Abstract
Determining the specific role(s) of proteases in cell migration and invasion will require high-resolution imaging of sites of protease activity during live-cell migration through extracellular matrices. We have designed a novel fluorescent biosensor to detect localized extracellular sites of protease activity and to test requirements for matrix metalloprotease (MMP) function as cells migrate and invade three-dimensional collagen matrices. This probe fluoresces after cleavage of a peptide site present in interstitial collagen by a variety of proteases including MMP-2, -9, and -14 (MT1-MMP) without requiring transfection or modification of the cells being characterized. Using matrices derivatized with this biosensor, we show that protease activity is localized at the polarized leading edge of migrating tumor cells rather than further back on the cell body. This protease activity is essential for cell migration in native cross-linked but not pepsin-treated collagen matrices. The new type of high-resolution probe described in this study provides site-specific reporting of protease activity and insights into mechanisms by which cells migrate through extracellular matrices; it also helps to clarify discrepancies between previous studies regarding the contributions of proteases to metastasis.
Keywords: Cell motility, Collagenase, Invasion, Metastasis, MMP, TIMP, MT1-MMP
1. Introduction
Because many proteases exist in both inactive and activated functional forms, clarifying the roles of proteases in cell migration and tumor cell invasion will require examining the localization and focal actions of protease activities rather than the mere presence of protease proteins. For example, even though certain tumor-associated matrix metalloproteases (MMPs)1 have been linked with cancer metastasis, the significance of these proteases for migration in physiologic tissue environments is controversial; data supporting metastasis as protease-driven, -dependent, -associated, or even irrelevant have been reported (Even-Ram and Yamada, 2005; Lopez-Otin and Matrisian, 2007; Mott and Werb, 2004; Nagase et al., 2006; Overall and Kleifeld, 2006; Sternlicht and Werb, 2001). Even the location of protease activity on the cell surface is uncertain: for example, a study with cells transfected with a FRET-based probe for the individual protease (MT1-MMP) reported localization of MT1-MMP activity at the leading edge of cells migrating on a flat (two-dimensional) substratum (Ouyang et al., 2008). In contrast, in a study of cells migrating in a three-dimensional collagen gel with analysis based on a DQ-collagen probe or antibody detection of a cleaved collagen epitope it was reported that proteolysis occurs much further back on the cell body; it was localized to a collagenolysis zone which was posterior to the leading edge/anterior cell attachment zone of MT1-MMP-transfected HT-1080 cells (Wolf et al., 2007).
Consequently, sensitive high-resolution protease-activity probes are needed to determine the precise localization of proteolysis by living stromal and invading cells, as well as to monitor the capacity of inhibitors to block this local cell-surface activity. This will allow rigorous evaluation of the role(s) and mechanisms of protease involvement in cell migration in three-dimensional settings. Existing probes for whole-cell protease activity (Hobson et al., 2006) and self-quenched probes for protease cleavage (Bremer et al., 2001; Horino et al., 2001; McIntyre et al., 2004; Ouyang et al., 2008; Sloane et al., 2006; Yang et al., 2007) have provided valuable approaches in cell culture and animals. However, an ideal protease probe would be based on a substrate with broad cell-surface protease susceptibility but with high spatial and temporal resolution, and it would be anchored to the extracellular matrix. It should also control for physical environmental effects and should not require transfection or other modifications of the cells being characterized.
We describe a newly designed fluorogenic protease substrate that provides high-resolution, live-cell images of focal protease activity on the surface of individual cells migrating on two-dimensional cell culture surfaces and through three-dimensional collagen matrices. Our goal was to design a probe to detect general matrix-degrading proteolytic activities. Using this novel probe, confocal microscopy, and time-lapse microscopy, we demonstrate localization of protease activity of cells migrating both on 2-dimensional gelatin substrata and in 3-dimensional collagen matrices. Additionally, we show inhibition of cell-surface protease activity and migration by physiologically relevant MMP inhibitors while examining differing conclusions in the literature concerning the cell-surface location of protease activity on cells migrating through three-dimensional extracellular matrices and the significance of matrix crosslinking in tumor cell invasion.
2. Results and Discussion
2.1. A novel protease probe
In order to explore the role(s) of proteases in tumor cell migration, we have designed and synthesized a new fluorogenic protease probe containing the peptide recognition/cleavage site GPLGIAG (cleaved between G and I) that is present in the α1 chain of interstitial collagen (Netzel-Arnett et al., 1991). We have characterized this probe in solution and immobilized it to detect protease activity associated with migrating tumor cells on 2-dimensional gelatin substrata and within 3-dimensional collagen gels.
