Abstract
Poliovirus initiates infection by binding to its cellular receptor (Pvr). We have studied this interaction by using cryoelectron microscopy to determine the structure, at 21-Å resolution, of poliovirus complexed with a soluble form of its receptor (sPvr). This density map aided construction of a homology-based model of sPvr and, in conjunction with the known crystal structure of the virus, allowed delineation of the binding site. The virion does not change significantly in structure on binding sPvr in short incubations at 4°C. We infer that the binding configuration visualized represents the initial interaction that is followed by structural changes in the virion as infection proceeds. sPvr is segmented into three well-defined Ig-like domains. The two domains closest to the virion (domains 1 and 2) are aligned and rigidly connected, whereas domain 3 diverges at an angle of ≈60°. Two nodules of density on domain 2 are identified as glycosylation sites. Domain 1 penetrates the “canyon” that surrounds the 5-fold protrusion on the capsid surface, and its binding site involves all three major capsid proteins. The inferred pattern of virus–sPvr interactions accounts for most mutations that affect the binding of Pvr to poliovirus.
To initiate infection, an animal virus must first attach to its cell-surface receptor and then deliver its nucleic acid to the appropriate cellular compartment. The study of viruses and their receptors has revealed that, in some cases, the receptor is simply a “hook” that concentrates virus on the cell surface, and genome release or “uncoating” is subsequently triggered by low pH or proteinases (1). For other viruses, the receptor is also an “unzipper,” in that it initiates conformational changes in the virion that prime it for uncoating. Poliovirus falls in the latter category. Its receptor (Pvr, also known as CD155) is a member of the Ig superfamily (2). Although the normal cellular function of Pvr is unknown, two close homologs have been shown to be cell-adhesion molecules that localize to adherens junctions (3). These proteins also serve as receptors for herpesviruses (4, 5). Knowledge of the three-dimensional structure of the viral capsid (6, 7) and the availability of cloned DNA encoding Pvr (2) make poliovirus a particularly attractive system for studying the mechanism of cell entry.
The poliovirus capsid is composed of 60 copies of 4 viral proteins (VP1, VP2, VP3, and VP4), arranged with icosahedral symmetry (6). The capsid surface has a corrugated topography: there is a prominent star-shaped peak (or “mesa”) at the 5-fold axis of symmetry, surrounded by a deep depression (the “canyon”), and another protrusion (the “propeller”) at the 3-fold axis. Beneath the canyon is a hydrocarbon-binding pocket (7). When poliovirus attaches to its receptor at physiological temperature, it undergoes a conformational alteration (to the 135S or A-particle state) that has been implicated in cell entry (8, 9).
Several genetic approaches have been used to study the poliovirus–Pvr interaction. Poliovirus mutants were selected for resistance to neutralization with a soluble form of Pvr (sPvr) (10) or for the ability to utilize mutant Pvr (11, 12). Others were generated by site-directed mutagenesis (13). Analysis of these mutants suggests that the principal contact site of Pvr on the capsid is the floor of the canyon, above the hydrocarbon-binding pocket, and on the outer (“south”) rim of the canyon. Other sites that modulate receptor utilization are located at or near the peak of the mesa. Mutagenesis of Pvr DNA has revealed that the binding site for poliovirus is contained in domain 1 of Pvr (d1), the membrane-distal domain (14–16). Mutations in the predicted C′C",CC′, DE, and EF loops, and the C"- and D-strands of this Ig-like domain disrupt virus binding (17–19).
To complement these genetic analyses, we have analyzed the complex of poliovirus type 1 bound to sPvr by cryoelectron microscopy. The resulting reconstruction revealed the manner in which domain 1 of Pvr inserts into the canyon. It also helped guide construction of a homology model of Pvr. This model, together with the known crystal structure of poliovirus (6), was used to identify specific interactions between the virus and receptor.
Materials and Methods
Purification.
