Abstract
Human dilated cardiomyopathy (DCM), a disorder of the cardiac muscle, causes considerable morbidity and mortality and is one of the major causes of sudden cardiac death. Genetic factors play a role in the etiology and pathogenesis of DCM. Disease-associated genetic variations identified to date have been identified in single families or single sporadic patients and explain a minority of the etiology of DCM. We show that a 600-kb region of linkage disequilibrium (LD) on 5q31.2-3, harboring multiple genes, is associated with cardiomyopathy in three independent Caucasian populations (combined P-value = 0.00087). Functional assessment in zebrafish demonstrates that at least three genes, orthologous to loci in this LD block, HBEGF, IK, and SRA1, result independently in a phenotype of myocardial contractile dysfunction when their expression is reduced with morpholino antisense reagents. Evolutionary analysis across multiple vertebrate genomes suggests that this heart failure-associated LD block emerged by a series of genomic rearrangements across amphibian, avian, and mammalian genomes and is maintained as a cluster in mammals. Taken together, these observations challenge the simple notion that disease phenotypes can be traced to altered function of a single locus within a haplotype and suggest that a more detailed assessment of causality can be necessary.
With the availability of a large catalog of single nucleotide polymorphisms (SNP) and high-throughput genotyping technologies, linkage disequilibrium (LD) mapping approaches are now frequently used to dissect the genetic basis of complex diseases. While LD mapping has proven to be effective in the identification of single genes or risk haplotypes for complex diseases (Botstein and Risch 2003), these genetic variants only represent pieces of the mosaic that determines disease. While LD on average extends blocks of ∼60 kb (1–100 kb) in Caucasians (Reich et al. 2001), several regions of the genome are characterized by long-range LD, for example, the peri-HLA region or the cytokine cluster on 5q31 with LD extending >500 kb. In addition, several of these regions have been repeatedly associated with common, complex diseases (Rioux et al. 2001; Dyment et al. 2004). The identification of disease susceptibility within blocks of long-range LD has generally been considered a misfortune, as it hampers molecular identification of the cause of disease.
There are several reasons to consider treating these evolutionarily conserved blocks as functional units, reflecting a more complex, organic, biological module. There is ample evidence that the order of genes along chromosomes in many eukaryotes is nonrandomly distributed (Lee and Sonnhammer 2003; Hurst et al. 2004), with similarities to the widespread, sometimes operon-driven, prokaryotic segregation of clustered genes that represent a functional unit (Lawrence 2002). A notable example is the complete genetic cosegregation of multiple enzymes in an antimicrobial defense pathway in oat plants (Qi et al. 2004). A recent study in yeast revealed that such gene clusters were formed through a set of genomic rearrangements under intense selective pressure (Wong and Wolfe 2005). In humans there is clear evidence for genomic clustering of genes within the same biological pathway (Lee and Sonnhammer 2003). In addition, Conrad and colleagues (Conrad et al. 2006) have shown that, although the extent of LD varies markedly across human populations, considerable sharing of haplotype structure exists in genomic architecture and inferred recombination hot spots generally match across ethnic groups. It is thus conceivable that clusters of genes reside within such conserved haplotype blocks as a consequence of natural selection to preserve them as functionally related units.
Human dilated cardiomyopathy (DCM) is a myocardial disease characterized by dilatation and impaired systolic function of the ventricles. DCM is the single largest cause of heart failure and cardiac transplantation (Towbin and Bowles 2006), with an annual incidence of 5–8 per 100,000 in the United States and in European populations (Karkkainen and Peuhkurinen 2007). The genetic background of DCM is heterogeneous, with both monogenic and multifactorial factors known to contribute to the disease. With respect to the monogenic background, up to 50% of all DCM cases reveal familial aggregation (Grunig et al. 1998), and mutations in >20 genes have been found to cause DCM (Franz et al. 2001; Ahmad et al. 2005). Almost half of these DCM causing mutations are located in genes that encode sarcomere proteins. The main sarcomeric filament proteins, actin (ACTC), myosin (MYH7), and titin (TTN) have been reported to harbor missense mutations (Olson et al. 1998; Kamisago et al. 2000; Daehmlow et al. 2002; Gerull et al. 2002, 2006; Itoh-Satoh et al. 2002; Karkkainen et al. 2004; Villard et al. 2005) and are restricted to a few families. In addition, DCM-causing mutations have been identified in genes encoding cytoskeletal proteins, nuclear proteins, ion channel proteins, and genes that regulate Ca2+ metabolism (Karkkainen and Peuhkurinen 2007). Mutations in the MYH7 gene were reported to account for ∼10% of DCM cases in a French study sample (Villard et al. 2005). Likewise, mutations in the lamin A/C gene (LMNA), encoding a matrix protein of the nuclear lamina, can be found in 5%–9% of DCM patients, depending on the study population (van Berlo et al. 2005). Taken together, known monogenic disease alleles account for 10%–20% of the cases across studies. Little is known about the polygenic basis of this complex disease. With the exception of association studies that identify candidate genes implicated in myocardial function, such as the alpha2C-adrenergic receptor (Regitz-Zagrosek et al. 2006) and the endothelin-A receptor gene (Herrmann et al. 2001), comprehensive association studies that study the complex genetic basis of DCM in a case-control setting are lacking.
There is accumulating evidence that inflammatory and autoimmune mechanisms may play a role in this idiopathic disease (Takeda 2003). For example, organ-specific auto-antibodies, inflammatory infiltrates, and proinflammatory cytokines have been observed in DCM patients (Maisch et al. 2005). In addition, increased levels of cytokines such as tumor necrosis factor have been shown to impair myocardial contractility (Matsumori 1996). Therefore, investigation of the inflammatory genetic background in DCM patients is a promising target for the identification of new susceptibility genes and new insights in disease pathogenesis.
