Abstract
Eight different angucyclinones have been produced in Streptomyces albus by combining three oxygenase genes together with the polyketide synthase and cyclases genes from the oviedomycin biosynthetic gene cluster from Streptomyces antibioticus ATCC 11891. Four of these compounds were fully characterized for the first time. Three of these angucyclinones—prejadomycin-2-carboxylate (2), 4a,12b-dehydro-UWM6 (5), and prejadomycin (3)—show a significant increase in their in vitro antitumor activity relative to oviedomycin (1). A hypothesis for the sequence of tailoring events catalyzed by these three oxygenases during oviedomycin biosynthesis is proposed. In this hypothesis OvmOII acts as a bifunctional oxygenase/dehydratase.
Keywords: angucycline, antitumor agents, combinatorial biosynthesis, polyketides, Streptomyces
Introduction
Oviedomycin (1) is an angucyclinone[1–4] polyketide produced by Streptomyces antibioticus ATCC 11891, which shows in vitro antitumor activity and induces apoptosis in cancer cell lines.
The structure elucidation and characterization of oviedomycin (ovm) and of its biosynthetic gene cluster has been reported previously.[5, 6] Oviedomycin possesses an unusually rich oxygen pattern, with seven oxygen atoms altogether. Particularly conspicuous is ring A, with its second quinone system generated by a specific oxygenase.[5, 6]
A biosynthetic pathway for oviedomycin has been proposed[6] that starts with the generation of a linear C-9 reduced polyketide of 20 C atoms through the action of a polyketide synthase (PKS) and ketoreductase (OvmPKST enzymes; Figure 1). This linear polyketide would undergo a series of cyclizations and aromatizations catalyzed by the corresponding aromatase (OvmA) and cyclase (OvmC).[6] Oviedomycin contains two keto groups (C-4 and C-12) and a hydroxy group (C-2) that should result from the actions of specific oxygenases. The ovm gene cluster contains three FAD-dependent oxygenase genes (ovmOI, ovmOII, and ovmOIII), which encode the enzymes that would be responsible for the introduction of those oxygens. OvmOI shows a high similarity to UrdE[7] (76% similarity, 67% identity), SimA7[8] (77 %, 67%), and LanE[9] (73 %, 61 %). Based on these sequence similarities, the expected function for OvmOI could be the introduction of a keto group at C-12 of the initial angucycline backbone, because these other proteins are thought to have similar functions. OvmOII shows a strong similarity to several other angucycline oxygenases involved in oxygenations at different positions in aglycons, such as LndM2[10] (75 %, 67%), responsible for C-6 hydroxylation in landomycin E, or Sim7[11] (71 %, 63%) and SimA8[8] (67 %, 60 %), thought to hydroxylate position C-12b in simocyclinone. These sequence data therefore do not allow a clear function to be ascribed to OvmOII. Finally, OvmOIII matches in databases correspond not to hydroxylation enzymes related to angucycline biosynthesis, but to compounds such as saframycin hydroxylase SfmO2[12] (50 %, 39%; saframycin is a tetrahydroisoquinoline resulting from the action of a PKS-NRPS system) and chromomycin A3 CmmOIV[13] (46 %, 38%; an aureolic acid polyketide). Since there is only one hydroxy group present in oviedomycin, OvmOIII could be involved in C-2 hydroxylation. Elucidation of the specific roles of each of these oxygenases could contribute to their use for combinatorial biosynthetic approaches. Additional oxygen functions often contribute to, and/or increase significantly, the biological activity of a compound.[ 4]
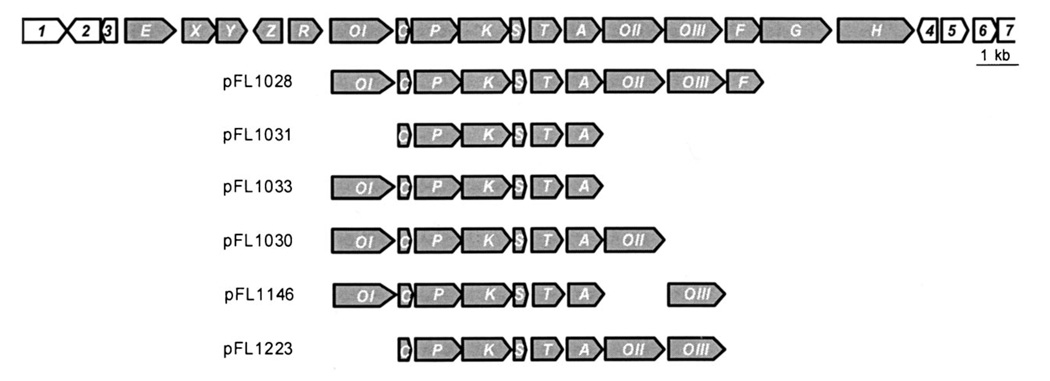
Figure 1.
Genetic organization of the oviedomycin (ovm) biosynthetic gene cluster; ovm genes are shown in gray. Functions for the corresponding enzymes are oxygenases (OvmOI, OvmOII, and OvmOIII), cyclase (OvmC), keto-acyl synthase α (OvmP), keto-acyl synthase β (OvmK), acyl carrier protein (OvmS), C9-ketoreductase (OvmT), aromatase (OvmA), and 4'-phosphopantetheinyl transferase (OvmF). Below: plasmids used in this work, showing the different gene compositions.
Here we report the heterologous expression of the minimum set of genes for oviedomycin biosynthesis in Streptomyces albus, as a starting point for the construction of different plasmids harboring different combinations of these FAD-dependent oxygenases together with the PKS, cyclase, aromatase, and reductase genes. These experiments led to the production of eight different compounds (four of them not previously described), which shed some light on the specific roles of these three oxygenases. Interestingly, three of these compounds showed significantly increased in vitro antitumor activity against human cancer cell lines relative to the parent oviedomycin.
