Abstract
Pyrethroid resistance in Anopheles funestus is a potential obstacle to malaria control in Africa. Tools are needed to detect resistance in field populations. We have been using a positional cloning approach to identify the major genes conferring pyrethroid resistance in this vector. A quantitative trait locus (QTL) named rp1 explains 87% of the genetic variance in pyrethroid susceptibility in two families from reciprocal crosses between susceptible and resistant strains. Two additional QTLs of minor effect, rp2 and rp3, were also detected. We sequenced a 120-kb BAC clone spanning the rp1 QTL and identified 14 protein-coding genes and one putative pseudogene. Ten of the 14 genes encoded cytochrome P450s, and expression analysis indicated that four of these P450s were differentially expressed between susceptible and resistant strains. Furthermore, two of these genes, CYP6P9 and CYP6P4, which are 25 and 51 times overexpressed in resistant females, are tandemly duplicated in the BAC clone as well as in laboratory and field samples, suggesting that P450 gene duplication could contribute to pyrethroid resistance in An. funestus. Single nucleotide polymorphisms (SNPs) were identified within CYP6P9 and CYP6P4, and genotyping of the progeny of the genetic crosses revealed a maximum penetrance value f2 = 1, confirming that these SNPs are valid resistance markers in the laboratory strains. This serves as proof of principle that a DNA-based diagnostic test could be designed to trace metabolic resistance in field populations. This will be a major advance for insecticide resistance management in malaria vectors, which requires the early detection of resistance alleles.
Anopheles funestus is one of the major malaria vectors in Africa. It is widely distributed from south of the Sahara desert to northern South Africa (Gillies and De Meillon 1968). The highly anthropophilic and endophilic behaviors of this mosquito make it an efficient vector of malaria, and, in many places, parasite infection rates of An. funestus exceed those of Anopheles gambiae. Malaria control relies extensively on the use of pyrethroid insecticides, either applied to insecticide-treated materials or as a residual spray inside houses. Resistance to pyrethroids is developing at an alarming rate in Anopheles mosquitoes in Africa, and resistance observed in An. funestus is disrupting malaria control in southern Africa (Hargreaves et al. 2000; Casimiro et al. 2006). There are fears that this resistance front could spread very rapidly and have a devastating impact on the many pyrethroid-based malaria control programs that are being widely implemented across the continent.
Elucidating the molecular basis of insecticide resistance greatly facilitates resistance management by enabling questions on the origin, spread, and impact of resistance to be assessed. The two major causes of insecticide resistance are alterations in the target sites and increases in the rate of insecticide metabolism. Three enzyme families, the cytochrome P450s, the esterases, and the glutathione S-transferases (GSTs), are primarily responsible for metabolic resistance (Hemingway and Ranson 2000). In An. funestus, pyrethroid resistance results from an increase in insecticide metabolism and is associated with increased activity of cytochrome P450s (Brooke et al. 2001), but the exact mechanism remains unclear. The appearance of P450-based pyrethroid resistance in southern African An. funestus had an immediate and significant effect on malaria transmission in South Africa, prompting a switch back to DDT in South Africa and the introduction of the carbamate bendiocarb in Mozambique (Hunt et al. 2005). However, resistance to these alternative insecticides has already developed in An. arabiensis, another malaria vector, in South Africa (Hargreaves et al. 2003), and hence effective resistance management strategies are imperative in this region. Identification and characterization of the specific genes involved in pyrethroid metabolism in resistant An. funestus is an important step toward this goal.
A preliminary study carried out using F2 progeny from crosses between a susceptible and a resistant strain of An. funestus identified a single quantitative trait locus (QTL) named rp1 located near the telomere of the 2R chromosome (Wondji et al. 2007b). However, the small sample sizes used for this study and the low recombination frequency at the F2 generation could have prevented the detection of additional QTLs. Moreover, the 15 cM confidence interval for the rp1 QTL precluded the identification of candidate genes. Advanced intercross lines (AIL) enable fine-scale mapping of QTLs by reducing the confidence interval surrounding QTLs (Darvasi and Soller 1995). AIL are produced from an F2 population by random intercrossing in each generation from F2 until advanced intercross generations are attained. In this method, many recombinational events required for fine-scale mapping of QTLs are accumulated in a single, relatively small population over the course of many generations rather than by producing and examining many progeny in a single large F2 or backcross generation (Darvasi and Soller 1995).
Here, we report the positional cloning of genes in the rp1 QTL that confer pyrethroid resistance in An. funestus, using the AIL approach with F6 and F8 generation progeny from reciprocal crosses between susceptible and resistant strains. We also report the identification of two additional QTLs not detected in the F2 generation.
Results
QTL mapping
χ2 goodness-of fit tests were used to identify loci significantly associated (P < 0.05) with pyrethroid resistance in progeny of reciprocal crosses between the resistant and susceptible families (Supplemental Fig. S1). Three sets of nine, four, and two loci, respectively, were significantly associated with pyrethroid resistance in Family 1. Seven loci from a single set were associated with resistance in Family 10. Each marker associated with resistance on the 2R chromosome has an additive effect in both families, with individuals homozygous for the resistant allele having a greater chance of surviving pyrethroid exposure than heterozygotes (Supplemental Fig. S2).
Analysis of the F6 progeny identified three QTLs in Family 1, with the major QTL located within the boundaries of the rp1 QTL identified previously using F2 samples (Wondji et al. 2007b). The two additional QTLs were named rp2 and rp3 for resistance to permethrin 2 and 3, respectively. The logarithm of the odd (LOD) scores of rp1, estimated by both interval mapping (IM) and composite interval mapping (CIM), was ∼40 for Family 1, which far exceeded the LOD scores of 4 and 3 for rp2 and rp3, respectively (Fig. 1A). Multiple interval mapping confirmed these three QTLs (see Supplemental Table S1 for parameters of each QTL). The genetic variance (σg2) explained by the three QTLs accounted for 97.4% of the phenotypic variance (σp2), with the vast majority of this (96%) contributed by rp1.
