Summary
A widely held view is that directional movement of tRNA in the ribosome is determined by an intrinsic mechanism and driven thermodynamically by transpeptidation. Here, we show that, in certain ribosomal complexes, the pretranslocation (PRE) state is thermodynamically favored over the posttranslocation (POST) state. Spontaneous and efficient conversion from the POST to PRE state is observed when EF-G is depleted from ribosomes in the POST state or when tRNA is added to the E site of ribosomes containing P-site tRNA. In the latter assay, the rate of tRNA movement is increased by streptomycin and neomycin, decreased by tetracycline, and not affected by the acylation state of the tRNA. In one case, we provide evidence that complex conversion occurs by reverse translocation (i.e., direct movement of the tRNAs from the E and P sites to the P and A sites, respectively). These findings have important implications for the energetics of translocation.
Introduction
During translation, the ribosome iteratively orchestrates three major events: decoding, peptidyl transfer, and translocation. Decoding is facilitated by elongation factor Tu (EF-Tu), which delivers aminoacyl-tRNA (aa-tRNA) to the A site as part of a ternary complex with GTP. Cognate codon-anticodon interaction in the 30S A site stimulates hydrolysis of GTP, release of aa-tRNA from EF-Tu, and movement of the acceptor end of aa-tRNA into the 50S A site (reviewed in Ogle and Ramakrishnan [2005]). Once aa-tRNA is in the A site of both subunits (termed the A/A state), the ribosome catalyzes transfer of the peptidyl group from P-site tRNA to the α-amino group of A-site aa-tRNA. Translocation of the tRNAs to their adjacent ribosomal sites is then catalyzed by elongation factor G (EF-G), which hydrolyzes GTP in the process. Translocation is believed to occur in a stepwise manner, in which the newly formed peptidyl-tRNA and deacylated tRNA move first with respect to the 50S subunit into hybrid A/P and P/E states and then move with respect to the 30S subunit (Blanchard et al., 2004; Moazed and Noller, 1989). Translocation results in a complex containing peptidyl-tRNA in the P site (P/P state), deacylated tRNA in the E site (E/E state), and a vacant A site.
More than 30 years ago, it was shown that ribosomes programmed with polyuridylic acid (poly-U) can synthesize polyphenylalanine (poly-Phe), albeit inefficiently, in the absence of elongation factors and GTP (Gavrilova and Spirin, 1971; Pestka, 1969). Based on these data, it was proposed that directional movement of tRNA is determined by a mechanism intrinsic to the ribosome and driven thermodynamically by transpeptidation (Spirin, 1978). The tRNA binding sites of the ribosome exhibit specificity with regard to the acylation state of tRNA, determined by interactions in the 50S subunit. Peptidyl transfer changes the acylation states of the bound tRNAs: the P-site tRNA becomes deacylated, and the A-site tRNA becomes peptidylated rather than aminoacylated. It was postulated that peptidyl transfer decreases the affinities of the tRNAs for their resident sites relative to their adjacent sites, making subsequent translocation thermodynamically favorable. Consistent with this prediction, efficient translocation of N-acetyl-(Phe)2-tRNAPhe in ribosomes programmed with poly-U was observed in the absence of EF-G (Bergemann and Nierhaus, 1983; Semenkov et al., 1992). This “thermodynamic gradient” model explained how a portion of the free energy liberated by transpeptidation could be coupled to translocation.
Chemical probing studies have shown that tRNA can move spontaneously with respect to the 50S subunit after peptidyl transfer, providing evidence that transpeptidation is coupled to tRNA movement (Moazed and Noller, 1989). The ability of the tRNAs to adopt the hybrid-state configuration appears to be important for rapid EF-G-dependent translocation (Dorner et al., 2006; Semenkov et al., 2000). However, spontaneous translocation with respect to the 30S subunit in a variety of ribosomal complexes is inefficient or not observed (Fredrick and Noller, 2003; Rodnina et al., 1997; Sharma et al., 2004), suggesting that transpeptidation is generally insufficient to promote translocation of the codon-anticodon helices. The inability to observe spontaneous translocation within the 30S subunit of these complexes can be explained by one or both of two possibilities: the energy barrier is too high to breach or the reaction is thermodynamically unfavorable.
EF-G is a five-domain GTPase that interacts with the intersubunit cleft on the A-site side of the ribosome (Agrawal et al., 1998; Wilson and Nechifor, 2004). In the 1970s, it was shown that EF-G can promote single- but not multiple-turnover translocation in the presence of nonhydrolyzable analogs of GTP (reviewed in Rodnina et al. [2001]). These data led to a model in which interaction of EF-G•GTP with the pretranslocation complex promotes tRNA-mRNA movement, and subsequent GTP hydrolysis facilitates release of EF-G from the complex. This model was refuted when rapid kinetic studies showed that GTP hydrolysis precedes tRNA-mRNA movement (Katunin et al., 2002; Rodnina et al., 1997). Hydrolysis of GTP by EF-G increased the rate of translocation by ∼ 50-fold, indicating that the free energy liberated by GTP hydrolysis is normally coupled to tRNA-mRNA movement. At the same time, these studies confirmed that EF-G can promote tRNA-mRNA movement in the absence of GTP hydrolysis, suggesting that binding of EF-G plays a major role in inducing directional movement of tRNA-mRNA. Movement of tRNA-mRNA and release of phosphate appear to be rate limited by a conformational rearrangement in the complex, which has been termed “unlocking” (Savelsbergh et al., 2003). Effects of mutations and antibiotics show that phosphate release and tRNA-mRNA movement are independent events, providing evidence that GTP hydrolysis and tRNA-mRNA movement are not directly coupled. Instead, GTP hydrolysis may drive the unlocking rearrangement and thereby indirectly accelerate tRNA-mRNA movement (Peske et al., 2004; Savelsbergh et al., 2005).
In this work, we investigate the relative stability of the pretranslocation (PRE) versus posttranslocation (POST) state in a number of different ribosomal complexes. We find that, in certain contexts, the PRE state is thermodynamically favored over the POST state. These findings challenge the idea that transpeptidation ensures the directionality of tRNA-mRNA movement in the ribosome and suggest instead that EF-G plays this role.
Results
Depletion of EF-G Causes Spontaneous Conversion of Ribosomes from the POST to PRE State
In the course of studying ribosome complexes by toeprinting (Hartz et al., 1989), we obtained evidence that removal of EF-G from ribosomes in the POST state resulted in spontaneous reformation of the PRE state (Figure 1). Binding of tRNAPhe to the P site of ribosomes containing message m617 resulted in a strong toeprint at +16, indicating positioning of the UUC codon in the P site (Figure 1C, lane 1). N-acetyl-Val-tRNAVal (AcVal-tRNAVal) was then added to pair with GUU in the A site, and a shift of the predominant toeprint to +17 was observed (Figure 1C, lane 2), characteristic of A-site binding (Jerinic and Joseph, 2000). Addition of EF-G-His6 and GTP resulted in translocation of AcVal-tRNAVal and GUU to the P site, as indicated by the toeprint at +19 (Figure 1C, lane 3). When EF-G-His6 was depleted from the reaction using Ni2+ agarose, the relative intensity of the +17 toeprint increased substantially (Figure 1C, lane 4), suggesting reformation of the PRE complex. Based on the toeprint intensities, the percentage of ribosomes in the POST state was reduced from 74% to 23% upon EF-G depletion. Addition of EF-G back to the reaction caused shifting of the toeprint to +19 (Figure 1C, lane 5), indicating that the +17 toeprint observed upon EF-G depletion corresponded to a functional PRE complex. These data provide evidence that, in this ribosomal complex, the PRE state is favored over the POST state at equilibrium.
