Abstract
Rap80 targets the breast cancer suppressor protein BRCA1 along with Abraxas and the BRCC36 deubiquitinating enzyme (DUB) to polyubiquitin structures at DNA double-strand breaks (DSBs). These DSB targeting events are essential for BRCA1-dependent DNA damage response-induced checkpoint and repair functions. Here, we identify MERIT40 (Mediator of Rap80 Interactions and Targeting 40 kD)/(C19orf62) as a Rap80-associated protein that is essential for BRCA1–Rap80 complex protein interactions, stability, and DSB targeting. Moreover, MERIT40 is required for Rap80-associated lysine63–ubiquitin DUB activity, a critical component of BRCA1–Rap80 G2 checkpoint and viability responses to ionizing radiation. Thus, MERIT40 represents a novel factor that links BRCA1–Rap80 complex integrity, DSB recognition, and ubiquitin chain hydrolytic activities to the DNA damage response. These findings provide new molecular insights into how BRCA1 associates with independently assembled core protein complexes to maintain genome integrity.
Keywords: C19orf62, HSPC142, MERIT40, BRCA1, Rap80, Abraxas, BRCC36
DNA lesions resulting from errors during DNA replication or from exogenous sources are normally repaired by the DNA damage response (DDR) system such that genome integrity is maintained. However, germline mutations in DNA repair and checkpoint control genes abrogate this response, which leads to genomic instability and a predisposition to neoplasia (Lengauer et al. 1998; DePinho 2000; Rouse and Jackson 2002; Venkitaraman 2002). The breast cancer suppressor protein BRCA1 is a key regulator of cellular responses to DNA lesions. Inactivating mutations in BRCA1 lead to deficiencies in G2- and S-phase cell cycle checkpoints, and a failure to repair DNA double-strand breaks (DSBs) (Moynahan et al. 1999; Xu et al. 2001; Yarden et al. 2002).
BRCA1 is at the core of a tumor suppressor network that forms mutually exclusive macromolecular complexes, each responsible for executing discrete processes in the DDR (Yu and Chen 2004; Greenberg et al. 2006). The most recently discovered BRCA1 complex (BRCA1–Rap80) is defined by new interactions with the BRCA1 C-terminal (BRCT) domain (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007). A phosphorylation-dependent interaction between the BRCA1 BRCT domain and the coiled-coiled domain protein Abraxas mediates interaction between BRCA1 and the Rap80 complex, consisting of Rap80, Abraxas, and the BRCC36 deubiquitinating enzyme (DUB) (Kim et al. 2007b; Liu et al. 2007; Wang et al. 2007). Following DNA damage, a series of PIKK (PI3 kinase-like kinase)-dependent phosphorylation events initiated on histone H2AX (γH2AX) lead to recruitment of the RNF8 E3 ubiquitin ligase along with Ubc13, a lysine63–ubiquitin (K63–Ub)-specific E2-conjugating enzyme. The E3/E2 pair RNF8/Ubc13 synthesizes K63-linked polyubiquitin chains on histones H2AX and H2A—and possibly other substrates—providing a DSB recognition platform (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007; Zhao et al. 2007). Rap80, through two N-terminal ubiquitin-interacting motifs (UIMs), specifically binds K63–Ub chains at sites of DNA damage (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007).
The central region of Rap80 (residues 233–399), termed the Abraxas-Interacting Region (AIR), mediates interaction with constituents of the BRCA1–Rap80 protein complex: BRCA1, BRCC36, BRCC45, and Abraxas (Sobhian et al. 2007; Wang and Elledge 2007). While the Rap80 UIMs are both necessary and sufficient for DSB localization (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007), the Rap80 AIR has also been proposed to play an adjunct role in DSB retention (Wang and Elledge 2007; Yan et al. 2007). It is currently unclear how the Rap80 AIR facilitates DSB localization, and how the entire repertoire of Rap80 AIR-associated proteins controls BRCA1–Rap80 complex DDR functions.
This study identifies a novel Rap80 AIR-associated protein, C19orf62/MERIT40 (Mediator of Rap80 Interactions and Targeting 40 kD), which is required for BRCA1–Rap80 complex integrity, DSB localization, and DUB activity. Each of these MERIT40-dependent functions is necessary for proper execution of the G2 checkpoint and for cellular resistance to ionizing radiation (IR). These findings provide new insights into how BRCA1 association with multicomponent protein complexes enables recognition and turnover of ubiquitin chains at DSBs for execution of essential DDR functions.
Results
Identification of MERIT40 as a novel component of the BRCA1–Rap80 complex that is necessary for BRCA1 DSB localization
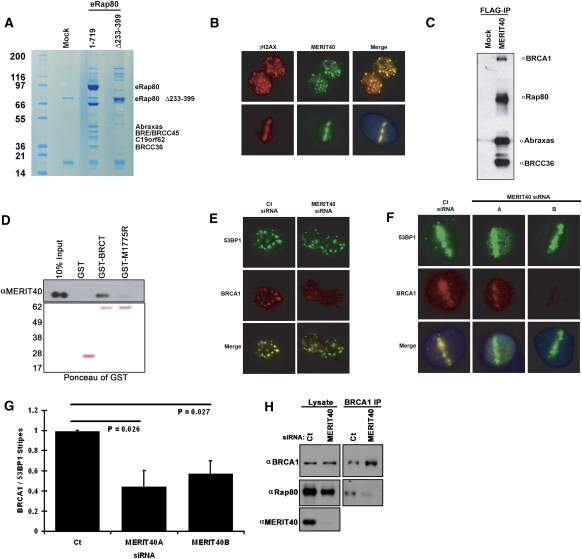
To gain insights into how the AIR might aid in Rap80 DSB recognition and repair properties, we performed tandem immunoaffinity purification from HeLa S3 nuclear extracts of epitope-tagged full-length Rap80 (1–719) versus Rap80 Δ233–399. The RAP80 (1–719) complex revealed association with four protein bands not present in the Rap80 Δ233–399 complex that migrate between ∼36 and 48 kD in a Coomassie-stained SDS–polyacrylamide gel (SDS-PAGE) (Fig. 1A). The four protein bands were excised and analyzed by mass spectrometry for definitive identification by numerous peptide sequence matches as Abraxas, BRE/BRCC45, BRCC36, and C19orf62/HSPC142 (Chromosome 19 ORF 62) (Fig. 1A; Supplemental Fig. S1). C19orf62, the gene product of a predicted ORF that migrated at ∼40 kD by SDS-PAGE (Fig. 1A), was shown previously to interact with BRE/BRCC45 by system-wide interactome-mapping experiments (Rual et al. 2005; Ewing et al. 2007). The putative C19orf62 gene product consists of 329 residues (∼36.5 kD), and protein sequence analysis revealed no known protein domains. A central region (93–320) is conserved in predicted genes from human and mouse to sea anemone and Arabidopsis, and is surrounded by stretches of acidic residues (Supplemental Fig. S2). Endogenous C19orf62, hereafter referred to as MERIT40, was confirmed to localize to γH2AX-positive DSBs by immunofluorescence (IF) following IR or laser microirradiation (Fig. 1B; Supplemental Fig. S3A,B; Rogakou et al. 1998).
Figure 1.