Figure 1.
Plot of LOD scores associated with pyrethroid resistance in Family 1 (A) and in Family 10 (B) using F6 progeny. The y-axis indicates LOD ratio scores, and the x-axis indicates chromosome positions. (Solid lines) LOD estimated by composite interval mapping, (dashed lines) LOD estimated by interval mapping. The straight line along the top of each graph represents the threshold value for LOD as determined by permutations. Names of markers are listed around QTL locations.
In the F6 progeny of Family 10, only rp1 was detected, with a LOD score close to that of Family 1 at 35 and 36 by IM and CIM, respectively (Fig. 1B). rp1 accounts for 87.7% of the genetic variance in susceptibility to pyrethroid in Family 10.
Additional fine-scale mapping using 400 F8 progeny from the same reciprocal cross families and additional loci indicated only a single QTL, rp1, in both families. In Family 1, none of the loci located in rp2 and rp3 showed a significant association with pyrethroid resistance, contrary to the results with F6 samples, suggesting that the higher recombination rate at the F8 generation has disrupted the linkage between these loci and the candidate genes or polymorphisms in these QTLs. This fine-scale mapping also reduced the boundaries of rp1, with only three out of nine loci linked to rp1 in the F6 generation remaining significantly associated with pyrethroid resistance at the F8 generation in Family 1 (Supplemental Fig. S2C) and in Family 10 (Supplemental Fig. S2D). In both families, the rp1 QTL boundaries of ≈8 cM were defined around SNPs within two P450 genes, CYP6P4 and CYP6P9 (Supplemental Table S1).
Identification of candidate genes in rp1 QTL
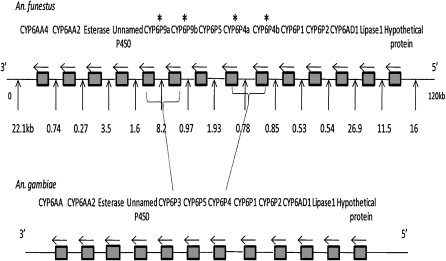
The high genetic variance in susceptibility to pyrethroids explained by rp1 strongly suggests that the major pyrethroid resistance genes in An. funestus are located in the genomic region encompassed by this QTL. We therefore screened an An. funestus BAC library, developed by the University of Notre Dame, to identify a BAC clone spanning this region. Four positive 384-well plates (Numbers 11, 29, 50, and 62) containing one or more of the loci spanning rp1 were identified. A single individual BAC clone (29F01) was sequenced and annotated. The 120-kb clone contained 14 putative protein coding genes, including a cluster of 11 P450 genes and one putative P450 pseudogene. The three additional genes are predicted to encode a carboxylesterase, a lipase precursor (lipase 1), and a hypothetical protein (Fig. 2). The full-length sequence of each of these genes has been submitted to GenBank (accession numbers EU852639–EU852649). The 11 cytochrome P450s all belong to the CYP6 family, and orthologs of these P450s are found in the same orientation and location on chromosome 2R, division 12D–13E, of An. gambiae (Fig. 2). However, in contrast to An. gambiae, tandem duplications of two of the P450 genes, CYP6P4 and CYP6P9, were observed in the An. funestus BAC clones. The duplication of CYP6P4 and CYP6P9 was confirmed by PCR amplification and sequencing of the 5′ flanking region of each copy of the two genes using genomic DNA from laboratory and field samples. The sequence similarity between the An. funestus and An. gambiae orthologs is given in Supplemental Table S2. There are 22 amino acid substitutions and a single indel at the carboxyl terminus between CYP6P4a (510 amino acids) and CYP6P4b (513 amino acids), resulting in 95% similarity (Supplemental Fig. S3A). CYP6P9a and CYP6P9b have 29 variant amino acids (94% similarity), and both have a predicted length of 509 amino acids (Supplemental Fig. S3B). The phylogenetic relationship between the cluster of An. funestus P450s contained in the BAC clone and their orthologs in An. gambiae and, where available, in An. mimimus, is shown in Supplemental Figure S4 and percentage similarities are given in Supplemental Table S2. As predicted from the phylogenetic relationship among these three species (Harbach 1994), the An. funestus P450s are more closely related to those of An. minimus, an Asian malaria vector, than to the African species An. gambiae.
Figure 2.
Schematic representation of gene organization of BAC 29F01 containing the rp1 QTL in An. funestus (top) compared with the organization of orthologous genes in An. gambiae (bottom). (Vertical arrows) Intergenic regions; (horizontal arrows) the 5′–3′orientation of each gene; (asterisks) duplicated genes.
Gene expression analysis
The transcript levels of each of nine P450 genes (the two copies of the duplicated genes being amplified together) from rp1 were determined for adult females from the resistant strain FUMOZ-R and the susceptible strain FANG in multiplex quantitative PCR (qPCR). We also studied the expression pattern of another set of 21 P450s located near the rp2 QTL (six) or elsewhere in the genome of An. funestus and one housekeeping gene, SP7, which served to normalize the data between genes. The amplification success of each gene (indicated by the presence or absence of a peak at the expected size on the GeXP expression profile) is indicated in Supplemental Table S3. No potential candidate genes were identified in the vicinity of rp3. The nearest coding genes are a microtubule binding gene (Accession number: BU038892) and a tubulin beta-3 chain gene (BU039021), both at division 39A of chromosome 3L (Sharakhov et al. 2002). Primers used are listed in Supplemental Table S3. For the duplicated CYP6P4 and CYP6P9 genes, primers were designed within nonvariant regions of the two gene copies in order to determine the combined transcript level of the two copies.
