruiting factors such as c-Jun and TEL at the promoter level.
A corepressor-based strategy for integration of multiple inflammatory signals
The induction, amplification, and ultimate resolution phases of inflammatory responses are regulated by a large number of proinflammatory and anti-inflammatory mediators that act at multiple levels within the cell. Here, we show that while LPS promotes the dismissal of NCoR and SMRT complexes from NCoR-specific, SMRT-specific, and NCoR/SMRT-dependent promoters, IFNγ and phorbol esters act selectively on SMRT and NCoR complexes, respectively. However, because of the coupled nature of NCoR and SMRT binding to NCoR/SMRT-dependent promoters, TPA is able to dismiss NCoR from both NCoR-specific and NCoR-SMRT-dependent genes, and IFNγ is able to mediate dismissal of SMRT from SMRT-specific and NCoR/SMRT-dependent genes. The cooperative recruitment of NCoR and SMRT to these promoters also provides an explanation for the unexpected finding that both NCoR and SMRT were required for transrepression of a large set of LXR-sensitive genes. Based on the ability of either LXRα or LXRβ to exert transrepression function (Joseph et al. 2003) and the expression of both receptors in macrophages (Supplemental Fig. S1), current efforts to develop LXRβ-selective ligands for use in prevention of atherosclerosis (Quinet et al. 2006) should result in compounds that not only reduce undesirable LXRα-mediated effects on serum triglycerides, but also retain anti-inflammatory activities. In concert, these studies provide evidence for a corepressor-based strategy for integration of inflammatory and anti-inflammatory signals that play essential roles in immunity and homeostasis.
Materials and methods
Reagents and plasmids
LPS, TPA (12-O-tetradecanoylphorbol-13-acetate), IFNγ, and PolyI:C were obtained from Sigma. Mouse TNFα was obtained from R&D Systems. GW3965 and Rosiglitazone were provided by GlaxoSmithKline. Point mutations on Mmp13-luc, Il12b-luc, and Ccl2-luc reporter vectors were made using the QuickChange site-directed mutagenesis kit (Stratagene).
Cell culture and transient transfection
Fetal liver-derived macrophages generated from embryonic day 14.5 embryo liver from NCoR−/− and SMRT−/− mice were plated and cultured in RPMI medium 1640 with 10% fetal bovine serum (HyClone; screened for low endotoxin levels) plus L cell media for 7 d (Jepsen et al. 2000; Jepsen et al. 2007). Thioglycollate-elicited macrophages were prepared as described previously (Ghisletti et al. 2007) from C57BL/6 mice (Charles River). For RNAi experiments in primary macrophages, cells were transfected with control or smart-pool siRNAs (100 nM; Dharmacon) directed against NCoR, SMRT, c-JUN, and TEL using lipofectamine 2000 (Invitrogen). Cells were used for experiments after 48 h incubation and target gene knockdown was validated by QPCR (Supplemental Fig. S13). RAW 264.7 cells were transiently transfected with Mmp13, Il12b, and Ccl2 promoters directing luciferase expression or MEKK1CA using Superfect reagent (Qiagen). A β-galactosidase expression vector was cotransfected as an internal control.
Expression array profiling
Total RNA (isolated by RNeasy kit, Qiagen) was prepared from liver-derived macrophages obtained from wild-type or knockout fetuses from the same mother. Purified RNA, 0.5 μg per sample, was labeled using the LRILAK PLUS, two-color low RNA input Linear Amplification kit and hybridized to a Mouse Whole Genome Microarray 4x44K 60 mer slide according to the manufacturer's instructions (Agilent). Two biological replicates were performed for each experimental condition.