In this study, we have identified a genomic region that harbors risk alleles for DCM via association studies coupled with a candidate gene approach enriched in proinflammatory mediators. We also provide some supporting evidence that multiple loci within the identified region may play a role in the disease and overall cardiac function. We identified a 600-kb LD block on chromosome 5q31.2-3, which harbors 16 genes and a gene cluster to be associated with cardiomyopathy in three independent human study samples. The association study is complemented by functional studies in zebrafish (Danio rerio), where cardiac phenotypes can be readily assessed through direct monitoring of the heart in the living animal (Driever and Fishman 1996). Gene knockdown experiments identified three genes orthologous to loci within this human region that cause a distinct cardiac phenotype in the zebrafish model. Comparative genomic analysis of the region across fish, amphibians, birds, and mammals provides evidence that the cluster was organized and became coexpressed during the course of mammalian evolution.
Results and Discussion
Exploration of the proinflammatory genetic background of DCM: Replicated association of a SNP in the 5q31.3 genomic region
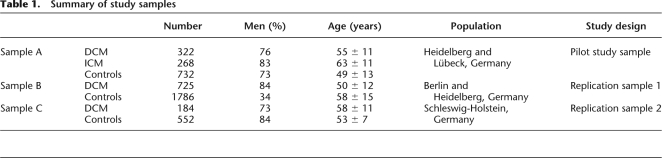
In a pilot study, we used a candidate gene approach to investigate the proinflammatory genetic background in a cardiomyopathy study sample comprising 590 patients (322 DCM patients, 268 ischemic cardiomyopathy [ICM] patients) and 732 healthy controls (Table 1, sample A) to identify common variants that are associated with myocardial dysfunction. Seventy-seven SNPs in 30 candidate genes (Supplemental Table 1), most of which orchestrate the inflammatory response, were selected for genotyping. Eight candidate genes were identified by association, most of which were exclusively associated with DCM (Supplemental Table 2). Independent replication studies were conducted and restricted to study samples consisting of DCM patients (Table 1, sample B with 725 DCM patients and 1786 controls, and sample C with 184 DCM patients and 552 controls) to validate the initial association signals. Only one SNP, rs2569193, located at the CD14 locus in a 600-kb block of long-range LD on chromosome 5q31.2-3 (www.hapmap.org), was consistently replicated and is associated with DCM across the three populations. Carriership for one or two copies of the minor allele A of rs2569193 (dominant inheritance model) reduced the risk for DCM (pilot study: odds ratio [OR] = 0.73, 95% confidence interval [CI] = 0.55–0.96, P = 0.024; first replication: OR = 0.81, 95% CI = 0.66–0.99, P = 0.039; second replication: OR = 0.64, 95% CI = 0.45–0.91, P = 0.012). The combined P-value for the repeated independent associations (n = 3) with DCM was 0.00087.
Table 1.
Summary of study samples
Fine-mapping of the 5q31.2-3 genomic region reveals one associated haplotype block harboring 16 genes and one gene cluster
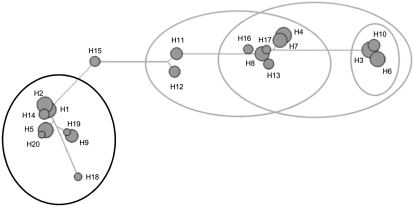
We genotyped 109 additional SNPs in the region of rs2569193 across the pilot study sample (Table 1, sample A) to (1) fine-map the underlying genomic region, (2) delineate the underlying haplotypes, and (3) define the most informative haplotype tagging SNPs for downstream analyses. These SNPs were selected from the HapMap project (www.hapmap.org) to capture the genetic variation within and beyond the main LD block, as well as known coding and promoter SNPs from genes in the region (Supplemental Table 3). These analyses confirmed a main LD block that spans 600 kb and harbors 16 genes and the protocadherin cluster (Supplemental Fig. 1, Supplemental Table 4). The two adjacent LD blocks were well separated by substantial recombination (Supplemental Fig. 1A). The association signal was confined to the main block, hereafter referred to as the CM cluster (Table 1; Supplemental Table 3). The CM cluster comprises 20 individual haplotypes with moderate to low frequencies but no common haplotype (Supplemental Fig. 1B), and thus exhibits a high degree of diversity. To capture the essential genetic variation in this LD block for further analyses we grouped the haplotypes according to their evolutionary relationship. Evolutionary relationships between the haplotypes were reconstructed using a median-joining network (Bandelt et al. 1999), a phylogenetic algorithm for recombination-free intraspecific data. Overall, the median-joining network placed the haplotypes into two branches, each of which accounts for about 50% of the haplotypes (Fig. 1). Thus, the identified haplotype groups are frequent in the general population and may explain a large proportion of the genetic variance contributing to DCM, not accounted for by the known genes of relevance in monogenic DCM.
Figure 1.
Median-joining network of the CM-cluster haplotypes. The tree shows a potential evolutionary path for the haplotypes from Supplemental Figure 1B. Circles represent distinct haplotypes and are scaled to approximate haplotype frequencies. Bars indicate the mutational distance between the haplotypes. The mutational distance between two sequences is defined as the fraction of sites where the residues differ in the alignment of these two sequences. In the case of haplotypes, this is the number of SNP sites that harbor not the same allelic variant on both haplotypes. The ellipses in gray mark the group of protective haplotypes (defined by SNP rs2569193, rs2240695, rs17286676), whereas the ellipse in black summarizes the risk associated group of haplotypes (defined by SNP rs6879217).