Expression and purification of sPvr, a recombinant form of the receptor, will be described in detail elsewhere (B.M., unpublished results). Briefly, DNA encoding the ectodomain of Pvr (residues 1 to 337) was fused to a C-terminal hexahistidine tag and stably expressed in 293T cells. sPvr was purified from cell supernatants by using Ni-agarose resin (Qiagen, Chatsworth, CA) followed by Q-Sepharose (Pharmacia) chromatography. Edman degradation revealed that the N terminus of purified sPvr is amino acid 28 of the unprocessed precursor, as previously reported (20). Virions were purified as described (9).
Cryoelectron Microscopy.
Thin films containing poliovirus complexed with sPvr were suspended over holey carbon films, vitrified, and examined by cryoelectron microscopy, essentially as described (21). Equal volumes of virions at 0.6 mg/ml and sPvr at 0.2 mg/ml were mixed, giving a sPvr-to-binding site ratio of ≈1.6:1 (60 binding sites per virus). Mixing was performed at 4°C to avoid converting virions to 135S particles (22, 23), and the specimens were frozen within 2 min of mixing to minimize aggregation. Focal pairs of micrographs were recorded on a Philips CM200-FEG electron microscope (FEI, Mahwah, NJ), equipped with a model 626 cryoholder (Gatan, Pleasanton, CA).
Image Reconstruction.
Density maps of the virion–sPvr complex were computed essentially as described (9). Particle images from 13 micrographs (six focal pairs plus one single micrograph) were combined. Focal settings ranged from 0.6 to 2.1 μm under focus. Our previous reconstruction of the virion (9) was the starting model for iterative model-based determination of particles' orientations and origins (24). The program ctfmix (25) was used to correct for contrast transfer function effects. After an initial reconstruction was computed from 280 images, we observed that the hand of the capsid had become inverted relative to the starting model. The original handedness was restored, but we also performed another more rigorous test. Orientations and origins were redetermined by using the same starting model but also by evaluating the alternative-handedness solution [corresponding to a 180° rotation of the image (26)] for each particle at each iteration. A particle was approved if its correlation coefficient was significantly higher (by ≥0.050) than that of the alternative-handedness solution (except for 2-fold orientations, which were rejected if the coefficients differed by >0.01). This second reconstruction, computed from 198 images, maintained the correct hand and was nearly identical to the first. The reconstruction presented is the average of these two maps. Resolution was assessed by correlation coefficient and R-factor (27) comparisons of sets of structure factors, each computed from half-data sets. A resolution of 21 Å was obtained by taking a correlation of 0.3 as cutoff (28) (25 Å if cutoff = 0.5). The R-factor criterion gave a similar result. After calibrating the size and density of the virion–sPvr reconstruction to that of the virion by comparing the radially averaged density (29) in the capsid region only, a difference map was computed.
Homology Modeling.
sPvr (residues 28–337) consists of three Ig-like β-sandwich domains: the N-terminal domain (d1) is V type, and d2 and d3 are C type (2). The sequences of all known atomic structures in the Protein Data Bank (PDB) were screened for homology with each of the Pvr domains by using iterated blast searches (30). Even the closest homologs in the PDB had low overall sequence identity (25% for 1NEU, 24% for 1IAO, 25% for 1BIH for d1, d2, and d3, respectively), which was insufficient to generate plausible models automatically. Accordingly, a structural alignment for each domain was constructed manually by superposition of three-dimensional structures (from three to four high-scoring “families” of molecules) and correlation of related sequences. Frequently, Pvr was most homologous to one known structure in one part of the molecule and to a different structure in another. The initial homology model for d1 was a hybrid of PDB entries 1NEU, chain A, which was most similar overall, and 8FAB, chain B, which was more similar around the A-, C-, E-, and G-strands. d2 was based on 1IAO, chain A, in strands A–C and on 1FAI, chain H, in strands D–G. d3 was based on 1BIH. After the domains were positioned in the density map (see below), the FG loop of d1 was shortened by two amino acids, and the G-strand of d1 was extended upward to achieve continuity with d2. Similarly, the N-terminal YYPP sequence of the A-strand of d3 was bent down to connect with the carboxyl end of d2.
Density Fitting.