Figure 1. Conversion of Ribosomal Complexes from the POST to PRE State by Depletion of EF-G-His6.
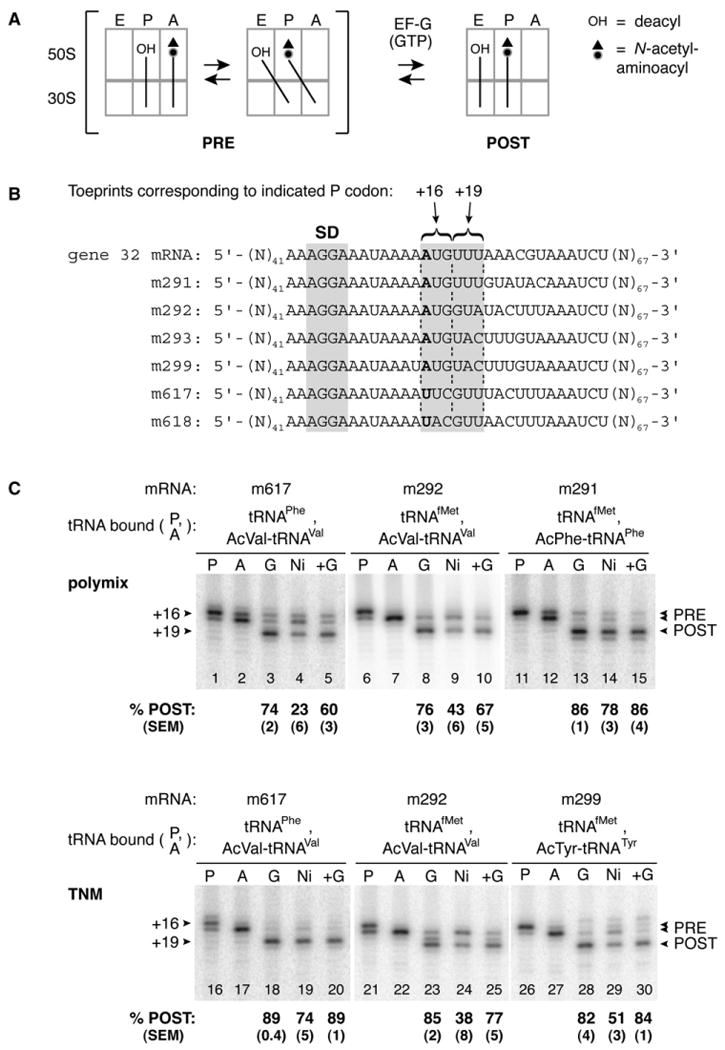
(A) Schematic of the stepwise movement of tRNA during translocation.
(B) Model mRNAs used in this study. Relevant codons and the Shine-Dalgarno element (SD) are indicated. Position +1 is shown in bold text.
(C) PRE complexes were formed by incubating programmed ribosomes with deacylated tRNA to fill the P site (P lanes) and then adding Ac-aa-tRNA to bind the A site (A lanes) in polymix or TNM buffer as indicated. Addition of EF-G-His6 and GTP resulted in translocation, evident by toeprints at position +19 (G lanes). Subsequent depletion of EF-G-His6 using Ni2+ agarose caused complex conversion from the POST to PRE state in several contexts (Ni lanes). Readdition of EF-G and GTP (+G lanes) converted complexes back to the POST state.
Reformation of the PRE complex upon EF-G depletion was observed in several complexes tested, but to varying degrees (Figure 1C and data not shown). Similar results were obtained when EF-G and GTP were removed using Sephacryl S-200 spin columns (data not shown). Notably, the POST complex containing AcPhe-tRNAPhe in the P site was particularly stable (Figure 1C, lane 14). This context, involving translocation of tRNAfMet and AcPhe-tRNAPhe, is most similar to those employed in several kinetics studies of translocation (Dorner et al., 2006; Savelsbergh et al., 2003; Studer et al., 2003). It was evident that several parameters (e.g., mRNA sequence, tRNA species, and buffer composition) influenced the relative stability of the POST state, making it difficult to assign a single experimental variable as most influential. Nevertheless, these data suggest that in certain complexes, the PRE state is thermodynamically favored over the POST state.
In these experiments, PRE complexes were typically made by nonenzymatic binding of N-acetyl-aminoacyl-tRNA (Ac-aa-tRNA) (an analog of peptidyl-tRNA) to the A site of ribosomes containing P-site-deacylated tRNA. Hence, we tested whether peptidyl transfer in the ribosome affected the relative stability of the POST complex (see Figure S1 in the Supplemental Data available with this article online). PRE complexes were made by EF-Tu-dependent loading of Val-tRNAVal into the A site of m617-programmed ribosomes containing P-site AcPhe-tRNAPhe. EF-G-catalyzed translocation resulted in a strong toeprint at +19, characteristic of the POST complex. When EF-G-His6 was then depleted from the reaction using Ni2+ agarose, reformation of the PRE complex was observed. The efficiency of complex conversion was ∼10% lower than when AcVal-tRNAVal was used (Figure 1C), indicating that the dipeptidyl group did stabilize the POST state. However, ∼60% conversion to the PRE state was still observed upon EF-G depletion (Figure S1), suggesting that the transpeptidation event itself does not act to ensure the directionality of tRNA-mRNA movement.
Conversion of Ribosomes from the POST to PRE State by Addition of E-Site tRNA
The EF-G depletion experiments suggested that in certain contexts translocation is reversible and the PRE state is more stable than the POST state. If these inferences were correct, we hypothesized that filling the E site of ribosomes containing P-site Ac-aa-tRNA in an appropriate context should result in formation of the PRE complex by reverse translocation. Indeed, we found that the PRE complex could be formed in this unconventional manner (Figure 2). When deacylated tRNA cognate for the E codon was added to ribosomes containing Ac-aa-tRNA in the P site, movement of mRNA in the 3′ direction indicated movement of the tRNAs into the 30S P and A sites, respectively. In the m292 context, movement of the tRNA from the P to A site was faster than its dissociation from the complex (see below), suggesting movement of the tRNAs directly to their adjacent sites. Using this assay, conversion of complexes from the POST to PRE state was monitored as a function of time, and in each case, the data fit well to a single exponential function. In polymix buffer, the observed rate (kobs) of m292 movement increased with the concentration of E-site tRNA, approaching a maximal rate of ∼0.14 min−1 at ∼10 μM (Figure 2C). By contrast, kobs was maximal in TNM buffer at 1 μM E-site tRNA, consistent with the ability of Mg2+ to increase the affinity of tRNA for the E site (Semenkov et al., 1996). Because polymix buffer mimics physiological conditions better, all further rate measurements were performed in polymix with 10 μM E-site tRNA unless stated otherwise.
Figure 2. Conversion of Ribosomes from the POST to PRE State by Addition of E-Site tRNA.