Based on the median-joining network, we selected four SNPs representing groups of evolutionary related haplotypes (Supplemental Fig. 1B; Fig. 1) for further analyses. First, the C allele of rs6879217 tags a haplotype group exhibiting low mutational distance, indicating that this group of haplotypes diverges at a few SNPs only and that the comprised haplotypes are closely related. Logistic regression analysis revealed that this haplotype group modestly increases disease risk when two haplotypes of this group are present in one individual (recessive inheritance model for the C allele of rs6879217: OR = 1.43, 95% CI = 1.02–2.00, P = 0.038). To analyze the remaining 50% of haplotypes, we identified three SNPs that tag overlapping groups of this haplotype branch with one of their alleles (A allele of rs17286676, A allele of rs2569193, C allele of rs2240695). The presence of haplotypes from these groups decreases the risk for cardiomyopathy (dominant inheritance model for the A allele of rs17286676: OR = 0.75, 95% CI = 0.57–0.99, P = 0.045; additive inheritance model for the A allele of rs2569193: OR = 0.73, 95% CI = 0.55–0.96, P = 0.024; dominant inheritance model for the C allele of rs2240695: OR = 0.77, 95% CI = 0.55–1.07, P = 0.115 [n.s.], respectively). This implies that this related haplotype group may either harbor protective variants or mirrors the observed overrepresentation of DCM susceptibility haplotypes.
Influence of associated CM-cluster SNPs on cardiac function phenotypes
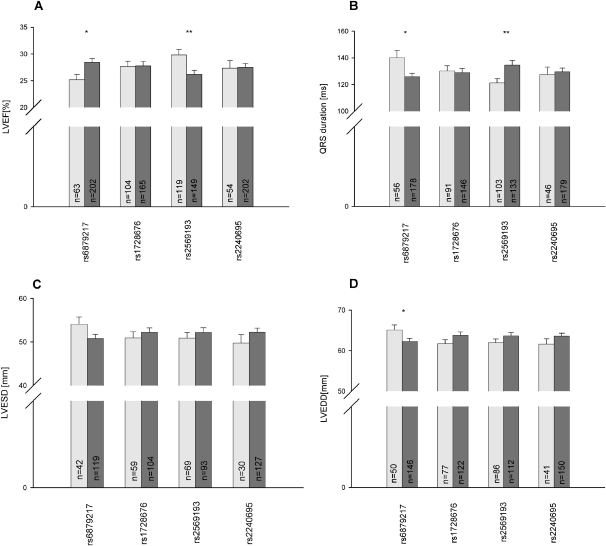
Apart from a general susceptibility to DCM, it is important to consider clinically relevant measures that determine the individual prognosis of DCM patients. Such clinically relevant subphenotypes comprise left ventricular ejection fraction (LVEF), left ventricular end-diastolic diameter (LVEDD), left ventricular end-systolic diameter (LVESD), and duration of the QRS complex, which reflects intraventricular conduction delay and is frequently increased in patients with DCM. Figure 2 shows mean values and standard errors of all four phenotypes in the DCM patients grouped for carriership of each of the haplotype group representative SNPs. For example, homozygous carriership of the minor C allele of rs6879217, which tags the group of DCM risk haplotypes, deteriorates almost all measures of heart function compared with carriers of only one or no copy (LVEF: 25.2 vs. 28.4%, P = 0.030; LVEDD: 65.1 vs. 62.3 mm, P = 0.048; LVESD: 54.1 vs. 50.8 mm, P = 0.086; QRS duration: 140.2 vs. 125.8 msec, P = 0.010). The fraction of the variance attributable to the DCM risk haplotype is estimated at 2%–3% depending on the phenotype (minor allele of rs6879217: LVEF 1.7%; LVEDD 2.0%; LVESD 0.18%; QRS time 2.84%). Notably, a similar difference in LVEF has been shown to translate into marked differences in prognosis in large clinical trials, for example, the Randomized Aldactone Evaluation Study (RALES) trial, where an increase in LVEF of 3% was associated with a reduction of mortality by 30% (Cicoira et al. 2002). Therefore, the presence of the SNP rs6879217, which is associated with a reduction of EF by 3.2%, may be important for the prognosis of DCM patients.
Figure 2.
Influence of associated CM-cluster SNPs on heart function phenotypes. Cardiomyopathy (DCM) patients were stratified into SNP carrier (dark gray bar) and noncarrier (light gray bar) groups. Mean values and standard errors are given for each subgroup for the following heart function phenotypes: left ventricular ejection fraction (LVEF) (A); duration of the QRS complex, a structure on the electrocardiogram that corresponds to the depolarization of the ventricles (QRS) (B); left ventricular end-systolic diameter (LVESD) (C); left ventricular end-diastolic diameter (LVEDD) (D). Student's t-test has been used to compare group means; *P < 0.05; **P < 0.01.
Knockdown of three CM-cluster genes, HBEGF, SRA1, and IK, independently cause heart failure in zebrafish
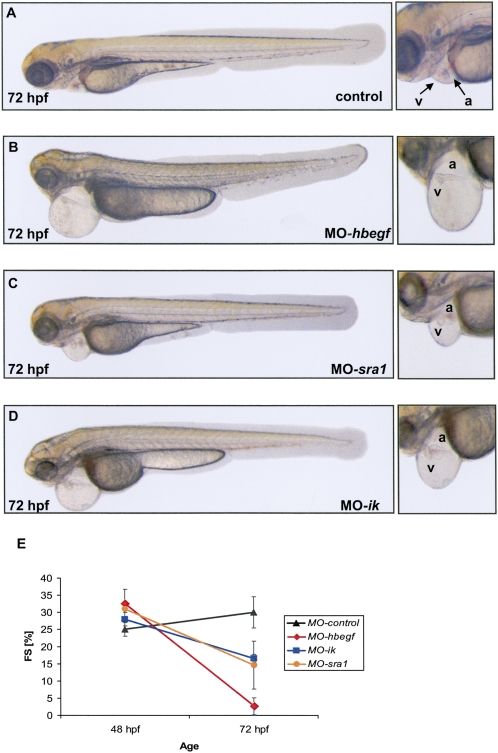
To gain insight into the functional activities of the CM-cluster genes, we assessed the function of their orthologs in zebrafish, an established model organism to study cardiac function in both forward and reverse genetics approaches. We were able to identify orthologous loci and conduct functional knockdown studies in zebrafish for eight genes in the region (orthology predictions for eight other loci as well as the protocadherin cluster could not be obtained). We performed Morpholino (MO) antisense knockdown experiments (Nasevicius and Ekker 2000) for zebrafish orthologous of HBEGF, ANKHD1, EIF4EBP3, SRA1, IK, WDR55, DND1, and ZMAT2 (see Supplemental Table 5A). Knockdown of three genes, HBEGF (heparin-binding epidermal growth factor [EGF]-like growth factor), IK (IK) cytokine, and SRA1 (steroid receptor RNA activator 1) independently resulted in impaired cardiac function phenotypes, with impaired contractility predominantly in ventricular heart chambers at 72 hr post fertilization (hpf) (Fig. 3; Supplemental Table 5B; Supplemental Movies 1–8). As observed in other zebrafish heart failure mutants (Rottbauer et al. 2005), MO-hbegf, MO-sra1, and MO-ik injected embryos also display a pericardial edema at 72 hpf.