The density map was modeled by manipulating four α-carbon models as rigid bodies. One of these corresponded to a protomer, consisting of one copy each of the four capsid proteins. Absence of movement of this body during automated refinement confirmed the correct size scale of the reconstruction. The other three bodies represented the domains of Pvr and began as homology models that were later modified slightly in their variable loops to optimize agreement with the density map. Models were fitted to the density by hand using the graphics program frodo (31) and then optimized automatically by using insout (9). Densities within 100.5 Å of the particle center were set to solvent level to mask out RNA-associated density.
During refinement, the agreement between the Fourier transforms of observed and model-based densities was optimized by minimizing the mean |F|-weighted absolute phase discrepancy, <Δφ>, in the resolution range of infinity to 20.2 Å. To ensure chemical plausibility, particularly of the connections between domains, complete atomic models were superimposed on the α-carbon models, adjusted by hand to improve stereochemistry, and energy-minimized together with poliovirus by using x-plor (32).
Results and Discussion
Visualization of sPvr Labeling.
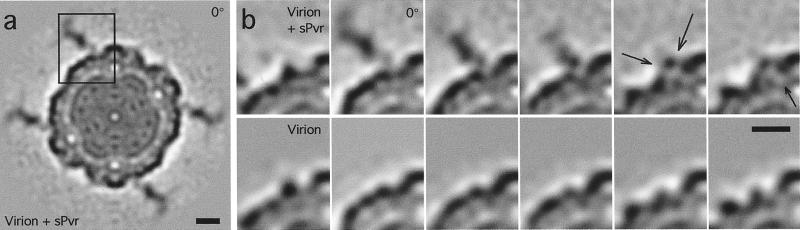
With or without sPvr, the most prominent feature in cryomicrographs of poliovirus is its roundish shape (Fig. 1a). Faint protrusions are discernible on labeled virions and represent attached sPvr molecules. However, the bound receptors are much more conspicuous after image reconstruction (see Fig. 1 b–d), which shows prominent protrusions, ≈115 Å long and 20–60 Å wide, extending outwards at an oblique angle relative to the capsid surface.
Figure 1.
(a) Cryomicrographs of poliovirus particles complexed with (Top) and without (Bottom) sPvr. Bar = 300 Å. Image reconstructions are shown of virion + sPvr [in stereo (b)] and, for comparison, of the virion (c) (from ref. 9). The two reconstructions were overlaid in d with the respective contour levels adjusted to clarify the interaction of sPvr with the virion. Bar = 100 Å. (e) Two views of a single sPvr molecule extracted from the difference map. Domain boundaries are marked. Bar = 25 Å.
Poliovirus Is Essentially Unchanged by sPvr Binding.
A comparison of corresponding sections through the virion–sPvr and virion density maps (Fig. 2) shows that the capsid is largely unchanged on binding receptor under these conditions, i.e., short incubations at 4°C. In particular, the sPvr-bound virion does not show the redistribution of RNA density characteristic of the 135S particle; see Fig. 2a of this paper and Fig. 3a of ref. 9. However, there are az few subtle changes. In particular, there is a small tunnel beneath the receptor-binding site on the floor of the canyon (Figs. 2 and 3b from this paper) that extends into the “pocket-factor” binding site in VP1 (7), possibly indicating that the pocket factor is expelled on Pvr binding.
Figure 2.
(a) A central section through the virion–sPvr reconstruction, normal to a 2-fold symmetry axis. The boxed region is shown in b. High density is shown as dark. The right edge of the box coincides with a 5-fold symmetry axis. (b) The virion–sPvr (Top) and virion (Bottom, from ref. 9) maps were oriented as in a rotated progressively about the 5-fold symmetry axis. (Left to Right) The rotation is −18°, 0°, 6°, 12°, 24°, and 30°. In each case, the boxed portion of the central section was extracted. Arrows indicate sPvr-related density that bridges the canyon (second panel from right) and the solvent-level density corresponding to the tunnel (rightmost panel). Bars = 50 Å.
Figure 3.