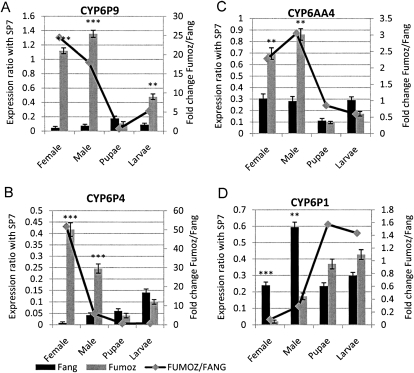
Four genes, CYP6P9, CYP6P4, CYP6AA4, and CYP6P1, were significantly differentially expressed (P < 0.001) between FUMOZ-R and FANG (Fig. 3A–D). Three of these were overexpressed in the resistant strain while one, CYP6P1, was overexpressed in the susceptible strain. The greatest fold changes were seen in the two duplicated genes, CYP6P9 (25-fold) and CYP6P4 (51-fold). The expression level of CYP6P9 was higher than that of CYP6P4 in both the susceptible (fivefold) and resistant strains (threefold). The overexpression of CYP6P9 in the FUMOZ-R strain has been reported earlier (Amenya et al. 2008), but the results from this multiplex qPCR show that two additional P450s within the boundaries of rp1 are also overexpressed. None of the genes within the region of the rp2 QTL or elsewhere in the genome was differentially expressed between the resistant and the susceptible strains (Supplemental Fig. S5A).
Figure 3.
Comparison of the patterns of gene expression of four candidate genes CYP6P9 (A), CYP6P4 (B), CYP6AA4 (C), and CYP6P1 (D) between the resistant strain FUMOZ and the susceptible FANG in different life stages. The normalized expression ratio of each gene against the SP7 gene is represented on the primary vertical axis, while the secondary vertical axis represents the fold change of each gene between FUMOZ and FANG. Significant differential expressions are indicated by asterisks: (***) P < 0.001, (**) P < 0.01.
The qPCR experiment was repeated using male adults, pupae, and larvae. Male adults generally presented a similar gene expression profile to that of females, with the same four genes differentially expressed between susceptible and resistant strains (Supplemental Fig. S5B). However, fold changes between the resistant and the susceptible strains were generally lower in males than in females. Indeed, apart from CYP6AA4, which presented a comparable level of expression between both sexes, the fold change of the three other genes was much lower for males than females. CYP6P9 was 18-fold overexpressed in resistant males compared with 25-fold for females, CYP6P4 was only fivefold overexpressed in males compared with 51 times in females, and CYP6P1, which was 12-fold overexpressed in susceptible females, was only three times overexpressed in susceptible males (Supplemental Fig. S5B). These results correlate with earlier studies showing that pyrethroid resistance is much higher in females than in males of An. funestus (Hunt et al. 2005). The expression levels of all the genes were significantly lower in pupae than in other life stages (Supplemental Fig. S5C), and none of the 30 genes studied were differentially expressed between the strains at the pupal stage. This could be explained by the fact that the pupal stage is a nonfeeding and metamorphic life stage during which most genes involved in detoxification or digestion are switched off or down-regulated in favor of those involved in metamorphosis. In the larval life stage, only one of the four genes that was differentially expressed in the adult stage, CYP6P9, showed differential expression between both strains, although it was a much lower fold change (fivefold compared with 18- and 25-fold in male and female adults, respectively [Supplemental Fig. S5D]). These results support the life stage specificity of pyrethroid resistance in the adult-selected FUMOZ-R strain, as suggested previously (Amenya et al. 2008). These gene expression results provide further support for a role of CYP6P9 and CYP6P4 in conferring pyrethroid resistance in An. funestus, with CYP6AA4 perhaps also being involved at a lesser extent. A comparative expression profile for some of these genes in all the life stages is presented in Supplemental Figure S5.
SNPs in CYP6P9 and CYP6P4 can predict the resistance status of An. funestus in the laboratory
To identify sequence variations associated with resistance to pyrethroids, we sequenced DNA fragments encompassing CYP6P4a and CYP6P9b from four resistant and four susceptible samples from the F6 generation of Family 1. CYP6P4a and CYP6P9b were chosen because they contain the SNPs used for the QTL mapping (Supplemental Fig. S2).
For CYP6P4a, a 2584-bp fragment (comprising the 5′ flanking region, coding sequences, and intron) was sequenced (accession number EU852651). CYP6P4a was highly polymorphic, with 103 polymorphic sites observed (Supplemental Table S4). Two indels, A/– and TT/–, were observed in the 5′ flanking region, with A and TT always deleted in resistant individuals. The distribution of polymorphic sites and the clustering of haplotypes in a phylogenetic tree correlated with the resistance status (Supplemental Fig. S6A) in the progeny of the crosses. Amino acid changes were clustered in the protein, with 17 polymorphic residues located between amino acids 50 and 100.
For CYP6P9b, a 2229-bp fragment (comprising the 5′ flanking region, coding sequences, and intron) was sequenced (accession number EU852650). Details of these polymorphisms are given in Supplemental Table S4. Two indels, CAA/– and TGT/–, were observed in the 5′ flanking region, with CAA and TGT being deleted in resistant individuals. The clustering of haplotypes according to the resistance status (Supplemental Fig. S6B) show that polymorphism at CYP6P9b is also closely associated with pyrethroid resistance.