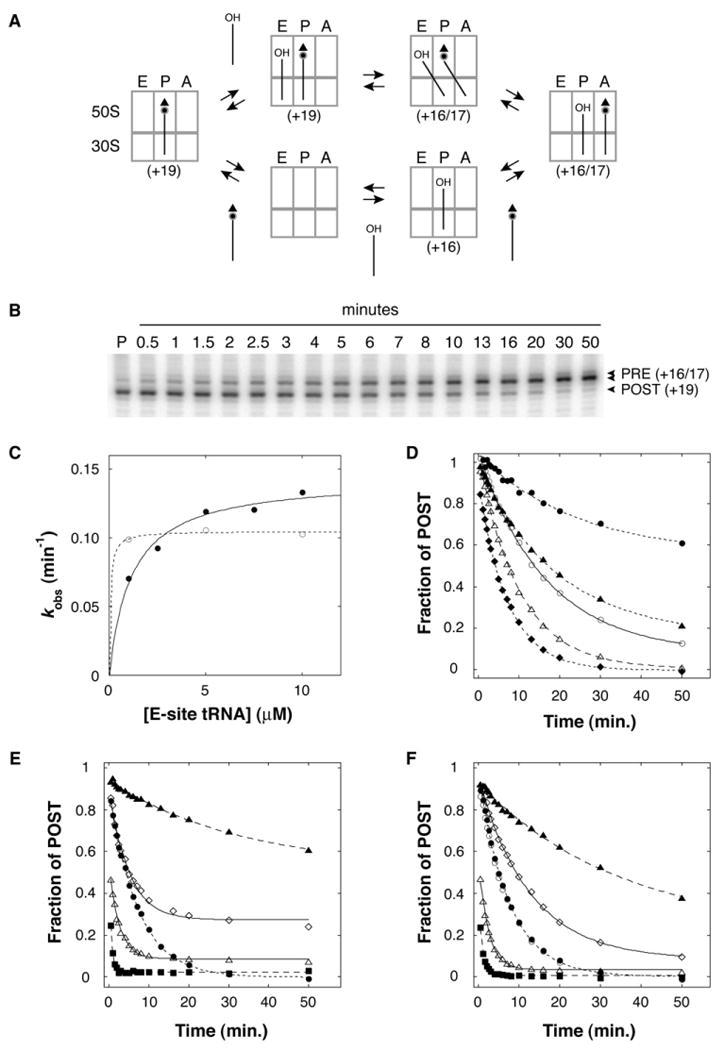
Addition of deacylated tRNA cognate for the E codon to ribosomes containing P-site Ac-aa-tRNA can result in movement of the mRNA in the 3′ direction, indicating formation of the PRE complex.
(A) Formation of the PRE complex by addition of E-site tRNA can occur by either of two pathways, which are not mutually exclusive. One pathway is reverse translocation, in which the tRNAs move directly to their adjacent sites (top). The other pathway involves complete dissociation of P-tRNA from the complex followed by de novo assembly of the PRE complex (bottom). The expected toeprint position(s) for each complex is indicated in parentheses.
(B) An example of POST-to-PRE conversion after addition of E-site tRNA. Ribosomes programmed with m617 were incubated with AcVal-tRNAVal to bind the P site (P lane), tRNAPhe was added at t = 0, and the position of mRNA in the complex was monitored as a function of time.
(C) The observed rate of POST-to-PRE conversion (kobs) after addition of tRNAfMet to m292-programmed ribosomes containing P-site AcVal-tRNAVal was determined in TNM (○) and polymix (●) buffer at several concentrations of E-site tRNAfMet. The data were fit to a hyperbolic function, giving maximum rates of 0.11 min−1 and 0.14 min−1 in TNM and polymix buffers, respectively.
(D) The rate (kobs) and amplitude (a) of POST-to-PRE conversion depended on the context (i.e., the mRNA and tRNA species bound). These parameters were determined for tRNAfMet and AcVal-tRNAVal paired to m292 (◆; kobs = 0.13 min−1, a = 1.0), tRNAPhe and AcVal-tRNAVal paired to m617 (△; kobs = 0.098 min−1, a = 0.99), tRNATyr andAcVal-tRNAVal paired to m618 (○; kobs = 0.060 min−1, a = 0.93), tRNAfMet and AcTyr-tRNATyr paired to m293 (▲; kobs = 0.052 min−1, a = 0.85), and tRNAfMet and AcPhe-tRNAPhe paired to m291 (●; kobs = 0.044 min−1, a = 0.43).
(E) POST-to-PRE conversion after addition of tRNAfMet to m292-programmed ribosomes containing P-site AcVal-tRNAVal in the absence of antibiotics (●) or in the presence of Neo (△), Str (■), Vio (◇), or Tet (▲).
(F) POST-to-PRE conversion after addition of tRNAfMet to m292-programmed ribosomes containing P-site-deacylated tRNAVal in the absence or presence of antibiotics, as indicated in (E). Time course when Val-tRNAVal was initially bound to the P site (○).
The observed rate of mRNA movement (kobs) depended on the particular complex (Figure 2D), and this context dependence was reminiscent of that observed in the experiments involving EF-G depletion. For example, conversion of complexes containing tRNAfMet and AcPhe-tRNAPhe paired to m291 was the slowest, and the POST state of this complex was the most stable after EF-G depletion. Complexes containing m292 and m617 exhibited the highest kobs values, and in these cases, the EF-G depletion experiments indicated that the POST state was less stable.
The Rate of POST-to-PRE Conversion Is Increased by Streptomycin and Neomycin and Decreased by Tetracycline
We screened several antibiotics for their ability to affect POST-to-PRE conversion (kobs) when added simultaneously with E-site tRNA. Streptomycin (Str), neomycin (Neo), and tetracycline (Tet) conferred obvious effects and were chosen for further investigation in two different contexts (Figure 2E, Table 1). In the m292 context, Neo and Str increased kobs by ∼3-fold and ∼14-fold, respectively. Str and aminoglycosides of the Neo class are known to increase miscoding, which may involve the ability of these antibiotics to influence conformational changes in the 30S subunit important for normal decoding (reviewed in Ogle and Ramakrishnan [2005]). One possibility is that Str and Neo promote a conformational change of the ribosome that facilitates POST-to-PRE conversion. Tet, whose primary target is the 30S A site (Brodersen et al., 2000; Moazed and Noller, 1987), decreased kobs by ∼4-fold and the extent of complex conversion by ∼60% (Figure 2E, Table 1). Bound Tet may occlude the 30S A site and thereby inhibit movement of tRNA from the P to A site. Viomycin (Vio), which binds at the subunit interface and strongly inhibits EF-G-catalyzed translocation (Rodnina et al., 1997; Yamada et al., 1978), increased kobs marginally and decreased the extent of complex conversion by ∼30% in this assay. Similar but less-marked effects of these antibiotics were observed in the m617 context (Table 1).
Table 1. Rates of mRNA Movement in the Complex and Dissociation of P-Site tRNA from the Complex.
| mRNA | m617 | m292 | |||
|---|---|---|---|---|---|
| P-Site tRNA | AcVal-tRNAVal | AcVal-tRNAVal | tRNAVal | tRNAVal | |
| E-Site tRNA Added | tRNAPhe | tRNAfMet | tRNAfMet | — | |
| kobs (min−1) | kobs (min−1) | kobs (min−1) | koff (min−1) | koff (min−1) | |
| Control | 0.098 | 0.13 | 0.13 ± 0.002 | 0.029 ± 0.001 | 0.018 ± 0.001 |
| Neomycin | 0.14 | 0.40 | 0.43 ± 0.02 | 0.0009 ± 0.0003 | 0.012 ± 0.002 |
| Streptomycin | 0.31 | 1.8 | 1.1 ± 0.2 | 0.0009 ± 0.0002 | 0.0051 ± 0.0003 |
| Viomycin | 0.20 | 0.20 | 0.072 ± 0.009 | 0.0027 ± 0.0002 | 0.018 ± 0.001 |
| Tetracycline | 0.026 | 0.030 | 0.033 ± 0.004 | 0.017 ± 0.002 | 0.026 ± 0.003 |
| Lincomycin | 0.12 | 0.12 | 0.11 | ND | ND |
Apparent rates of mRNA movement (kobs) upon addition of E-site tRNA were determined in the absence or presence of antibiotic in different contexts as indicated. Dissociation rates of [3′-32P]-tRNAVal (koff) from ribosomes in the presence or absence of E-site tRNA were determined in the m292 context (mean ± SEM). ND, not determined.