Results and Discussion
Two cosmid clones from an S. antibioticus gene library have been sequenced. Both include the oviedomycin gene cluster and are able to direct the biosynthesis of this angucyclinone in S. albus. These cosmids also contain several putative primary metabolism genes at their 5'- and 3'-ends (Figure 1).[6] Furthermore, for two genes of the oviedomycin gene cluster (ovmX, ovmZ) no specific function could be assigned.[6]
With the aim of establishing the minimum set of genes necessary for oviedomycin biosynthesis, we cloned several fragments from the cosmid cosAB4 into the bifunctional E. coli/Streptomyces shuttle vector pEM4A. In this vector, the cloned genes were placed under the control of the constitutive promoter of the erythromycin resistance gene ermE, which allows efficient expression of genes under its control in Streptomyces. The construct containing the minimum set of genes able to produce oviedomycin in S. albus (Figure 2B) was pFL1028 (Figure 1). This plasmid contains, under the control of the ermE promoter, the genes ovmOI, ovmC, ovmP, ovmK, ovmS, ovmT, ovmA, ovmOII, ovmOIII, and ovmF. Gene ovmF codes for a 4'-phosphopantetheinyl transferase, and is present in this construction only for DNA cloning reasons; ovmF is not essential for oviedomycin production since other constructs lacking this gene (see below) still direct the biosynthesis of the corresponding polyketides. According to this result, all other genes present in the cluster were unnecessary for oviedomycin biosynthesis in S. albus.
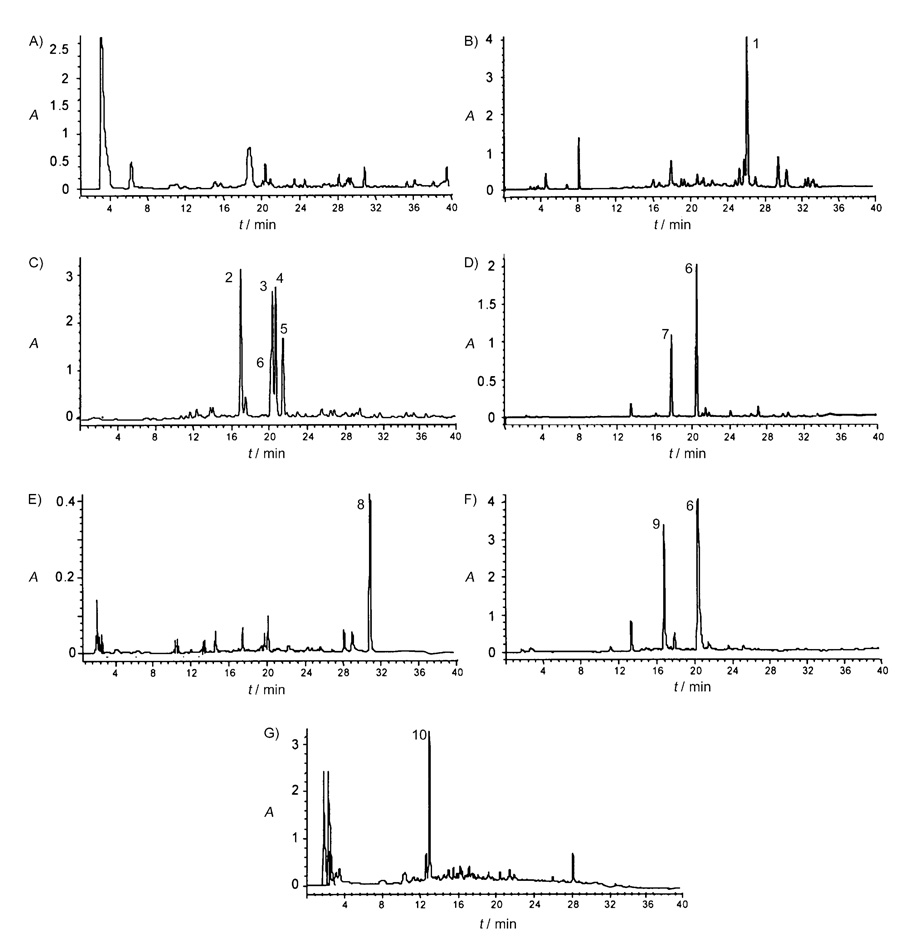
Figure 2.
HPLC chromatograms from cell culture extracts of Streptomyces albus strains containing plasmids A) pEM4A, B) pFL1028, C) pFL1031, D) pFL1033, E) pFL1030, F) pFL1146, and G) pFL1223.
Consequently, the minimum set of genes required for oviedomycin biosynthesis was that encoding the PKS, a ketoreductase, an aromatase, a cyclase, and three oxygenases. To establish the specific contribution of each of the three oxygenases to the tailoring modifications in oviedomycin at C-12 (ring C quinone), C-4 (ring A quinone), and C-2 (ring A hydroxy group), we generated several constructs in pEM4A, containing the oviedomycin ovmCPKSTA (cyclase, PKS, ketoreductase, and aromatase) genes alone (pFL1031; see the Experimental Section) or with various oxygenase gene combinations: pFL1033 (plus ovmOI), pFL1030 (plus ovmOIOII), pFL1146 (plus ovmOIOIII), and pFL1223 (plus ovmIIOIII; Figure 1).
When S. albus was transformed with pFL1031, the simplest set of oviedomycin genes, which is a construct lacking all three oxygenase genes, five peaks that had been absent in the control strain S. albus (pEM4A; Figure 2A) were detected in HPLC chromatograms of ethyl acetate extracts of the corresponding cultures (Figure 2C). These five compounds were isolated, and their structures were elucidated and found to correspond to the angucyclinones prejadomycin-2-carboxylate (2; 18.1 min), prejadomycin (3; 20.5 min), UWM6 (4; 20.7 min), 4a,12b-dehydro-UWM6 (5; 21.2 min), and rabelomycin (6; 20.4 min), this last being a minor compound.