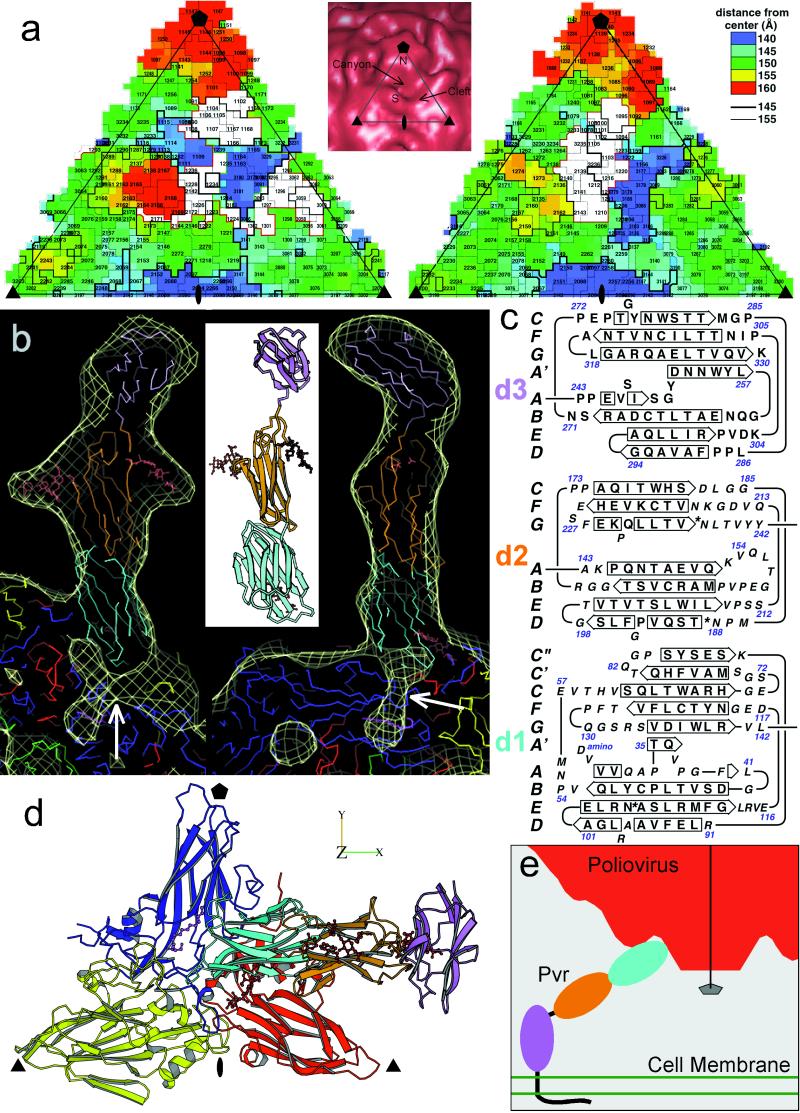
(a) “Road map” representations (36, 37) of poliovirus (Left) and rhinovirus-14 [Right; (33)]. The corresponding triangular area of the capsid surface, bounded by a 5-fold and two 3-fold icosahedral symmetry axes, is marked (Inset). The radial distances of surface residues from the virion center are color coded and contoured [see key (Top Right)]. Nomenclature: 3145, residue 145 of VP3. The receptor footprints are shown in white. (b) A ribbon diagram (38) of the sPvr model is flanked by two views (39) of a single sPvr molecule as portrayed in the cryoelectron microscopy density map (white cage), enclosing the model of the three sPvr domains, d1 (cyan), d2 (orange), and d3 (violet). Carbohydrates attached to d2 [to N188 (Left) and N237 (Right)] and possibly to d1 are shown in brown. Also shown are the capsid proteins VP1 (blue), VP2 (yellow), VP3 (red), and VP4 (green). The tunnel beneath the sPvr-binding site is evident (white arrows). “Pocket factor” is magenta. (c) The sPvr sequence is mapped onto secondary structural elements of the homology model. Asn residues thought to be glycosylated are marked with asterisks. (d) Ribbon diagram (38) showing the docking of the sPvr model onto the capsid surface. Same color conventions as in b. The axes allow this view to be related to Fig. 4. (e) Schematic diagram showing a possible binding configuration of poliovirus with intact membrane-bound Pvr.