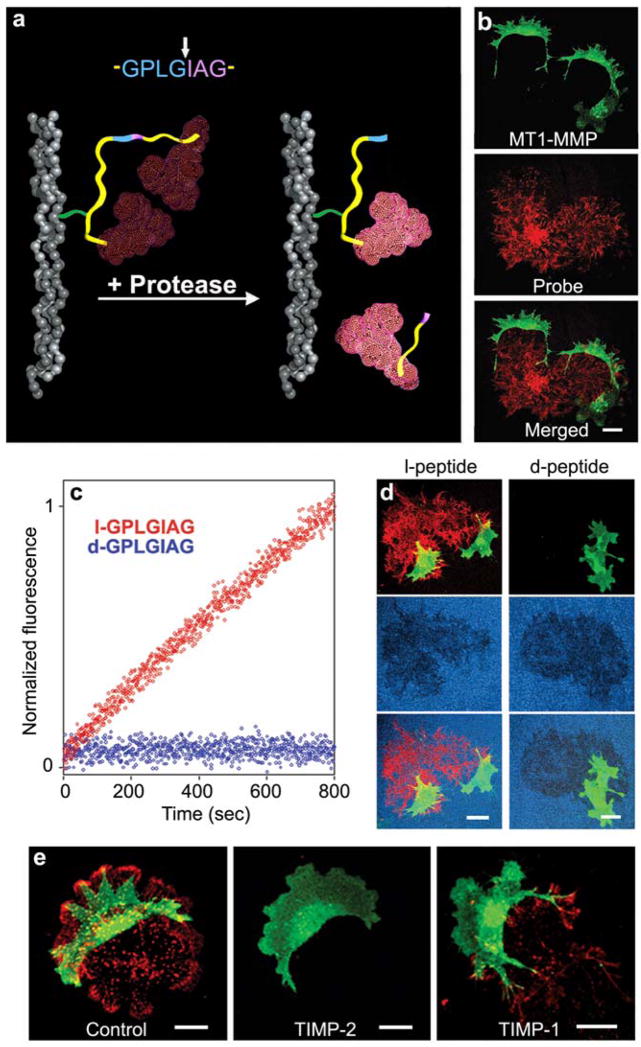
The self-quenched fluorescence probe was immobilized on a solid support such as triple-helical collagen as shown in Figure 1a or gelatin via chemical crosslinking. Contact with cell-associated protease activity produces cleavage and substantially increased local fluorescence at the site of activity (Fig. 1b). The red fluorescence patterns in this figure result from cleavage of the GPLGIAG peptide immobilized to the substratum, and they show the migratory paths taken by cells (green) as the latter traverse a 2-dimensional gelatin matrix.
Fig. 1.
Fluorogenic probe for detection of protease activity in solution, on a 2-dimensional substratum, or in a 3-dimensional extracellular matrix. (a) The GPLGIAG cleavage site from interstitial collagen is incorporated into a peptidyl backbone (yellow) with constrained conformation (Packard and Komoriya, 2008). The 18 amino acid peptide is then homodoubly labeled with rhodamine 6G fluorophores (red) that self-quench by formation of an intramolecular H-type excitonic dimer (Packard et al., 1996). After cleavage of the GPLGIAG site (between blue and purple in the diagram), two fragments are generated, which separate (one fragment remains immobilized and the other diffuses away), thereby disrupting the intramolecular quenching to generate fluorescence (bright red). The probe can be used to detect protease activities in solution or localized proteolytic activity if immobilized with a biotin-streptavidin linker (green) on a surface such as collagen (gray triple helix). (b) Migrating A2 cells [a clone of breast carcinoma MDA-MD-231 cells expressing MT1-MMP with an enhanced GFP marker] (green) produce fluorescence patterns (red) resulting from cleavage of the probe immobilized on a gelatin substratum. (c) Solution kinetics of cleavage by MT1-MMP show stereospecificity of the probe containing all l-amino acids (red) compared to the control with all d-amino acids (blue). (d) Stereospecificity of l- versus d-peptide is retained after immobilization. Cleavage and fluorescence of the probe (red) by A2 cells (green) can be seen in the left column, whereas the right column shows the refractoriness to cleavage of the control (d-amino acids) peptide. Degradation of the gelatin substratum by migrating cells detected by loss of fluorescence (dark areas in blue background) is equivalent. (e) Cleavage of probe (red) by A2 cells (green) on a gelatin substratum is inhibited strongly by 100 nM TIMP-2 compared with untreated control and 100 nM TIMP-1, but migratory morphology is unaffected. Scale bars, 20 μm.
The protease and d/l isoform specificity of probes were characterized in solution by fluorometry. Figure 1c shows kinetics of cleavage by the MT1-MMP (MMP-14) catalytic domain, confirming that the l-isomer peptide is readily digested whereas the d-isomer peptide is totally refractory to cleavage. This difference in protease susceptibility between the l- and d-peptides was confirmed for cellular proteases of live cells by immobilizing each peptide in a gelatin substratum-cleavage assay for focal proteolysis by structures termed invadopodia (Aoyama and Chen, 1990; Artym et al., 2006) (Fig. 1d). Tracks of cleaved probe were detected (left panel) coinciding with regions of gelatin degradation (dark areas in center panel). In contrast, the d-peptide control probe was not cleaved by cell-associated proteases, even though the labeled gelatin substrate itself remained susceptible to protease degradation (Fig. 1d, right panels). Consequently, we routinely used the l-peptide as the protease probe and the d-peptide as the direct control for physical environmental effects. Taken together, these studies demonstrate that a combination of the classical gelatin degradation assay with this new high-resolution protease activity assay should provide further insights into the regulation of the earliest proteolytic sites of attack.
As expected, the GPLGIAG probe is susceptible to cleavage by a variety of MMPs and other proteases, providing a general biosensor for cell-associated proteases including collagen-degrading enzymes such as collagenases MMP-1, -8, -13, and -14 (MT1-MMP), cathepsin K, gelatinases MMP-2 and -9, and elastase (Table 1).
Table 1.