Compounds 3, 4, and 6 (Scheme 1) were easily identified as the known angucyclinones prejadomycin, UWM6, and rabelomycin respectively, by comparison of their NMR data with those of authentic samples and the reported data in the literature.[7, 14, 15]
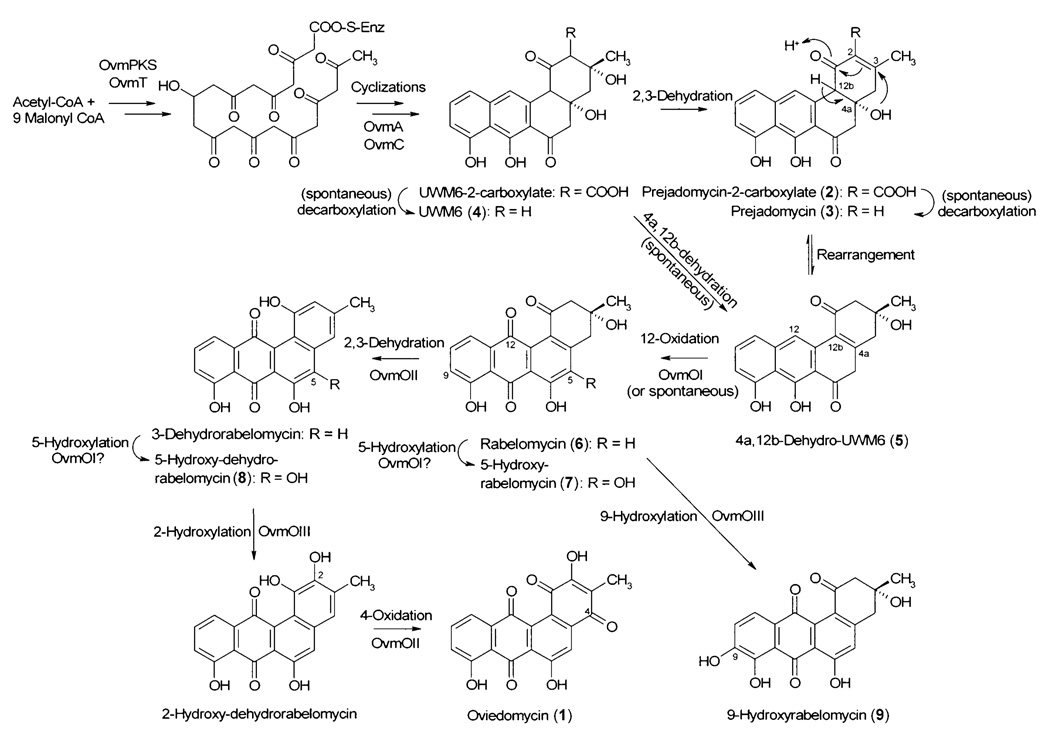
Scheme 1.
Proposed biosynthetic pathway for oviedomycin; all numbered compounds were isolated and characterized. It is proposed that compounds 7, 8, and 9 are shunt products.
Rabelomycin (6) had been isolated as an early shunt product from several other biosynthetic routes, such as those for jadomycin and urdamycin and a silent angucycline gene cluster.[16–20] Rabelomycin (6) and UWM6[16, 21, 22] have previously been identified by Hutchinson et al. in several mutants from the jadomycin producer S. venezuelae or in a strain of S. lividans that expresses a group of jadomycin genes. Cultures of a S. lividans strain containing the minimal PKS, ketoreductase, aromatase, and cyclase (JadABCEDI) from the jadomycin pathway accumulated UWM6 (4) and rabelomycin (6).[16] UWM6 (4) is an unstable compound, which is spontaneously converted into rabelomycin (6) through dehydration and oxidation steps.[16] Prejadomycin (3; also known as 2,3-dehydro-UWM6),[16–20, 23] along with its homologue, was first isolated from a mutant of the gilvocarcin pathway in which oxygenase GilOI was inactivated,[20] and then from the jadomycin producer lacking the jadomycin oxygenase JadH.[17–20, 23] In both cases 3 was shown to be a real intermediate of gilvocarcin M and jadomycin B biosyntheses, respectively, and not a shunt product.[18, 20] It was also shown that rabelomycin (6) is a shunt product,[17–20, 23] while UWM6 (4) is a biosynthetic intermediate of jadomycin biosynthesis, because it is converted, although in low yield, into jadomycins.[18] Recently, prejadomycin (3) was also found to be an intermediate of the landomycin biosynthetic pathway.[19]
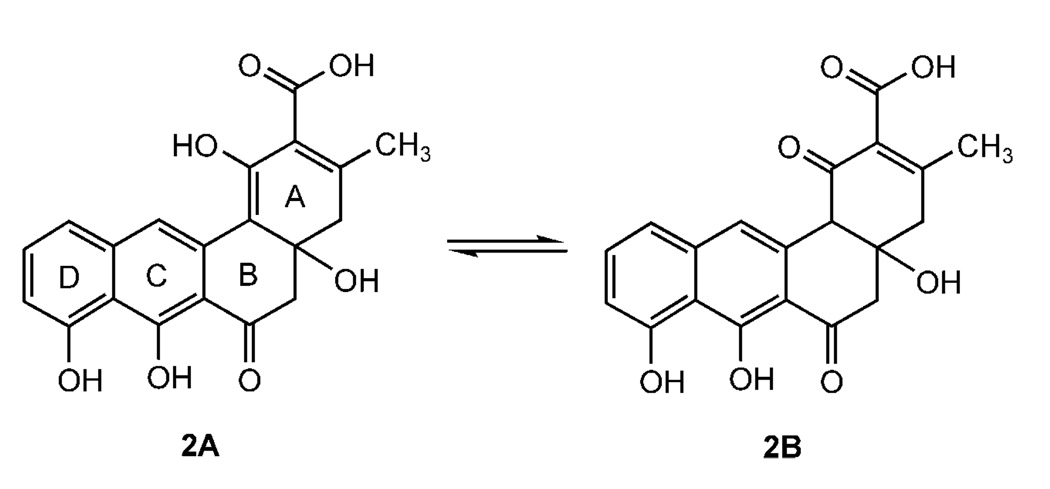
Compound 2 showed UV absorption maxima at 266 and 407 nm. The APCI mass spectrum of 2 displayed signals at m/z 369 [M+H]+ in the positive ion mode and m/z 367 [M−H]− in the negative ion mode, thus suggesting a molecular weight of 368. The molecular formula of 2 was confirmed as C20H16O7 by HREI-MS (m/z 368.0834, 10%; calcd. 368.0896). The 1H NMR spectrum of 2 showed characteristic signals at δ = 15.54 and 9.42 for the 8- and 7-OH groups, respectively,[16] as well as a broad singlet at δ=9.85. Three aromatic protons were coupled together in an ABC system at δ=7.56 (8.2 Hz), 7.25 (8.2 Hz), and 6.88 (8.2 Hz), and one singlet at δ = 6.85 was found as well. Similar signals had previously been described for prejadomycin (3).[17] In the aliphatic region, two pairs of methylene protons and one methyl group showing an HSQC correlation to δ=43.8, 40.6, and 21.6, respectively, were observed. A major difference from the spectra of 3 was the lack of the olefinic proton at C-2 of 3 in ring A at δ = 5.95, and the appearance of a carboxylic acid carbonyl at δ = 175.2. The molecular weight of 2 in the (−)-APCI-MS was 44 mass units greater than that of 3, suggesting the presence of an extra COOH group at C-2 in compound 2, which would also be expected from the biosynthetic pathway. The structure of compound 2 was confirmed by its 1D and 2D NMR spectra (see the Supporting Information) and by comparison of its spectral data with those of the known prejadomycin (3). Compound 2 was concluded to be an analogue of 3 in which ring A has a COOH group and was therefore named prejadomycin-2-carboxylate. Prejadomycin-2-carboxylate (2) exists as a tautomeric mixture as shown in Scheme 2, with 2A being the major tautomer, as has been shown by NMR.