Toeprinting maps the position of mRNA with respect to the ribosome and thereby allows the position of tRNA within the 30S subunit to be inferred. To independently assess the topology of tRNAs in these complexes, puromycin reactivity of Ac-[14C]-Val-tRNAVal was assayed before and after a 10 min incubation with E-site tRNA, employing conditions analogous to those used in toeprinting. In these experiments, puromycin (1 mM) was added to each complex and incubated for 10 s prior to extraction of Ac-[14C]-Val-puromycin into ethyl acetate. Under these conditions, the amount of puromycin product should reflect the amount of AcVal-tRNAVal in the P/P state (Semenkov et al., 1992; Sharma et al., 2004). We found that the level of puromycin reactivity correlated with the fraction of POST complex estimated by toeprinting (Figure 3). These data indicate that Ac-[14C]-Val-tRNAVal is moving out of the P/P state in these complexes and that toeprinting is a reliable indicator of tRNA binding topology.
Figure 3. Movement of mRNA in the 3′ Direction with Respect to the 30S Subunit after Addition of E-Site tRNA Correlates with Movement of Ac-aa-tRNA Out of the P/P State, as Determined by Loss of Puromycin Reactivity.
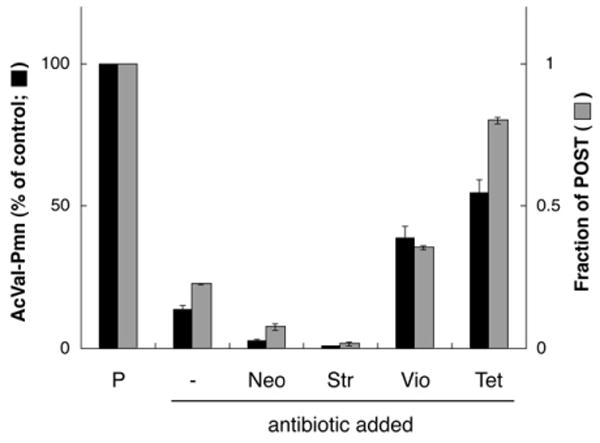
Ribosomes programmed with m292 and containing Ac-[14C]-Val-tRNAVal bound to the P site (P column) were incubated for 10 min in the presence of tRNAfMet alone (−) or in the additional presence of antibiotics as indicated. In these complexes, the level of puromycin reactivity (black bars) correlated with the fraction of ribosomes in the POST state inferred by toeprinting (gray bars). Data represent mean ± SEM.
Acylation State of tRNAVal Does Not Affect the Rate of POST-to-PRE Conversion
Aa-tRNA binds the A site with higher affinity than either the deacylated or peptidyl form (Fahlman et al., 2004; Fahlman and Uhlenbeck, 2004; Semenkov et al., 2000). Hence, it was of interest to test whether the acylation state of tRNA influenced the rate of POST-to-PRE conversion. Addition of tRNAfMet to the E site of m292-programmed ribosomes containing P-site AcVal-tRNAVal, Val-tRNAVal, or tRNAVal resulted in virtually identical rates of complex conversion (Figures 2E and 2F, Table 1). When antibiotics were added with tRNAfMet to ribosomes containing P-site tRNAVal, kobs values were comparable to the AcVal-tRNAVal case. The only exception was Vio, where kobs was ∼2-fold lower in the tRNAVal case. The fact that the peptidyl group did not appreciably affect kobs in the absence or presence of various antibiotics suggests that complex conversion occurs by a similar mechanism in both cases.
Complex Conversion Cannot Be Easily Explained by Dissociation and Reassociation of tRNA from the Ribosome
Conversion of ribosomes from the POST to PRE state could be explained by two independent pathways, which are not mutually exclusive (Figure 2A). One pathway, reverse translocation, involves movement of the tRNAs directly to their adjacent sites. The second pathway involves complete dissociation of P-site tRNA from the complex followed by de novo assembly of the PRE complex. To investigate which pathway was predominant in the m292 context, we compared the rate of P-site [3′-32P]-tRNAVal dissociation from the complex (koff) to the rate of mRNA movement (kobs) (Figure 4, Table 1). Without antibiotic, koff was 0.018 min−1 and 0.029 min−1 in the absence and presence of E-site tRNA, respectively. These rates are considerably slower than mRNA movement (kobs), suggesting that conversion of the complex is primarily due to movement of the tRNAs directly to their adjacent sites. The small difference in koff due to the presence of E-site tRNA may reflect a slightly higher dissociation rate of [3′-32P]-tRNAVal from the A site than from the P site. When Neo, Str, or Vio was included at t = 0, the difference between kobs and koff increased: kobs was faster than koff by a factor of ∼500, ∼1000, and ∼30 in the presence of Neo, Str, and Vio, respectively (Figure 4, Table 1). Thus, at least in these cases, complex conversion does not seem to involve dissociation of tRNAVal from the complex. The simplest explanation is reverse translocation (i.e., direct movement of the codon-anticodon helices from the E and P sites to the P and A sites, respectively). In the presence of Tet, kobs decreased to 0.033 min−1, a rate similar to koff (0.017 min−1) (Table 1). The effect of Tet might be explained by its ability to occlude the 30S A site, which would be expected to inhibit reverse translocation.
Figure 4. Complex Conversion Cannot Be Explained by Complete Dissociation and Reassociation of tRNA.
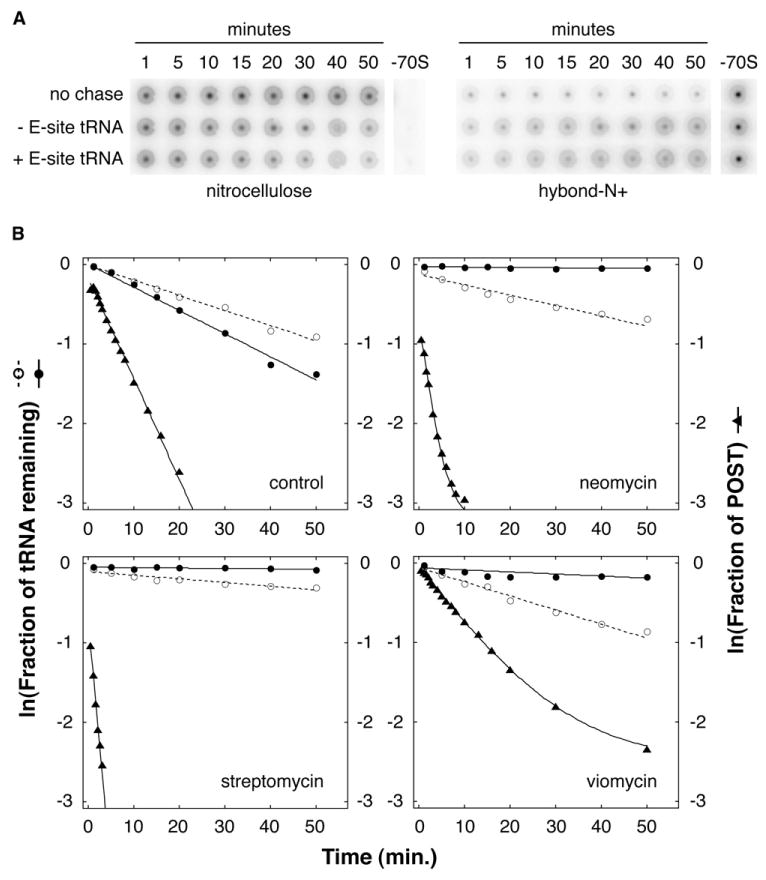
Dissociation rate (koff) of P-site [3′-32P]-tRNAVal from ribosomes programmed with m292 was determined after addition of excess unlabeled tRNAVal (chase) in the absence or presence of E-site tRNAfMet.