Figure 3.
CM-cluster zebrafish knockdown phenotypes. Zebrafish embryos were injected with either control Morpholino (control) (A) or 2 ng of HBEGF Morpholino (MO-hbegf) (B), 4 ng of SRA1 Morpholino (MO-sra1) (C), or 2 ng of IK Morpholino (MO-ik) (D), and images were recorded at 72 hpf with a lateral view (head to the left, tail to the right). In contrast to the control-injected zebrafish embryos, hbegf, sra1, and ik morphants display severe pericardial edema. On the right, corresponding higher magnified views detailing cardiac chamber structure (a, atrium; v, ventricle) are shown for each morphant dysfunction. (E) Fractional shortening (FS) of the ventricular chamber was measured at different time points after injection (48 hpf and 72 hpf). Whereas in control-injected embryos, FS of the ventricle slightly increases from 48 hpf to 72 hpf, a dramatic decrease of FS in hbegf, sra1, and ik morphant ventricles can be observed by 72 hpf.
For one of these genes, the growth factor HBEGF, a role in cardiomyopathies has been established previously, whereas the literature evidence for the role of SRA1 and IK in cardiac function is more novel. HBEGF null mice have been shown to develop severe dilated cardiomyopathy with enlarged ventricular chambers and diminished contractile function due to impaired phosphorylation of ERBB2/B4 tyrosine kinase receptors (Iwamoto et al. 2003). It is also of note that in humans, cardiac side effects are reported in breast cancer patients upon treatment with Herceptin (Force et al. 2007), a monoclonal antibody for the ERBB2 receptor tyrosine kinase, which acts through disruption of EGF ligand signaling (Iwamoto et al. 2003). IK encodes a cytokine known to down-regulate expression of HLA class II antigens (Krief et al. 1994). Down-regulation of HLA class II antigens in CD34+ hematopoietic progenitor cells by IK cytokine has been shown to be a prerequisite for their proliferation and differentiation (Cao et al. 1997). It has been reported that therapeutic administration of CD34+ cells has been shown to improve LVEF after experimental myocardial infarction (Kocher et al. 2001). SRA1 is also known to stimulate proliferation as well as apoptosis in vivo (Lanz et al. 2003). Taken together, all three genes appear to be important for cell proliferation or act as cell survival mediators and provide a novel mechanism contributing to heart failure.
Organization of the CM-cluster genes during vertebrate evolution
Our findings indicate that at least three genes in this disease-associated LD block play important roles in cardiac function and may contribute to disease risk, since targeted intervention of multiple loci within this region yielded cardiovascular phenotypes in the zebrafish model. This result challenges the otherwise reasonable assumption that changes in phenotype or disease risk can be traced to a single functional variant within an allele. To our knowledge, this is the first report on multiple genes causing a severe heart failure phenotype in zebrafish while cosegregating on a single haplotype block in humans. However, the exact polyploid nature and balancing of risk contribution of all genes in the CM cluster requires further in-depth testing.
It is conceivable that clusters of genes reside within such conserved haplotype blocks as a consequence of natural selection to preserve them as a functionally related unit. Several studies addressed the evolution of this gene cluster (Lipovich et al. 2001). The genes HBEGF, SLC4A9, ANKHD1, and FGF1 (which is adjacent to the CM cluster) are located in an ancient gene cluster that was spread across the genome through repeated duplication events. Altogether, seven paralogous EGF-SLC4A-ANK-FGF clusters are present in the human genome. The evolutionary history of these clusters involves multiple rounds of local and genome-wide duplication events. Conrad et al. (2006) showed that the LD block that harbors the CM cluster is largely intact in all four populations (European [CEU], Chinese [CHB], Japanese [JPT], and African [YRI]) of the HapMap database (www.hapmap.org). This supports the notion of considerable sharing of haplotypes and inferred recombination points across ethnicities. Furthermore, the CM cluster is preserved in syntenic blocks on mouse and rat chromosomes 18, implying that the cluster formation probably took place prior to the divergence of humans and rodents.