Scheme 2.
Prejadomycin-2-carboxylate (2) exists as a tautomeric mixture (data for the minor tautomer 2b are not listed in Table S1 in the Supporting Information).
The other new compound of this experiment—compound 5—gave a quasimolecular ion at m/z 325 [M+H]+ by (+)-APCI-MS, confirmed by EIMS ([M]+, m/z 324), thereby identifying 5 as possessing a molecular weight of 324. High-resolution measurements (HREI-MS) of 5 showed a molecular ion at m/z 324.0909 (calcd 324.0998), consistent with the molecular formula C19H16O5. The molecular weight of 5 is 18 units lower than that of UWM6 (4). The implication that compound 5 differs from 4 in the loss of H2O was supported by the NMR data (Table S1 in the Supporting Information). The UV spectrum of 5 recorded in MeOH exhibited UV absorption bands at 241 and 403 nm, which closely resemble those reported for UWM6 (4).[16] The 1H and 13C NMR spectra of 5 showed that compound 5 differs from UWM6 (4) only at the C-4a and C-12b positions. A methine signal at δ = 3.84 in the 1H NMR spectrum of 4 had disappeared in that of 5. In addition, the 13C NMR spectrum of 5 showed the appearance of two quaternary carbon atoms at δ = 127.6 and 142.8 at the C-12b and C-4a positions in place of one CH carbon at δ = 58.9 and one quaternary carbon at δ = 74.0 in 4. Direct comparison of the spectroscopic data for 5 with those for UWM6 (4)[16] and interpretation of the 2D NMR data thus allowed 5 to be assigned as 4a,12b-dehydro-UWM6. This compound might correspond to an angucycline found by Hutchinson et al.,[16] which was not characterized because of its instability. This compound was also recently found to exist in equilibrium with prejadomycin (3) through an intramolecular Michael rearrangement reaction.[19]
The new compound prejadomycin-2-carboxylate (2) is the only compound out of the five isolated that still contains the COOH group originating from the incorporation of the last malonyl-CoA extender unit. This COOH group at C-2 was previously believed to be highly unstable and to undergo a nonenzymatic decarboxylation early during the biosynthesis of this class of decaketides once ring A had been formed.[1–4] Prejadomycin-2-carboxylate (2) is accumulated along with the previously known compounds 3 and 4. These three compounds are most likely biosynthetic intermediates of the oviedomycin pathway. Rearrangement would generate 4a,12b-dehydro- UWM6 (5), and its (spontaneous) 12-oxidation would lead to rabelomycin (6; Scheme 1). Alternatively, the sequence of events could be 12-oxidation of 4, followed by 4a,12b-dehydration to provide 6. A less likely pathway would be the spontaneous 4a,12b-dehydration of 4 followed by 12-oxidation (Scheme 1).
Transformation of S. albus with pFL1033 (Figure 1), a construct containing the same genes as pFL1031 but also including the ovmOI oxygenase gene, led to the production of two compounds (Figure 2D): rabelomycin (6), now as the major product, and its derivative 5-hydroxyrabelomycin (7, HPLC retention time 17.7 min) as the minor product.[24] The presence of the oxygenase OvmOI had thus induced a drastic change in the type and amount of accumulated products in relation to the previous experiment, with S. albus (pFL1031), in which rabelomycin (6) was only a minor product. The two compounds generated in this experiment each contain a quinone system in ring C, so OvmOI is the enzyme that introduces the oxygen into C-12, and likely the first acting oxygenase during oviedomycin biosynthesis. This assignment is in agreement with the fact that, at the amino acid sequence level, OvmOI is highly similar to UrdE, the oxygenase believed to carry out this 12-oxygenation during urdamycin biosynthesis.[7] Since compounds 6 and 7 are both dehydrated at the 4a,12b-bond, in contrast to compounds 2, 3, and 4, OvmOI could also possess a dehydratase activity, as has been described for various oxygenases acting on early angucyclinone intermediates, such as the oxygenases GilOI, GilOIV, JadH, and JadF.[17–20, 23] This possibility has been reinforced by the fact that the presence of OvmOI in other constructs (see below) always results in compounds containing that double bond. Compound 7 contains an extra hydroxy group, not found in oviedomycin (1), at C-5, so compound 7 is a shunt product, the generation of which is caused either by an oxygenase from the host strain S. albus or by a second, normally suppressed, reaction of OvmOI. In this context, a recent analysis of the oxygenases involved in the gilvocarcin and jadomycin biosyntheses[7, 19, 23] showed that these were multienzyme complexes, disruption of which can cause the introduction of an O atom at a position not normally affected. It is noticeable that the simultaneous presence of OvmOI and OvmOIII abolishes the formation of C-5 hydroxylated derivatives (see below). This supports the existence of a possible interaction between OvmOI and OvmOIII, as has also been suggested for some oxygenases in other systems. Most probably, the correct formation of oviedomycin requires the presence of all three oxygenases for their right function and for the correct channeling of intermediates from one oxygenase to the following one.