(A) A phosphorimager scan of nitrocellulose and Hybond-N+ membranes is shown as an example of the data used to quantify the fraction of bound [3′-32P]-tRNAVal.
(B) Dissociation of P-site tRNAVal in the absence (○) or presence (●) of E-site tRNAfMet was compared to mRNA movement (▲) in the absence (control) or presence of antibiotics as indicated.
Several Antibiotics Convert Ribosomes from the POST to PRE State in the Presence of EF-G and GTP
The EF-G depletion experiments indicated that in several contexts the continual presence of EF-G was necessary to keep the population of ribosomes in the POST state. We reasoned that addition of antibiotics that inhibit EF-G-dependent translocation might also reveal the labile nature of the POST state. Indeed, reformation of the PRE complex was observed when several antibiotics were added subsequent to EF-G and GTP (Figure 5). Addition of Neo, Par, Str, and Vio to ribosomes in the POST state caused their conversion to the PRE state despite the presence of EF-G and GTP. These antibiotics were shown previously to decrease the rate and/or extent of translocation (Cabanas et al., 1978; Fredrick and Noller, 2003; Modolell and Vazquez, 1977; Peske et al., 2004; Rodnina et al., 1997; Studer et al., 2003). PRE reformation induced by these antibiotics was observed in several different complexes including those containing dipeptidyl-tRNA, although the dipeptidyl group did attenuate POST-to-PRE conversion, consistent with the EF-G depletion experiments (Figure S1). In certain contexts, thiostrepton and fusidic acid had a similar but less-marked effect, while blasticidin S, carbomycin, chloramphenicol, erythromycin, kasugamycin, lincomycin, sparsomycin, spectinomycin, and tetracycline did not promote reformation of the PRE complex in this assay (Figure 5 and data not shown). Interestingly, in the presence of Str and EF-G(GTP), we observed an additional toeprint at position +18, between the characteristic PRE (+16/17) and POST (+19) toeprints. This intermediate +18 toeprint was not observed in experiments lacking EF-G, and a very prominent +18 toeprint was observed in the presence of fusidic acid and EF-G (data not shown). Because fusidic acid is known to stabilize EF-G•GDP on the ribosome, the +18 toeprint observed in the presence of Str may result from an increased association of EF-G with the complex.
Figure 5. Conversion of Ribosomal Complexes from the POST to PRE State by Addition of Antibiotics.
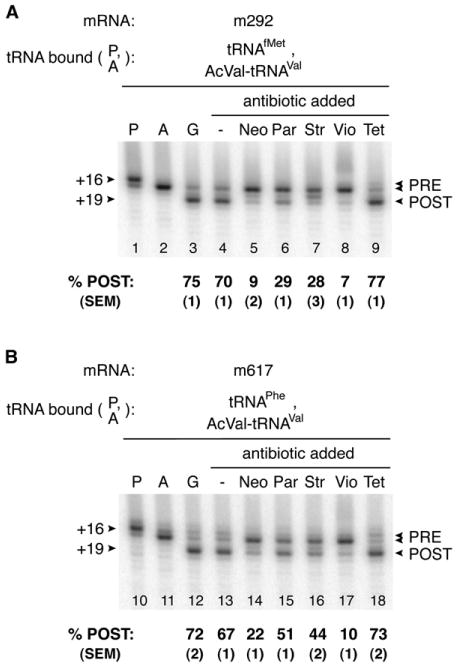
The PRE complex was formed in polymix buffer by incubating ribosomes with m292 (A) or m617 (B) and deacylated tRNA to fill the P site (P lanes) and then adding Ac-aa-tRNA to bind the A site (A lanes) as indicated. EF-G(GTP) was then added to form the POST complex (G lanes), which was further incubated in the absence (−) or presence of various antibiotics as indicated.
To further explore the effects of these antibiotics, we assayed puromycin reactivity in analogous complexes formed by addition of antibiotics after EF-G(GTP) (Figure 6A). Addition of Par and Vio decreased puromycin reactivity of AcVal-tRNAVal and %POST (determined by toeprinting) to similar degrees. In contrast, puromycin reactivity of AcVal-tRNAVal in complexes formed by addition of Neo and Str was considerably higher than expected based on toeprinting. For example, toeprinting indicated that Neo decreased %POST by 8-fold, whereas puromycin reactivity of AcVal-tRNAVal decreased by only ∼30%. Similarly, Str converted more than half the ribosomes to the PRE state as indicated by toeprinting, yet puromycin reactivity of AcVal-tRNAVal in the complex was unchanged by the antibiotic. These data might be explained if the antibiotic can stabilize an A/P-like state of AcVal-tRNAVal that remains highly puromycin reactive (Borowski et al., 1996) or if the complex is dynamic in the presence of EF-G, allowing AcVal-tRNAVal to sample the P/P state with high enough frequency to account for the level of puromycin reactivity observed. Interestingly, addition of Tet after EF-G(GTP) increased %POST by ∼10% and puromycin reactivity by ∼60%, suggesting that Tet can stabilize the POST state.
Figure 6. Comparison between the Puromycin Reactivity of Ac-[14C]-Val-tRNAVal and the Percentage of Ribosomes in the POST State as Determined by Toeprinting.
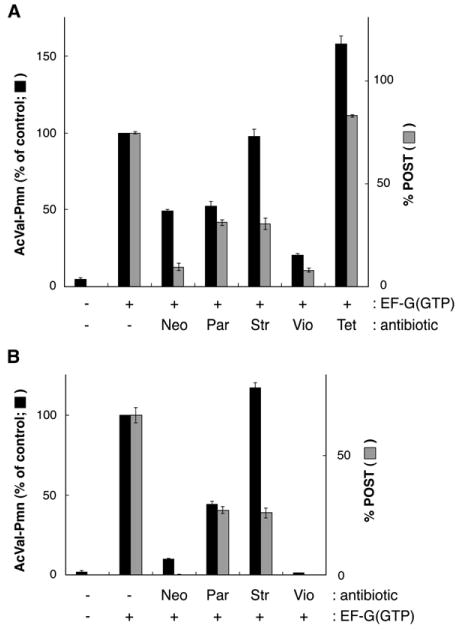
First, the PRE complex (−, −) was formed by incubating ribosomes with m292 and tRNAfMet to bind the P site and then adding Ac-[14C]-Val-tRNAVal to bind the A site. To the PRE complex, EF-G(GTP) was added before (A) or after (B) antibiotics as indicated. Data represent mean ± SEM.
For comparison, we performed an analogous experiment in which the antibiotic was added before EF-G(GTP) (Figure 6B). With Par and Str, the data were highly similar, suggesting that these reactions had reached equilibrium in both experiments. When Neo and Vio were added before EF-G(GTP), no translocation was detected by toeprinting, consistent with earlier studies (Peske et al., 2004; Rodnina et al., 1997; Studer et al., 2003). In the case of Vio, no increase in puromycin reactivity was observed either. With Neo, some puromycin product was formed (∼10% of the control) after EF-G(GTP) addition despite no evidence for the POST complex by toeprinting.
Discussion
In this study, we provide evidence that the PRE state is more stable than the POST state in several ribosomal complexes. Spontaneous conversion of complexes from the POST to PRE state was observed by depletion of EF-G or by binding tRNA to the E site of ribosomes containing P-site tRNA. These findings challenge the generalized idea that the POST state of the ribosome is intrinsically more stable than the PRE state. Although transpeptidation may be sufficient to promote directional movement of tRNA in the 50S subunit (Moazed and Noller, 1989), transpeptidation is insufficient to bias movement in the 30S subunit in several contexts.