ChIP assays
ChIP assays were performed in primary macrophages, liver-derived macrophages, and RAW 264.7 cells transfected with various reporter plasmids. After stimulation, cells were cross-linked by adding directly to the medium formaldehyde to a final concentration of 1% at room temperature. After 10 min, ice-cold PBS was added and plates were placed on ice, washed extensively with PBS, and scraped. After centrifugation, cells were lysed in Lysis buffer (50 mM Tris at pH 8.0, 5 mM EDTA, 1% SDS) supplemented with proteases inhibitors. Chromatin was sheared by sonication (Bioruptor UCD-200 ultrasound sonicator from Diagenode), resulting in DNA fragments between 150 and 200 bp in size. After centrifugation to remove cell debris, 5% of the sample was kept as INPUT and then diluted 10 times in dilution buffer (50 mM Tris at pH 8.0, 0.5% Triton X-100, 0.1 M NaCl, 2 mM EDTA) supplemented with protease inhibitors. Extracts were precleared for 2 h at 4°C using 2 μg of sheared salmon sperm DNA (Invitrogen, Inc.) and 45 μL of protein A-Sepharose (50% slurry in dilution buffer) (Sigma). Immunoprecipitations were carried out overnight at 4°C with 1 μg of each antibody. In parallel, supernatants were incubated with normal rabbit serum as controls. Immune complexes were collected with protein A-Sepharose and washed three times (5 min each) with low salt buffer (20 mM Tris at pH 8.0, 0.1% SDS, 1% Triton, 2 mM EDTA, 200 mM NaCl) and three times with TE. Immune complexes were extracted with 1% SDS (v/v), 0.1 M NaHCO3, and heated overnight at 65°C to reverse the cross-linking. After proteinase K digestion (100 μg, 1 h at 50°C), DNA fragments were purified on QIAquick Spin columns (Qiagen) in 50 μL of EB (elution buffer) and 1 μL was used in each QPCR. Sequences of promoter-specific primers are available in Supplemental Table S2 and were designed to specifically amplify proximal promoter regions containing the binding sites for c-Jun and TEL discussed in the text. For ReChIP assay, after the first immunoprecipitation, beads were eluted in 10 mM DTT and diluted with ReChIP dilution buffer (50 mM Tris/HCl at pH 7.4, 150 mM NaCl, 1% Triton, 2 mM EDTA, protease inhibitors) and subjected to a second immunoprecipitation. Anti-NCoR (Affinity Bioreagents), anti-SMRT (Affinity Bioreagents), anti-TEL (Santa Cruz Biotechnologies), anti-IRF2 (Santa Cruz Biotechnologies), anti-METS (Klappacher et al. 2002), anti-p50 (Santa Cruz Biotechnologies), p65 (Santa Cruz Biotechnologies), c-Rel (Santa Cruz Biotechnologies), c-Jun (Santa Cruz Biotechnologies), c-Fos (Santa Cruz Biotechnologies), and LXR (Santa Cruz Biotechnologies) antibodies were used. Data are presented as enrichment of the precipitated target sequence as compared to input DNA.
RNA isolation and QPCR
Total RNA (isolated by RNeasy kit, Qiagen) was prepared from macrophages. One microgram of total RNA was used for cDNA synthesis, and 1 μL of cDNA was used for real-time PCR using inflammatory gene-specific primers. Primer sequences can be found in Supplemental Table S2. PCR (SYBERgreen) analysis was performed on an Applied Biosystems 7300 real-time PCR system. Values are normalized with GAPDH content.
Western blotting
Nuclear and cytoplasmic extracts were prepared from RAW 264.7 cells, liver-derived macrophages, and thioglycollate-elicited macrophages using a commercial kit (NE-PER; Pierce). Extracts were resolved by SDS-PAGE and immunoblotted using anti-NCoR, anti-SMRT (Affinity Bioreagents), anti-GAPDH (Abcam), and anti-lamin (Cell Signaling) antibodies.
Co-IP assay
HeLa cells were transfected with Flag-SMRT or Flag-NCoR and Ha-TEL or Ha-cJUNand lysed in high-salt lysis buffer. Flag-SMRT or Flag-NCoR was immunoprecipitated using anti-Flag beads (Sigma) and Ha-Tel and cJun was detected using anti-Ha (Covance) or anti-cJun antibody (Santa Cruz Biotechnologies).