The three structural differences between rabelomycin (6) and oviedomycin (1) are located in ring A: absence of the 2-hydroxy group and the keto group at C-4, and the lack of a 2,3-enoyl bond. The two other oviedomycin oxygenases (OvmOII and OvmOIII) should thus be responsible for these modifications. In order to clarify these aspects, we decided to study the effect of the simultaneous presence of OvmOI with OvmOII or OvmOIII. When S. albus was transformed with plasmid pFL1030 (Figure 1), which contains all the necessary genes for oviedomycin biosynthesis except ovmOIII, this strain produced one single compound with a retention time of 31.1 min (Figure 2E). Structure elucidation of this compound revealed that it corresponded to the new compound 5-hydroxy-dehydrorabelomycin (8). As in the case of 5-hydroxy-rabelomycin (7), this hydroxylation at C-5 could be caused by a chromosomally encoded S. albus oxygenase or by OvmOI. Thus, the main effect caused by the introduction of oxygenase OvmOII is the conversion of the rabelomycin scaffolds 6 and 7 (the only products generated in the presence of OvmOI alone) into the dehydrorabelomycin derivative 8.
We anticipated that the addition of OvmOII in S. albus (pFL1030) should add to the oxygenation pattern in ring A, but the 4-oxo and 2-hydroxy groups were still absent in 8 (Scheme 1). OvmOII therefore does not carry out the second oxygenation event that takes place after the introduction of the 12-oxygen by OvmOI during oviedomycin biosynthesis, possibly because OvmOII cannot act in the absence of OvmGOIII. However, the presence of OvmOII in a system that already contains the first tailoring oxygenase OvmOI apparently causes the formation of a 2,3-enoyl bond. This suggests that OvmOII has a 2,3-dehydratase activity, like those found for GilOIV and JadF,[23] leading to the formation of 8 from 7.
Another experiment consisted of the generation of a system containing the oxygenase OvmOI together with OvmOIII. S. albus was transformed with pFL1146 (Figure 1), which contains all the necessary genes for oviedomycin biosynthesis except ovmOII. This recombinant strain accumulated two products in the culture broths. These compounds turned out to be rabelomycin (6) and the new 9-hydroxyrabelomycin (9, 16.8 min; Figure 2 F). The result clearly reasserted the role of oxygenase OvmOII as a 2,3-dehydratase (2-CH2 and 3-OH are still present in 6, 7, and 9). Moreover, the presence of oxygenase OvmOIII also causes the introduction of a hydroxy group at C-9, an aromatic C atom ortho to a phenolic hydroxy group. This might indicate that OvmOIII could be involved in the generation of the 2-hydroxy group found in ring A of oviedomycin (1), which is also consistent with its sequence comparison, in which hydroxylases are its closest homologues. Since ring A is not (yet) aromatic in the absence of OvmOII, OvmOIII is probably unable to carry out a hydroxylation at the saturated C-2.
As a final experiment, S. albus was transformed with a system containing ovmOII and ovmOIII, but lacking ovmOI (pFL1223, Figure 1). Analysis of the cultures from this recombinant strain showed the production of a main compound with an absorption spectrum completely different from those of oviedomycin and its intermediates or shunt products (Figure 2 G). Mass analysis of this compound revealed a molecular weight 82 units greater than oviedomycin. From these data, we concluded that this compound would be an aberrant cycled shunt product and so it was not further characterized. This result was further confirmation that OvmOI is the oxygenase that acts first during oviedomycin biosynthesis.
In summary, several important conclusions relating to the biosynthesis of oviedomycin can be deduced from all these experiments (Scheme 1). Firstly, oxygenation at C-12 is an early event (like in landomycin and urdamycin biosynthesis, but unlike in jadomycin and gilvocarcin biosyntheses), which in the oviedomycin pathway is catalyzed by OvmOI. In addition, OvmOI appears to have a dehydratase activity, which would initially generate a 2,3-double bond in ring A and eventually (after rearrangement) the 4a,12b-double bond, because all the compounds formed in the presence of OvmOI contain this 4a,12b-enoyl bond. The next events of oviedomycin biosynthesis are probably the C-2 decarboxylation followed by the (re)-generation of the 2,3-double bond in ring A, which requires the 2,3-dehydratase activity of OvmOII, because this 2,3-elimination of water occurs only in the presence of OvmOII. However, the expression of OvmOII does not produce any additional oxygenation event. There are two possible explanations for this. The oxygenase activity of OvmOII (most likely introduction of the oxygen at C-4 and generation of the quinone system in ring A of 1) requires the simultaneous presence of the oxygenase OvmOIII. Alternatively, and more probably, the order of reactions is 2,3-dehydration by OvmOII, then introduction of the oxygen at C-2 through OvmOIII, and, finally, introduction of the oxygen at C-4 by OvmOII, followed by dehydrogenation to the p-quinone system found in ring A of oviedomycin (1). In absence of an aromatic ring A, OvmOIII is unable to introduce an oxygen atom at C-2 and hydroxylates the C-9 instead.
Antitumor activity has been described in the literature for several angucycline and angucyclinone polyketides: the landomycins,[ 24] urdamycins,[25] and 6-hydroxytetrangulol,[26] for example. The last of these compounds induced apoptosis in U937 leukemia cells by activating the protease caspase-3,[26] though the related compound tetrangulol was inactive. Oviedomycin shares the presence of this 6-hydroxy group, and we therefore tested its antitumor activity against three human cancer cell lines: MDA-MB-231 (breast cancer), A549 (non-small- cell lung cancer), and HT29 (colon cancer). The same in vitro antitumor activity tests were also carried out with the eight compounds isolated from these experiments. Surprisingly, although most of the compounds showed antitumor activity similar to that of oviedomycin, three of the compounds—prejadomycin-2-carboxylate (2), prejadomycin (3), and 4a,12b-dehydro-UWM6 (5)—showed activities ten times greater than that of oviedomycin (Table 1). This result implies that C-12 oxygenation is not necessarily a key biosynthetic step with respect to the generation of compounds with antitumor activity, and that incompletely tailored intermediates or shunt products can be even more active than the final compound. These three active derivatives share the presence of an extra hydroxy group (at C-4a in 2 and 3, or at C-3 in 5). However, the simultaneous presence of both hydroxy groups (C-4a and C-3), as in UWM6 (4), abolishes the increase in activity. The presence of the C-2 carboxy group in 2 does not seem to be important for target binding, because the activity of this compound is about the same as that of its decarboxylated counterpart 3. Therefore, elimination of all oxygenases has contributed to the generation of three more active compounds (2, 3, and 5), which could serve as scaffolds for further tailoring.
Table 1.