The idea that the POST state is thermodynamically favored over the PRE state stems largely from the observation that synthesis of poly-Phe can occur in the absence of EF-G and GTP (Spirin, 1978). However, it should be kept in mind that factor-independent translation is observed in reactions containing excess aa-tRNA. Binding of aa-tRNA to the A site of ribosomes in the POST state results in peptidyl transfer, an exergonic and effectively irreversible step in protein synthesis. Thus, factor-independent translation would be predicted to occur as long as the rate of spontaneous (forward) translocation is high enough, regardless of whether the PRE or POST state is thermodynamically favored. Our study predicts that the rate of spontaneous (forward) translocation in certain contexts is extremely slow, which may explain why translation of heteropolymeric mRNA in the absence of translation factors has not been observed.
An implication of our study is that the directionality of codon-anticodon movement during translocation is determined by EF-G. It has been proposed that EF-G acts as a Brownian motor, akin to other molecular motors that couple NTP hydrolysis to directional movements (Wintermeyer et al., 2004). According to this model, interaction of EF-G•GTP and hydrolysis of GTP induce conformational changes that unlock the PRE complex, allowing Brownian movement of tRNA to take place. Insertion of extended domain 4 of EF-G into the 30S A site then prevents reverse movement of tRNA and thereby biases movement of tRNA-mRNA in the forward direction. Here, we show that movement of tRNA can occur in the reverse direction, lending support to the Brownian motor model.
Ribosomal Complexes in Which the PRE State Is Favored Over the POST
The difference in free energy between the PRE and POST state has been estimated previously based on binding affinities of various forms of tRNAPhe for the A, P, and E sites (Lill et al., 1986). In this case, the transition from PRE to POST state was predicted to be exergonic, but only by ∼1 kcal/mol. Here, we studied the relative stability of the PRE versus POST state using toeprinting, which allows the fraction of ribosomes in each state to be directly assessed. For several complexes, the observed rate of POST-to-PRE conversion was higher than rates of spontaneous (forward) translocation measured previously. For example, kobs for POST-to-PRE conversion in the m292 case (0.13 min−1) was nearly 10-fold higher than the rate of spontaneous translocation of N-acetyl-(Phe)2-tRNAPhe (0.014 min−1) (Semenkov et al., 1992). When induced by addition of E-site tRNA, POST-to-PRE conversion was highly efficient in several contexts (a ≈ 1; Figure 2D), suggesting that the EF-G-His6 depletion experiments generally underestimated the relative stability of the PRE state. This was not unexpected, as any residual EF-G contamination would promote POST complex formation. To further assess the relative stability of the PRE state, we tried to compare the rates of PRE-to-POST and POST-to-PRE conversions under otherwise identical conditions in three different mRNA contexts (Figure S2). In the cases of m292 and m293, efficient conversion from POST to PRE was readily observed, but we could not detect spontaneous conversion from the PRE to POST state even after prolonged incubation. In the case of m291, spontaneous PRE-to-POST conversion was detected, although it was extremely slow (6 × 10−4 min−1). These data provide further evidence that the transition from PRE to POST state can be thermodynamically “uphill.” It is plausible that the PRE-to-POST transition in other ribosomal complexes is similarly endergonic, which may explain why efficient spontaneous translocation was not observed (Fredrick and Noller, 2003).
Mechanism of POST-to-PRE Conversion
Conversion of ribosomes from the POST to PRE state can be explained by reverse translocation or by a dissociation/reassociation pathway (Figure 2A). In the m292 context, we showed that the rate of mRNA movement (kobs) was ∼4-fold faster than the rate of dissociation of P-site 3′-[32P]-tRNAVal (koff) from the complex. In the presence of Neo, Str, or Vio, the difference in these rates increased to ∼500-fold, ∼1000-fold, and ∼30-fold, respectively. These data suggest that the predominant pathway of POST-to-PRE conversion, at least in this complex, involves reverse translocation.
We were unable to measure koff in complexes containing AcVal-tRNAVal because the amount of chase AcVal-tRNAVal necessary for the experiment was prohibitive for practical reasons. Thus, we cannot rule out the possibility that POST-to-PRE conversion in this case occurs primarily via a dissociation/reassociation pathway. However, there are several observations to suggest that POST-to-PRE conversion is similar regardless of whether tRNAVal carries an N-acetyl-Val group. First, kobs is identical regardless of the acylation state of tRNAVal (Figures 2E and 2F, Table 1). Second, the effects of antibiotics are similar in both cases (Figures 2E and 2F, Table 1). Third, it has been shown that aminoacylation of a number of tRNAs does not increase their dissociation rate from the P site (Fahlman et al., 2004; Fahlman and Uhlenbeck, 2004). Although those studies analyzed the effect of an aminoacyl group, it seems unlikely that acetylation of the α-amino group of aa-tRNA would destabilize its interaction with the P site, based on earlier studies and the fact that ample space is available for the peptidyl analog in the 50S P site (Hansen et al., 2002; Lill et al., 1986).
Possible Relevance for the Allosteric Three-Site Model
Our findings provide an alternative explanation for data supporting the allosteric three-site model, which posits negative cooperativity between the A and E sites (Rheinberger, 1991). An observation presented in support of this model comes from experiments analogous to those of Figure 2. Binding of tRNA cognate for the E codon to ribosomes containing P-site tRNA generated a complex in which the A site was blocked (Gnirke et al., 1989). We propose that POST-to-PRE conversion (by reverse translocation or a dissociation/reassociation mechanism), rather than allosteric conformational changes, occluded the A site in those experiments. Occupation of the A site with tRNA might also explain the observation that various aminoglycosides and Vio promoted occlusion of the A site specifically when a sufficient amount of tRNA was present to fill at least two ribosomal sites (Hausner et al., 1988). To reconcile the effects of these antibiotics in the framework of the allosteric three-site model, it was postulated that aa-tRNA selection in the A site is fundamentally different in initiation (“i-type”) and elongation (“e-type”) complexes and that aminoglycosides and Vio inhibit A-site tRNA binding in e-type complexes specifically (Hausner et al., 1988). However, this explanation conflicts with evidence that several of these antibiotics stabilize tRNA in the A site (Ogle et al., 2002; Peske et al., 2004). Indeed, our data may hold relevance for numerous studies in which the topology of tRNA binding was not recurrently verified.
Effects of Antibiotics on POST-to-PRE Conversion
We found that addition of Neo, Par, Vio, or Str subsequent to EF-G(GTP) caused conversion of ribosomes from the POST to PRE state. Neo, Par, and Vio have been shown to dramatically decrease the rate of EF-G-dependent translocation (Peske et al., 2004; Rodnina et al., 1997; Studer et al., 2003). It was proposed that Par and Vio inhibit translocation by stabilizing the PRE complex and thereby increasing the energy barrier for tRNA-mRNA movement (Peske et al., 2004). Consistent with this model, our data indicate that these antibiotics stabilize the PRE state of the ribosome. Neo, which differs from Par by only a single functional group, also stabilizes the PRE complex in our assay. Unlike the other antibiotics, Str was shown to confer little effect on the rate of EF-G-dependent translocation (Peske et al., 2004). However, Str did stabilize peptidyl-tRNA interaction in the PRE complex by 45-fold, consistent with evidence from our study that Str can stabilize the PRE state of the ribosome. The fact that Str had little effect on the rate of EF-G-dependent translocation was puzzling because the energy barrier should increase due to stabilization of the PRE state (Peske et al., 2004). The authors proposed that Str can induce a conformational change in the 30S subunit that facilitates tRNA-mRNA movement, thereby decreasing the activation energy for translocation and compensating for stabilization of the PRE state. The idea that Str can promote a conformation of the 30S subunit that facilitates tRNA-mRNA movement is supported by our observation that Str can stimulate spontaneous reverse translocation by 14-fold (Table 1).
