Abstract
The natural history of cancers associated with virus exposure is intriguing, since only a minority of human tissues infected with these viruses inevitably progress to cancer. However, the molecular reasons why the infection is controlled or instead progresses to subsequent stages of tumorigenesis are largely unknown. In this article, we provide the first complete DNA methylomes of double-stranded DNA viruses associated with human cancer that might provide important clues to help us understand the described process. Using bisulfite genomic sequencing of multiple clones, we have obtained the DNA methylation status of every CpG dinucleotide in the genome of the Human Papilloma Viruses 16 and 18 and Human Hepatitis B Virus, and in all the transcription start sites of the Epstein-Barr Virus. These viruses are associated with infectious diseases (such as hepatitis B and infectious mononucleosis) and the development of human tumors (cervical, hepatic, and nasopharyngeal cancers, and lymphoma), and are responsible for 1 million deaths worldwide every year. The DNA methylomes presented provide evidence of the dynamic nature of the epigenome in contrast to the genome. We observed that the DNA methylome of these viruses evolves from an unmethylated to a highly methylated genome in association with the progression of the disease, from asymptomatic healthy carriers, through chronically infected tissues and pre-malignant lesions, to the full-blown invasive tumor. The observed DNA methylation changes have a major functional impact on the biological behavior of the viruses.
Epigenetics encompasses a large number of mechanisms underlying embryonic development, differentiation, and cell identity including DNA methylation and histone modifications and is increasingly recognized as being involved in human diseases such as cancer and imprinting disorders, among others (Bernstein et al. 2007; Esteller 2007; Feinberg 2007; Jirtle and Skinner 2007; Jones and Baylin 2007). For the first time, it is possible to define whole epigenomes, which represent all epigenetic marks in a given cell type, thanks to the development of new powerful genomic technologies (Bernstein et al. 2007; Esteller 2007; Feinberg 2007; Jirtle and Skinner 2007; Jones and Baylin 2007). In contrast to the human genome, human epigenomes vary between tissues, among individuals, and between healthy and disease states. Under these circumstances, distinct epigenomes might explain why the same genotypes generate different phenotypes as occurs in the Agouti mice (Michaud et al. 1994), cloned animals (Humpherys et al. 2001), and monozygotic twins (Fraga et al. 2005).
Determining the complete DNA methylome entails the description of all the methylated nucleotides in an organism. The gold standard technique for analyzing the methylation state of individual cytosines is bisulfite sequencing in which unmethylated cytosines are converted to uracils and read as thymines, while methylated cytosines are protected from conversion. Bisulfite sequencing yields precise nucleotide resolution data, but to date, this method has been limited to relatively small genome coverage (Rakyan et al. 2004; Eckhardt et al. 2006; Frigola et al. 2006). Alternative approaches involve the isolation of methylated fractions of the genome by methylation-sensitive restriction (Lippman et al. 2005), or immunoprecipitation with a methylcytosine-specific antibody (Weber et al. 2005, 2007; Keshet et al. 2006), combined with hybridization to genomic microarrays. This is exemplified by the recent DNA methylation analyses of the Arabidopsis genome (Zhang et al. 2006; Vaughn et al. 2007; Zilberman et al. 2007), further expanded by using sequencing-by-synthesis technology (Lister et al. 2008). We decided to combine these latter two approaches, extensive analyses of DNA methylation by bisulfite genomic sequencing of multiple clones and the use of model organisms, to obtain the complete DNA methylomes of double-stranded DNA viruses associated with human cancer. The chosen option recalls the U.S. Department of Energy's establishment in 1994 of the Microbial Genome Program (MGP) as a companion to its Human Genome Program. From the very start, the MGP achieved remarkable success, and microbial genomics has become one of the most exciting and high-profile fields in biology today. Thus, the complete description of the DNA methylomes of model organisms, such as double-stranded DNA viruses, might have an enormous impact on the success of the human epigenome project (Jones and Martienssen 2005; Brena et al. 2006).
Worldwide, it has been estimated that viral infections are etiologically linked to 15% of cancer cases, accounting for nearly 1.5 million new cases and 1 million deaths annually (Butel 2000; Gatza et al. 2005). Three well-studied oncogenic viruses are the human papilloma virus (HPV) (Papillomaviridae; Alphapapillomavirus; Human Papillomavirus), the hepatitis B virus (HBV) (Hepadnaviridae; Orthohepadnavirus; Hepatitis B virus), and the Epstein-Barr Virus (EBV) (Herpesviridae; Gammaherpesvirinae; Lymphocryptovirus; Human Herpesvirus 4). All of these are double-stranded DNA viruses, which are not only involved in the development of infectious diseases (such as infectious mononucleosis and hepatitis), but also in the formation of cervical, liver, and nasopharyngeal tumors and lymphomas, respectively (Butel 2000; Gatza et al. 2005). It is known that DNA methylation is present in some of these viral sequences—for example, in HPV16 (Thain et al. 1996; Kim et al. 2003) and EBV (Ambinder et al. 1999; Minarovits 2006)—but the complete coverage of their genomes and the biological consequences have not been systematically addressed. Herein, we present the complete DNA methylomes of the HPV16, HPV18, and HBV viruses and the DNA methylation analyses of all the transcription start sites of EBV obtained by bisulfite genomic sequencing of multiple clones. We also analyze the dynamic changes in the viral DNA methylome and their functional relevance in the natural history of the disease.