Antitumor activities of compounds.
| Cell line | GI50 [µm] | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| MDA-MB-231 (breast cancer) |
5.6 | 0.38 | 0.24 | 24.8 | 0.64 | 6.8 | 13.3 | 2.56 | 7.06 |
| A549 (NSCL) | 8.7 | 0.59 | 0.29 | >100 | 0.77 | 5.32 | 7.62 | 6.24 | 3.10 |
| HT29 (colon cancer) |
6.9 | 1.17 | 1.29 | >100 | 1.85 | 10.9 | 12.4 | 8.03 | 14.1 |
Gl50: 50% growth inhibition.
Cloning of ovm PKS genes together with several combinations of ovm oxygenases genes has allowed the characterization of four new angucyclines and the generation of three compounds ten times more active than oviedomycin. These results shed light on the enzymatic activity of each oviedomycin oxygenase, establishing a scheme for the biosynthesis of this polyketide antitumor agent.
Experimental Section
Microorganisms, culture conditions, and plasmids
Streptomyces albus J1074 (ilv-1, sal-2)[27] was used as host for expression of ovie-domycin genes. E. coli DH10B (Invitrogen) was used as host for subcloning. Cultures of E. coli were grown in TSB medium (Merck). R5A medium[28] was used for oviedomycin production. R5 medium was used for protoplast regeneration after plasmid transformation.[ 29] Benett medium was used for sporulation of S. albus.[29] When plasmid-containing clones were grown, the medium was supplemented with the appropriate antibiotics: ampicillin (100 µgmL−1), kanamycin (25 µgmL−1), apramycin (25 µgmL−1), and thiostrepton (50 µgmL−1). pUC18,[30] pUK21,[31] pCRBlunt (Invitrogen), pIAGO,[28] and pEM4A[32] were used for subcloning.
DNA manipulation techniques
Plasmid DNA preparations, restriction endonuclease digestions, alkaline phosphatase treatments, DNA ligations, and other DNA manipulations were performed by standard techniques for Escherichia coli[33] and Streptomyces.[29] Preparation of S. albus protoplasts, transformation, and selection of transformants were carried out as described.[29]
Plasmid constructions
pFL1028
A 10906 bp DNA fragment from cosmid cosAB4[5, 6] was obtained by digestion with restriction enzymes BfuI (position 7826 in the accession number AJ632203) and SgfI (position 18732), and blunt ended. This DNA band was subcloned in the blunt-ended EcoRI site of pEM4A, generating pFL1028. In this construct, genes ovmOI, ovmC, ovmP, ovmK, ovmS, ovmT, ovmA, ovmOII, ovmOIII, and ovmF are placed under the control of ermE* promoter.
pFL1030
A 8966 bp DNA fragment from cosAB4 was obtained by digestion with restriction enzymes BfuI (position 7826) and KpnI (position 16 792). This DNA band was blunt ended and cloned in the blunt-ended EcoRI site of pEM4A. In this construction, genes ovmOI, ovmC, ovmP, ovmK, ovmS, ovmT, ovmA, and ovmOII are placed under the control of ermE* promoter.
pFL1031
A 5487 bp DNA fragment from pFL1028 was obtained by digestion with restriction enzymes SphI (position 9143) and XhoI (position 14 630). This DNA band was blunt ended and cloned in the blunt-ended EcoRI site of pEM4A. In this construction, genes ovmC, ovmP, ovmK, ovmS, ovmT, and ovmA are placed under the control of ermE* promoter.
pFL1033
A 6804 bp DNA fragment from cosAB4 was obtained by digestion with restriction enzymes BfuI (position 7826) and XhoI (position 14 630). This DNA band was blunt ended and cloned in the blunt-ended EcoRI site of pEM4A. In this construction, genes ovmOI, ovmC, ovmP, ovmK, ovmS, ovmT, and ovmA are placed under the control of ermE* promoter.
pFL1146
A 2541 bp DNA fragment from pFL1028 was obtained by digestion with restriction enzyme SalI (positions 15633 and 18174). This DNA band containing the gene ovmOIII was cloned in the SalI site of pUC18, generating plasmid pFL1142. From this plasmid, ovmOIII gene was obtained as an XbaI and blunt-ended HindIII fragment, which was subcloned in the XbaI and bluntended PstI restriction sites of vector pIAGO, generating pFL1144, in which ovmOIII gene is placed under the control of ermE promoter. Finally, a blunt-ended EcoRI/HindIII fragment from pFL1144 containing ermE promoter and ovmOIII was subcloned into the HindIII restriction site (previously blunt ended) of pFL1029 (a thiostrepton version of pFL1033) polylinker, which is located just upstream of ermE* promoter, generating pFL1146. This plasmid contains the genes ovmOI, ovmC, ovmP, ovmK, ovmS, ovmT, and ovmA placed under the control of ermE* promoter and gene ovmOIII placed under the control of ermE promoter.
pFL1223
A 5232 bp AvrII/SgfI DNA fragment containing the genes ovmOII and ovmOIII was obtained from pFL1028 and subcloned in pWHM3, generating pFL1222, where these two genes are under the control of ermE promoter. These two genes and this promoter were rescued from pFL1222 as a HindIII/EcoRI fragment and blunt ended prior to subcloning in the blunt-ended HindIII site of pFL1031, giving rise to pFL1223. This construction contains all the genes for oviedomycin biosynthesis (except ovmOI) under the control of two divergent ermE promoters.
DNA sequencing
Sequencing of the different constructs was performed on double-stranded templates derived from pUC18 clones by use of the dideoxynucleotide chain termination method[34] and the Cy5 AutoCycle Sequencing Kit (Amersham Pharmacia Biotech). DNA was sequenced with primers supplied in the kits (18-mer) with the aid of an ALF-express automatic DNA sequencer (Amersham Pharmacia Biotech).
HPLC analyses for oviedomycin and derivatives
HPLC-MS analyses were carried out with chromatographic equipment coupled to a ZQ4000 mass spectrometer (Waters-Micromass), with acetonitrile and trifluoroacetic acid (TFA; 0.1%) in water as HPLC solvents and on a reversed-phase column (Symmetry C18, 2.1 × 150 mm, Waters). Samples were eluted with acetonitrile (10%) over the first 4 min, followed by a linear gradient from 10 to 88% acetonitrile over 26 min, at a flow rate of 0.25 mLmin−1. When required, an additional 5 min elution with 100% acetonitrile was performed at the end of the gradient. Detection and spectral characterization of peaks were carried out with a photodiode array detector and Empower software (Waters). MS analyses were carried out by electrospray ionization in the positive mode, with a capillary voltage of 3 kV and cone voltages of 20 and 100 V. The same conditions were used for HPLC analysis, except for the column size (4.6×250 mm) and flow rate (1 mLmin−1).