Detection of protease activity in solution by the GPLGIAG probe using fluorometry
| Protease | Cleavage rate (fmol/sec) |
|---|---|
| MMP-1 | 63.2 |
| MMP-2 | 337.0 |
| MMP-3 | 67.4 |
| MMP-7 | 5.6 |
| MMP-8 | 35.2 |
| MMP-9 | 447.8 |
| MMP-13 | 15.3 |
| MT1-MMP | 99.8 |
| MT2-MMP | 22.8 |
| MT3-MMP | 135.0 |
| ADAMTS13 | 0.3 |
| Cathepsin B | 13.2 |
| Cathepsin K | 342.2 |
| Elastase | 163.0 |
| Granzyme B | 0.8 |
| Urokinase | 0.7 |
2.2. Monitoring protease inhibition in solution and in living cells in 2D culture
Matrix-degrading activity is modulated by the endogenous tissue inhibitors of metalloproteases, the TIMPs. TIMPs are 22–28 kDa proteins that form stoichiometric inhibitory complexes with MMPs. In solution-phase protease activity studies using enzymes and TIMP inhibitors at equimolar concentrations (50 nM) (Table 2), the GPLGIAG probe reported the inhibition of MMPs by TIMP-1 and TIMP-2 consistent with known TIMP activities as follows: stoichiometric levels of TIMP-2 inhibited the three MMPs that are membrane-bound under normal physiological conditions as well as MMP-2 by more than 95% and showed considerable inhibition of MMP-9 activity. In contrast, TIMP-1 was unable to inhibit any of the transmembrane MMPs but did inhibit MMP-2 strongly and MMP-9 to a lesser extent. Cleavage of the GPLGIAG probe by elastase, a serine protease with known collagenolytic activity, was unaffected by either TIMP-1 or TIMP-2. Thus, this new probe together with its control substrate can faithfully report the presence of cell-associated protease activities and their differential inhibition by TIMPs. Identification of the specific proteases involved in the total proteolytic activity detected by this probe and their roles in biological processes such as migration could be facilitated by using other inhibitors, e.g. a spectrum of pharmacological, peptide, and antibody inhibitors plus RNA interference approaches to determine the contribution of individual proteases.
Table 2.
Comparisons of protease inhibitory activities of TIMP-1 and TIMP-2
| Protease | Inhibition by TIMP-1 | Inhibition by TIMP-2 |
|---|---|---|
| MT-1 MMP | 0 | 97.1 ± 1.6% |
| MT-2 MMP | 0 | 98.0 ± 1.1% |
| MT-3 MMP | 0 | 99.9 ± 1.1% |
| MMP-2 | 98.3 ± 1.4% | 97.1 ± 1.2% |
| MMP-9 | 49.3 ± 1.0% | 86.0 ± 1.4% |
| Elastase | 0 | 0 |
Values indicate mean ± S.D.
When the GPLGIAG probe was tested with live tumor cells and TIMP inhibitors, the probe conjugated to a 2-dimensional gelatin matrix (Fig. 1e) showed strong fluorescence signals that documented proteolysis where cells had migrated; in contrast to control cells (left panel), migrating tumor cells exposed to TIMP-2 showed no fluorogenic signal (middle panel). TIMP-1 treated cells continued to show fluorescence (right panel), consistent with substantially less inhibition of endogenous proteolytic activity. The migratory morphology of cells in all three 2-dimensional conditions remained similar with or without MMP inhibition.
2.3. Location of protease activity on tumor cells in a 3-dimensional collagen matrix
Since the most abundant and structurally important molecular component of stromal matrices in vivo is collagen, we immobilized the GPLGIAG probe on 3-dimensional type I collagen matrices to characterize the location and function of cell-surface protease activity. The concentration of 3-dimensional collagen gels for routine analyses was chosen to be 2.5 mg/ml based on preliminary studies in which we found that even though the velocity of tumor cell migration was inversely proportional to collagen concentration/density (e.g., see Fig. 2f), 2.5 mg/ml gels supported rapid cell migration similar to a lower concentration gel, but with less cell-to-cell variation in migration speeds.
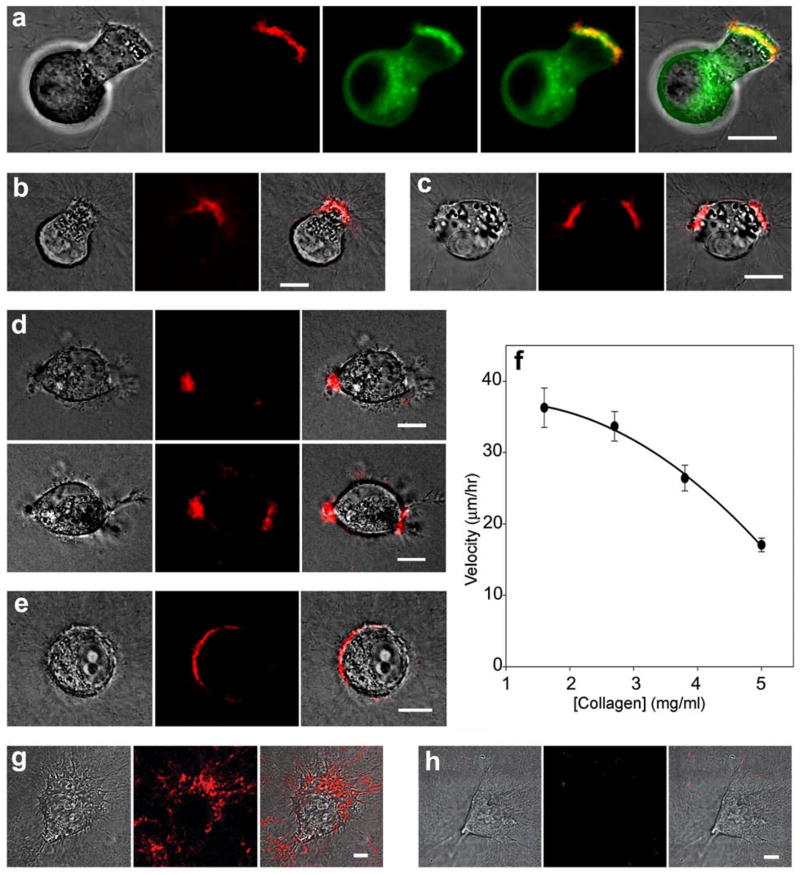
Fig. 2.