Two SNPs, one from CYP6P4a and one from CYP6P9b, were selected to see if they could be used to predict the resistance status of the progeny from the laboratory crosses. For CYP6P9b, we chose the A/G mutation at position 1480, and for CYP6P4a, we chose the A/G mutation at position 2099 located in the intron and always showing a consistent association with the phenotype in the eight samples sequenced above. These two SNPs were genotyped using the pyrosequencing method in 325 resistant and 325 susceptible individual mosquitoes from F6 and F8 generations of Family 1 and Family 10 used above for QTL mapping. Primers were designed to specifically amplify the corresponding fragment of CYP6P4a and CYP6P9b containing these two SNPs. The results presented in Table 1 show a very strong correlation between the genotypes at these two SNPs and pyrethroid resistance. For the total sample, a maximum value of 1 for f2 penetrance (probability of being resistant if having two copies of the resistant allele, which is G in both genes) was obtained for the G/G genotype at both SNPs for CYP6P9b and CYP6P4a. The opposite observation was made for f0 penetrance (probability of being resistant if having zero copies of the resistant allele G) with only an average probability of 1.2% and 1.1%, respectively, for CYP6P4a and CYP6P9b. An intermediate result was observed for f1 penetrance (probability of being resistant if having one copy of the resistant allele G) with probabilities of 61% and 72%, respectively, for CYP6P4a and CYP6P9b, indicating that resistance is additive at both genes. On average, the risk of being resistant for an individual with the G/G genotype r2 (Genotype Relative Risk) compared with one with the A/A genotype is 83.3 and 90.9 times for CYP6P4 and CYP6P9, respectively, while the heterozygote A/G is respectively 50.8 and 65.4 times more likely to be resistant than an A/A mosquito. When comparing results from F6 and F8 generations, we observed that there is a decrease of the genotype relative risk from F6 to F8 samples for both SNPs (see Table 1), indicating that recombination occurring between the F6 and F8 generations has probably reduced the association between the two SNPs and the causative mutation at both genes. However, the fact that f2 penetrance remained unchanged indicates that these two SNPs are very close to the causative mutations. The strong association observed between these two SNPs and pyrethroid resistance serves as proof of principle that a DNA-based marker could be identified in order to design a diagnostic test to trace this metabolic resistance in field populations.
Table 1.
Penetrance and genotype relative risk (GRR) of two mutations in CYP6P9b and CYP6P4a strongly associated with pyrethroid resistance in An. funestus
The parameters f0, f1, and f2 are the probabilities of being resistant in individuals with zero, one, and two copies of the resistant allele (G allele for both CYP6P4 and CYP6P9), respectively, while r2 and r1 are a quantification of the risk for a mosquito to be resistant if it is homozygous at the resistance locus (G/G for both genes) or heterozygous (A/G for both genes), respectively, compared with a susceptible mosquito (A/A at both genes).
Sus, susceptible; Res, resistant.
Discussion
The long-term objective of this work is to develop a DNA-based assay to monitor for the presence of pyrethroid resistance in individual An. funestus mosquitoes. Current monitoring for resistance requires the use of bioassays on large numbers of live insects and requires trained entomologists. The use of DNA-based assays would simplify resistance monitoring and, as the assays can be performed on dried mosquitoes, would enable a much wider geographical area to be sampled. Using DNA-based assays also enables resistance to be detected when it first appears in a population, which is the stage at which preventative action must be taken. Here, we report significant progress toward this goal by identifying candidate genes that are tightly linked to the resistance phenotype and demonstrating that simple SNP assays can be used to predict resistance status in laboratory strains.
Three QTLs associated with resistance to permethrin were identified, but one of these, rp1, explains the vast majority of the genetic variance. However, given that the boundaries of rp2 and rp3 are currently much larger than rp1, further fine-scale mapping of these two minor QTLs could result in higher LOD scores for these two loci. rp2 is also located within the vicinity of a cluster of P450s. Around 20 P450s are located in the corresponding position in An. gambiae (chromosome 3R) (Ranson et al. 2002), and five P450s were genetically mapped to the rp2 region in An. funestus in this study. However, none of these rp2 P450s was found to be overexpressed in the resistant strain. No candidate genes associated with pyrethroid resistance were identified for rp3. None of the three QTLs are linked to the target site of pyrethroids, the voltage-gated sodium channel gene. Mutations in this gene, found on chromosome 3R in An. funestus, are linked with a resistance phenotype known as knockdown resistance (kdr). kdr mutations are widespread in An. gambiae populations (Martinez-Torres et al. 1998; Ranson et al. 2000) but have not been reported in An. funestus to date. Instead, metabolic resistance, caused by increased activity of cytochrome P450s, appears to be the major resistance mechanism both in laboratory and field populations of An. funestus (Brooke et al. 2001; Amenya et al. 2008). Earlier reports have identified CYP6P9 as being highly overexpressed in the pyrethroid-resistant FUMOZ populations. Here, we confirm this result but also identify additional P450 genes, clustered with CYP6P9, that are overexpressed and reveal two gene duplications of two of the candidate genes. The fact that these gene duplication events are present only in An. funestus suggests that they occurred after the divergence of An. gambiae and An. funestus 36–80 Mya (Krzywinski et al. 2006). However, these gene duplications are probably fairly ancient given that the two copies show relatively extensive sequence variation (94% nucleotide sequence similarity for CYP6P4a and b and 95% similarity for CYP6P9a and b) and would certainly predate any selection by pyrethroids. Nevertheless, it is intriguing that these main candidate genes associated with resistance to pyrethroids are both duplicated. Duplication of P450 genes involved in metabolic resistance has also been observed in Drosophila species. The Rst(2)DDT locus conferring resistance to DDT in Drosophila melanogaster encompasses a cluster of six P450 genes, among which two duplicated P450 genes, CYP6G1 and CYP12D1, are the only genes up-regulated in the resistant strain (Brandt et al. 2002). Indeed, CYP12D1 is present in two copies, products of a very recent gene duplication event on chromosome 2R in D. melanogaster, while CYP6G1, confirmed to confer resistance to DDT in D. melanogaster (Daborn et al. 2002), was recently found to be also duplicated (Emerson et al. 2008). These cases of P450 gene duplication associated with insecticide resistance suggest that P450 gene duplication could potentially generate genes that are more metabolically active against insecticides. This is also supported by the observation made in Drosophila that both copies of CYP12D1 need to be overexpressed to confer a resistant phenotype (Daborn et al. 2007). In humans, duplication of CYP2D6 results in an ultra-rapid metabolism of its substrates (Agundez et al. 2001). Similarly, it is possible that the duplication of CYP6P9 and CYP6P4 increases pyrethroid metabolism in resistant An. funestus mosquitoes. To further investigate the potential role of these P450 gene duplication events in pyrethroid resistance, we would need to measure copy number polymorphisms in a wider population of An. funestus. Further work is also needed to verify that both copies of the genes are functional and are capable of metabolizing pyrethroids.