For purification purposes, all strains were grown by a two-step culture method, as previously described.[28] In the production step, eight 2-liter Erlenmeyer flasks, each containing medium (400 mL), were incubated for 5 days. The cultures were centrifuged and filtered, and the broth was solid-phase extracted.[35] In the case of strain S. albus (pFL1031), the broth was acidified to pH 4.0 with formic acid before extraction. The fractions obtained were analyzed by HPLC, and those containing the desired compounds were dried in vacuo. The extracts were redissolved and chromatographed by use of a µBondapak C18 radial compression cartridge (PrepPak Cartridge, 25 × 100 mm, Waters) or a preparative column (SunFire Prep C18, 10 × 250 mm, Waters). Isocratic elutions with mixtures of acetonitrile or methanol and trifluoroacetic acid in water (0.1%) were optimized for every compound to be purified. In all cases, after every purification step, the collected compounds were diluted fourfold with water and were desalted and concentrated by solidphase extraction, followed by lyophilization. The following yields were obtained: S. albus (pFL1030) 5-hydroxy-dehydrorabelomycin (8, 2.0 mg, 0.625 mg L−1); S. albus (pFL1031) prejadomycin-2-carboxylate (2, 5.7 mg, 1.78 mg L−1), rabelomycin (6, 4.2 mg, 1.31 mg L−1), prejadomycin (3, 17.8 mg, 5.56 mg L−1), UWM6 (4, 21.5 mg, 6.71 mg L−1), 4a,12b-dehydro-UWM6 (5, 3.7 mg, 1.15 mg L−1); S. albus (pFL1033) 5-hydroxyrabelomycin (7, 7.3 mg, 2.28 mg L−1), rabelomycin (6, 37.3 mg, 11.65 mg L−1); S. albus (pFL1146) 9-hydroxyrabelomycin (9, 13.8 mg, 4.31 mg L−1), rabelomycin (6, 33.0 mg, 10.31 mg L−1).
Structure elucidation and characterization
The structures of the isolated compounds were elucidated by nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry. The ESI mass spectra (ESI-MS) were acquired with a Finnigan LCQ mass spectrometer. The electron impact (EI) ionization mass spectra were measured with a Finnigan PolarisQ mass spectrometer. The high-resolution EI ionization mass spectra were recorded at 25 eV on a JEOL JMS-700T M station (magnetic-sector instrument) at a resolution greater than 10 000. All NMR data were recorded in [D6]DMSO with either a Varian Mercury 300 or a Varian Inova 400 MHz spectrometer. All NMR assignments were confirmed by heteronuclear single quantum correlation and heteronuclear multiple bond correlation spectra, allowing unambiguous assignment of all signals.
Physicochemical properties of new compounds 2, 5, 8, and 9
Prejadomycin-2-carboxylate (2)
Yellow solid; 1.8 mg L;−1 Rrel = 18.1 min; UV maxima (from HPLC-diode array): λ = 266 (69 %), 280 (100), 407 nm (29%); (−)-APCI-MS: m/z 367 [M−H]− (100); (+)-APCI-MS: m/z (%): 369 [M+H]+ (100); HREI-MS: m/z 368.0834; calcd for C20H16O7: 368.0896; for 1H and 13C NMR data see the Supporting Information.
4a,12b-Dehydro-UWM6 (5)
Yellow solid; 1.2 mg L−1; Rrel = 21.2 min; UV maxima (from HPLC-diode array): λ=241 (45 %), 259 (89 %), 403 nm (16%); (−)-APCI-MS: m/z (%): 323 [M−H]− (100); (+)-APCIMS: m/z (%): 325 [M+H]+ (100); HREI-MS: m/z 324.0909; calcd for C19H16O5 : 324.0998; 1H and 13C NMR data: see the Supporting Information.
5-Hydroxy-dehydrorabelomycin (8)
Green solid; 0.6 mg L−1; Rrel = 31.1 min; UV maxima (from HPLC-diode array): λ = 224 (44 %), 345 (67 %), 456 nm (15%); (−)-APCI-MS: m/z (%): 335 [M−H]− (100); (+)-APCI-MS: m/z (%): 337 [M+H]+ (100); HREI-MS: m/z 336.0642; calcd for C19H12O6 : 336.0634; 1H and 13C NMR data: see the Supporting Information.
9-Hydroxyrabelomycin (9)
Yellow solid; 4.3 mg L;−1 Rrel = 16.8 min; UV maxima (from HPLC-diode array): λ=271 (75 %), 286 (100 %), 437 nm (20%); (−)-APCI-MS: m/z (%): 353 [M−H]− (100); (+)-APCI-MS: m/z (%): 355 [M+H]+ (100); HREI-MS: m/z 354.0714; calcd for C19H14O7: 354.0739; 1H and 13C NMR data: see the Supporting Information.
Antitumor activity tests
Different concentrations of the corresponding compounds were used for testing of their antitumor activities against three human cancer cell lines (MDA-MB-231, A549, and HT29). Quantitative measurement of cell growth and viability was carried out by colorimetric sulforhodamine B assay.[36]
Acknowledgements
We thank Leire Peña and Patricia Oro for helpful technical assistance. This research was supported by grants from the Spanish Ministry of Education and Science (BMC2005-04115 and PET2005–0401 to C.M.), from Red Temática de Investigación Co-operativa de Centros de Cáncer (Ministry of Health, Spain; ISCIII-RETIC RD06/0020/0026), and from the US National Institute of Health (CA 102102 to J.R.). We thank Pharmamar S.A. for helping us with the antitumor assays. We also thank Obra Social Cajastur for financial support to F.L., and the NMR and MS core facilities of the University of Kentucky for use of their instruments.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.200800425.