Imaging of protease activity on the surface of tumor cells migrating in a 3-dimensional collagen matrix. (a) Although eGFP-MT1-MMP (green) is expressed not only at the polarized leading edge of an A2 cell invading through the collagen matrix, most protease activity (red) localizes to the leading edge. (b and c) A2 cells migrating through 3-dimensional collagen matrices labeled with the protease probe show strong fluorescence (red) and accumulation of intracellular vesicles at leading edges. (d) BT549 breast carcinoma cells migrating through a 3-dimensional collagen matrix show a similar localization to the leading edge. In the top row both proteolysis and accumulation of intracellular vesicles are on the left side of the cell whereas 105 minutes later (bottom row) both have relocated to the right. (e) PC-3 prostate carcinoma cells invading a collagen gel also show localization to the leading edge of the migrating cell. (f) Effects of collagen density/concentration on tumor cell motility. HT-1080 fibrosarcoma cells were embedded in type I crosslinked collagen at different collagen concentrations (1.6 to 5.0 mg/ml). After 48 hours, migration of cells was measured at 10 minute intervals for an additional 48 hour period. Note the inverse correlation between average cell velocity and collagen concentration. However, there was no significant difference between cell velocities in 1.6 and 2.5 mg/ml collagen (P > 0.05 by Student-Newman-Keuls Multiple Comparisons Test), but larger differences in concentration affected migration velocity (P<0.001 for 1.6 versus 3.8 mg/ml; P<0.01 for 2.7 versus 5.0 mg/ml). (g) HT-1080 cells in 3-dimensional collagen matrices labeled with the l-peptide probe show extensive non-polarized extracellular proteolysis consistent with their high, relatively random protrusiveness. (h) HT-1080 cells in collagen matrices labeled with the d-isomer control show no detectable fluorescence. Scale bars, 10 μm.
We first characterized the location of protease activity in tumor cells migrating within 3-dimensional 2.5 mg/ml collagen gels. Imaging of breast carcinoma MDA-MD-231 cells transfected with the MT1-MMP collagenase with an eGFP marker on the cytoplasmic tail (A2 cells) revealed discrepancies between the location of total protease protein and local proteolytic activity (Fig. 2a). Although the eGFP-MT1-MMP molecule (green) is expressed throughout the cell, the collagenase activity (red) is highly localized. Specifically, proteolytic activity is confined to the polarized leading edge of a cell invading through the collagen matrix; this region is characterized by a zone of local cell surface protrusions and intracellular accumulation of vesicles (e.g., see DIC images at the left of Fig. 2a,b,c).
Because the use of cells transfected with a chimeric GFP-protease might affect protease activity or the subcellular localization of a transfected protein might not be physiological, we examined the localization of protease activity on several different types of tumor cells that had not been transfected. Similar localization at the leading edge is observed in BT549 cells (Fig. 2d) and PC-3 cells (Fig. 2e). Notably, as the area of proteolysis moved from left (top left panel of Fig. 2d) to right (bottom left panel of Fig. 2d), the intracellular vesicles also relocalized. In contrast, HT-1080 cells often change polarity and directions of migration rapidly, and they show a broad pattern of proteolysis consistent with the multiple directions of migration (Fig. 2g). For all cell types in 3-dimensional gels, the protease activity was stereo-specific for the l-peptide GPLGIAG probe (Fig. 2g) but not its direct d-peptide control (Fig. 2h), providing a control to rule out non-specific physical effects such as aggregation or tension on the matrix-bound probe. These findings differ from the reported localization of collagenolytic activity by MT1-MMP overexpressing HT-1080 cells to a proteolysis zone which is posterior to the leading edge (Wolf et al., 2007). These significant differences suggest that the sites of measurable proteolysis can differ depending on the cell type, experimental over-expression of a protease, and/or the assay used. Nevertheless, our data suggest that proteolysis is most prominent at the polarized leading edge of non-transfected tumor cells using our detection method for protease activity that has low background at sites of collagen tension and aggregation; local aggregation can produce spurious signals near the cell body of migrating and invading cells.
2.4. Direct comparisons with DQ-collagen in a 3D collagen matrix
We compared protease localization using the GPLGIAG probe with the localization patterns using DQ-collagen, a frequently used reagent in which collagen is directly labeled with multiple fluorescein molecules at a density sufficient to quench fluorescence substantially; release of fluorescence from DQ-collagen is used to detect collagenolytic enzyme activities (Sloane et al., 2006). Because migrating tumor cells can rearrange and locally cluster collagen fibrils, we also examined fluorescence patterns of collagen directly pre-labeled with Alexa 647 succinimidyl ester. PC-3 prostate carcinoma cells showed fluorescence from the GPLGIAG probe that was localized to the leading edge (Fig. 3a). In contrast, the fluorescence signal from DQ-collagen around the same cell was found not only at the leading edge, but it was also present in substantial amounts elsewhere in the matrix (compare green DQ-collagen signal with the DIC pattern of collagen and the merged image). The localization of DQ-collagen fluorescence around the BT549 cell in the matrix in Fig. 3b is very similar to that of collagen directly labeled with Alexa-647, suggesting that aggregated and/or mechanically disrupted collagen can also produce a DQ-collagen-type signal. This phenomenon can also be seen very weakly with the GPLGIAG probe. Consequently, we suggest that the use of fluorescent probes of protease activity should include controls of directly labeled matrix (e.g., Alexa-labeled collagen) and l-/d-isomer controls to rule out effects due to interactions of the probe with the local physical environment.