Structure of sPvr.
The structural invariance of the virion on receptor binding allowed us to obtain a representation of sPvr from the difference map (Fig. 1e). Three domains are apparent, whose shapes and dimensions are consistent with those of Ig-like domains. We equate them with domains d1, d2, and d3 (2), in order of increasing distance from the capsid surface. d1 makes contact with the capsid. d2 has two prominent nodules and is connected to d1 by a narrow region. The long axes of these two domains are aligned, and they appear to be firmly connected because there is little diminution of density in d2, as would arise from a flexible hinge. The distal domain, d3, is oriented quite differently, with an angle of ≈60° between its long axis and that of d2. Its density is lower (see Fig. 2), suggesting that d2 and d3 may be more loosely joined.
Contact Region on the Virus.
sPvr binds at a glancing angle such that its d1 domain penetrates the canyon and makes contact with the capsid surface near the center of the icosahedral asymmetric unit bounded by a 5-fold axis and two 3-fold axes (Fig. 3a). The major contact points are in a cleft on the “south rim” of the canyon and on the side of the mesa on the “north rim,” so that the receptor appears to bridge the canyon (see Figs. 1 c and d, 2b, and 3a). The binding aspect of sPvr to poliovirus differs substantially from that of the rhinovirus receptor, intercellular adhesion molecule-1, to rhinovirus (33).
By overlaying the virion–sPvr reconstruction and the difference map on the atomic-resolution coordinates of the virion, we found that the sPvr-binding site includes many residues in VP1 (102–108, 166–169, 213–214, 222–236, 293–297, 301–302), a few in VP2 (140–144, 170–172), and several in VP3 (58–62, 93, 182–186). The footprint of sPvr on the capsid surface consists of three distinct patches. Two of them are similar to the footprint of intercellular adhesion molecule-1 on rhinovirus (33); cf. Fig. 3a. The third patch in the southeast corner is unique.
Pseudoatomic Model of sPvr.
The homology models for d1, d2, and d3 were fitted into the reconstruction (Fig. 3b). Because the density map exhibits constrictions between the domains, determining the placement of the domains was mainly a matter of fixing their orientations about their long axes. The d1 model could be fitted into the density map in either of two orientations, 180o apart. One orientation was entirely consistent with mutational data implicating the C′C" and DE loops of Pvr, and the EF (166–169) and GH (213–236) loops of VP1, the EF loop of VP2 (140–144), and the GH loop of VP3 (182–186) as important interaction sites; the other was inconsistent with these data. The orientations of d2 and d3 were unambiguous. β-Strand and loop assignments in the final model (Fig. 3b Inset) are given in Fig. 3c.
The d1 model (residues 29–142) fits the reconstructed density well and exhibits notable complementarity with the virus surface. Adaptation of the initial homology model to fit the density map required major changes in only one large loop (CC′). This loop, which projects laterally from the side and top of the β-sandwich in all close-sequence homologs, would protrude significantly through the envelope in any plausible orientation of d1 unless the loop were folded closer to the body of the domain. There is additional density between the virus surface near the B- and C-strands of VP1 and the FG loop of the receptor as presently modeled, suggesting that the FG loop may adopt a different conformation than in the homologs. There is some extra density between the receptor and the virus surface in the vicinity of N105, which is a potential glycosylation site (Fig. 3 b–d). However, this feature appears too small to accommodate a full-length glycosyl modification. Interestingly, the mutant N105A, which is not glycosylatable at this site, exhibits increased virus binding and infectivity (19). There is no evidence for glycosylation at N120.
The orientation of d2 (residues 143–242) was constrained by several factors. (i) Two of the glycosylation sites should coincide with the prominent nodules that extend laterally on either side of the domain; (ii) the convex shape of one face and concave shape of the opposite face of the domain should match between the density map and the model (see Fig. 3b); (iii) the C terminus of the G-strand should be located near d3. In the preferred orientation, the glycosylation sites at N188 and N237 account for the lateral protrusions. The close proximity of N218 to N237 makes it difficult to ascertain whether N218 is glycosylated as well.