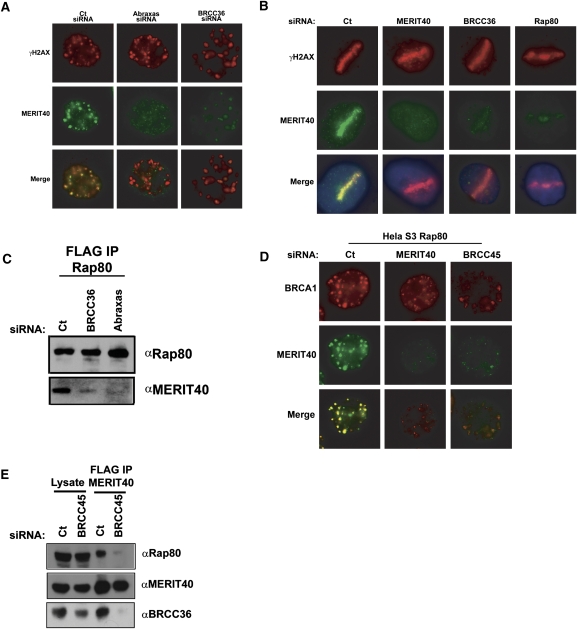
Identification of MERIT40 as a novel BRCA1–Rap80 complex constituent that is required for BRCA1 DSB localization. (A) Coomassie-stained gel representing eRap80 (1–719 and Δ233–399) complexes after consecutive Flag and HA affinity-purification steps from HeLa S3 nuclear extracts. The identities of both eRap80 species and the four Coomassie-stained bands that are uniquely present in the eRap80 wild-type complex are indicated as Abraxas, BRE/BRCC45, BRCC36, and C19orf62. (B) IF for endogenous MERIT40 in HeLa cells at 4 h following IR (top panel) and 30 min after laser microirradiation (bottom panel), demonstrating MERIT40 colocalization with γH2AX at DSBs. (C) Immunoblot (IB) of the eMERIT40 complex after Flag IP from HeLa S3 cells. BRCC36, Abraxas, Rap80, and BRCA1 were detected as MERIT40-associated proteins. (D) MERIT40 interacts with the BRCA1 BRCT domain, but not a clinical BRCT domain mutant. Nuclear extracts of HeLa S3 cells expressing eMERIT40 were incubated with recombinant GST, GST-BRCT, or GST-M1775R proteins bound to GST beads. (Top) IB with anti-HA antibody was performed to detect eMERIT40. (Bottom) The Ponceau-stained membrane is displayed showing the amount of each GST fusion protein. (E,F) MERIT40 is required for BRCA1 DSB localization. IF was performed for BRCA1 following Ct or MERIT40 siRNA treatment in IR-treated (E) and laser-microirradiated (F) HeLa cells. MERIT40 siRNA A and B target different sequences in the coding region of the MERIT40 mRNA. (G) Quantification of BRCA1 stripes as a percentage of 53BP1 stripes displayed graphically from F. At least 200 cells were counted in triplicate for the analysis. Error bars represent standard deviation (SD) and P values were calculated by the Student's t-test. (H) MERIT40 is required for BRCA1 association with Rap80. BRCA1 IP was performed in HeLa S3 cells following transfection with MERIT40 or Ct siRNA. IB was performed on the BRCA1 IP eluates as indicated.
MERIT40 interaction with BRCA1, RAP80, Abraxas, and BRCC36 was confirmed by coimmunoprecipitation (co-IP) (Fig. 1C). Tandem affinity purification of MERIT40 complexes revealed strong associations with Rap80, Abraxas, BRCC45, and BRCC36, and a substantially less than stoichiometric association with BRCA1 as assessed by Coomassie staining of the purified complex (data not shown). MERIT40 interacted with BRCT repeats of wild-type BRCA1 but not a cancer-causing BRCT mutant, M1775R, known to disrupt BRCA1 DSB localization to IR-induced foci (IRIF) (Fig. 1D; Scully et al. 1999). Thus, similar to Rap80, Abraxas, and BRCC36, MERIT40 specifically associates with an intact BRCT domain capable of mediating BRCA1 DSB localization and DDR activities (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007).
Interaction with the BRCT domain of BRCA1 defines multiple distinct BRCA1 protein complexes via mutually exclusive protein interactions (Yu and Chen 2004; Greenberg et al. 2006). Rap80 is required for BRCA1 IRIF formation and maximal accumulation at laser-generated DSBs by targeting BRCA1 to γH2AX–MDC1-dependent ubiquitin chains on chromatin domains flanking DSBs (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007). The remaining BRCA1 complexes, containing BRCA2, Brip1, or CtIP, are thought to recognize the nonnucleosomal regions more proximal to DSBs (Bekker-Jensen et al. 2006; Sartori et al. 2007; Sobhian et al. 2007). BRCA1 IRIF localization was strongly reduced with knockdown of MERIT40 using siRNA (Fig. 1E; Supplemental Fig. S4C). Furthermore, BRCA1 localization to laser-microirradiated stripes was reduced in intensity and frequency following MERIT40 knockdown (Fig. 1F,G), suggesting that MERIT40 contributes an essential component of the Rap80 complex for BRCA1 DSB recognition. Indeed, as BRCA1 resides in at least four different complexes, it appears that, like H2AX, Rap80, and RNF8 (Celeste et al. 2003; Mailand et al. 2007; Sobhian et al. 2007), MERIT40 does not influence DSB recognition for the entire set of BRCA1 complexes, explaining the diminished, albeit still present, BRCA1 localization at stripes in MERIT40-depleted cells. Finally, MERIT40 knockdown largely diminished the association of Rap80 with BRCA1, implying the requirement for MERIT40 in BRCA1 DSB localization occurs vis-à-vis maintenance of BRCA1 association with the Rap80 complex (Fig. 1H).
MERIT40 localizes to RNF8-synthesized ubiquitin structures at DSBs in a manner dependent on its interaction with Rap80 and independent of interaction with BRCA1
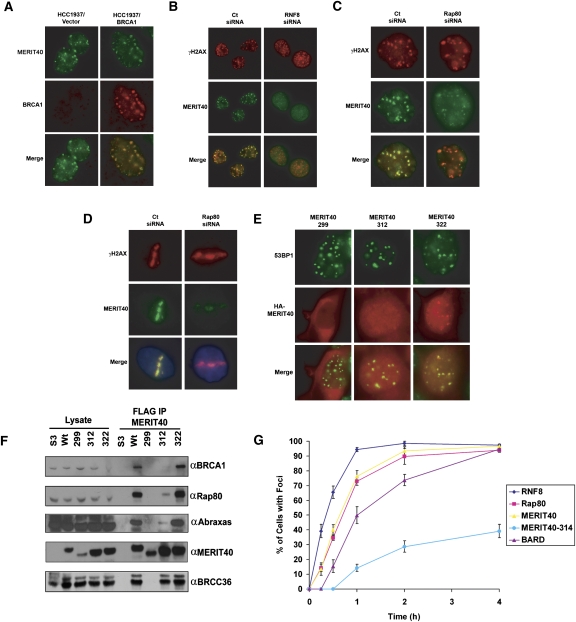
Although MERIT40 is necessary for BRCA1 localization to DNA damage, this requirement is not reciprocated. MERIT40 forms IRIF in BRCA1 BRCT-mutated HCC1937 cells (Fig. 2A). This would suggest that MERIT40, like Rap80, Abraxas, and BRCC36, is upstream of BRCA1 recruitment to sites of DNA damage. Each of these BRCA1–Rap80 complex members is dependent on RNF8–Ubc13-directed ubiquitination of chromatin adjacent to DSB sites (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007; Zhao et al. 2007). MERIT40 localization to IRIF also depends on these activities (Figs. 2B; Supplemental S4A,B). Consequently, as with the other members of this complex, the recruitment of MERIT40 to DSBs is downstream from chromatin ubiquitination by Ubc13/RNF8.
Figure 2.