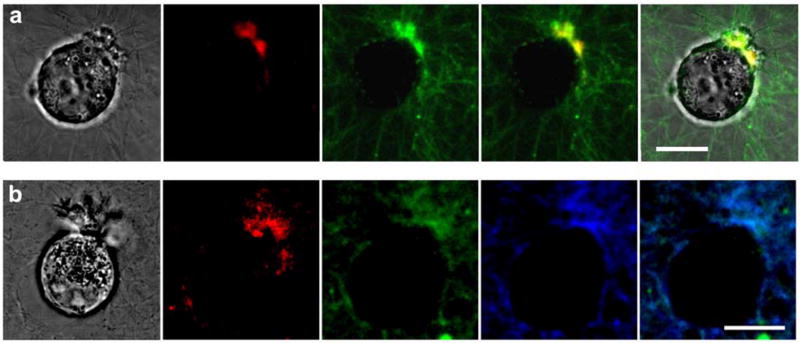
Fig. 3.
Fluorescence from GPLGIAG probe cleavage versus DQ- and Alexa 647-labeled collagen. (a) PC-3 cells display a slight asymmetry during migration with intracellular vesicles (see DIC) and fluorescence from cleavage of the GPLGIAG probe (red) at the polarized leading edge. In contrast, the DQ-collagen cleavage signal (green) is present at both the leading edge and in substantial amounts elsewhere in the matrix. The fourth panel represents merged images of GPLGIAG probe (red) and DQ-collagen (green); the fifth panel shows merged DIC (gray), GPLGIAG probe (red), and DQ-collagen (green). (b) Fluorescence from cleavage of the GPLGIAG probe (red) at the site of matrix invasion by a BT549 cell while DQ-collagen (green) and collagen directly labeled with Alexa 647 (blue) co-localize throughout the matrix. Fifth panel shows merged DQ-collagen (green) and Alexa 647 (blue). Scale bars, 10 μm.
2.5. Role of protease activity in tumor cell invasion through 3-dimensional collagen gels
We applied the GPLGIAG probe to study migration and invasion in real time. As a tumor cell migrates through a 3-dimensional matrix, it leaves behind a record of its passage by removal of matrix (Sabeh et al., 2004) and cleavage products, as exemplified by the PC3 cell in Figure 4a invading a 3-dimensional collagen gel of 2.5 mg/ml type I collagen derivatized with the GPLGIAG probe. In comparison, the migration/invasion paths taken by HT-1080 cells are often substantially more convoluted due to the frequent changes in direction of cell migration by this cell type (Fig. 4b).
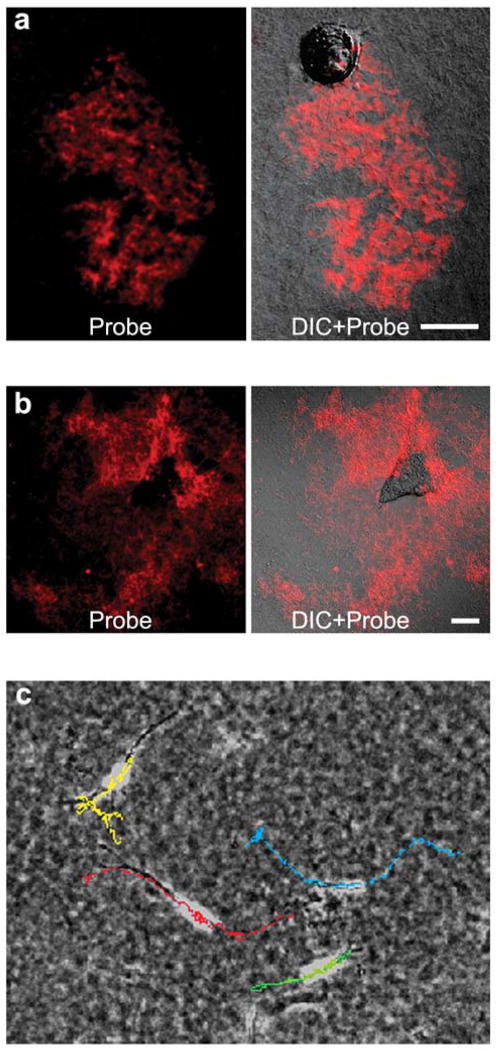
Fig. 4.
Invasion of tumor cells in 3-dimensional collagen gels. (a) Cleavage and fluorescence of the GPLGIAG probe documents proteolysis of the collagen matrix where a PC-3 cell migrated in zig-zag fashion over a 4 day period. (b) Over a four day time course, similar protease activity patterns are observed for an HT-1080 cell. (c) Video time-lapse tracking of migration paths of HT-1080 cells during 48 hours in a 3-dimensional collagen sandwich gel. Scale bars, 20 μm.