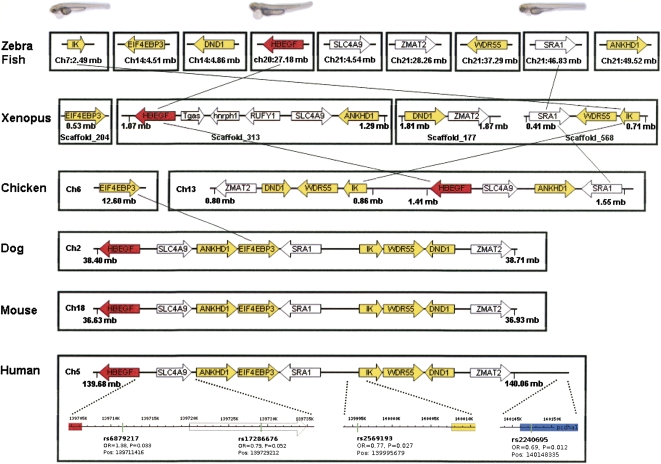
Therefore, we conducted evolutionary analyses to address how these susceptibility genes accumulated in one cosegregating genomic segment. First, we performed a comparative genomics analysis to trace the emergence of the CM cluster across multiple vertebrate genomes (Fig. 4). The eight orthologs, successfully identified in the zebrafish genome (excluding 8 additional loci and the protocadherins), are not clustered but scattered across chromosomes 7, 14, 20, and 21. First genomic rearrangements become apparent in Xenopus tropicalis, where HBEGF, ANKHD1, and SLC4A9 cocluster onto Scaffold_313, while IK and SRA1 cocluster onto Scaffold_568. In Gallus gallus, most of the CM-cluster genes already aggregate on chromosome 13, with EIF4EBP3 being the only ortholog not yet integrated. In Canis familiaris, EIF4EBP3 is finally integrated into the cluster, and the cluster is then preserved in the mammalian lineage. Notably, these genomic rearrangements coincide with the evolution of heart anatomy, from the two-chamber fish heart to the three-chamber amphibian heart to the four-chamber avian heart (Fishman and Olson 1997). It is conceivable that the CM cluster emerged as a consequence of selective pressure challenged by these anatomical changes. Similar adaptive genomic rearrangements were observed in the evolution of yeast strains, where the birth of a metabolic gene cluster coincided with a biochemical reorganization of functionally associated genes under a strong selective pressure (Wong and Wolfe 2005).
Figure 4.
Organization of the CM-cluster genes during vertebrate evolution. CM-cluster orthologs were mapped to their chromosomal positions using ENSEMBL and BLAST analysis. Gene size and chromosome length were not drawn to scale. Colored genes were analyzed for expression coregulation; genes illustrated in the same color were found to be coordinately expressed (Supplemental Table 6). Heart failure phenotypes in zebrafish are shown on the top, and SNPs associated with human cardiomyopathy at the bottom.
In silico analysis has also demonstrated that HBEGF and SLC4A9 may share a bidirectional promoter, further supporting the notion that clustered genes might be coordinately regulated (Lipovich et al. 2001). Therefore, we next performed an mRNA correlation study via K-nearest neighbor (KNN) analysis (Massart et al. 1988) across a large subset of the National Center for Biotechnology Information (NCBI) GEO (Gene Expression Omnibus) database. KNN analysis for all genes residing within the CM cluster, with the exception of SRA1, which is not present on the Affymetrix HG-U133A chip, revealed a significant degree of mRNA expression coregulation of IK with four additional genes in the region: ANKHD1, EIF4EBP3, WDR55, and DND1 (Supplemental Table 6). The correlated expression of CM-cluster genes adds further evidence to support the conservation and function of this region as a unit.
Our findings support the notion that the emergence and maintenance of LD in the genome can reflect clusters of functionally cooperating genes, which jointly determine a complex trait. These types of clusters have been well documented in human metabolic pathways (Lee and Sonnhammer 2003). The conservation of some recombination “hot spots” across diverse human populations also supports this type of selection (Guryev et al. 2006). Our observations may be of general importance and suggest that areas of LD identified as contributing to complex disease risk will need to be rigorously evaluated to understand the functional role each gene may play in determining risk. Finally, a joint examination of genes in such clusters may more holistically identify dysregulated pathways as well as shed light on the general architecture and function of the genome.
Methods
Study samples
Patients were referred for cardiac evaluation to the Divisions of Cardiology at the German Heart Institute Berlin, Germany; the University Hospital of Lübeck, Germany; the University Hospital of Heidelberg, Germany; and the University Hospital of Kiel, Germany. Only patients with DCM according to World Health Organization (WHO) criteria and definitions of cardiomyopathies (Richardson et al. 1996) were enrolled in the study. Coronary angiography and left heart catheterization were used to identify potential study subjects exhibiting a marked reduction of left ventricular ejection fraction (LVEF) ≤50% unexplained by coronary artery disease (maximal luminal stenosis of 50% or less of an epicardial coronary artery). Patients with hypertensive heart disease, congenital or valvular heart disease, primary pulmonary hypertension, as well as inflammatory or metabolic (e.g., thyroid dysfunction, electrolyte disturbance) heart disease were excluded. At the time of entry into the study, the patients' past and current medical history, current medications, symptoms, and physical examination findings were recorded. Baseline variables included New York Heart Association (NYHA) functional class, routine blood chemistry, and 12-lead electrocardiogram (ECG). Patients were scheduled for regular clinical follow-up evaluations every six months in the ambulatory care center. Two-dimensional transthoracic echocardiograms were recorded on videotapes and analyzed off-line by an independent observer. Left atrial size, LVEDD, and LVESD were measured from M-mode tracings of parasternal long- and short-axis views. LVEF was derived from apical two- and four-chamber views according to the modified Simpson rule (Schiller et al. 1989). All measurements were performed in triplicate and averaged. The study was approved by the local ethics committees. All patients gave written informed consent. The investigation conformed to the principles outlined in the Declaration of Helsinki.
Control individuals were randomly identified through official population registries of the county of Schleswig-Holstein in northern Germany. They were screened by use of a health questionnaire and recruited through the population-based PopGen Biobank (Krawczak et al. 2006). The PopGen Catchment Area, Northern Schleswig-Holstein, is home to ∼1.1 million people. There is no historical, demographic, or genetic evidence suggesting that etiological factors relevant in the present context differ substantially between this and other regions of Germany (Steffens et al. 2006).
SNP selection
Candidate gene approach
The 77 SNPs for the primary candidate gene approach were selected in 2002, based on the information on SNP variation in the human genome at that time, to tag our candidate genes, which were selected as likely pathophysiological contributors to cardiomyopathies.
Fine mapping of 5q31 genomic region
Haplotype tagging SNPs were selected from the CEPH (Centre d'Etude du Polymorphisme Humain) population of the HapMap project (www.hapmap.org). Coding and promoter SNPs were selected using SNPper database (http://snpper.chip.org/) and NCBI database (http://www.ncbi.nlm.nih.gov/SNP/). We selected all SNPs within the first 500 bases from the first exon for genotyping as these might be possible promoter SNPs. Chromosome positions for SNPs refer to the reference assembly as annotated in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/).