Acknowledgments
We thank Dr. Martel-Pellettier for providing Mmp13-luc, Dr. R. Natajaran for Ccl2-luc, and Dr. S. Smale for Il12b-luc plasmid. We thank Lynn Bautista for assistance with preparation of the manuscript. These studies were supported in part by NIH grants DK063491, CA052599, GM069338, DK074868, DK3949, HL088093, and HL065445, and a LeDucq Foundation Transatlantic Network Grant. M.G.R. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1773109.
Supplemental material is available at http://www.genesdev.org.
References
- Akira S., Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Angel P., Imagawa M., Chiu R., Stein B., Imbra R.J., Rahmsdorf H.J., Jonat C., Herrlich P., Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- Baek S.H., Ohgi K.A., Rose D.W., Koo E.H., Glass C.K., Rosenfeld M.G. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Blaschke F., Takata Y., Caglayan E., Collins A., Tontonoz P., Hsueh W.A., Tangirala R.K. A nuclear receptor corepressor-dependent pathway mediates suppression of cytokine-induced C-reactive protein gene expression by liver X receptor. Circ. Res. 2006;99:e88–e99. doi: 10.1161/01.RES.0000252878.34269.06. [DOI] [PubMed] [Google Scholar]
- Bradley M.N., Zhou L., Smale S.T. C/EBPβ regulation in lipopolysaccharide-stimulated macrophages. Mol. Cell Biol. 2003;23:4841–4858. doi: 10.1128/MCB.23.14.4841-4858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S.R., Nucifora G. The leukemia-associated gene TEL encodes a transcription repressor which associates with SMRT and mSin3A. Biochem. Biophys. Res. Commun. 1999;264:871–877. doi: 10.1006/bbrc.1999.1605. [DOI] [PubMed] [Google Scholar]
- Chen J.D., Evans R.M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- de la Torre P., Diaz-Sanjuan T., Garcia-Ruiz I., Esteban E., Canga F., Munoz-Yague T., Solis-Herruzo J.A. Interleukin-6 increases rat metalloproteinase-13 gene expression through Janus kinase-2-mediated inhibition of serine/threonine phosphatase-2A. Cell. Signal. 2005;17:427–435. doi: 10.1016/j.cellsig.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Ghisletti S., Huang W., Ogawa S., Pascual G., Lin M.E., Willson T.M., Rosenfeld M.G., Glass C.K. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARγ. Mol. Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C.K., Ogawa S. Combinatorial roles of nuclear receptors in inflammation and immunity. Nat. Rev. Immunol. 2006;6:44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- Gri G., Savio D., Trinchieri G., Ma X. Synergistic regulation of the human interleukin-12 p40 promoter by NFκB and Ets transcription factors in Epstein-Barr virus-transformed B cells and macrophages. J. Biol. Chem. 1998;273:6431–6438. doi: 10.1074/jbc.273.11.6431. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Lane W.S., Fischle W., Verdin E., Lazar M.A., Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes & Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guidez F., Petrie K., Ford A.M., Lu H., Bennett C.A., MacGregor A., Hannemann J., Ito Y., Ghysdael J., Greaves M., et al. Recruitment of the nuclear receptor corepressor N-CoR by the TEL moiety of the childhood leukemia-associated TEL-AML1 oncoprotein. Blood. 2000;96:2557–2561. [PubMed] [Google Scholar]
- Hermanson O., Jepsen K., Rosenfeld M.G. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- Hester K.D., Verhelle D., Escoubet-Lozach L., Luna R., Rose D.W., Glass C.K. Differential repression of c-myc and cdc2 gene expression by ERF and PE-1/METS. Cell Cycle. 2007;6:1594–1604. doi: 10.4161/cc.6.13.4336. [DOI] [PubMed] [Google Scholar]