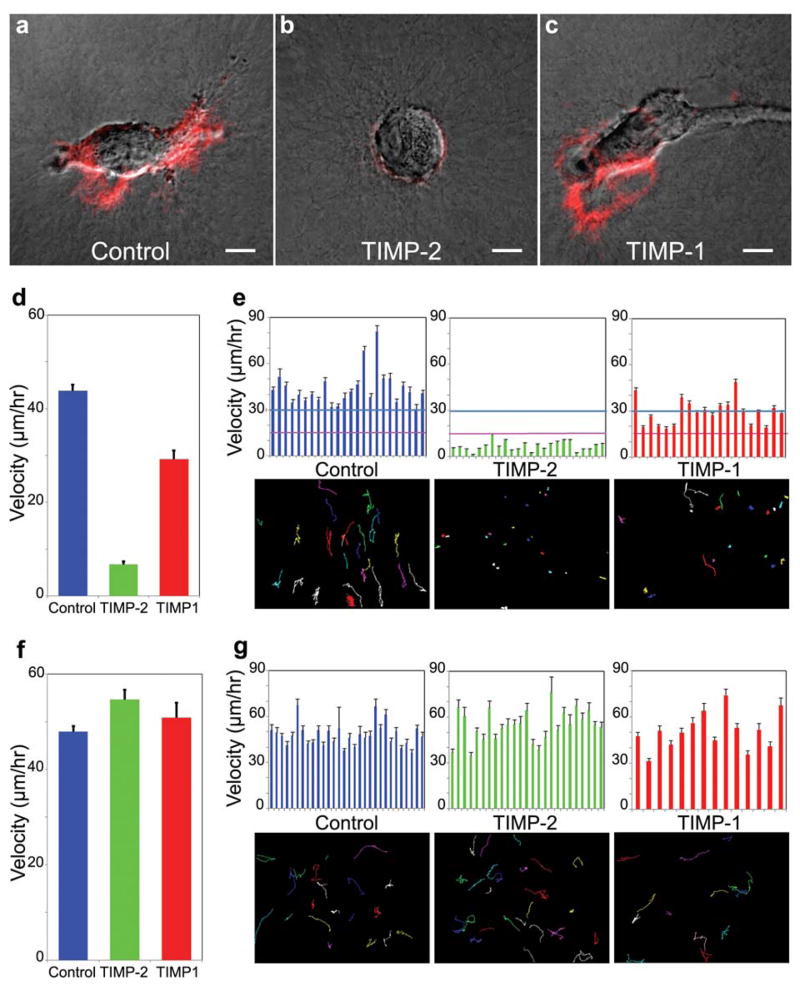
We quantified rates of tumor cell migration using video time-lapse microscopy (Fig. 4c) and cell tracking with MetaMorph software. For inhibiting migration-associated proteases, we used TIMPs and avoided non-physiological chemical inhibitors such as the Zn++ chelating inhibitors BB-94 and GM6001. Preliminary tests identified the effective concentrations needed for TIMP inhibition of probe proteolysis: 100 nM TIMP-2 was sufficient to inhibit proteolysis by the A2 and PC-3 cells, but HT-1080 cells required an increase of TIMP-2 concentration to 200 nM for effective suppression of proteolysis according to GPLGIAG probe-derivatized collagen gels (Fig. 5b and data not shown; see also Fig. 1e). In contrast to TIMP-2, TIMP-1 was substantially less effective as determined by fluorescence intensity (Fig. 5c; see also Fig. 1e). The effects of these inhibitors on probe-detected activity reflected those on tumor cell migration/invasion through native crosslinked type I collagen gels as follows (Fig. 5d, e): TIMP-2 inhibited individual HT-1080 cell migration by 85% (P <0.001). The lesser inhibition of collagenolytic activity by TIMP-1 was reflected in a velocity reduction of ~33% (P >0.001). Specifically, tracking of individual cells between 48 and 96 hours of culture showed the following average velocities ± standard errors: 43.8 ± 11.5 μm/hr for control cells (n=24), 6.73 ± 3.04 μm/hr for TIMP-2-treated (n=24), and 29.2 ± 8.3 μm/hr for TIMP-1-treated (n=20) cells (Fig. 5d). Further analyses showed that the velocity of 100% of TIMP-2-treated cells was less than 15 μm/hr whereas 100% of both control and TIMP-1-treated cells exceeded this value (Fig. 5e). Although TIMP-1 was not as strong an inhibitor of cell surface-associated protease activity and migration/invasion as TIMP-2, the velocity of 60% of cells exposed to this inhibitor was less than 30 μm/hr, whereas all control cells exceeded this rate.
Fig. 5.
Effect of MMP inhibitors on proteolysis and velocity of HT-1080 cells. (a) Control showing protease activity (red). (b) TIMP-2 (200 nM) inhibited cleavage of the GPLGIAG probe (red) accompanied by rounder cell morphology. (c) TIMP-1 (200 nM) was a substantially less effective inhibitor as determined by fluorescence intensity, except at some leading lamellae. (d) Comparison of effects of TIMP-1 and TIMP-2 on tumor cell migration/invasion through acid-extracted, native crosslinked type I collagen gels. (e) Comparisons of velocity (top row) and migration paths (bottom) of individual cells though crosslinked collagen. (f) Comparison of effects of TIMP-1 and TIMP-2 on tumor cell migration/invasion through non-crosslinked, type I pepsinized collagen gels. (g) Comparisons of velocity (top row) and migration pattern (bottom) of individual cells through this telopeptide-free non-crosslinked collagen. Scale bars, 10 μm.