SNP genotyping
The pilot study and the replication were performed by standard TaqMan allelic discrimination method using an ABI 7900 sequence detector (Applied Biosystems). For high-density LD mapping of the CM cluster we used the SNPlex Genotyping System (Applied Biosystems). In brief, DNA samples were evaluated before genotyping by gel electrophoresis for the presence of high-molecular-weight DNA and adjusted to 20–30 ng/μL DNA content using the Picogreen fluorescent dye (Molecular Probes–Invitrogen). One microliter of genomic DNA was amplified by the GenomiPhi (Amersham) whole-genome amplification system and fragmented at 99°C for 5 min. One hundred nanograms of DNA were dried overnight in TwinTec hardshell 384-well plates (Eppendorf) at room temperature. Genotyping was performed with these plates using an automated platform, employing TECAN Freedom EVO and 96-well and 384-well TEMO liquid handling robots (TECAN). Genotypes were generated by automatic calling using the Genemapper 4.0 software (Applied Biosystems). All genotypes were additionally reviewed manually and call rates >95% required. All process data were logged into, and administered by, a database-driven LIMS 58. Genotyping results obtained with the SNPlex system were confirmed on the genotyping platform by random choice using a TaqMan assay (Applied Biosystems), which resulted in 99.8% genotype concordance, thus excluding artifacts due to technological problems.
Statistical analysis
Hardy–Weinberg equilibrium in cases and controls was tested using the exact test as implemented in the program Haploview 3.2 (Barrett et al. 2005) at a significance level of 0.05. LD between two markers, as well as inference of haplotypes, was also calculated with Haploview 3.2. The Network 4.1 program (Bandelt et al. 1999) was used to reconstruct haplotype phylogenies. Logistic regression was used to model the relationship between a binary outcome variable and one or more predictor variables. All ORs for genotype variables refer to the minor allele of the SNP and were adjusted for age and gender of study participants. Logistic regression analyses were performed using STATA statistical package (version 9); P-values are given according to the Wald statistic. We calculated combined P-values for determining the overall significance of the observed independent association findings using Fisher's method (Fisher 1946). For subphenotype analyses, mean values of the quantitative phenotypes in patient groups stratified according to their genetic risk were compared using two-tailed unpaired Student's two-sample t-test. Since there is no consensus on what represents a sufficiently conservative P-value for studies that test multiple hypotheses, and since the tested SNPs are in LD and the clinical subphenotypes are correlated, P-values from the t-test were not adjusted for multiple testing. Instead, we performed two independent replication studies, since we believe that replication of results is a powerful argument toward the validity of a finding.
Identification of orthologs
5q31.2-3 CM-cluster orthologs were identified by performing TBLASTN analysis against genomic databases of Danio rerio, Xenopus tropicalis, Gallus gallus, Canis familiaris, Mus musculus, and Homo sapiens (The Institute for Genomic Research, NCBI, Sanger), as well as comparative genomics analysis using Ensembl databases. Genes with the best sequence identity to human proteins (minimum BLAST2 alignment of 55% positives on the amino acid level and minimum score of 100 bits) were considered orthologs.
Morpholino injection procedures
Total amount of 2, 4, and 8 ng of Morpholino modified antisense oligonucleotides (Gene-Tools) complementary to the translational start site or splice site of the zebrafish orthologs (Supplemental Table 5A) and control Morpholino were microinjected into the yolks of one- to two-cell stage wild-type embryos. For control injections the standard Gene-Tools control Morpholino (CCTCTTACCTCAGTTACAATTTATA) was used. Phenotypes were examined by digital photo and video imaging at 24, 48, and 72 hpf.
Methods for correlated expression calculation (KNN)
We collected 393 human tissue and cell line samples from 21 different projects within the NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) utilizing the Affymetrix HG-U133A chip (Affymetrix Inc.). Primary tumor samples were excluded from the collection to minimize the impact of genetic and epigenetic changes specific to cancer on the results. The Affymetrix CEL files were analyzed with the MAS 5.0 algorithm from Affymetrix and standardized using quantile normalization (Bolstad et al. 2003), making use of the publicly available Bioconductor tools (www.bioconductor.org). To perform the expression correlation search, these GEO data were further standardized by converting each probe's intensity levels across all samples within a GEO project into a standard distribution. Euclidean distances between all probes in the 393-dimensional sample space were then calculated. Neighbors of a query probe were reported in units of standard deviation from the mean distance of the query probe to others. All values were multiplied by −1.
Acknowledgments
This work was mainly supported by grant 01GS0420 (project NHK-S19T12) of the National Genome Research Network (www.ngfn.de) funded by the Bundesministerium für Bildung und Forschung (BMBF), Germany. We are indebted to Jens Boos for his assistance in the recruitment of patients and to Stefan Kirov, Milan Hiersche, and Christoph Preuss for their assistance in bioinformatics analyses concerning the conservation of the CM cluster. We are grateful to Howard J. Jacob for the helpful suggestions and critical appraisal of the paper.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.076653.108.