The d3 model (residues 243–330) fits its portion of the envelope well (Fig. 3b), despite the weakness of this density compared with d2 and d1. The orientation of d3 is constrained by the requirements to position its A-strand on the lower surface of d3, and to fit the large CD loop into a bulge on the top surface of the envelope. Thus positioned, the d3 model is clearly shape- similar to its envelope (Fig. 3b). There is no compelling evidence for glycosylation at any of d3's three potential sites.
One implication of the 60° turn between the axes of d2 and d3 is illustrated in Fig. 3e. If d3 were to emerge from the cell membrane with its long axis normal to the plane of membrane, then d1 and d2 would be inclined at an angle of ≈30° relative to this plane. Given the binding aspect of Pvr to the virus (Fig. 1c), this configuration would orient a 5-fold axis perpendicular to the membrane, thus facilitating multiple symmetry-related attachment of receptors around this axis. Moreover, it would bring the mesa at this axis close to the membrane. This juxtaposition is relevant to the proposal that a continuous channel is formed along the 5-fold axis of the capsid and through the membrane, as a mechanism for membrane attachment and RNA release (9, 34).
Overview of the Binding Site.
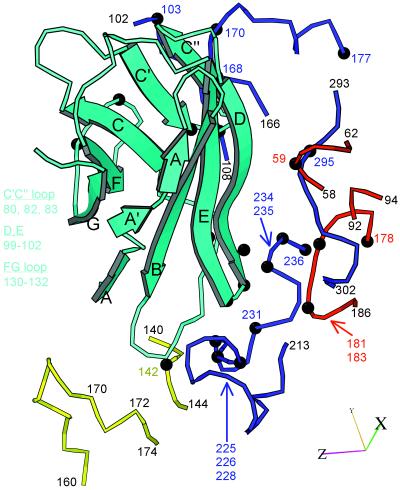
All contacts of the receptor with the virus involve domain d1, which approaches the capsid surface from the right as shown on Fig. 3d (east in Fig. 3a) with its face containing strands A, B, E, and D pointing downward, toward the virus surface. The majority of interactions with the virus involve the BC, C′C", DE, and FG loops (Fig. 4). The BC loop inserts in a groove defined by the smaller EF loop of VP2 (138–142), residues at the distal end of the GH loop of VP1, and residues in the bulge at the C-terminal end of the GH loop of VP1(234–235). The DE loop contacts the residues at the N-terminal end of the GH loop of VP1 (214), the distal end of the GH loop of VP1 (224–226), and the GH loop of the 5-fold related copy of VP3 (182–183). The FG loop contacts the C-strand of VP1, and the C′C" loop of the receptor contacts the EF loop of VP1 (166–169). The latter contacts are the only ones made with residues on the north wall of the canyon.
Figure 4.
Ribbon diagram (38) of the capsid-binding d1 domain of Pvr (cyan), with β-strands labeled, juxtaposed with segments of the capsid proteins with which it is inferred to interact. The viral segments are shown as tubes, with VP1, blue; VP2, yellow; and VP3, red. Residue numbers are provided as landmarks. Black balls and colored numbers denote amino acids implicated by genetic analysis in receptor binding. Similarly, Pvr residues shown by mutation to be important for virus binding are listed (Left). The axes allow this view to be related to Fig. 3d.
In addition to contacts involving loops at the N-terminal end of d1, the N-terminal end of the D-strand and the C-terminal end of the E-strand (which are just below the d1-d2 junction) contact the hairpin “knob” proximal to the B-strand of VP3 (58–60) from the 5-fold related protomer and residues near the C terminus of VP1 (293–297) from the adjacent protomer (Fig. 4). These resides are located in the “southeast” portion of the receptor footprint (Fig. 3a).
Correlation with Mutational Data.