Results
The DNA methylome of HPV16
There are more than 100 different papilloma family members, and, in humans, these are responsible for a variety of benign proliferations. However, infection with two specific high-risk HPVs—HPV-16 and HPV-18—is associated with ∼90% of uterine cervical cancers, >50% of other anogenital tumors, and a small percentage of head and neck tumors (zur Hausen 2002). HPV particles consist of circular DNA molecules, ≈8000 bp long, wrapped into a protein shell that is composed of two molecules (L1 and L2). The genome has the coding capacity for these two proteins and at least six so-called early proteins (E1, E2, E4–E7) that are necessary for the replication of the viral DNA and for the assembly of newly produced virus particles within the infected cells. Both sets of genes are separated by an upstream regulatory region (URR) of ∼1000 bp that does not code for proteins but contains cis-elements required for regulation of gene expression, replication of the genome, and its packaging into virus particles (Butel 2000; zur Hausen 2002; Gatza et al. 2005). Two of the viral proteins—E6 and E7—are essential for HPV-mediated cellular transformation (Wise-Draper and Wells 2008). These interfere with many cellular processes, overriding signaling pathways and cell cycle control by altering the function of basal transcription factors TP53 (also known as p53) and RB1 (also known as Rb), among other target proteins (Wise-Draper and Wells 2008).
The natural history of the disease is intriguing, since only a minority of cervical tissues infected with HPV inevitably progress to cancer. Given that around 291 million women throughout the world are carriers of HPV DNA (de Sanjose et al. 2007), the incidence of cervical cancer is relatively low (zur Hausen 2002). Thus, the development of cervical cancer occurs in a few women who cannot resolve their infection and who maintain persistent active infection for years or decades following initial exposure. However, the molecular reasons why the infection is controlled or instead progresses to subsequent stages of tumorigenesis are largely unknown. The complete HPV DNA methylomes obtained in our study might provide important clues to help us understand the described process.
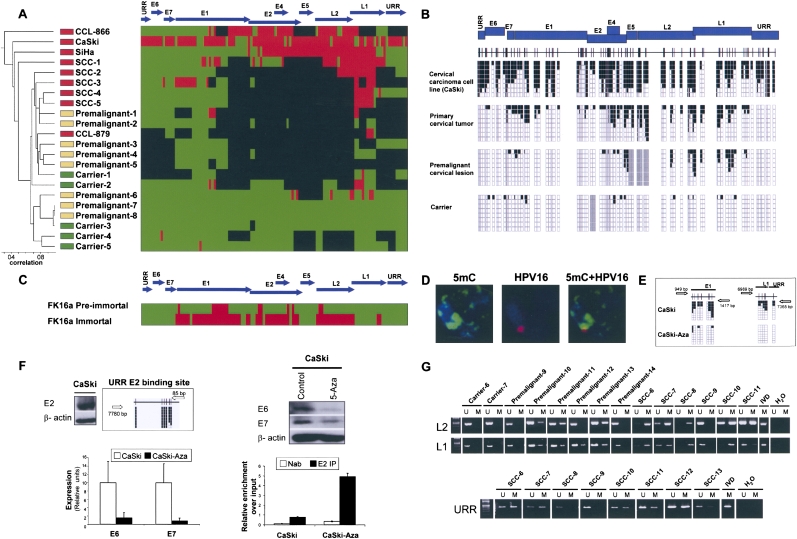
First, we sequence the whole DNA methylome of the HPV16 virus (110 CpGs in 7904 nt) in a collection of human cervical samples corresponding to the different progressive stages of the disease: asymptomatic carriers (n = 5), pre-malignant disease (the so-called Cervical Intraepithelial Neoplasia, CIN; n = 8), and the primary cervical carcinomas (n = 5). We have also completed the HPV16 DNA methylome of four established cervical cancer cell lines (CaSki, SiHa, CCL-866, and CLL-879). We discovered that the HPV16 DNA methylome undergoes a progressive increase in its DNA methylation content from tissue of women who are carriers of the virus but without any symptom of clinical disease, through early pre-tumorigenic lesions, to full-blown primary cervical carcinomas (Fig. 1A). In this sequence of events, the HPV16 DNA methylomes of the invasive cervical cancer cell lines CaSki and SiHa are the ones demonstrating the higher DNA methylation levels. We developed an unsupervised clustering analysis for the 18 HPV16 DNA methylomes obtained that also distinguished the four different biological groups: carriers, pre-malignancies, primary tumors, and cancer cell lines (Fig. 1A). The DNA methylation patterns and deletions of the HPV genomes are both responsible in the establishment of the clustering. Examples of the bisulfite genomic sequencing analysis of multiple clones of the HPV16 virus genome are shown in Figure 1B. To examine the HPV16 DNA methylomes, please go to http://ubio.bioinfo.cnio.es/Methylyzer/main/index.html.