- Hoberg J.E., Yeung F., Mayo M.W. SMRT derepression by the IκB kinase α: A prerequisite to NF-κB transcription and survival. Mol. Cell. 2004;16:245–255. doi: 10.1016/j.molcel.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Honda K., Taniguchi T. IRFs: Master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Hong S.H., Privalsky M.L. The SMRT corepressor is regulated by a MEK-1 kinase pathway: Inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol. Cell. Biol. 2000;20:6612–6625. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein A.J., Naar A.M., Heinzel T., Torchia J., Gloss B., Kurokawa R., Ryan A., Kamei Y., Soderstrom M., Glass C.K., et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Hermanson O., Onami T.M., Gleiberman A.S., Lunyak V., McEvilly R.J., Kurokawa R., Kumar V., Liu F., Seto E., et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Solum D., Zhou T., McEvilly R.J., Kim H.J., Glass C.K., Hermanson O., Rosenfeld M.G. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Gleiberman A.S., Shi C., Simon D.I., Rosenfeld M.G. Cooperative regulation in development by SMRT and FOXP1. Genes & Dev. 2008;22:740–745. doi: 10.1101/gad.1637108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas B.A., Privalsky M.L. SMRT and N-CoR corepressors are regulated by distinct kinase signaling pathways. J. Biol. Chem. 2004;279:54676–54686. doi: 10.1074/jbc.M410128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S.B., Castrillo A., Laffitte B.A., Mangelsdorf D.J., Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat. Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- Karin M., Greten F.R. NF-κB: Linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- Klappacher G.W., Lunyak V.V., Sykes D.B., Sawka-Verhelle D., Sage J., Brard G., Ngo S.D., Gangadharan D., Jacks T., Kamps M.P., et al. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell. 2002;109:169–180. doi: 10.1016/s0092-8674(02)00714-6. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Kim J.H., Lee Y.C., Cheong J., Lee J.W. Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-κB, and serum response factor. J. Biol. Chem. 2000;275:12470–12474. doi: 10.1074/jbc.275.17.12470. [DOI] [PubMed] [Google Scholar]
- Li Q., Verma I.M. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Lutterbach B., Westendorf J.J., Linggi B., Patten A., Moniwa M., Davie J.R., Huynh K.D., Bardwell V.J., Lavinsky R.M., Rosenfeld M.G., et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S., Sumita K., Shen H., Kanoh M., Xu X., Sato M., Matsumoto M., Shinomiya H., Asano Y. Identification of IFN regulatory factor-1 binding site in IL-12 p40 gene promoter. J. Immunol. 2003;170:997–1001. doi: 10.4049/jimmunol.170.2.997. [DOI] [PubMed] [Google Scholar]
- Masumi A., Ozato K. Coactivator p300 acetylates the interferon regulatory factor-2 in U937 cells following phorbol ester treatment. J. Biol. Chem. 2001;276:20973–20980. doi: 10.1074/jbc.M101707200. [DOI] [PubMed] [Google Scholar]
- Mavrothalassitis G., Ghysdael J. Proteins of the ETS family with transcriptional repressor activity. Oncogene. 2000;19:6524–6532. doi: 10.1038/sj.onc.1204045. [DOI] [PubMed] [Google Scholar]
- Murphy T.L., Cleveland M.G., Kulesza P., Magram J., Murphy K.M. Regulation of interleukin 12 p40 expression through an NF-κ B half-site. Mol. Cell. Biol. 1995;15:5258–5267. doi: 10.1128/mcb.15.10.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S., Lozach J., Jepsen K., Sawka-Verhelle D., Perissi V., Sasik R., Rose D.W., Johnson R.S., Rosenfeld M.G., Glass C.K. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc. Natl. Acad. Sci. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S., Lozach J., Benner C., Pascual G., Tangirala R.K., Westin S., Hoffmann A., Subramaniam S., David M., Rosenfeld M.G., et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill L.A., Bowie A.G. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- Pascual G., Fong A.L., Ogawa S., Gamliel A., Li A.C., Perissi V., Rose D.W., Willson T.M., Rosenfeld M.G., Glass C.K. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendas A.M., Balbin M., Llano E., Jimenez M.G., Lopez-Otin C. Structural analysis and promoter characterization of the human collagenase-3 gene (MMP13) Genomics. 1997;40:222–233. doi: 10.1006/geno.1996.4554. [DOI] [PubMed] [Google Scholar]