The role of CYP6P9 and CYP6P4 in conferring pyrethroid resistance is supported by their high overexpression level in the resistant strain FUMOZ and also by the correlation observed between polymorphisms at these genes and the resistance phenotype in advanced intercross lines. Preliminary work has also indicated that enzymes from both genes are able to metabolize pyrethroids in vitro (B. Stevenson and C. Wondji, unpubl.). The causative mutations responsible for the overexpression of these P450 genes are still unknown. Analysis of the 5′ promoter region of each gene did not reveal major genomic changes such as has been observed in the CYP6G1 gene in Drosophila where a long terminal repeat (LTR) retrotransposon known as Accord is inserted in the 5′ promoter region (Daborn et al. 2002). However, the short indels and polymorphisms in the 5′ promoter region of both copies of CYP6P9 and CYP6P4 may influence the transcription of these genes, as observed in D. melanogaster (CYP6A8) (Maitra et al. 2002) and in the house fly Musca domestica (CYP6D3) (Kasai and Scott 2001). Further promoter analysis and genotype–phenotype association studies using field populations are needed to identify the functional polymorphism responsible for the increased expression of these P450 genes.
Methods
Mosquito crosses
Reciprocal crosses between the FANG and FUMOZ-R strains (Hunt et al. 2005) were set up as described previously (Wondji et al. 2007b). F2 progeny of each isofemale line were randomly intercrossed until the F6 and F8 generations. Bioassays on 1- to 3-d-old adults using the standard WHO bioassay procedure (WHO 1998) were carried out on the F6 and F8 generations. Mosquitoes were exposed to 0.75% permethrin at two different exposure times of 30 min and 2 h and mortality was recorded 24 h post-exposure. Specimens killed after the short exposure of 30 min (time empirically determined to kill 10% of the population or LT10) were considered susceptible while those that survived the long exposure of 2 h (LT90) were considered resistant. This approach minimizes the level of phenotype misclassification and increases the power of QTL detection because specimens from the two extreme phenotypes contribute more linkage information than the other specimens (Lander and Botstein 1989). F6 and F8 progeny from each family were stored on silica gel for subsequent DNA extraction and genotyping.
Genotyping of molecular markers
Progeny from two of the 11 families used to detect rp1 in the F2 generation in a previous study (Wondji et al. 2007b) were maintained for a further six generations in this study. Family 1 originated from a cross between a female from the susceptible strain FANG and a male from the resistant strain FUMOZ-R, while family 10 originated from the reciprocal cross between a FUMOZ-R female and a FANG male. In each family equal numbers of surviving and dead progeny were genotyped (75 dead and 75 alive for family 1, 48 dead and 48 alive for family 10). DNA was extracted using the LIVAK method (Collins et al. 1987), and the microsatellite loci were genotyped as described previously (Wondji et al. 2005). A total of 72 microsatellite markers was scored in the parental P1 and F1 samples, and the F6 progeny were genotyped for informative markers in each family using the Beckman CEQ8000 fragment analysis program.
Single nucleotide polymorphism (SNP) genotyping
SNP markers chosen from a set of SNPs previously identified in An. funestus (Wondji et al. 2005, 2007a) were genotyped in simplex and multiplex pyrosequencing reactions. These were scored in the parents (P1) and in F1 for the two families, and informative SNPs were genotyped for F6 and F8 samples. Multiplex assays were designed using the Pyrosequencing SNP Multiplex Entry software (Pyrosequencing AB), which allows the simultaneous genotyping of three SNPs. Sequences to be analyzed for each of the three SNPs were entered in the software and a dispensation order was obtained that allowed nonambiguous genotyping of each of the three SNPs. Multiplex PCR was carried out using three different primer pairs specific for each SNP to be genotyped. Forward and biotinylated reverse primers (10 pmol) were added to a PCR reaction containing 1× HotStarTaq buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 1 U of HotStarTaq (Qiagen), and 30 ng of genomic DNA. Amplification was performed with the following conditions: one cycle for 5 min at 95°C; 50 cycles for 20 sec at 94°C, 30 sec at 57°C, and elongation for 20 sec at 72°C; followed by one cycle for 5 min at 72°C. Three different sequencing primers were used to simultaneously genotype the three SNPs of each multiplex assay. The pyrosequencing reaction was carried out as described by Wondji et al. (2007b) according to the manufacturer's instructions using the PSQ 96 SNP Reagent Kit (Biotage AB), and the genotype was determined using SNP Software (Biotage AB).