MERIT40 DSB localization depends on the RNF8 E3 ligase and interaction with Rap80. (A) IF for MERIT40 and BRCA1 in irradiated HCC1937 cells reconstituted with vector (left panels) or wild-type BRCA1 (right panels), demonstrating MERIT40 localization to IRIF occurs independently of wild-type BRCA1 status. (B) IF for MERIT40 and γH2AX in IR-treated HeLa cells following Ct or RNF8 siRNA treatment, demonstrating dependence on RNF8 for MERIT40 IRIF. (C,D) IF for MERIT40 and γH2AX in irradiated (C) and laser-microirradiated (D) HeLa cells following Ct or Rap80 siRNA treatment demonstrates reduced MERIT40 DSB localization in Rap80-depleted cells. (E) IF detection by α-HA antibodies of ectopically expressed forms of Flag-HA-MERIT40 following IR. (Left panel) MERIT40-299 (residues 1–299) is cytoplasmic. (Middle panel) MERIT40-312 (residues 1–312) is nuclear and weakly colocalizes with 53BP1. (Right panel) MERIT40-322 (residues 1–322) strongly colocalizes with 53BP1, akin to full-length MERIT40 (residues 1–329). (F) IB of lysates and Flag IP elutions from HeLa S3 cells expressing epitope-tagged MERIT40 truncations (299, 312, 322, and wild type). MERIT40-299 loses all interactions whereas MERIT40-312 demonstrates reduced interaction with Rap80, Abraxas, and BRCC36. MERIT40-322 interacts with Rap80, Abraxas, and BRCC36. (G) Kinetics of Flag-HA-tagged RNF8, Rap80, MERIT40, MERIT40-314, and BARD IRIF formation in HeLa S3 cells following 10 Gy IR, showing MERIT40 localization occurs more slowly than RNF8 but faster than BARD IRIF. At least 200 cells were counted in triplicate for the analysis. Error bars represent SD. Representative cells for each time point are shown in Supplemental Figure S5.
Depletion of Rap80 by siRNA knockdown also resulted in strongly reduced accumulation of MERIT40 at IRIF and laser-induced DSBs (Fig. 2C,D), indicating that Rap80 is necessary for MERIT40 recruitment to sites of DNA damage. Consistent with these findings, a MERIT40 allelic series displayed a strong correlation between Rap80 binding and DSB localization (Fig. 2E,F; Supplemental Fig. S4D–F). A series of MERIT40 C-terminal truncations was examined for foci formation and interaction with Rap80, Abraxas, and BRCC36. Loss of the seven C-terminal amino acids in MERIT40 (MERIT40-1–322) had no discernable effect on either foci formation or interaction with Rap80, Abraxas, or BRCC36. However, truncation of the C-terminal 30 amino acids (MERIT40-1–299) prevented nuclear accumulation, IRIF formation, and Rap80 association. Truncation to residue 312 reduced both IRIF and Rap80 interaction to similar extents. Finer mapping of the MERIT40 C terminus revealed that amino acids 310–314 were important for robust foci formation, whereas residues 317–321 were completely dispensable (Supplemental Fig. S4F). Thus, a strong correlation exists for MERIT40 between Rap80 binding and DSB localization, consistent with Rap80 targeting MERIT40 to RNF8-dependent ubiquitin structures at DSBs. These data would predict that RNF8 precedes both Rap80 and MERIT40 at DSBs, and that Rap80 and MERIT40 accumulate at DSBs along with and independently of BRCA1–BARD1. Consistent with this hypothesis, HeLa S3 cells that were engineered to express either Flag-HA-tagged RNF8, BARD1, Rap80, MERIT40, or MERIT40-1–314 demonstrated more rapid accumulation of RNF8 at IRIF, followed by almost identical accumulation of MERIT40 and Rap80, and slower foci formation for BRCA1 (Fig. 2G; Supplemental Fig. S5). MERIT40-1–314 displayed reduced IRIF at all time points.
MERIT40 is required for Rap80 complex integrity and DSB localization
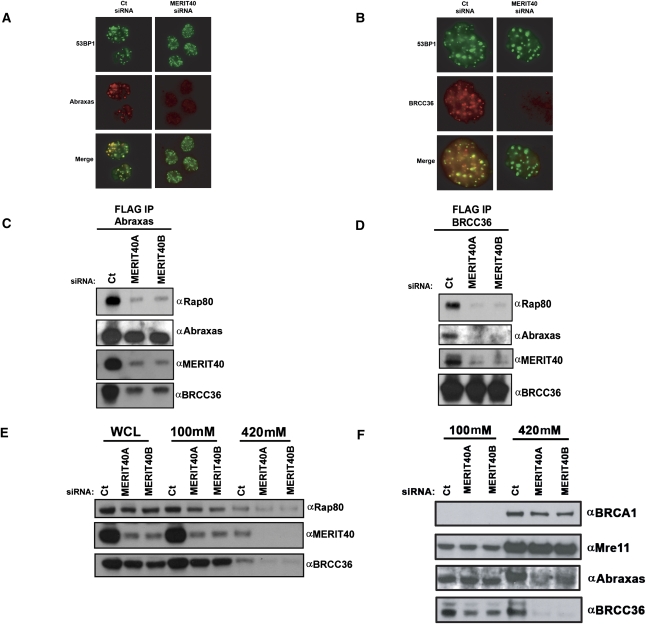
The preceding data reveal that MERIT40 is a key member of the BRCA1–Rap80 complex that mediates BRCA1 DSB recruitment via association with Rap80. Following MERIT40 depletion, both Abraxas and BRCC36 IRIF were also strongly reduced, raising the possibility that Abraxas and BRCC36 fail to bind Rap80 in the absence of MERIT40 (Fig. 3A,B). Indeed, IP of epitope-tagged Abraxas, BRCC36, and Rap80 individually from HeLa S3 cell lines at 48 h following transfection of two different MERIT40 siRNAs resulted in strongly decreased interaction of Rap80 with both Abraxas and BRCC36 (Fig. 3C,D; Supplemental Fig. S6A). In addition, MERIT40 knockdown reduced the Abraxas–BRCC36 interaction, revealing that MERIT40 is essential to the integrity of multiple interactions within the BRCA1–Rap80 complex. Like Abraxas, CtIP associates with the BRCA1 BRCT domain in a phosphorylation-dependent manner (Yu and Chen 2004). Mass spectrometry of Rap80 or MERIT40 complexes did not reveal the presence of CtIP. Correspondingly, MERIT40 knockdown did not impair CtIP foci formation (Supplemental Fig. S7), indicating that MERIT40 does not influence all BRCA1-dependent activities at DSBs. In sum, MERIT40 is required for the interaction of Rap80 with other members of the complex, potentially explaining its requirement for localization of each complex constituent to DSBs.
Figure 3.
MERIT40 is required for DSB targeting and chromatin association of Abraxas and BRCC36. (A,B) MERIT40 knockdown reduces Abraxas (A) and BRCC36 (B) DSB localization. IF was performed for Abraxas and BRCC36 in cells transfected with Ct or MERIT40 siRNA at 4 h after 10 Gy IR. Images are representative of >200 cells counted per condition in three independent experiments. (C,D) HeLa S3 cells expressing eAbraxas (C) and eBRCC36 (D) were immunoprecipitated with anti-Flag-M2 Agarose beads at 72 h after transfection with Ct or either MERIT40A- or MERIT40B-targeted siRNAs. IB of Flag elutions was performed as indicated, demonstrating that both Abraxas and BRCC36 require MERIT40 for interaction with Rap80. (E,F) MERIT40 depletion results in reduced levels of Rap80 and BRCC36 (E) and Abraxas (F) in the high-salt extractable fractions, as shown by IB of sequential 100 mM and 420 mM NaCl-containing lysates.
Rap80 has been shown to be necessary for BRCC36 association with chromatin (Sobhian et al. 2007). The requirement for MERIT40 for Abraxas and BRCC36 DSB localization and interaction with Rap80 would predict that MERIT40 is necessary for the subcellular localization of members of the Rap80 complex. In agreement with this idea, Rap80, BRCC36, and Abraxas displayed increased extractability from chromatin and/or nuclear matrix-rich fractions in MERIT40-depleted cells (Fig. 3E,F). Notably, MERIT40 knockdown reduced total protein levels of Abraxas and BRCC36 as well as Rap80 (Fig. 3E,F), suggesting an additional mechanism whereby MERIT40 could influence the integrity and DSB localization of BRCA1–Rap80 complex members.