The interactions described above are consistent with mutational analyses (17–19) in that the majority of mutations that affect receptor binding map to the virion–sPvr interface (Fig. 4). Specifically, mutations in the C′C" loop, the C terminus of the D-strand, and the DE and FG loops of the receptor have been associated with alterations in virus binding and the ability of the receptor to support infections (17–19). However, five single-site mutations in the BC loop do not affect binding, perhaps because none of these residues is essential (19). Similarly, several mutations within the receptor footprint on the virus have been shown to alter the ability to be neutralized by sPvr, bind wild-type receptor, or initiate infection with mutated receptors (refs. 10–13; V.R., unpublished data). These include mutations in the C-strand, EF loop, and C terminus of VP1; in the N-terminal end of the smaller EF loop of VP2; and in the β-bend (residues 58–60) and GH loop of VP3.
Not all of the capsid residues identified in the genetic studies cited above are involved in contacts with Pvr. Those that are not—with one exception—are buried in interfaces between subunits or on the inner surface of the capsid. The exception involves mutations in the BC loop of VP1 that have been associated with mouse adaptation, ability to use mutated receptor, and ability to establish persistent infection (reviewed in ref. 1). The BC loop is well outside the footprint of the receptor in the road map (Fig. 3a), and viruses in which it is replaced by a variety of heterologous sequences are viable. Alterations in the BC loop are associated with significant changes in thermal stability and the ability of the virus to undergo receptor-mediated conversions. We therefore suggest that these mutations affect cell-entry steps downstream from receptor binding (8).
A similar explanation has been proposed for mutations in residues that are buried in interfaces, in the drug-binding pocket in VP1, or on the inner surface of the protein shell (8). Indeed, most of these mutations have been shown to alter thermal stability or the ability to undergo receptor-mediated structural alterations. However, mutations in several nonexposed residues (including residues 178 of VP3 and 177, 231, and 241 of VP1) result in significant reductions in affinity for receptor, in competition assays with wild-type virus (10). These data suggest that tight receptor binding may require minor conformational changes in the virion. This suggestion is further supported by the recent demonstration that the ability to bind receptor is ablated by capsid-binding drugs, such as WIN51711, at low temperature but not at 37°C (V.R., unpublished data). The imputed conformational changes may be related to the “breathing” of the virus under physiological conditions (35). Because the complexes studied here were formed by brief incubations at 4°C, we infer that they represent the initial state of receptor binding.
Acknowledgments
We thank Simon Tsang for purified virus and James Conway for helpful suggestions and acknowledge support from National Institutes of Health Grants AI20566 (to J.H.) and AI20017 (to V.R.).
Note added in Proof. After this paper was submitted, a paper was published describing the structures of two human rhinoviruses with two- and five-domain fragments of the rhinovirus receptor, the intercellular adhesion molecule-1 (40).
Abbreviations
- VP1
viral protein 1
- Pvr
poliovirus receptor
- d1
domain 1 of Pvr. Known loops and strands in capsid proteins are denoted by capitals
- putative loops and strands in receptor domains are in bold italic capitals.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Racaniello V R. Structure (London) 1996;4:769–773. doi: 10.1016/s0969-2126(96)00083-4. [DOI] [PubMed] [Google Scholar]
- 2.Mendelsohn C L, Wimmer E, Racaniello V R. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, et al. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 5.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 6.Hogle J M, Chow M, Filman D J. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 7.Filman D J, Syed R, Chow M, Macadam A J, Minor P D, Hogle J M. EMBO J. 1989;8:1567–1579. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wien M W, Chow M, Hogle J M. Structure (London) 1996;4:763–767. doi: 10.1016/s0969-2126(96)00082-2. [DOI] [PubMed] [Google Scholar]