QTL mapping
The JoinMap 2.0 package (Stam and Van Ooijen 1995) was used to build a genetic linkage map for each of the two families using the same parameters as described previously (Wondji et al. 2007b). DrawMap 1.1 (van Ooijen 1994) software was then used to plot the genetic map. Associations between genotypes at each locus and the resistance phenotype were assessed using a contingency χ2 analysis. The null hypothesis was that susceptibility to permethrin is equal in each genotype class. For loci with a significant χ2, we analyzed the inheritance of the alleles at these loci. The a priori hypothesis was that a higher mortality rate would occur among F6 and F8 individuals with one or both alleles inherited from the susceptible parent. The JoinMap linkage map and the genotype/phenotype data were entered into Windows QTL Cartographer 2.5 (Wang et al. 2005). Interval mapping (IM) (Lander and Botstein 1989), composite-interval mapping (CIM) (Zeng 1994), and multiple-interval mapping (MIM) (Zeng et al. 1999) procedures were implemented for each family using the parameters described previously (Wondji et al. 2007b).
Sequencing of the BAC clone containing the rp1 QTL
A BAC library for An. funestus was recently constructed by The Institute for Genomic Research (TIGR) using genomic DNA fragments from the FUMOZ colony (N. Lobo, pers. comm.). The library was screened by PCR using the following primers located within the boundaries of the rp1 QTL as described in Hong et al. (2003):
CYP6P9 (forward: GCGCCTTAGACAAGAGATCA; reverse: AAGGGATGTCGCTTCTTCTC),
CYP6P4 (forward: GTACGAGACTGGCAAAGAAT; reverse: AAGGAAGACGTATGGATGG),
BU85 (forward: AAAAAGAATGGC CGGAAAGT; reverse: CTCATCGCCCAGAATTTCAT).
DNA of whole 384-well plates was pooled and a PCR was carried out for each plate using 10 pmol of each primer and 30 ng of BAC DNA as template in 25-μL reactions containing 1× Hot StarTaq buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 1 U of Hot StarTaq (Qiagen). The cycle parameters were: 1 cycle for 15 min at 95°C; 35 cycles of 30 sec at 94°C, 30 sec at 57°C, and elongation for 30 sec at 72°C; followed by 1 cycle of 10 min at 72°C. The positive plates after electrophoresis of PCR products on a 1.5% agarose gel were then subdivided into six column-pools and four row-pools and the PCR screen was repeated. Finally, individual colonies from the set of 16 identified from the pooled column and row screen were used as a template to identify the individual clone containing the markers of interest. The BAC clone was grown overnight at 37°C, harvested in a glycerol solution, and stored at −80°C. The size of the BAC clone was estimated after a restriction digestion using the BamH1 restriction enzyme to separate the insert from the vector. The BAC clone was then fragmented by sonication into shorter, random sequences of ∼2–5 kb. These small fragments were cloned into plasmid vectors (pC31) and sequenced using an ABI DNA Sequencer. The reads were trimmed using Seqman (http://www.dnastar.com/web/r3.php), which automatically removes known vector sequences from the end of reads. Read sequences were later assembled using the computer program phrap (P. Green unpubl.; http://www.genome.washington.edu/UWGC/analysistools/Phrap.cfm). The program Genemark.hmm version 2.2 (Lukashin and Borodovsky 1998) was used to locate genes in the assembled sequence of the BAC clone. Putative genes were annotated by using the BLASTX (http://www.ncbi.nlm.nih.gov/blast/) program of GenBank and using the Ensembl Mosquito BlastView. Further detailed annotation of the P450 genes was aided by the P450 site (http://p450.sophia.inra.fr/).
Quantitative PCR
The expression pattern of each gene was compared between the resistant strain FUMOZ-R and the susceptible strain FANG using the GenomeLab GeXP genetic analysis system from Beckman Coulter. This system utilizes a highly multiplexed quantitative PCR approach to analyze the expression of up to 30 genes simultaneously. Total RNA was extracted from three batches (biological replicates) of 15 mosquitoes from each life stage (1-d-old female and male adults, pupae, and fourth-instar larvae) using a PicoPure RNA isolation kit (Arkturis) according to the manufacturer's instructions. Total RNA quantity and quality were assessed using a Nanodrop spectrophotometer (Nanodrop Technologies). The GenomeLab eXpress Profiler program was used for automated primer design, calculation of relative gene expression values, data checking, and data analysis. The quantitative PCR reaction was carried out using the GenomeLab GeXP Start Kit (Beckman and Coulter) according to the protocol provided. First, a reverse transcription (RT) reaction was carried out in a 20-μL total volume containing 1× RT buffer, the reverse primers mix at 50 nM each, 20 U of reverse transcriptase, 5 μL of the Kanr RNA (positive control), and 50 ng of the sample RNA. The final volume was brought to 20 μL with DNase/RNase-free H2O. This reaction mixture was incubated with the following program: 1 min at 48°C, 5 min at 37°C, 1 h at 42°C, 5 min at 95°C. Secondly, a PCR reaction was carried out with the mixture containing, at the final concentration, 1× PCR buffer, 1.25 mM MgCl2, the forward primers mix at 20 nM each, 3.5 U of the thermo-Start DNA polymerase (ABgene AB-0908/A), and 9.3 μL of the cDNA from the RT reaction. The cycle parameters were 10 min at 95°C, 35 cycles of 30 sec at 94°C, 30 sec at 55°C, 1 min at 68°C. The resulting samples were diluted 10 times and added in a 96-well microplate with the DNA size standard-400 and run on the GenomeLab GeXP genetic analysis system. The expression level of the SP7 ribosomal gene was used to normalize for variation in total cDNA concentration.