References
- Ahmad F., Seidman J.G., Seidman C.E. The genetic basis for cardiac remodeling. Annu. Rev. Genomics Hum. Genet. 2005;6:185–216. doi: 10.1146/annurev.genom.6.080604.162132. [DOI] [PubMed] [Google Scholar]
- Bandelt H.J., Forster P., Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Botstein D., Risch N. Discovering genotypes underlying human phenotypes: Past successes for mendelian disease, future approaches for complex disease. Nat. Genet. 2003;33(Suppl.):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- Cao L.X., Le Bousse-Kerdiles M.C., Clay D., Oshevski S., Jasmin C., Krief P. Implication of a new molecule IK in CD34+ hematopoietic progenitor cell proliferation and differentiation. Blood. 1997;89:3615–3623. [PubMed] [Google Scholar]
- Cicoira M., Zanolla L., Rossi A., Golia G., Franceschini L., Brighetti G., Marino P., Zardini P. Long-term, dose-dependent effects of spironolactone on left ventricular function and exercise tolerance in patients with chronic heart failure. J. Am. Coll. Cardiol. 2002;40:304–310. doi: 10.1016/s0735-1097(02)01965-4. [DOI] [PubMed] [Google Scholar]
- Conrad D.F., Jakobsson M., Coop G., Wen X., Wall J.D., Rosenberg N.A., Pritchard J.K. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- Daehmlow S., Erdmann J., Knueppel T., Gille C., Froemmel C., Hummel M., Hetzer R., Regitz-Zagrosek V. Novel mutations in sarcomeric protein genes in dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2002;298:116–120. doi: 10.1016/s0006-291x(02)02374-4. [DOI] [PubMed] [Google Scholar]
- Driever W., Fishman M.C. The zebrafish: Heritable disorders in transparent embryos. J. Clin. Invest. 1996;97:1788–1794. doi: 10.1172/JCI118608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyment D.A., Ebers G.C., Sadovnick A.D. Genetics of multiple sclerosis. Lancet Neurol. 2004;3:104–110. doi: 10.1016/s1474-4422(03)00663-x. [DOI] [PubMed] [Google Scholar]
- Fisher R.A. Statistical methods for research workers. Oliver and Boyd; London, UK: 1946. [Google Scholar]
- Fishman M.C., Olson E.N. Parsing the heart: Genetic modules for organ assembly. Cell. 1997;91:153–156. doi: 10.1016/s0092-8674(00)80397-9. [DOI] [PubMed] [Google Scholar]
- Force T., Krause D.S., Van Etten R.A. Molecular mechanisms of cardiotoxicity of tyrosine kinase inhibition. Nat. Rev. Cancer. 2007;7:332–344. doi: 10.1038/nrc2106. [DOI] [PubMed] [Google Scholar]
- Franz W.M., Muller O.J., Katus H.A. Cardiomyopathies: From genetics to the prospect of treatment. Lancet. 2001;358:1627–1637. doi: 10.1016/S0140-6736(01)06657-0. [DOI] [PubMed] [Google Scholar]
- Gerull B., Gramlich M., Atherton J., McNabb M., Trombitas K., Sasse-Klaassen S., Seidman J.G., Seidman C., Granzier H., Labeit S., et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002;30:201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- Gerull B., Atherton J., Geupel A., Sasse-Klaassen S., Heuser A., Frenneaux M., McNabb M., Granzier H., Labeit S., Thierfelder L. Identification of a novel frameshift mutation in the giant muscle filament titin in a large Australian family with dilated cardiomyopathy. J. Mol. Med. 2006;84:478–483. doi: 10.1007/s00109-006-0060-6. [DOI] [PubMed] [Google Scholar]
- Grunig E., Tasman J.A., Kucherer H., Franz W., Kubler W., Katus H.A. Frequency and phenotypes of familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 1998;31:186–194. doi: 10.1016/s0735-1097(97)00434-8. [DOI] [PubMed] [Google Scholar]
- Guryev V., Smits B.M.G., van de Belt J., Verheul M., Hubner N., Cuppen E. Haplotype structure is conserved across mammals. PLoS Genet. 2006;2:e121. doi: 10.1371/journal.pgen.0020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann S., Schmidt-Petersen K., Pfeiffer J., Perrot A., Bit-Avragim N., Eichhorn C., Dietz R., Kreutz R., Paul M., Osterziel K.J. A polymorphism in the endothelian-A receptor genes predicts survival in patients with idiopathic dilated cardiomyopathy. Eur. Heart J. 2001;20:1948–1953. doi: 10.1053/euhj.2001.2626. [DOI] [PubMed] [Google Scholar]
- Hurst L.D., Pal C., Lercher M.J. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Itoh-Satoh M., Hayashi T., Nishi H., Koga Y., Arimura T., Koyanagi T., Takahashi M., Hohda S., Ueda K., Nouchi T., et al. Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2002;291:385–393. doi: 10.1006/bbrc.2002.6448. [DOI] [PubMed] [Google Scholar]
- Iwamoto R., Yamazaki S., Asakura M., Takashima S., Hasuwa H., Miyado K., Adachi S., Kitakaze M., Hashimoto K., Raab G., et al. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc. Natl. Acad. Sci. 2003;100:3221–3226. doi: 10.1073/pnas.0537588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisago M., Sharma S.D., DePalma S.R., Solomon S., Sharma P., McDonough B., Smoot L., Mullen M.P., Woolf P.K., Wigle E.D., et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N. Engl. J. Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- Karkkainen S., Peuhkurinen K. Genetics of dilated cardiomyopathy. Ann. Med. 2007;39:91–107. doi: 10.1080/07853890601145821. [DOI] [PubMed] [Google Scholar]
- Karkkainen S., Helio T., Jaaskelainen P., Miettinen R., Tuomainen P., Ylitalo K., Kaartinen M., Reissell E., Toivonen L., Nieminen M.S., et al. Two novel mutations in the β-myosin heavy chain gene associated with dilated cardiomyopathy. Eur. J. Heart Fail. 2004;6:861–868. doi: 10.1016/j.ejheart.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Kocher A.A., Schuster M.D., Szabolcs M.J., Takuma S., Burkhoff D., Wang J., Homma S., Edwards N.M., Itescu S. Neovascularization of ischemic myocardium by human bone-marrow-derived angioblasts prevents cardiomyocyte apoptosis, reduces remodeling and improves cardiac function. Nat. Med. 2001;7:430–436. doi: 10.1038/86498. [DOI] [PubMed] [Google Scholar]