MERIT40 is required for stability of Rap80 complex members
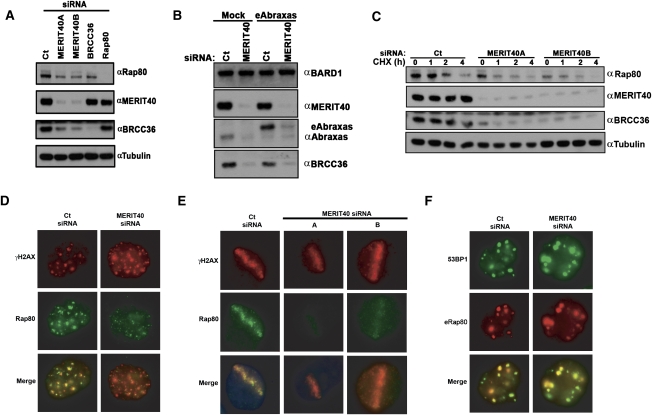
MERIT40 is essential for BRCA1–Rap80 complex integrity, raising the possibility that maintenance of protein–protein interactions within the BRCA1–Rap80 complex is necessary for the stability of each complex component. In support of this hypothesis, MERIT40 knockdown resulted in reduced protein levels for Rap80, BRCC36, and Abraxas (Fig. 4A,B), but did not reduce BRCA1 protein levels (Supplemental Fig. S6C). Conversely, Rap80 or BRCC36 depletion did not appear to influence endogenous MERIT40 protein levels (Fig. 4A), indicating that MERIT40 has a greater influence over BRCA1–Rap80 protein complex stability than either Rap80 or BRCC36. Consistent with this assertion, Rap80 and BRCC36 demonstrated reduced protein stability following cycloheximide (CHX) treatment in MERIT40-depleted cells (Fig. 4C), and ectopic expression of either Abraxas or BRCC36 reduced the levels of their respective endogenous protein (Fig. 4A; Supplemental Fig. S6D), while ectopic MERIT40 did not affect endogenous MERIT40 levels (Supplemental Fig. S6E). A plausible explanation is that ectopic Abraxas and BRCC36 displace the respective endogenous protein from interaction with MERIT40 and the other core complex members, resulting in reduced stability.
Figure 4.
MERIT40 mediates the stability of the BRCA1–RAP80 complex at DSBs. (A) MERIT40 depletion results in decreased Rap80 and BRCC36 protein levels. IB was performed on HeLa cell lysates at 72 h following siRNA transfection as indicated. (B) MERIT40 is required for endogenous Abraxas stability (left) and ectopic Abraxas in an eAbraxas cell line (right). IB was performed for Abraxas in nontransduced (mock) or eAbraxas-expressing HeLa cells at 72 h after Ct or MERIT40 siRNA. Note that eAbraxas expression causes reduced levels of endogenous Abraxas protein. (C) MERIT40 depletion reduces Rap80 and BRCC36 protein levels and half-life. HeLa cells were transfected with Ct or MERIT40 siRNAs. Untreated (0) or treated cells were lysed at the indicated time following the addition of CHX (60 μg/mL). IB was performed as indicated. (D,E) MERIT40 knockdown reduces Rap80 DSB localization. IF was performed for Rap80 following Ct or MERIT40 siRNA treatment at IR-induced (D) or laser-induced (E) DSBs as indicated. Images are representative of >200 cells for each condition in three independent experiments. (E) Quantification of Rap80 colocalization with γH2AX at stripes is shown in Supplemental Figure S8A. (F) HeLa S3 cells containing stably expressed eRap80 were transfected with Ct- or MERIT40-targeted siRNA and subsequently examined for IRIF with α-HA and α-53BP1 antibodies. eRap80 is still able to localize to IRIF following MERIT40 depletion.
The tandem UIM domains of Rap80 are both necessary and sufficient for DSB localization (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007), while the Rap80 AIR has been proposed to play an adjunct role in DSB recognition (Wang and Elledge 2007; Yan et al. 2007). To determine if MERIT40 contributed to this role, MERIT40 knockdown was performed and Rap80 localization to IRIF and laser-induced DSBs was examined. In both cases, MERIT40 depletion strongly reduced endogenous Rap80 localization to sites of DNA damage (Fig. 4D,E; Supplemental Fig. S8A). Conversely, ectopic Rap80 (eRap80) localized to IRIF in Ct or MERIT40 siRNA-transfected cells (Fig. 4F), as did ectopic Rap80Δ233–399 (Supplemental Fig. S8B), indicating that MERIT40 largely influences Rap80 DSB localization through its effects on Rap80 protein stability, rather than by providing an essential DSB recognition signal.
Interdependency of the BRCA1–Rap80 complex for DSB localization and interaction with Rap80
Abraxas and BRCC36 have also been reported to strongly contribute to Rap80 IRIF formation (Wang and Elledge 2007). A plausible mechanism is that each of these proteins is necessary for MERIT40 to interact with Rap80. In this event, knockdown of Abraxas or BRCC36 would strongly reduce MERIT40 foci formation and interaction with Rap80. This proved to be accurate, as MERIT40 DSB localization at IRIF and laser stripes was strongly reduced in either Abraxas- or BRCC36-depleted cells (Fig. 5A,B; Supplemental Fig. S4B). Similarly, BRCC36 and Abraxas knockdown disrupted MERIT40 interaction with Rap80 (Fig. 5C), suggesting interdependencies among members of the BRCA1–Rap80 complex for interaction with the Rap80 AIR and for DSB recruitment. BRCC45 was present in the Rap80 (1–719) complex at similar levels to the other components, and has been reported to form a binary interaction with C19orf62/MERIT40 by yeast two-hybrid assay (Rual et al. 2005). BRCC45 was also originally identified in complex with BRCC36, and knockdown of BRCC36 or BRCC45 was determined to have similar effects on G2 checkpoint and viability responses to IR (Dong et al. 2003). We thus interrogated the biochemical function of BRCC45 in this complex to determine if it was also required for MERIT40 DSB retention and interaction with Rap80. BRCC45 knockdown strongly diminished MERIT40 and BRCA1 foci and, correspondingly, prevented MERIT40 interaction with Rap80 and BRCC36, implicating it as a critical interacting partner for BRCA1–Rap80 complex integrity (Fig. 5D,E; Supplemental Fig. S4B).
Figure 5.
Interdependence of the BRCA1–Rap80 complex for DSB localization and protein interactions. (A) IF for MERIT40 and γH2AX at 4 h after 10 Gy IR in HeLa cells following Ct (left panel), Abraxas (middle panel), or BRCC36 (right panel) siRNA treatment demonstrates reduced MERIT40 DSB localization. Results are quantified and displayed graphically in Supplemental Figure S4B. (B) IF for MERIT40 and γH2AX in laser-irradiated HeLa cells following Ct, MERIT40, BRCC36, or Rap80 siRNA treatment demonstrates reduced MERIT40 DSB localization. Images are representative of >200 cells for each condition in three independent experiments. (C) Requirement for BRCC36 and Abraxas in Rap80 interactions with MERIT40. Flag IP was performed on HeLa S3 cells expressing ectopic Rap80 at 72 h after transfection with Ct-, BRCC36-, or Abraxas-targeted siRNAs. IB of Flag elutions was performed as indicated, demonstrating that both Abraxas and BRCC36 are required for Rap80 interaction with MERIT40. (D) IF for BRCA1 and MERIT40 following Ct (left panel), MERIT40 (middle panel), or BRCC45 (right panel) siRNA treatment in HeLa S3 cells stably expressing ectopic Flag-HA tagged Rap80. Depletion of MERIT40 or BRCC45 reduces BRCA1 IRIF. Images are representative of >200 cells for each condition in three independent experiments. (E) Requirement for BRCC45 in MERIT40 interaction with Rap80 and BRCC36. Flag IP was performed on HeLa S3 cells expressing ectopic MERIT40 at 72 h after transfection with Ct- or BRCC45-targeted siRNAs. IB of Flag elutions was performed as indicated, demonstrating that BRCC45 is required for MERIT40 interaction with Rap80 and BRCC36.