Sequencing of candidate genes
The P450 genes CYP6P9b and CYP6P4a containing the SNPs with the highest correlation with pyrethroid resistance were amplified from four susceptible and four resistant individuals from the F6 generation of family 1 and sequenced to identify potential polymorphisms associated with pyrethroid resistance. The following primers were used:
CYP6P4a (forward: CGCACAATGCTCGCATGTTTACC; reverse: GACGCCTCGATCATGAAACAC), CYP6P9b (forward: CATACTCATAATAACTAGACGCG; reverse: TACACTGCCGACACTACGAAG).
Sequences were aligned using ClustalW (Thompson et al. 1994). Haplotypes and amino acid translations were generated using DnaSP 4.50 (Rozas et al. 2003). Phylogenetic trees were constructed using MEGA 4 (Tamura et al. 2004). The penetrance and Genotype Relative Risk (GRR) for two SNPs in CYP6P4a and CYP6P9b were estimated. The penetrance estimates f0, f1, and f2 are the probability of being resistant in individuals with zero, one, and two copies of the resistant allele, respectively. fi = Pr(resistant | i copies of resistant allele) (i = 0, 1, 2), where f indicates penetrance and Pr indicates probability (Schaid and Sommer 1993). GRR (AA) = r2 = f2/f0; GRR(Aa) = r1 = f1/f0, r2 being the genotype relative risk of the homozygote resistant allele, while r1 is for the heterozygote genotype (Schaid and Sommer 1993).
Acknowledgments
This work was supported by the National Institute of Health (NIH) grant IU01 A1058271-01 and by a Wellcome Trust Research Career Development Fellowship to C.S.W.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.gov/Genbank/) under accession nos. EU852639–EU852651.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.087916.108.
References
- Agundez J.A., Gallardo L., Ledesma M.C., Lozano L., Rodriguez-Lescure A., Pontes J.C., Iglesias-Moreno M.C., Poch J., Ladero J.M., Benitez J. Functionally active duplications of the CYP2D6 gene are more prevalent among larynx and lung cancer patients. Oncology. 2001;61:59–63. doi: 10.1159/000055354. [DOI] [PubMed] [Google Scholar]
- Amenya D.A., Naguran R., Lo T.C., Ranson H., Spillings B.L., Wood O.R., Brooke B.D., Coetzee M., Koekemoer L.L. Over expression of a cytochrome P450 (CYP6P9) in a major African malaria vector, Anopheles funestus, resistant to pyrethroids. Insect Mol. Biol. 2008;17:19–25. doi: 10.1111/j.1365-2583.2008.00776.x. [DOI] [PubMed] [Google Scholar]
- Brandt A., Scharf M., Pedra J.H., Holmes G., Dean A., Kreitman M., Pittendrigh B.R. Differential expression and induction of two Drosophila cytochrome P450 genes near the Rst(2)DDT locus. Insect Mol. Biol. 2002;11:337–341. doi: 10.1046/j.1365-2583.2002.00344.x. [DOI] [PubMed] [Google Scholar]
- Brooke B.D., Kloke G., Hunt R.H., Koekemoer L.L., Temu E.A., Taylor M.E., Small G., Hemingway J., Coetzee M. Bioassay and biochemical analyses of insecticide resistance in southern African Anopheles funestus (Diptera: Culicidae) Bull. Entomol. Res. 2001;91:265–272. doi: 10.1079/ber2001108. [DOI] [PubMed] [Google Scholar]
- Casimiro S., Coleman M., Mohloai P., Hemingway J., Sharp B. Insecticide resistance in Anopheles funestus (Diptera: Culicidae) from Mozambique. J. Med. Entomol. 2006;43:267–275. doi: 10.1603/0022-2585(2006)043[0267:iriafd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Collins F.H., Mendez M.A., Rasmussen M.O., Mehaffey P.C., Besansky N.J., Finnerty V. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am. J. Trop. Med. Hyg. 1987;37:37–41. doi: 10.4269/ajtmh.1987.37.37. [DOI] [PubMed] [Google Scholar]
- Daborn P.J., Yen J.L., Bogwitz M.R., Le Goff G., Feil E., Jeffers S., Tijet N., Perry T., Heckel D., Batterham P., et al. A single p450 allele associated with insecticide resistance in Drosophila. Science. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- Daborn P.J., Lumb C., Boey A., Wong W., Ffrench-Constant R.H., Batterham P. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem. Mol. Biol. 2007;37:512–519. doi: 10.1016/j.ibmb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Darvasi A., Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J.J., Cardoso-Moreira M., Borevitz J.O., Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- Gillies M.T., De Meillon B. The anophelinae of Africa South of the Sahara. The South African Institute for Medical Research; Johannesburg: 1968. [Google Scholar]
- Harbach R.E. Review of the internal classification of the genus Anopheles (Diptera: Culicidae): The foundation for comparative systematics and phylogenetic research. Bull. Entomol. Res. 1994;84:331–342. [Google Scholar]
- Hargreaves K., Koekemoer L.L., Brooke B.D., Hunt R.H., Mthembu J., Coetzee M. Anopheles funestus resistant to pyrethroid insecticides in South Africa. Med. Vet. Entomol. 2000;14:181–189. doi: 10.1046/j.1365-2915.2000.00234.x. [DOI] [PubMed] [Google Scholar]
- Hargreaves K., Hunt R.H., Brooke B.D., Mthembu J., Weeto M.M., Awolola T.S., Coetzee M. Anopheles arabiensis and An. quadriannulatus resistance to DDT in South Africa. Med. Vet. Entomol. 2003;17:417–422. doi: 10.1111/j.1365-2915.2003.00460.x. [DOI] [PubMed] [Google Scholar]