2.6. Direct comparison of protease function in crosslinked versus non-crosslinked collagen gels
These data establishing the sensitivity of HT-1080 motility to inhibition of MMP activity contradict data published using the same tumor cell line reporting insensitivity to inhibition of MMPs and other proteases (Wolf et al., 2003). The conflict was resolved when we re-evaluated cell migration of HT-1080 cells using the same non-crosslinked, pepsinized type I collagen used by Wolf et al.: there is a striking absence of any inhibition of motility by either TIMP-1 or TIMP-2 at 200 nM (Fig. 5f, g; control: 47.9 ± 1.2 μm/hr (n=30); TIMP-2: 65.6 ± 2.1 μm/hr (n=25); TIMP-1: 50.8 ± 3.2 μm/hr (n=14)). Another study showed that tumor cell tunneling down into collagen gels was sensitive to MMP inhibition in crosslinked rat tail collagen but not in pepsinized collagen gels (Sabeh et al., 2004). Taken together, all of these studies indicate that collagenolytic activity is necessary for tumor cell invasion in a 3-dimensional collagen gel, and that this requirement can be abrogated by using collagen that has been modified with pepsin to remove the telopeptides and crosslinking. Consequently, collagenase activity is required for HT-1080 cell migration/invasion only in endogenously crosslinked collagen gels, highlighting the crucial importance of the type of 3-dimensional assay system used in the analysis.
2.7. Conclusions
By imaging living cells using a new probe for protease activity, we have established the following: (1) This novel probe containing a known collagen cleavage site can serve as a high-resolution, specific reporter of cell-associated protease activity. (2) This protease activity is localized to the leading edge of migrating tumor cells. (3) The path of proteolytic cleavage by a migrating cell can be visualized in 2- and 3-dimensional matrices. (4) Cell-derived protease activity detected by the probe is required for tumor cell migration in a cross-linked collagen matrix, but not if the collagen is non-crosslinked. (5) The GPLGIAG probe can be used to determine inhibitor concentrations needed to suppress cell-surface protease activity, and (6) TIMP-2 at 1–2×10−7 M inhibits both protease activity and migration. The same approach could be used to study a single protease by substituting a protease-specific target peptide sequence. Use of different fluorophores and sequences would then permit simultaneous analysis of multiple proteases with the same high spatial and temporal resolution for further analyses of the mechanisms of tumor cell invasion, inflammation, embryonic development, and other forms of tissue remodeling.
3. Experimental procedures
3.1. Chemicals
Dulbecco’s modified Eagle’s medium (DMEM) and RPMI-1640 medium plus fetal calf serum (FCS) were purchased from HyClone. NuSerum IV and acid-extracted type I collagen with endogenous crosslinking from rat tails were from BD Biosciences, pepsinized triple-helical bovine type I collagen termed Nutragen (formerly Vitrogen) from Inamed BioMaterials, bovine gelatin from Sigma, NeutrAvidin and biotinamidohexanoic acid 3-sulfo-N-hydroxysuccinimide ester, sodium salt from Pierce, and 6-carboxyrhodamine 6G-succinimidyl ester, Alexa 647-succinimidyl ester, and DQ-collagen from Molecular Probes. Human MMP-2, MMP-9, catalytic domains of MT1-MMP, MT2-MMP, and MT3-MMP, and TIMP-1 were from EMD Biosciences; human neutrophil elastase was from MP Biochemicals.
3.2. Cell lines and transfectants
PC-3 and HT-1080 cell lines were from ATCC and Lombardi Cancer Center, Washington, DC, respectively. A clone of the MDA-MB-231 line stably transfected with carboxy terminal enhanced green fluorescent protein (eGFP) MT1-MMP (eGFP-MT1-MMP) (Artym et al., 2006) named A2 was generated by serial dilution. The PC-3 line was cultured in 1:1 DMEM:RPMI-1640 plus 10% FCS and 5% NuSerum IV; HT-1080 cells were cultured in DMEM containing 10% FCS; and the A2 clone was cultured in DMEM with 10% FCS plus 500 μg/ml G418.
3.3. Protease biosensor
Octadecapeptides with the sequence KDPC5GPLGIAGIGC5PKGY (where C5 is 6-aminohexanoic acid) with the collagenase cleavage sequence GPLGIAG (cleavage site between LG and IA) were synthesized by standard FMOC solid phase chemistry and purified by reverse phase HPLC. Each purified peptide was homodoubly labeled with rhodamine 6G at a molar ratio of 3:1 dye:peptide and characterized as previously described (Packard et al., 1996). For peptide immobilization, the amino terminus of each homodoubly labeled peptide was covalently linked by N-hydroxysuccinimidyl chemistry to biotin, purified by reverse phase chromatography, and then complexed at a molar ratio of 3 biotinylated peptides to 1 NeutrAvidin molecule. For 3-dimensional studies, the NeutrAvidin-labeled peptide complex was added to collagen covalently labeled with biotin, incubated 18 hours at room temperature, washed extensively with phosphate buffered saline (PBS) and stored in 0.02 N HCl.
3.4. Protease activity studies in solution
Homodoubly labeled substrates were analyzed at a concentration of 2 μM in 0.1 M NaCl, 0.01 M CaCl2, 0.1 M Tris, pH 7.5 or in 50 mM MES, 5 mM DTT, 2.5 mM EDTA, pH 5.5 for cathepsin K at 37°C using a Photon Technology International (PTI) fluorometer operating with FeliX™ software. Excitation was set at 530 ± 2 nm with a polarizer at 54.7° and emission at 555 ± 2 nm with a polarizer at 90°.