Whether POST-to-PRE conversion induced by Neo, Par, Vio, and Str in the presence of EF-G(GTP) occurs by reverse translocation or via a pathway of dissociation/reassociation remains unclear. Moreover, these antibiotics may differentially promote tRNA movement through one pathway or the other. Interestingly, differences in the activities of these antibiotics were apparent when puromycin reactivity was assayed in these complexes. Addition of Par or Vio to ribosomes in the POST state decreased puromycin reactivity of AcVal-tRNAVal and the fraction of ribosomes in the POST state similarly. On the other hand, puromycin reactivity of AcVal-tRNAVal after addition of Neo and Str was substantially higher than expected based on toeprinting. These data may be explained by either of two hypotheses. One is that the antibiotic stabilizes an A/P-like state of AcVal-tRNAVal that remains highly puromycin reactive. Precedence for this hypothesis comes from the ability of a variant of EF-G lacking domain I to promote movement of peptidyl-tRNA into such a state (Borowski et al., 1996). Another hypothesis is that the complex is dynamic in the presence of EF-G(GTP) and the antibiotic, allowing AcVal-tRNAVal to enter the P/P state with high enough frequency to account for the level of puromycin reactivity. EF-G is believed to catalyze translocation by inducing a rearrangement that unlocks the ribosome, allowing tRNA-mRNA movement to occur (Savelsbergh et al., 2003). If, under certain conditions (e.g., in the presence of Str), EF-G•GTP can unlock the POST complex, it is conceivable that EF-G can catalyze the movement of tRNA-mRNA in both directions. Clearly, further work is necessary to clarify the effects of these antibiotics on tRNA-mRNA movement.
Karimi and Ehrenberg showed that Str and Neo can destabilize peptidyl-tRNA in the P site and stabilize peptidyl-tRNA in the A site (Karimi and Ehrenberg, 1994, 1996), consistent with our findings that these antibiotics promote POST-to-PRE conversion. They also analyzed a number of ram and restrictive mutations known to increase and decrease miscoding, respectively. Interestingly, the ram mutations conferred effects similar to Neo and Str (destabilization of P-site peptidyl-tRNA and stabilization of A-site peptidyl-tRNA), while the restrictive mutations conferred the opposite effects (stabilization of P-site peptidyl-tRNA and destabilization of A-site peptidyl-tRNA). It was proposed that Str, Neo, and ram mutations promote a conformation of 16S rRNA that enhances dissociation of peptidyl-tRNA from the P site, while restrictive mutations promote an alternative conformation that inhibits P-tRNA dissociation (Karimi and Ehrenberg, 1996). In line with their model, we suggest that, by inducing conformational changes in the ribosome, Str and Neo destabilize the POST complex and thereby decrease the activation energy for reverse translocation (Figure 4). It should also be mentioned, though, that the experiments of Karimi and Ehrenberg suggest that Str and Neo promote dissociation of P-site peptidyl-tRNA from the ribosome, consistent with the possibility that conversion of complexes containing Ac-aa-tRNA in our study may involve a dissociation/reassociation mechanism.
Physiological Relevance of Reverse Translocation
The fastest rates of reverse translocation measured here are about four orders of magnitude slower than the rate of EF-G-dependent translocation. Therefore, reverse translocation is not predicted to influence translation elongation unless the reaction is catalyzed. In an independent, concurrent study, it was shown that LepA, a highly conserved bacterial protein homologous to EF-G, can catalyze reverse translocation (Qin et al., 2006). The presence of LepA in an in vitro transcription/translation system increased the fraction of active translational products substantially. These data suggest that LepA may regulate translation elongation to improve the accuracy of translation and/or facilitate correct folding of the nascent polypeptide. A number of other EF-G homologs exist in E. coli that have yet to be well characterized (Caldon and March, 2003). It will be interesting to see whether these or other unrelated proteins can similarly regulate translation elongation by promoting reverse translocation.
Experimental Procedures
Ribosomes from E. coli MRE600, factors, and mRNAs were purified as described (Boon et al., 1992; Fredrick and Noller, 2002, 2003). Purified tRNAs (Sigma) were charged and acetylated or [32P] labeled as described (Fredrick and Noller, 2002; McGarry et al., 2005).
The position of mRNA in the ribosome was determined by toeprinting (Hartz et al., 1989). For EF-G-His6 depletion experiments in polymix buffer (Ehrenberg et al., 1990), [32P]-labeled primer was annealed to mRNA (0.5 μM) in 5 mM potassium phosphate (pH 7.3) and 95 mM KCl by heating to 60°C and placing on ice. Polymix salts (5 mM Mg[OAc]2, 0.5 mM CaCl2, 5 mM NH4Cl, 8 mM putriscine, and 1 mM spermidine), DTT (1 mM), ribosomes (0.7 μM), and tRNA (1 μM) were added (final concentrations indicated throughout) and incubated at 37°C for 20 min to bind the P site. Next, Ac-aa-tRNA (1 μM) was added and incubated at 37°C for 10 min to bind the A site. Then, EF-G-His6 (1 μM) and GTP (300 μM) were added and incubated at 37°C for 10 min. (The POST complex in TNM buffer (50 mM Tris-HCl [pH 7.6], 100 mM NH4Cl, 20 mM MgCl2, 6 mM β-mercaptoethanol) was formed similarly as described (Fredrick and Noller, 2003).) A portion of the POST complex (50 μl) was added to 500 μl of Ni-NTA agarose (Qiagen) pre-equilibrated with either polymix or TNM, incubated at 25°C for 5 min, filtered, and incubated at 37°C for 10 min. Finally, GTP (300 μM) and EF-G-His6 (1 μM) were added to the flowthrough, incubated at 37°C for 10 min. At each stage of the experiment, a 2 μl aliquot was removed and added to 10 μl of extension mix for primer extension (PE) analysis (Fredrick and Noller, 2003). The percentage of POST complexes was calculated as %POST = 100 × (post)/(pre + post), where post and pre correspond to toeprint intensities from equivalent-sized areas of the gel centered at position +19 (+18 to +20) and position +16 (+15 to +17), respectively.
For toeprinting experiments shown in Figures 5 and 6A, the POST complex was diluted by 10% in the absence or presence of Neo (0.1 mM), Par (0.1 mM), Str (1 mM), or Vio (1 mM) and incubated at 37°C for 10 min. For experiments in Figure 6B, the PRE complex was incubated with antibiotics at 37°C for 3 min prior to addition of EF-G(GTP). Aliquots were removed for PE analysis as described above.
To measure puromycin reactivity of AcVal-tRNAVal, ribosome complexes were formed as described for toeprinting except that radio-labeled primer was excluded and the concentration of Ac-[14C]-Val-tRNAVal was 0.3 μM. Based on filter binding, the fraction of Ac-[14C]-Val-tRNAVal bound was 0.4 (3.6 pmol). At each step of the experiment, an equivalent amount of complex was removed, incubated at 37°C for 10 s with or without 1 mM puromycin, and immediately extracted for 1 min with ethyl acetate. The amount of Ac-[14C]-Val-puromycin (AcVal-Pmn) in the ethyl acetate phase was determined by liquid scintillation counting. In Figures 3, 6A, and 6B, 100% AcVal-Pmn corresponds to 3.0, 1.0, and 1.5 pmol, respectively.
To measure the rate of reverse translocation, the P-site binding complex was formed as described above. Reverse translocation was initiated by adding deacylated tRNA to bind the E site at t = 0, and the reaction was incubated at 37°C. Neo (0.1 mM), Str (10 mM), Vio (1 mM), Tet (0.1 mM), and lincomycin (10 mM) were also added at t = 0 if indicated. A 2 μl aliquot was removed at each time point and immediately added to 10 μl of prewarmed PE mix and further incubated at 37°C for 2 min. The fraction of POST complex at time t, F(t), was determined by calculating F(t) = {(post)/(pre + post)}/FP, where FP is the equivalent value derived from the P lane. The data were then plotted as a function of time and fit to the single exponential function F(t) = F0{1 − a × exp(−kobs × t)}, where F0 corresponds to the fraction of POST complex at t = 0, a is the amplitude, and kobs is the observed rate.