- Hemingway J., Ranson H. Insecticide resistance in insect vectors of human disease. Annu. Rev. Entomol. 2000;45:369–389. doi: 10.1146/annurev.ento.45.1.371. [DOI] [PubMed] [Google Scholar]
- Hong Y.S., Hogan J.R., Wang X., Sarkar A., Sim C., Loftus B.J., Ren C., Huff E.R., Carlile J.L., Black K., et al. Construction of a BAC library and generation of BAC end sequence-tagged connectors for genome sequencing of the African malaria mosquito Anopheles gambiae. Mol. Genet. Genomics. 2003;268:720–728. doi: 10.1007/s00438-003-0813-7. [DOI] [PubMed] [Google Scholar]
- Hunt R.H., Brooke B.D., Pillay C., Koekemoer L.L., Coetzee M. Laboratory selection for and characteristics of pyrethroid resistance in the malaria vector Anopheles funestus. Med. Vet. Entomol. 2005;19:271–275. doi: 10.1111/j.1365-2915.2005.00574.x. [DOI] [PubMed] [Google Scholar]
- Kasai S., Scott J.G. Expression and regulation of CYP6D3 in the house fly, Musca domestica (L.) Insect Biochem. Mol. Biol. 2001;32:1–8. doi: 10.1016/s0965-1748(01)00073-x. [DOI] [PubMed] [Google Scholar]
- Krzywinski J., Grushko O.G., Besansky N.J. Analysis of the complete mitochondrial DNA from Anopheles funestus: An improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol. Phylogenet. Evol. 2006;39:417–423. doi: 10.1016/j.ympev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin A.V., Borodovsky M. GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra S., Price C., Ganguly R. Cyp6a8 of Drosophila melanogaster: Gene structure, and sequence and functional analysis of the upstream DNA. Insect Biochem. Mol. Biol. 2002;32:859–870. doi: 10.1016/s0965-1748(01)00174-6. [DOI] [PubMed] [Google Scholar]
- Martinez-Torres D., Chandre F., Williamson M.S., Darriet F., Berge J.B., Devonshire A.L., Guillet P., Pasteur N., Pauron D. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol. Biol. 1998;7:179–184. doi: 10.1046/j.1365-2583.1998.72062.x. [DOI] [PubMed] [Google Scholar]
- Ranson H., Jensen B., Vulule J.M., Wang X., Hemingway J., Collins F.H. Identification of a point mutation in the voltage-gated sodium channel gene of Kenyan Anopheles gambiae associated with resistance to DDT and pyrethroids. Insect Mol. Biol. 2000;9:491–497. doi: 10.1046/j.1365-2583.2000.00209.x. [DOI] [PubMed] [Google Scholar]
- Ranson H., Claudianos C., Ortelli F., Abgrall C., Hemingway J., Sharakhova M.V., Unger M.F., Collins F.H., Feyereisen R. Evolution of supergene families associated with insecticide resistance. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- Rozas J., Sanchez-DelBarrio J.C., Messeguer X., Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schaid D.J., Sommer S.S. Genotype relative risks: Methods for design and analysis of candidate-gene association studies. Am. J. Hum. Genet. 1993;53:1114–1126. [PMC free article] [PubMed] [Google Scholar]
- Sharakhov I.V., Serazin A.C., Grushko O.G., Dana A., Lobo N., Hillenmeyer M.E., Westerman R., Romero-Severson J., Costantini C., Sagnon N., et al. Inversions and gene order shuffling in Anopheles gambiae and A. funestus. Science. 2002;298:182–185. doi: 10.1126/science.1076803. [DOI] [PubMed] [Google Scholar]
- Stam P., Van Ooijen J.W. JoinMap (tm) version 2.0: Software for the calculation of genetic linkage maps. CPRO-DLO; Wageningen, The Netherlands: 1995. [Google Scholar]
- Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen J.W. DrawMap: A computer program for drawing genetic linkage maps. J. Hered. 1994;85:66. [PubMed] [Google Scholar]
- Wang S., Basten C.J., Zeng Z.-B. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University; Raleigh, NC: 2005. [Google Scholar]
- WHO. Test procedures for insecticide resistance monitoring in malaria vectors, bio-efficacy and persistence of insecticides on treated surfaces. World Health Organization; Geneva, Switzerland: 1998. [Google Scholar]
- Wondji C.S., Hunt R.H., Pignatelli P., Steen K., Coetzee M., Besansky N., Lobo N., Collins F.H., Hemingway J., Ranson H. An integrated genetic and physical map for the malaria vector Anopheles funestus. Genetics. 2005;171:1779–1787. doi: 10.1534/genetics.105.044800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji C.S., Hemingway J., Ranson H. Identification and analysis of Single Nucleotide Polymorphisms (SNPs) in the mosquito Anopheles funestus, malaria vector. BMC Genomics. 2007a;8:5. doi: 10.1186/1471-2164-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji C.S., Morgan J.C., Coetzee M., Hunt R., Steen K., Black W.C., Hemingway J., Ranson H. Mapping a Quantitative Trait Locus conferring pyrethroid resistance in the African malaria vector Anopheles funestus. BMC Genomics. 2007b;8:34. doi: 10.1186/1471-2164-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z.B. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z.B., Kao C.H., Basten C.J. Estimating the genetic architecture of quantitative traits. Genet. Res. 1999;74:279–289. doi: 10.1017/s0016672399004255. [DOI] [PubMed] [Google Scholar]