Interdependency among multiple protein constituents for Rap80 complex integrity
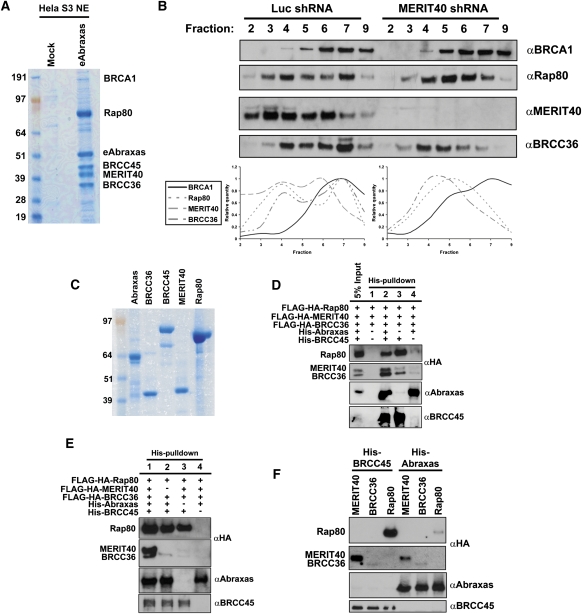
The Rap80 complex appears to contain relatively equal amounts of five components (Fig. 1A). To determine if comparable levels of associations found in the Rap80 complex would be maintained regardless of which constituent was purified, we performed tandem immunoaffinity chromatography with Flag-HA-Abraxas (eAbraxas) from HeLa S3 nuclear extracts (Fig. 6A). As for the Rap80 complex, eAbraxas purification revealed five strong Coomassie-stained bands and numerous substoichiometric interactions, strongly suggesting that Rap80 forms near-stoichiometric interactions in vivo with Abraxas, BRCC45, MERIT40, and BRCC36. To test the hypothesis that MERIT40 plays a scaffolding role for the integrity of this complex, we performed sucrose gradient sedimentation on nuclear extracts derived from HeLa S3 cells expressing shRNA to Luciferase (Luc) or to MERIT40 (Fig. 6B). MERIT40, Rap80, and BRCC36 displayed a biphasic migration through the gradient in Luc shRNA extracts. MERIT40 migrated with Rap80 and BRCC36 throughout the sucrose gradient and also appeared in an earlier fraction than either Rap80 or BRCC36, consistent with it being stable in the absence of binding to other components of the assembled complex. The MERIT40-depleted nuclear extracts demonstrated a more monophasic distribution of Rap80 and BRCC36, with each protein shifting its distribution to smaller complexes in less dense regions of the gradient. Thus, MERIT40 depletion seems to prevent the complete assembly of the Rap80 complex. BRCA1 migration was not qualitatively affected by MERIT40, consistent with it being assembled into several alternative complexes (Greenberg et al. 2006).
Figure 6.
Biochemical reconstitution of the Rap80 complex in vitro. (A) Coomassie-stained gel representing eAbraxas complexes after consecutive Flag and HA affinity-purification steps from HeLa S3 nuclear extracts (NE). The predominant interacting proteins are readily observed in strong Coomassie-stained bands for Rap80, eAbraxas, BRCC45, MERIT40, and BRCC36. BRCA1 was detected substoichiometrically as an Abraxas-associated protein. (B) HeLa S3 nuclear extracts derived from cells expressing shRNA to either luciferase (luc) or MERIT40 were centrifuged through a 5%–22.5% sucrose gradient, and individual fractions were probed by IB as indicated. MERIT40 depletion shifts peak protein levels of Rap80 and BRCC36 to lower fractions. Quantification of bands was performed using ImageJ software from the NIH. (C) Coomassie-stained gel representing purified proteins used for in vitro binding assays. Recombinant Abraxas and BRCC45 are His-NusA- and His-ZZ-tagged, respectively; were bacterially derived; and are estimated to be ∼90% pure. Recombinant BRCC36, MERIT40, and Rap80 derived from baculovirus-infected Sf9 cells are Flag-HA-tagged and are estimated to be ∼95% pure. (D,E) IB of in vitro binding assays was performed as indicated. BRCC36, MERIT40, and Rap80 were baculovirally expressed and mixed in equal molar concentrations with both Abraxas and BRCC45 or combinations thereof. HisPur Cobalt resin was used for coprecipitation of recombinant proteins. (F) IB of in vitro binding assays was performed as indicated. His-tagged BRCC45 or His-tagged Abraxas were mixed in equal molar concentration with Rap80, MERIT40, or BRCC36 individually and coprecipitated using HisPur Cobalt resin.
The interdependence of each member of the Rap80 complex for DSB localization and protein–protein associations provides evidence for a series of interdependent interactions that regulate Rap80 complex integrity and DSB localization. The data supporting this model were derived exclusively from in vivo experiments (Figs. 1–5), precluding a more precise analysis of how the Rap80 core complex is assembled at a biochemical level. Complicating this analysis is the observation that protein stability of each component of the complex was dependent on MERIT40 expression.
Therefore, we set forth to determine the requirements for Rap80 complex assembly in vitro using recombinant protein. Flag-HA-tagged Rap80, MERIT40, and BRCC36 were expressed and purified as soluble protein from baculovirus-infected Sf9 cells, while BRCC45 and Abraxas were expressed and purified from Escherichia coli as his-NusA or his-ZZ-N-terminal fusion proteins, respectively. Purified protein for each component is documented in Figure 6C. The Rap80 complex was assembled in a stepwise fashion by combining all of the components in binding buffer, followed by precipitation of His-Abraxas and His-BRCC45 with Co2+-agarose beads (Fig. 6D,E). This demonstrated coprecipitation of Rap80, MERIT40, and BRCC36, while control pull-downs lacking any His-tagged protein did not bring down detectable levels of any of the Flag-HA-tagged proteins (Fig. 6D,E). Removing an individual component of the complex recapitulated the in vivo interdependencies that were observed for association with the Rap80 complex in prior experiments (Figs. 3–5). The absence of MERIT40 strongly reduced BRCC36 association with the complex (Fig. 6E). Similarly, removal of Abraxas reduced association of both MERIT40 and BRCC36 with BRCC45 and Rap80 (Fig. 6D). A striking finding from this analysis was that the absence of Abraxas did not reduce BRCC45 association with Rap80, while removing BRCC45 strongly diminished Abraxas association with Rap80 and other components, suggesting that BRCC45 plays an essential role in mediating association between the Rap80 AIR and other components of the complex (Fig. 6D,E). We next attempted to identify binary interactions within the complex by incubating each component individually with either BRCC45 or Abraxas. BRCC45 maintained association with Rap80 and MERIT40, and weaker association with BRCC36. Abraxas displayed reduced association with Rap80 in comparison with BRCC45, and also coprecipitated MERIT40 and BRCC36 (Fig. 6F). In sum, both in vitro and in vivo mapping approaches demonstrate similar interdependencies for the assembly of the Rap80 complex, with new evidence that BRCC45 provides a critical link to the Rap80 AIR.