- 9.Belnap D M, Filman D J, Trus B L, Cheng N, Booy F P, Conway J F, Curry S, Hiremath C N, Tsang S K, Steven A C, et al. J Virol. 2000;74:1342–1354. doi: 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colston E, Racaniello V R. EMBO J. 1994;13:5855–5862. doi: 10.1002/j.1460-2075.1994.tb06930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colston E M, Racaniello V R. J Virol. 1995;69:4823–4829. doi: 10.1128/jvi.69.8.4823-4829.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao S, Racaniello V. J Virol. 1997;71:9770–9777. doi: 10.1128/jvi.71.12.9770-9777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harber J, Bernhardt G, Lu H H, Sgro J Y, Wimmer E. Virology. 1995;214:559–570. doi: 10.1006/viro.1995.0067. [DOI] [PubMed] [Google Scholar]
- 14.Koike S, Ise I, Nomoto A. Proc Natl Acad Sci USA. 1991;88:4104–4108. doi: 10.1073/pnas.88.10.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selinka H C, Zibert A, Wimmer E. Proc Natl Acad Sci USA. 1991;88:3598–3602. doi: 10.1073/pnas.88.9.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison M E, Racaniello V R. J Virol. 1992;66:2807–2813. doi: 10.1128/jvi.66.5.2807-2813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison M E, He Y J, Wien M W, Hogle J M, Racaniello V R. J Virol. 1994;68:2578–2588. doi: 10.1128/jvi.68.4.2578-2588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki J, Koiki S, Ise I, Sato-Yoshida Y, Nomoto A. J Biol Chem. 1994;269:8431–8438. [PubMed] [Google Scholar]
- 19.Bernhardt G, Harber J, Zibert A, deCrombrugghe M, Wimmer E. Virology. 1994;203:344–356. doi: 10.1006/viro.1994.1493. [DOI] [PubMed] [Google Scholar]
- 20.Bibb J A, Bernhardt G, Wimmer E. J Gen Virol. 1994;75:1875–1881. doi: 10.1099/0022-1317-75-8-1875. [DOI] [PubMed] [Google Scholar]
- 21.Zlotnick A, Cheng N, Conway J F, Booy F P, Steven A C, Stahl S J, Wingfield P T. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 22.Gomez Yafal A, Kaplan G, Racaniello V R, Hogle J M. Virology. 1993;197:501–505. doi: 10.1006/viro.1993.1621. [DOI] [PubMed] [Google Scholar]
- 23.Arita M, Koike S, Aoki J, Horie H, Nomoto A. J Virol. 1998;72:3578–3586. doi: 10.1128/jvi.72.5.3578-3586.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker T S, Cheng R H. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 25.Conway J F, Steven A C. J Struct Biol. 1999;128:106–118. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- 26.Belnap D M, Olson N H, Baker T S. J Struct Biol. 1997;120:44–51. doi: 10.1006/jsbi.1997.3896. [DOI] [PubMed] [Google Scholar]
- 27.Winkelmann D A, Baker T S, Rayment I. J Cell Biol. 1991;114:701–713. doi: 10.1083/jcb.114.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyazawa A, Fujiyoshi Y, Stowell M, Unwin N. J Mol Biol. 1999;288:765–786. doi: 10.1006/jmbi.1999.2721. [DOI] [PubMed] [Google Scholar]
- 29.Belnap D M, Grouchulski W D, Olson N H, Baker T S. Ultramicroscopy. 1993;48:347–358. doi: 10.1016/0304-3991(93)90110-j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones T A. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- 32.Brunger A T. x-plor: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 33.Olson N H, Kolatkar P R, Oliveira M A, Cheng R H, Greve J M, McClelland A, Baker T S, Rossmann M G. Proc Natl Acad Sci USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rueckert R R. In: Fields Virology. 3rd Ed. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 609–654. [Google Scholar]
- 35.Li Q, Yafai A G, Lee Y M-H, Hogle J, Chow M. J Virol. 1994;68:3965–3970. doi: 10.1128/jvi.68.6.3965-3970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossmann M G, Palmenberg A C. Virology. 1988;164:373–382. doi: 10.1016/0042-6822(88)90550-8. [DOI] [PubMed] [Google Scholar]
- 37.Chapman M S. Protein Sci. 1993;2:459–469. doi: 10.1002/pro.5560020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 39.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Kolatkar P R, Bella J, Olson N H, Bator C M, Baker T S, Rossmann M G. EMBO J. 1999;18:6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]