- Krawczak M., Nikolaus S., von Eberstein H., Croucher P.J., El Mokhtari N.E., Schreiber S. PopGen: Population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- Krief P., Augery-Bourget Y., Plaisance S., Merck M.F., Assier E., Tanchou V., Billard M., Boucheix C., Jasmin C., Azzarone B. A new cytokine (IK) down-regulating HLA class II: Monoclonal antibodies, cloning and chromosome localization. Oncogene. 1994;9:3449–3456. [PubMed] [Google Scholar]
- Lanz R.B., Chua S.S., Barron N., Soder B.M., DeMayo F., O'Malley B.W. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol. Cell. Biol. 2003;23:7163–7176. doi: 10.1128/MCB.23.20.7163-7176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.G. Shared strategies in gene organization among prokaryotes and eukaryotes. Cell. 2002;110:407–413. doi: 10.1016/s0092-8674(02)00900-5. [DOI] [PubMed] [Google Scholar]
- Lee J.M., Sonnhammer E.L. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 2003;13:875–882. doi: 10.1101/gr.737703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovich L., Lynch E.D., Lee M.K., King M.C. A novel sodium bicarbonate cotransporter-like gene in an ancient duplicated region: SLC4A9 at 5q31. Genome Biol. 2001;2:RESEARCH0011. doi: 10.1186/gb-2001-2-4-research0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisch B., Richter A., Sandmoller A., Portig I., Pankuweit S. Inflammatory dilated cardiomyopathy (DCMI) Herz. 2005;30:535–544. doi: 10.1007/s00059-005-2730-5. [DOI] [PubMed] [Google Scholar]
- Massart D.L., Vandeginste B.G.M., Deming S.N., Michotte Y., Kaufman L. Chemometrics: A Textbook (Data Handling in Science and Technology) Vol. 2. Elsevier; New York: 1988. The K-nearest neighbor method; pp. 395–397. [Google Scholar]
- Matsumori A. Cytokines in myocarditis and cardiomyopathies. Curr. Opin. Cardiol. 1996;11:302–309. doi: 10.1097/00001573-199605000-00011. [DOI] [PubMed] [Google Scholar]
- Nasevicius A., Ekker S.C. Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Olson T.M., Michels V.V., Thibodeau S.N., Tai Y.S., Keating M.T. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- Qi X., Bakht S., Leggett M., Maxwell C., Melton R., Osbourn A. A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. 2004;101:8233–8238. doi: 10.1073/pnas.0401301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regitz-Zagrosek V., Hocher B., Bettmann M., Brede M., Hadamek K., Gerstner C., Lehmkuhl H.B., Hetzer R., Hein L. α2C-Adrenoceptor polymorphism is associated with improved event-free survival in patients with dilated cardiomyopathy. Eur. Heart J. 2006;27:454–459. doi: 10.1093/eurheartj/ehi659. [DOI] [PubMed] [Google Scholar]
- Reich D.E., Cargill M., Bolk S., Ireland J., Sabeti P.C., Richter D.J., Lavery T., Kouyoumjian R., Farhadian S.F., Ward R., et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Richardson P., McKenna W., Bristow M., Maisch B., Mautner B., O'Connell J., Olsen E., Thiene G., Goodwin J., Gyarfas I., et al. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation. 1996;93:841–842. doi: 10.1161/01.cir.93.5.841. [DOI] [PubMed] [Google Scholar]
- Rioux J.D., Daly M.J., Silverberg M.S., Lindblad K., Steinhart H., Cohen Z., Delmonte T., Kocher K., Miller K., Guschwan S., et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat. Genet. 2001;29:223–228. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- Rottbauer W., Just S., Wessels G., Trano N., Most P., Katus H.A., Fishman M.C. VEGF-PLCγ1 pathway controls cardiac contractility in the embryonic heart. Genes & Dev. 2005;19:1624–1634. doi: 10.1101/gad.1319405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller N.B., Shah P.M., Crawford M., DeMaria A., Devereux R., Feigenbaum H., Gutgesell H., Reichek N., Sahn D., Schnittger I., et al. Recommendations for quantitation of the left ventricle by two-dimensional echocardiography. American Society of Echocardiography Committee on Standards, Subcommittee on Quantitation of Two-Dimensional Echocardiograms. J. Am. Soc. Echocardiogr. 1989;2:358–367. doi: 10.1016/s0894-7317(89)80014-8. [DOI] [PubMed] [Google Scholar]
- Steffens M., Lamina C., Illig T., Bettecken T., Vogler R., Entz P., Suk E.K., Toliat M.R., Klopp N., Caliebe A., et al. SNP-based analysis of genetic substructure in the German population. Hum. Hered. 2006;62:20–29. doi: 10.1159/000095850. [DOI] [PubMed] [Google Scholar]
- Takeda N. Cardiomyopathy: Molecular and immunological aspects (review) Int. J. Mol. Med. 2003;11:13–16. [PubMed] [Google Scholar]
- Towbin J.A., Bowles N.E. Dilated cardiomyopathy: A tale of cytoskeletal proteins and beyond. J. Cardiovasc. Electrophysiol. 2006;17:919–926. doi: 10.1111/j.1540-8167.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- van Berlo J.H., de Voogt W.G., van der Kooi A.J., van Tintelen J.P., Bonne G., Yaou R.B., Duboc D., Rossenbacker T., Heidbuchel H., de Visser M., et al. Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: Do lamin A/C mutations portend a high risk of sudden death? J. Mol. Med. 2005;83:79–83. doi: 10.1007/s00109-004-0589-1. [DOI] [PubMed] [Google Scholar]
- Villard E., Duboscq-Bidot L., Charron P., Benaiche A., Conraads V., Sylvius N., Komajda M. Mutation screening in dilated cardiomyopathy: Prominent role of the beta myosin heavy chain gene. Eur. Heart J. 2005;26:794–803. doi: 10.1093/eurheartj/ehi193. [DOI] [PubMed] [Google Scholar]
- Wong S., Wolfe K.H. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat. Genet. 2005;37:777–782. doi: 10.1038/ng1584. [DOI] [PubMed] [Google Scholar]