MERIT40 links Rap80 complex-associated DUB activity to the G2 checkpoint and cellular resistance to IR
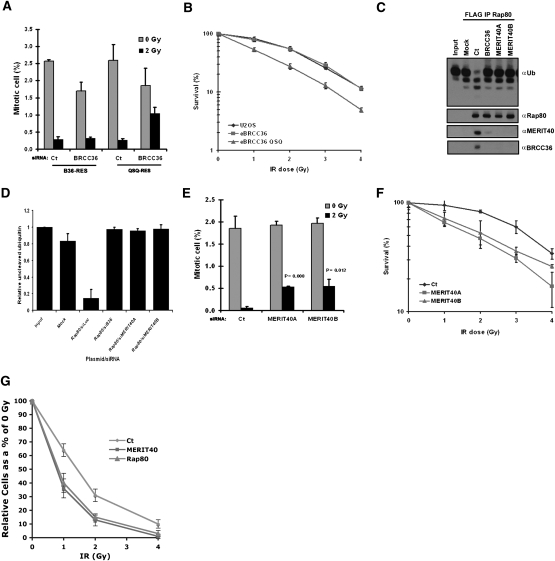
Each member of the BRCA1–Rap80 complex is required for checkpoint and viability responses to IR. Given their interdependencies for complex integrity, DSB localization, and DDR function, a unifying hypothesis is that the BRCA1–Rap80 complex executes its DDR functions by delivering ubiquitin recognition and ubiquitin chain-modifying enzymatic activities to DSBs. To specifically test if DUB activity was responsible for the reported BRCC36 DDR activities (Dong et al. 2003), we engineered U2OS cell lines that stably express siRNA-resistant forms of ectopic BRCC36 (B36-RES) or a BRCC36 QSQ (QSQ-RES) DUB mutant that replaces the Zn2+-binding histidine residues with glutamines (BRCC36H122H124 to BRCC36Q122Q124). These residues are required for JAMM (JAB1/MPN/Mov34 metalloenzyme) domain DUB activity (Ambroggio et al. 2004; Sobhian et al. 2007), but did not affect BRCC36 association with Rap80 or DSB recognition (Supplemental Fig. S9). We examined each BRCC36 line for BRCA1–Rap80 complex-dependent responses to IR. Expression of the siRNA-resistant wild-type BRCC36 maintained G2 checkpoint activity following endogenous BRCC36 knockdown, while cells reconstituted with the DUB-inactive BRCC36 QSQ mutant remained deficient for the IR-induced G2 checkpoint (Fig. 7A). Similarly, expression of BRCC36-QSQ conferred a dose-dependent IR supersensitivity compared with expression of BRCC36 wild type or a vector control in U2OS cells (Fig. 7B), indicating that DUB activity is essential for both checkpoint and IR resistance responses.
Figure 7.
MERIT40 and BRCC36 DUB contributions to the DDR. (A) Knockdown of endogenous BRCC36 in U2OS cells that ectopically express siRNA-resistant forms of either B36 wild type or B36 QSQ reveals that BRCC36 DUB activity is required for the G2 checkpoint. Mitotic cells were detected by FACS with an antibody against phosphohistone H3. Error bars represent SD. (B) BRCC36 DUB deficiency increases IR sensitivity as measured by colony formation in U2OS cells that express eBRCC36 wild type or eBRCC36 QSQ. Each group was plated at low density and exposed to escalating doses of IR as indicated. Cells were maintained in culture for 12 d following IR and subsequently Giemsa-stained. Colonies were counted and normalized as a percentage of colonies formed at 0 Gy. Each point represents the average of three independent experiments. Error bars indicate SD. (C) Knockdown with either BRCC36 or MERIT40 siRNA reduces Rap80-associated K63-Ub DUB activity. eRap80 complexes were purified from nuclear extracts of HeLa S3 cells at 48 h after transfection with the indicated siRNAs. Mock represents Flag IP from HeLa S3 cells that do not express ectopic protein. Flag-purified complexes were incubated with hexa-K63–Ub, and the products were detected by IB with antibody to Ub. The figure is representative of three independent experiments. (D) Quantification of C using ImageJ software from the NIH. Relative uncleaved ubiquitin represents the relative intensity of the uppermost K63–Ub band in the IB normalized to input. Error bars represent SD. (E) MERIT40 is required for the G2 checkpoint. The G2 checkpoint assay was performed as in A following Ct or MERIT40 siRNAs as indicated. The percentage of mitotic cells is graphically displayed. Error bars represent SD and P values were calculated by the Student's t-test for each MERIT40 siRNA compared with Ct siRNA. (F) Knockdown of endogenous MERIT40 in HeLa cells reveals increased IR sensitivity. Colony formation assay was performed as in B following Ct or MERIT40 siRNAs. Cells were treated with the indicated doses of IR at 48 h after siRNA transfection. Error bars represent SD. (G) MERIT40 and Rap80 contribute to IR resistance in BRCA1 mutated cells. HCC1937 cells were exposed to escalating doses of IR 48 h after transfection with the indicated siRNAs. Relative cell number at each dose of IR is displayed graphically. Error bars represent SD.
As MERIT40 depletion abrogates BRCC36 IRIF (Fig. 3B) and interactions with Rap80 and Abraxas (Fig. 3C, D), we thus reasoned that MERIT40 influences DDR events by enabling Rap80 to target BRCC36 K63–DUB activity to DSBs. To investigate this possibility, we performed an in vitro DUB assay of K63–Ub with Rap80 complexes that were purified from HeLa S3 nuclear extracts at 72 h after transfection with Ct, BRCC36, or either of two different MERIT40 siRNAs. BRCC36 knockdown quantifiably reduced Rap80-associated DUB activity to levels similar to a mock control (Fig. 7C,D). Depletion of MERIT40 resulted in loss of Rap80 association with BRCC36 and loss of Rap80-associated DUB activity comparable with BRCC36 depletion (Fig. 7C,D). These findings (Fig. 7A–D) are compatible with a model in which translocation of BRCC36 DUB activity to DSBs is an essential element of the BRCA1–Rap80 complex in the DDR (Sobhian et al. 2007). Consistent with this hypothesis, MERIT40 depletion resulted in both a partial G2 checkpoint deficit (Fig. 7E) and increased IR sensitivity (Fig. 7F).
Each member of the Rap80 complex localizes to DSBs independently of its interaction with BRCA1 and also appears to be present at many DSB sites prior to wild-type BRCA1 (Fig. 2A,G), raising the possibility that members of the Rap80 core complex may have, at least in part, BRCA1-independent functions in the DDR. We thus determined if MERIT40 and Rap80 influenced the DDR in BRCA1 mutant cells. This entailed knockdown of either MERIT40 or Rap80 in HCC1937 cells and examining IR sensitivity in comparison with Ct siRNA-transfected cells (Fig. 7G). Both MERIT40- and Rap80-transfected HCC1937 cells demonstrated a dose-dependent IR supersensitivity compared with Ct-transfected cells, suggesting that in addition to targeting BRCA1 to DSBs, MERIT40 and Rap80 at least partially contribute to the DDR independent of their association with BRCA1.
Discussion
The cellular response to DSBs resulting from both endogenous and exogenous genotoxic stress involves the recruitment of distinct protein complexes containing the tumor suppressor protein BRCA1. The BRCA1–Rap80 complex, defined by its phosphorylation-dependent interaction between the BRCA1 BRCT domain and Abraxas in association with Rap80 and BRCC36, is one of these complexes that exerts its functions at ubiquitinated chromatin regions adjacent to DSBs. MERIT40 is a new member of the BRCA1–Rap80 complex and contributes an essential role in G2 checkpoint control and IR resistance functions.
MERIT40 does not contain obvious sequence homology with known protein domains or enzymatic activities. Instead, MERIT40 may play a scaffolding role for the BRCA1–Rap80 complex by maintaining the interactions, stability, and DSB targeting for each component. Consequently, depletion of MERIT40 abrogates both interactions and DSB-targeted activities for the entire BRCA1–Rap80 complex. The importance of MERIT40 in maintaining the stability and DSB targeting of the BRCA1–Rap80 complex cannot be overlooked. Protein levels of Abraxas and BRCC36 as well as Rap80 were reproducibly reduced to a greater extent by MERIT40 knockdown than by siRNA against several of the other constituents (Figs. 3E,F, 4A–C). The requirement of protein–protein interactions for protein stability has been commonly observed, with codependency of BRCA1–BARD1 interaction being a notable example (Joukov et al. 2001).
While the Rap80 AIR has been postulated to play an adjunct role in DSB recognition (Wang and Elledge 2007; Yan et al. 2007), the mechanistic basis for this observation has remained unclear. Our results indicate that interactions with Abraxas, BRCC36, BRCC45, and MERIT40 serve to stabilize Rap80 protein levels, and that this may explain, at least in part, the importance of the AIR for DSB localization. Indeed, ectopic Rap80 or ectopic Rap80 Δ233–399 localize to IRIF independently of MERIT40 or BRCC45 (Fig. 4F; Supplemental Fig. S8), indicating that overexpression of Rap80 species containing intact UIM domains is sufficient to bypass a requirement for MERIT40 interaction. Conversely, MERIT40 knockdown abrogated IRIF formation for ectopic BRCC36 or Abraxas and also disrupted both the in vivo and in vitro association of these proteins with Rap80, suggesting that Rap80 is still the dominant DSB recognition element for the entire complex.