3.5. 2- and 3-dimensional motility and imaging studies
Two-dimensional gelatin matrices were prepared as described (Artym et al., 2006) except that the NeutrAvidin-biotin-peptide complex was incubated with the substratum for 15 minutes followed by four PBS washes; assays were performed using 1 × 104 cells. Three-dimensional collagen matrices were prepared using 1/10 volume 10X DMEM with either rat-tail collagen or Nutragen. For imaging collagenase activity, equal amounts of collagen and collagen labeled with the fluorogenic probe via biotin were mixed. Sandwich matrices were prepared by spreading 50 μl collagen on the 14 mm glass insert of a MatTek Petri dish with a plastic micropipet tip, 30 minute incubation at 37°C, addition of 1 × 104 cells, and 30 minute incubation at 37°C. The medium above the attached cells was removed, and 300 μl collagen solution was layered over the cells and incubated 30 minutes at 37°C. For single collagen layer studies, the 300 μl layer was omitted.
3.6. Imaging and motility studies
Cell movement was monitored using Zeiss Axiovert 25 microscopes at 37°C using A-Plan 5X objectives at 10 minute intervals with Infinity 2 CCD video cameras from Lumenera. Positions of cell nuclei were tracked using MetaMorph (Molecular Devices) software. Confocal images were collected using a Zeiss 510 laser scanning confocal microscope with Plan-Apochromat 40X or 63X objectives at 37°C. Fluorescence and differential interference contrast (DIC) images were acquired as stacks of single optical sections ranging from 0.4 to 1.0 μm in thickness with pinhole photodetector apertures set equal to 1 Airy unit and analyzed using MetaMorph and Volocity (Improvision).
Acknowledgments
This work was supported in part by the intramural research program of the NIDCR, NIH. We thank Thomas Bugge for helpful suggestions on the manuscript.
Footnotes
Abbreviations: MMP, matrix metalloprotease; MT1-MMP, membrane-type matrix metalloprotease-1 (MMP-14); TIMP, tissue inhibitor of metalloprotease; A2, clone of MDA-MD-231 cells expressing MT1-MMP with eGFP marker
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoyama A, Chen WT. A 170-kDa membrane-bound protease is associated with the expression of invasiveness by human malignant melanoma cells. Proc Natl Acad Sci U S A. 1990;87:8296–8300. doi: 10.1073/pnas.87.21.8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66:3034–3043. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- Bremer C, Bredow S, Mahmood U, Weissleder R, Tung CH. Optical imaging of matrix metalloproteinase-2 activity in tumors: feasibility study in a mouse model. Radiology. 2001;221:523–529. doi: 10.1148/radiol.2212010368. [DOI] [PubMed] [Google Scholar]
- Even-Ram S, Yamada KM. Cell migration in 3D matrix. Curr Opin Cell Biol. 2005;17:524–532. doi: 10.1016/j.ceb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Hobson JP, Liu S, Rono B, Leppla SH, Bugge TH. Imaging specific cell-surface proteolytic activity in single living cells. Nat Methods. 2006;3:259–261. doi: 10.1038/nmeth862. [DOI] [PubMed] [Google Scholar]
- Horino K, Kindezelskii AL, Elner VM, Hughes BA, Petty HR. Tumor cell invasion of model 3-dimensional matrices: demonstration of migratory pathways, collagen disruption, and intercellular cooperation. FASEB J. 2001;15:932–939. doi: 10.1096/fj.00-0392com. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- McIntyre JO, Fingleton B, Wells KS, Piston DW, Lynch CC, Gautam S, Matrisian LM. Development of a novel fluorogenic proteolytic beacon for in vivo detection and imaging of tumour-associated matrix metalloproteinase-7 activity. Biochem J. 2004;377:617–628. doi: 10.1042/BJ20030582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JD, Werb Z. Regulation of matrix biology by matrix metalloproteinases. Curr Opin Cell Biol. 2004;16:558–564. doi: 10.1016/j.ceb.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562–573. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Netzel-Arnett S, Fields GB, Birkedal-Hansen H, Van Wart HE. Sequence specificities of human fibroblast and neutrophil collagenases. J Biol Chem. 1991;266:6747–6755. [PubMed] [Google Scholar]
- Ouyang M, Lu S, Li XY, Xu J, Seong J, Giepmans BN, Shyy JY, Weiss SJ, Wang Y. Visualization of polarized membrane type 1 matrix metalloproteinase activity in live cells by fluorescence resonance energy transfer imaging. J Biol Chem. 2008;283:17740–17748. doi: 10.1074/jbc.M709872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall CM, Kleifeld O. Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- Packard BZ, Komoriya A. Intracellular protease activation in apoptosis and cell-mediated cytotoxicity characterized by cell-permeable fluorogenic protease substrates. Cell Res. 2008;18:238–247. doi: 10.1038/cr.2008.17. [DOI] [PubMed] [Google Scholar]
- Packard BZ, Toptygin DD, Komoriya A, Brand L. Profluorescent protease substrates: intramolecular dimers described by the exciton model. Proc Natl Acad Sci U S A. 1996;93:11640–11645. doi: 10.1073/pnas.93.21.11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, Lopez-Otin C, Shapiro S, Inada M, Krane S, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167:769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloane BF, Sameni M, Podgorski I, Cavallo-Medved D, Moin K. Functional imaging of tumor proteolysis. Annu Rev Pharmacol Toxicol. 2006;46:301–315. doi: 10.1146/annurev.pharmtox.45.120403.095853. [DOI] [PubMed] [Google Scholar]
- Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang Z, Lin J, Lu J, Liu BF, Zeng S, Luo Q. Detection of MMP activity in living cells by a genetically encoded surface-displayed FRET sensor. Biochim Biophys Acta. 2007;1773:400–407. doi: 10.1016/j.bbamcr.2006.11.002. [DOI] [PubMed] [Google Scholar]