- Perissi V., Aggarwal A., Glass C.K., Rose D.W., Rosenfeld M.G. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Plevy S.E., Gemberling J.H., Hsu S., Dorner A.J., Smale S.T. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: Evidence of functional synergy between C/EBP and Rel proteins. Mol. Cell. Biol. 1997;17:4572–4588. doi: 10.1128/mcb.17.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet E.M., Savio D.A., Halpern A.R., Chen L., Schuster G.U., Gustafsson J.A., Basso M.D., Nambi P. Liver X receptor (LXR)-β regulation in LXRα-deficient mice: Implications for therapeutic targeting. Mol. Pharmacol. 2006;70:1340–1349. doi: 10.1124/mol.106.022608. [DOI] [PubMed] [Google Scholar]
- Roy S.K., Hu J., Meng Q., Xia Y., Shapiro P.S., Reddy S.P., Platanias L.C., Lindner D.J., Johnson P.F., Pritchard C., et al. MEKK1 plays a critical role in activating the transcription factor C/EBP-β-dependent gene expression in response to IFN-γ. Proc. Natl. Acad. Sci. 2002;99:7945–7950. doi: 10.1073/pnas.122075799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S., Hoffmann A., Liou H.C., Baltimore D., Smale S.T. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc. Natl. Acad. Sci. 2000;97:12705–12710. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S., Williams K.J., Saccani S., Zhou L., Hoffmann A., Ghosh G., Gerondakis S., Natoli G., Smale S.T. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes & Dev. 2005;19:2138–2151. doi: 10.1101/gad.1329805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis-Herruzo J.A., Rippe R.A., Schrum L.W., de La Torre P., Garcia I., Jeffrey J.J., Munoz-Yague T., Brenner D.A. Interleukin-6 increases rat metalloproteinase-13 gene expression through stimulation of activator protein 1 transcription factor in cultured fibroblasts. J. Biol. Chem. 1999;274:30919–30926. doi: 10.1074/jbc.274.43.30919. [DOI] [PubMed] [Google Scholar]
- Tardif G., Pelletier J.P., Dupuis M., Hambor J.E., Martel-Pelletier J. Cloning, sequencing and characterization of the 5′-flanking region of the human collagenase-3 gene. Biochem. J. 1997;323:13–16. doi: 10.1042/bj3230013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hiebert S.W. TEL contacts multiple co-repressors and specifically associates with histone deacetylase-3. Oncogene. 2001;20:3716–3725. doi: 10.1038/sj.onc.1204479. [DOI] [PubMed] [Google Scholar]
- Wang J., Hoshino T., Redner R.L., Kajigaya S., Liu J.M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.M., Contursi C., Masumi A., Ma X., Trinchieri G., Ozato K. An IFN-γ-inducible transcription factor, IFN consensus sequence binding protein (ICSBP), stimulates IL-12 p40 expression in macrophages. J. Immunol. 2000;165:271–279. doi: 10.4049/jimmunol.165.1.271. [DOI] [PubMed] [Google Scholar]
- Yang C.H., Wei L., Pfeffer S.R., Du Z., Murti A., Valentine W.J., Zheng Y., Pfeffer L.M. Identification of CXCL11 as a STAT3-dependent gene induced by IFN. J. Immunol. 2007;178:986–992. doi: 10.4049/jimmunol.178.2.986. [DOI] [PubMed] [Google Scholar]
- Zhang J., Kalkum M., Chait B.T., Roeder R.G. The N-CoR–HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- Zhu C., Rao K., Xiong H., Gagnidze K., Li F., Horvath C., Plevy S. Activation of the murine interleukin-12 p40 promoter by functional interactions between NFAT and ICSBP. J. Biol. Chem. 2003;278:39372–39382. doi: 10.1074/jbc.M306441200. [DOI] [PubMed] [Google Scholar]