In vivo analysis of the BRCA1–Rap80 complex suggests interdependent interactions between Abraxas, BRCC36, and MERIT40 for association with Rap80. In vitro reconstitution with recombinant members of the complex confirms this model. These studies also revealed the importance of BRCC45 for association of each component of the complex with the Rap80 AIR. This study provides the first evidence for how BRCC45 may function in the DDR, by modulating assembly of the BRCA1–Rap80 complex at the Rap80 AIR. Furthermore, the MERIT40–BRCC45 interaction likely plays a role in the scaffolding function of MERIT40, as BRCC45 protein levels were strongly reduced following MERIT40 knockdown (Supplemental Fig. S10). This function of BRCC45 appears critical for in vivo DDR responses as well, as BRCC45 knockdown prevented MERIT40 and BRCA1 IRIF and MERIT40 association with Rap80 (Fig. 5D,E).
These experiments thus provide novel molecular insights into how BRCA1 functions in the DDR. We propose a model whereby interchangeable core protein complexes associate with the BRCA1 BRCT domain to target BRCA1 for different checkpoint and repair functions at the site of DNA damage. Each of these core complexes would also assemble and associate with DSBs in the absence of BRCA1 interaction and have some BRCA1-independent DNA repair functions. This is consistent with recent reports documenting that CtIP associates with Mre11 independently of BRCA1 (Sartori et al. 2007), and that Brip1/FANCJ interacts with MLH1 and MS2 independently of BRCA1 and contributes to DNA cross-link repair even in the absence of BRCA1 interaction (Bridge et al. 2005; Peng et al. 2007). Our findings suggest a similar scenario for the Rap80 complex in that (1) BRCA1 displays a substoichiometric association with the core complex, (2) Rap80 and MERIT40 localize to DSBs independently of BRCA1 interaction and are present at more IRIF than BRCA1 in cells expressing wild-type BRCA1, and (3) MERIT40 or Rap80 knockdown confers increased sensitivity to IR in BRCA1 mutated HCC1937 cells. These results set the stage for studying synthetic lethal interactions that target BRCT-associated core complexes in BRCA1-deficient cancers. BRCC36 DUB activity represents one such potential target.
A second insight gained from this study is the importance of delivery of BRCC36 DUB activity to DSBs for the BRCA1–Rap80-dependent DDR. While BRCC36 had been implicated in G2 checkpoint and viability responses to IR (Dong et al. 2003), the present study is the first to demonstrate that BRCC36 DUB activity is essential for these DDR functions. Thus, BRCC36 DUB activity is a key component in G2 checkpoint control and viability responses to IR. Indeed, the USP1 and USP28 DUBs have been reported to have roles in the DDR (Nijman et al. 2005; Huang et al. 2006; Zhang et al. 2006; Bassermann et al. 2008), although it is unclear if they function at the site of DNA damage or elsewhere in the nucleus. Regardless, an emerging concept from these independent studies is that, similar to kinase/phosphatase cascades, regulation of ubiquitination and deubiquitination is an important aspect of the DDR (Huang and D'Andrea 2006).
In summary, we identified MERIT40, a novel provider of BRCA1–Rap80 complex integrity and DDR function. Given the central importance of BRCA1 in genome integrity control and tumor suppression, these findings raise the possibility that MERIT40, like other essential BRCA1-associated factors, may be worthy of investigation as a target of disease-causing mutations.
Materials and methods
Cells
Adherent and S3 HeLa cells and U2OS cells were cultured in DMEM (GIBCO) with 10% calf serum. HCC1937 cells were cultured in RPMI (GIBCO) with 10% calf serum. Phoenix A retroviral packaging lines were used to produce eRap80, eRap80 Δ233–399, eAbraxas, eBARD, eBRCC36, and MERIT40 retrovirus. Cell lines infected with retrovirus were selected on anti-IL2 receptor antibody-coated (Upstate Biotechnologies) DynabeadsR (Invitrogen). Transient transfections were performed on 293T cells using lipofectamine (Invitrogen).
Purification of Rap80-containing complexes
Rap80 complexes were purified from nuclear extracts of HeLa S3 cells stably expressing N-terminal Flag- and HA epitope-tagged eRap80 or eRap80 Δ233–399 by a standard two-step affinity chromatography method (Nakatani and Ogryzko 2003). Purified material was subsequently electrophoresed on a 4%–12% Bis-Tris SDS gel (Invitrogen), and Coomassie-stained bands were excised and submitted for tandem mass spectrometry analysis at the University of Pennsylvania Proteomics Facility.
G2 checkpoint assay
The G2 checkpoint assay was performed by assessing the percentage of mitotic cells at 0 Gy and at 1 h after 2 Gy IR as described previously (Xu et al. 2001). A rabbit polyclonal antibody (Ab) against phosphorylated histone H3 (Upstate Biotechnologies) was used to detect mitotic cells.
DNA damage induction
An MDS Nordion Gammacell fixed source Cs-137 irradiator was used to treat cells with 10 Gy of radiation. Cells were incubated for 15 min, 30 min, 1 h, 2 h, or 4 h at 37°C, and fixation and IF were performed as described previously (Greenberg et al. 2006).
A PALM MicroBeam laser microdissection system (Carl Zeiss MicroImaging, Inc.) was used to induce DNA DSBs as described previously (Bekker-Jensen et al. 2006; Sobhian et al. 2007). Cells were grown on coverslips for 36 h in standard media containing 10 μM BrdU (Sigma-Aldrich). Laser stripes were performed on at least 70 cells per coverslip with the PALM Microbeam laser (λ = 337 nm) at 62% power on the 40× objective. Cells were incubated for 30 min at 37°C, and fixation and IF were performed as described previously (Greenberg et al. 2006).
Antibodies
A MERIT40 rabbit polyclonal Ab was generated against a GST-MERIT40 fusion protein and used for immunoblot (IB) at 1:500 and IF at 1:50 dilution. γH2AX was detected by IF with mouse monoclonal Ab clone JBW301 (Upstate Biotechnologies) at 1:2500 dilution. 53BP1 was detected by IF with a rabbit polyclonal Ab (Novus) at 1:250 dilution. BRCA1 was detected by IB with mouse monoclonal Ab MS110 at a 1:10 dilution (Scully et al. 1997), and by IF with mouse monoclonal antibody SC-6954 (Santa-Cruz Biotechnologies) at 1:50. A Rap80 rabbit polyclonal antibody (Sobhian et al. 2007) was used for IB at 1:500 and IF at 1:100 dilutions. BRCC36 was detected for IB with a rabbit polyclonal Ab (Sobhian et al. 2007). An Abraxas rabbit polyclonal Ab was generated against a His-ZZ-Abraxas fusion protein and used for IB at 1:500 dilution. BRCC45 was detected for IB by a rabbit polyclonal Ab raised against human BRCC45 (Zymed Corp.). HA-tagged proteins were detected by IB at 1:1000 and for IF at 1:1000 dilutions using mouse monoclonal Ab HA.11 (Covance).
DUB activity assay
Flag-peptide-eluted Rap80 complexes were incubated with K63-linked hexa-ubiquitin (Boston Biochem) in DUB buffer at 37°C as described previously (Sobhian et al. 2007).
RNAi
Transfections were performed using either oligofectamine or lipofectamine RNAiMax (Invitrogen) per the manufacturer's instructions.