The dissociation rate of P-site tRNA from the complex was measured by filter binding (Fahlman and Uhlenbeck, 2004). Typically, ∼0.1 μM [3′-32P]-tRNAVal (∼4 × 104 cpm/pmol), 0.5 μM m292, and 0.7 μM ribosomes were incubated in polymix buffer at 37°C for 20 min to bind the P site. Dissociation of [3′-32P]-tRNAVal from the complex was monitored after adding unlabeled tRNAVal (to 11 μM) at t = 0. If indicated, antibiotics and/or E-site tRNAfMet (to 10 μM) was also added at t = 0. A 2 μl aliquot was removed at each time point, diluted into 15 μl of buffer (10 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 60 mM NH4Cl, 6 mM β-mercaptoethanol), filtered at room temperature, and washed with 200 μl of the same buffer. The fraction of [3′-32P]-tRNAVal bound to the complex was calculated from the fraction of radiolabel on the nitrocellulose membrane, where the spots without chase tRNA and those without ribosomes were used for background correction. koff was determined by fitting the data to the single exponential function F(t) = F0{1 − exp(−koff × t)}, where F(t) is the fraction of [3′-32P]-tRNAVal bound at time t and F0 is the fraction of [3′-32P]-tRNAVal bound at t = 0.
Supplementary Material
Acknowledgments
We thank B. Kraal for the strain to overexpress EF-Tu-His6, K. McGarry for PheRS, H. Roy for ATP(CTP):tRNA nucleotidyltransferase, K. Nierhaus for suggestions and sharing unpublished data, and M. Ibba and J. Cate for comments on the manuscript. This work was supported by NIH grant GM072528.
Footnotes
Supplemental Data: Supplemental Data include two figures and can be found with this article online at http://www.molecule.org/cgi/content/full/24/6/931/DC1/.
References
- Agrawal RK, Penczek P, Grassucci RA, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome. Proc Natl Acad Sci USA. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergemann K, Nierhaus KH. Spontaneous, elongation factor G independent translocation of Escherichia coli ribosomes. J Biol Chem. 1983;258:15105–15113. [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K, Vijgenboom E, Madsen LV, Talens A, Kraal B, Bosch L. Isolation and functional analysis of histidine-tagged elongation factor Tu. Eur J Biochem. 1992;210:177–183. doi: 10.1111/j.1432-1033.1992.tb17406.x. [DOI] [PubMed] [Google Scholar]
- Borowski C, Rodnina MV, Wintermeyer W. Truncated elongation factor G lacking the G domain promotes translocation of the 3′ end but not of the anticodon domain of peptidyl-tRNA. Proc Natl Acad Sci USA. 1996;93:4202–4206. doi: 10.1073/pnas.93.9.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- Cabanas MJ, Vazquez D, Modolell J. Inhibition of ribosomal translocation by aminoglycoside antibiotics. Biochem Biophys Res Commun. 1978;83:991–997. doi: 10.1016/0006-291x(78)91493-6. [DOI] [PubMed] [Google Scholar]
- Caldon CE, March PE. Function of the universally conserved bacterial GTPases. Curr Opin Microbiol. 2003;6:135–139. doi: 10.1016/s1369-5274(03)00037-7. [DOI] [PubMed] [Google Scholar]
- Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M, Bilgin N, Kurland CG. Design and use of a fast and accurate in vitro translation system. In: Spedding G, editor. Ribosomes and Protein Synthesis—A Practical Approach. Oxford: IRL Press; 1990. pp. 101–129. [Google Scholar]
- Fahlman RP, Uhlenbeck OC. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry. 2004;43:7575–7583. doi: 10.1021/bi0495836. [DOI] [PubMed] [Google Scholar]
- Fahlman RP, Dale T, Uhlenbeck OC. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- Gavrilova LP, Spirin AS. Stimulation of “non-enzymic” translocation in ribosomes by p-chloromercuribenzoate. FEBS Lett. 1971;17:324–326. doi: 10.1016/0014-5793(71)80177-1. [DOI] [PubMed] [Google Scholar]
- Gnirke A, Geigenmuller U, Rheinberger HJ, Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle: Analysis with a heteropolymeric mRNA. J Biol Chem. 1989;264:7291–7301. [PubMed] [Google Scholar]
- Hansen JL, Schmeing TM, Moore PB, Steitz TA. Structural insights into peptide bond formation. Proc Natl Acad Sci USA. 2002;99:11670–11675. doi: 10.1073/pnas.172404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Hausner TP, Geigenmuller U, Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle: new insights into the inhibition mechanisms of aminoglycosides, thiostrepton, and viomycin. J Biol Chem. 1988;263:13103–13111. [PubMed] [Google Scholar]
- Jerinic O, Joseph S. Conformational changes in the ribosome induced by translational miscoding agents. J Mol Biol. 2000;304:707–713. doi: 10.1006/jmbi.2000.4269. [DOI] [PubMed] [Google Scholar]
- Karimi R, Ehrenberg M. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur J Biochem. 1994;226:355–360. doi: 10.1111/j.1432-1033.1994.tb20059.x. [DOI] [PubMed] [Google Scholar]
- Karimi R, Ehrenberg M. Dissociation rates of peptidyl-tRNA from the P site of E. coli ribosomes. EMBO J. 1996;15:1149–1154. [PMC free article] [PubMed] [Google Scholar]
- Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- Lill R, Robertson JM, Wintermeyer W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry. 1986;25:3245–3255. doi: 10.1021/bi00359a025. [DOI] [PubMed] [Google Scholar]
- McGarry KG, Walker SE, Wang H, Fredrick K. Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol Cell. 2005;20:613–622. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Modolell J, Vazquez D. The inhibition of ribosomal translocation by viomycin. Eur J Biochem. 1977;81:491–497. doi: 10.1111/j.1432-1033.1977.tb11974.x. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- Pestka S. Studies on the formation of transfer ribonucleic acid-ribosome complexes. VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. J Biol Chem. 1969;244:1533–1539. [PubMed] [Google Scholar]
- Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. The highly conseved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- Rheinberger HJ. The function of the translating ribosome: allosteric three-site model of elongation. Biochimie. 1991;73:1067–1088. doi: 10.1016/0300-9084(91)90149-u. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribsosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Semenkov YP, Savelsbergh A, Katunin VI, Peske F, Wilden B, Wintermeyer W. Mechanism of tRNA translocation on the ribosome. Mol Biol. 2001;35:559–568. [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenkov YP, Shapkina TG, Kirillov SV. Puromycin reaction of the A site bound peptidyl tRNA. Biochimie. 1992;74:411–417. doi: 10.1016/0300-9084(92)90080-x. [DOI] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc Natl Acad Sci USA. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat Struct Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin AS. Energetics of the ribosome. Prog Nucleic Acid Res Mol Biol. 1978;21:39–62. doi: 10.1016/s0079-6603(08)60266-4. [DOI] [PubMed] [Google Scholar]
- Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- Wilson KS, Nechifor R. Interactions of translation factor EF-G with the bacterial ribosome before and after mRNA translocation. J Mol Biol. 2004;337:15–30. doi: 10.1016/j.jmb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina M. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Yamada T, Mizugichi Y, Nierhaus KH, Wittman HG. Resistance to viomycin conferred by RNA of either ribosomal subunit. Nature. 1978;275:460–461. doi: 10.1038/275460a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