References
- 1.Rohr J, Thiericke R. Nat. Prod. Rep. 1992;9:103–137. doi: 10.1039/np9920900103. [DOI] [PubMed] [Google Scholar]
- 2.Krohn K, Rohr J. Top. Curr. Chem. 1997;188:127–195. [Google Scholar]
- 3.Rawlings BJ. Nat. Prod. Rep. 1999;16:425–484. doi: 10.1039/a900566h. [DOI] [PubMed] [Google Scholar]
- 4.Rix U, Fischer C, Remsing LL, Rohr J. Nat. Prod. Rep. 2002;19:542–580. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- 5.Méndez C, Künzel E, Lipata F, Lombó F, Cotham W, Walla M, Bearden DW, Braña AF, Salas JA, Rohr J. J. Nat. Prod. 2002;65:779–782. doi: 10.1021/np010555n. [DOI] [PubMed] [Google Scholar]
- 6.Lombó F, Braña AF, Salas JA, Méndez C. ChemBioChem. 2004;5:1181–1187. doi: 10.1002/cbic.200400073. [DOI] [PubMed] [Google Scholar]
- 7.Faust B, Hoffmeister D, Weitnauer G, Westrich L, Haag S, Schneider P, Decker H, Künzel E, Rohr J, Bechthold A. Microbiology. 2000;146:147–154. doi: 10.1099/00221287-146-1-147. [DOI] [PubMed] [Google Scholar]
- 8.Trefzer A, Bechthold A. Antimicrob. Agents Chemother. 2002;46:1174–1182. doi: 10.1128/AAC.46.5.1174-1182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westrich L, Domann S, Faust B, Bedford D, Hopwood DA, Bechthold A. FEMS Microbiol. Lett. 1999;170:381–387. doi: 10.1111/j.1574-6968.1999.tb13398.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhu L, Ostash B, Rix U, Nur-E-Alam M, Mayers A, Luzhetskyy A, Mendez C, Salas JA, Bechthold A, Fedorenko V, Rohr J. J. Org. Chem. 2005;70:631–638. doi: 10.1021/jo0483623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galm U, Schimana J, Fiedler HP, Schmidt J, Li SM, Heide L. Arch. Microbiol. 2002;178:102–114. doi: 10.1007/s00203-002-0429-z. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Deng W, Song J, Ding W, Zhao QF, Peng C, Song WW, Tang GL, Liu W. J. Bacteriol. 2008;190:251–263. doi: 10.1128/JB.00826-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menéndez N, Mohammad N, Braña AF, Rohr J, Salas JA, Méndez C. Chem. Biol. 2004;11:21–32. doi: 10.1016/j.chembiol.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Krishnamurthy VV, Russell DJ, Hadden CE, Martin GE. J. Magn. Reson. 2000;146:232–239. doi: 10.1006/jmre.2000.2141. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds WF, Enriquez RG. J. Nat. Prod. 2002;65:221–244. doi: 10.1021/np010444o. [DOI] [PubMed] [Google Scholar]
- 16.Kulowski K, Wendt-Pienkowski E, Han L, Yang K, Vining LC, Hutchinson CR. J. Am. Chem. Soc. 1999;121:1786–1794. [Google Scholar]
- 17.Rix U, Wang C, Chen Y, Lipata FM, Remsing Rix LL, Greenwell LM, Vining LC, Yang K, Rohr J. ChemBioChem. 2005;6:838–845. doi: 10.1002/cbic.200400395. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Wang C, Greenwell L, Rix U, Hoffmeister D, Vining LC, Rohr J, Yang K. J. Biol. Chem. 2005;280:22508–22514. doi: 10.1074/jbc.M414229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baig I, Kharel M, Kobylyankyy A, Zhu L, Rebets Y, Ostash B, Luzhetskyy A, Bechthold A, Fedorenko VA, Rohr J. Angew. Chem. 2006;118:8006–8010. doi: 10.1002/anie.200603176. Angew. Chem. Int. Ed. 2006, 45, 7842-7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu T, Fischer C, Beninga C, Rohr J. J. Am. Chem. Soc. 2004;126:12262–12263. doi: 10.1021/ja0467521. [DOI] [PubMed] [Google Scholar]
- 21.Rix U, Zheng J, Remsing Rix LL, Greenwell L, Yang K, Rohr J. J. Am. Chem. Soc. 2004;126:4496–4497. doi: 10.1021/ja031724o. [DOI] [PubMed] [Google Scholar]
- 22.Metsä-Ketelä M, Palmu K, Kunnari T, Ylihonko K, Mäntsälä P. Antimicrob. Agents Chemother. 2003;47:1291–1296. doi: 10.1128/AAC.47.4.1291-1296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kharel MK, Zhu L, Liu T, Rohr J. J. Am. Chem. Soc. 2007;129:3780–3781. doi: 10.1021/ja0680515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu L, Luzhetskyy A, Luzhetska M, Mattingly C, Adams V, Becht-hold A, Rohr J. ChemBioChem. 2007;8:83–88. doi: 10.1002/cbic.200600360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drautz H, Zähner H, Rohr J, Zeeck A. J. Antibiot. 1986;39:1657–1669. doi: 10.7164/antibiotics.39.1657. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita N, Harada T, Shin-ya K, Seto H. J. Antibiot. 1998;51:79–81. doi: 10.7164/antibiotics.51.79. [DOI] [PubMed] [Google Scholar]
- 27.Chater KF, Wilde LC. J. Gen. Microbiol. 1980;116:323–334. doi: 10.1099/00221287-116-2-323. [DOI] [PubMed] [Google Scholar]
- 28.Fernández E, Weissbach W, Reillo CS, Braña AF, Méndez C, Rohr J, Salas JA. J. Bacteriol. 1998;180:4929–4937. doi: 10.1128/jb.180.18.4929-4937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich: The John Innes Foundation; 2000. [Google Scholar]
- 30.Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 31.Vieira J, Messing J. Gene. 1991;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]
- 32.Blanco G, Perez-Patallo E, Braña AF, Rohr J, Mendez C, Salas JA. Chem. Biol. 2001;8:253–263. doi: 10.1016/s1074-5521(01)00010-2. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, a Laboratory Manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson AR. Proc. Natl. Acad. Sci. USA. 1992;24:104–108. [Google Scholar]
- 35.Sánchez C, Butovich IA, Braña AF, Rohr J, Méndez C, Salas JA. Chem. Biol. 2002;9:519–531. doi: 10.1016/s1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]
- 36.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. J. Natl. Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]