The following siRNA target sequences were used: Abraxas, CGUUUAGAGAGAGGCUGCUUCACAA; BRCC36, AACAUCAACAUGUGAAGGCCTT; BRCC45, GGUGCAGUACGUGAUUCAA; Ct, UCGAAGUAUUCCGCGUACGTT; MERIT40A, AGCUUUGGCAGUGACAUUUCCUCUG; MERIT40B, CCCAUAUUUCUUCUUUGACGUUGUU; Rap80, CCAGUUGGAGGUUUAUCAA; RNF8, GGACAAUUAUGGACAACAATT; Ubc13, GGACUAGGCUAUAUGCCAU.
GST pull-down assay
Lysate from cells expressing Flag-HA tagged MERIT40 was mixed with equal amounts of recombinant GST, GST-Brca1 BRCT, or GST-Brca1 BRCT M1775R protein that was bound to glutathione–Sepharose beads. After incubation overnight at 4°C, the beads were washed three times with PBS. The bound proteins were released by boiling in SDS loading buffer and were detected by IB with α-HA Ab.
CHX chase
Forty-eight hours to 72 h after siRNA knockdown, HeLa cells were treated with CHX (60 μg/mL) or vehicle for the time indicated. Cells were harvested, lysed in RIPA buffer (50 mM NaCl, 50 mM Tris at pH 7.4, 2 mM EDTA, 1% Nonidet P-40, 0.1% SDS, proteinase inhibitor cocktail), and subjected to IB.
Purification of recombinant proteins and protein–protein interaction assays
His-tagged versions of Abraxas and BRCC45 were expressed in E. coli using autoinduction media. Cells were resuspended in a buffer containing 25 mM Tris (pH 8.5), 250 mM NaCl, 25 mM imidazole, and 10% glycerol. Lysozyme, PMSF, and 1% Tween20 were added prior to brief sonication. After high-speed centrifugation, the lysate was applied to a Ni-NTA (Qiagen) column and washed extensively. Recombinant protein was eluted using 250 mM imidazole, 25 mM Tris 8.5, 250 mM NaCl, and 10% glycerol and run on a Superdex 75 gel filtration column. The purity of the final protein was estimated to be ∼90% by Coomassie staining (see Fig. 6C).
Flag-HA-tagged Rap80, BRCC36, and MERIT40 were cloned in pVL1393 and expressed using the BaculoGold baculoviral expression system (BD Biosciences). High-titer recombinant virus was generated and used to infect Sf9 cells using standard methods. Two days to 3 d after infection, cells were harvested and lysed in a buffer containing 50 mm Tris (pH 8.0), 120 NaCl, 0.5% NP-40 and PMSF, and 5 mM β-mercaptoethanol, and the recombinant protein was purified using α-Flag bead IP (Sigma) and Flag peptide elution. Purity was estimated to be ∼95% by Coomassie staining (Fig. 6C). All recombinant protein was quantified using the Coomassie Plus reagent (Pierce).
For the binding assays, 30 μM concentrations of each protein were incubated in a binding buffer containing 50 mM Tris (pH 8.0), 100 mM NaCl, 30 mM imidazole, 10% glycerol, 1 mM DTT, and 0.1 mg/mL BSA for 2–3 h at 4°C. Pull-downs were performed by incubating protein complex with HisPur Cobalt resin (Pierce) for 30 min and washing three times with binding buffer. Protein was eluted from the cobalt resin using 200 mM imidazole and run on a 4%–12% bis-tris gradient gel (Invitrogen). After transferring to a nylon membrane (Whatman), protein was detected using mouse α-HA (Covance), rabbit α-Abraxas, and HRP-linked secondary antibodies, and was visualized using the ECL plus reagent (Perkin-Elmer).
Sucrose gradients
Five percent to 22.5% sucrose gradients in KETENG-100 buffer (100 mM KCl, 0.5mM EDTA, 20mM Tris at pH 7.4, 0.1% NP-40, 1.5mM MgCl2, 10% glycerol) were prepared in 14 × 89-mm polyclear tubes (Seton Scientific) using a Gradient Master (BioComp) using the following settings: time, 2.25 min; angle, 81.5°; speed 15 rpm. Samples to be analyzed were loaded on the gradient and centrifuged at 35,000 rpm for 15 h in an SW41 rotor (Beckman Coulter). Fractions of the gradient (0.5 mL) were collected using a piston gradient fractionator (BioComp) with a speed setting of 0.3. Fractions were analyzed by Western blotting.
Acknowledgments
We thank J. Chen for reviewing results prior to publication, J.A. Diehl for helpful discussions, and L. Pontano and M. Romero for advice on baculoviral transduction and protein purification in Sf9 cells. This work was supported by K08 award 1K08CA106597-01 from the National Cancer Institute, the Sidney Kimmel Foundation Scholar Award, The Mary Kay Ash Foundation Translational Innovation Award, and funds from the Abramson Family Cancer Research Institute to R.A.G.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1739609.
Supplemental material is available at http://www.genesdev.org.
References
- Ambroggio X.I., Rees D.C., Deshaies R.J. JAMM: A metalloprotease-like zinc site in the proteasome and signalosome. PLoS. 2004;2:e2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassermann F., Frescas D., Guardavaccaro D., Busino L., Peschiaroli A., Pagano M. The Cdc14B–Cdh1–Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–267. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge W.L., Vandenberg C.J., Franklin R.J., Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- Celeste A., Fernandez-Capetillo O., Kruhlak M.J., Pilch D.R., Staudt D.W., Lee A., Bonner R.F., Bonner W.M., Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- DePinho R.A. The age of cancer. Nature. 2000;408:248–254. doi: 10.1038/35041694. [DOI] [PubMed] [Google Scholar]
- Dong Y., Hakimi M.A., Chen X., Kumaraswamy E., Cooch N.S., Godwin A.K., Shiekhattar R. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell. 2003;12:1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- Ewing R.M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M.D., O'Connor L., Li M., et al. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg R.A., Sobhian B., Pathania S., Cantor S.B., Nakatani Y., Livingston D.M. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes & Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.T., D'Andrea A.D. Regulation of DNA repair by ubiquitylation. Nat. Rev. Mol. Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., D'Andrea A.D. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006;4:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V., Chen J., Fox E.A., Green J.B., Livingston D.M. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci. 2001;98:12078–12083. doi: 10.1073/pnas.211427098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Chen J., Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007a;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kim H., Huang J., Chen J. CCDC98 is a BRCA1–BRCT domain-binding protein involved in the DNA damage response. Nat. Struct. Mol. Biol. 2007b;14:710–715. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K.W., Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Liu Z., Wu J., Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat. Struct. Mol. Biol. 2007;14:716–720. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Moynahan M.E., Chiu J.W., Koller B.H., Jasin M. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Nakatani Y., Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Huang T.T., Dirac A.M., Brummelkamp T.R., Kerkhoven R.M., D'Andrea A.D., Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Peng M., Litman R., Xie J., Sharma S., Brosh R.M., Jr, Cantor S.B. The FANCJ/MutLα interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007;26:3238–3249. doi: 10.1038/sj.emboj.7601754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rouse J., Jackson S.P. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Chen J., Ochs R.L., Keegan K., Hoekstra M., Feunteun J., Livingston D.M. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Scully R., Ganesan S., Vlasakova K., Chen J., Socolovsky M., Livingston D.M. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- Sobhian B., Shao G., Lilli D.R., Culhane A.C., Moreau L.A., Xia B., Livingston D.M., Greenberg R.A. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman A.R. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Wang B., Elledge S.J. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., Elledge S.J. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Kim St., Kastan M.B. Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 2001;21:3445–3450. doi: 10.1128/MCB.21.10.3445-3450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Kim Y.S., Yang X.P., Li L.P., Liao G., Xia F., Jetten A.M. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;67:6647–6656. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden R.I., Pardo-Reoyo S., Sgagias M., Cowan K.H., Brody L.C. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 2002;30:285–289. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- Yu X., Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Zaugg K., Mak T.W., Elledge S.J. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- Zhao G.Y., Sonoda E., Barber L.J., Oka H., Murakawa Y., Yamada K., Ikura T., Wang X., Kobayashi M., Yamamoto K., et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]