Figure 1.
The DNA methylome of HPV16. (A) Unsupervised clustering analysis of the HPV16 DNA methylome in asymptomatic carriers of the virus, pre-malignant lesions (CIN), primary tumors (SCCs), and cancer cell lines (CaSki, SiHA, CCL-879, and CCL-866). (Red) Methylated, (green) unmethylated, and (black) deleted sequences. (Top) The HPV16 genome. (B) Example of bisulfite genomic sequencing analysis of multiple clones for the HPV16 genome. (Black squares) Methylated and (white squares) unmethylated CpG dinucleotides; (gray squares) deleted genome sequences. (C) Bisulfite sequencing showing how the mostly unmethylated HPV16 DNA methylome from pre-immortal keratinocytes undergo hypermethylation in immortalized cells. (D) Confocal studies for fluorescence in situ hybridization for HPV16 and 5-methylcytosine DNA staining demonstrate the colocalization of the integrated viral genome in a human chromosome and the DNA methylation signal in CaSki cells. (E) The use of a DNA demethylating agent in CaSki cells causes DNA hypomethylation of the HPV16 genome. (F) The expression of HPV16 E6 and E7 oncoproteins is associated with a loss of binding of the E2 repressor to the URR methylated sites. (Top left) The expression of E2 and the methylated status of the URR binding site in CaSki. (Top right) DNA demethylating treatment reduces E6 and E7 expression in Western blot and (bottom left) qRT-PCT in association with a recruitment of the E2 protein to the demethylated URR E2-binding site, (bottom right) shown by q-ChIP. (G) Methylation-specific PCR analysis of the HPV16 L2, L1, and URR sequences in cervical tumorigenesis. The presence of a band under the U or M lanes indicates unmethylated or methylated sequences. In vitro methylated DNA (IVD) is shown as a positive control.
The dynamic changes experienced by the viral DNA methylome during the tumorigenic process were highlighted when we studied cultured primary human foreskin keratinocytes transfected with the entire genome of HPV16 that integrates in the host genome (Steenbergen et al. 1996; Van Tine et al. 2004). The HPV16 DNA methylome from the pre-immortal keratinocytes was almost completely unmethylated, while the immortal descendant cells featured a densely methylated viral genome (Fig. 1C). Most importantly, genetic changes in the HPV16 genome were not observed. This observation was a universal feature of our studies: Because our DNA methylation analysis by bisulfite genomic sequencing also allows putative nucleotide changes (such as point mutations, insertions, or deletions) to be identified, we were able to assess the contribution of these genetic changes in the virus itself to the progression of the disease (with the exception of T or C nucleotide changes that could be caused by failed bisulfite conversion). We did not observe any particular viral nucleotide change associated with the natural history of cervical tumorigenesis.
The HPV16 genome does not code for any gene of the DNA methylation machinery, such as DNA methyltransferases (DNMTs). Thus, the viral genome is methylated by the host human cellular DNMTs. The integration of HPV16 in the human genome occurs in most cervical tumors and a significant proportion of pre-malignant lesions, and so it is reasonable to suggest that human DNMTs might methylate these sequences (Bernstein et al. 2007; Esteller 2007; Feinberg 2007; Jirtle and Skinner 2007; Jones and Baylin 2007). A list of the known integration sites for the studied viral genomes is provided in Supplemental Table S1. Indeed, in our study, we have confirmed by FISH analysis that the HPV16 genome is integrated within the host cell DNA in CaSki and SiHa cells (Supplemental Fig. S1). A correlation between a higher number of integrated copies and a higher DNA methylation level might exist, and it could be the focus of future research in the area. A list of the known copy numbers for the studied viral genomes is provided in Supplemental Table S2. Most importantly, confocal studies demonstrated the colocalization of the integrated viral genome and 5-methylcytosine DNA staining (Fig. 1D). The use of a DNA demethylating agent (5-aza-2′-deoxycytidine) in CaSki cells did, indeed, cause DNA hypomethylation of the HPV16 genome (Fig. 1E). Interestingly, the HPV16 genome is not merely a passive spectator of the process but might actively participate by recruiting DNMTs using the viral oncoprotein E7 (Burgers et al. 2007).
The HPV16 DNA methylomes described here also reflect the proposed expression patterns of the virus in cervical tumorigenesis, which are characterized by a significant overexpression of E6 and E7 in the carcinomas (Butel 2000; zur Hausen 2002; Gatza et al. 2005; Wise-Draper and Wells 2008). Our sequencing effort shows that, for a subset of cases spanning the range of stages, there are genomic viral regions that are deleted, particularly the E1/E2 region. The protein E2 inhibits cell proliferation by regulating the viral upstream regulatory region (URR), repressing the E6 and E7 oncoproteins, and, ultimately, causing cell cycle arrest at G2 (Frattini et al. 1997). Thus, the genomic loss of E2 could contribute to the progression of the disease. However, the HPV16 DNA methylomes described here show that there are many cases in which there are not any ruptures at the E2 locus, and, accordingly, E2 is expressed (Fig. 1F). We observed instead methylation of the E2-binding sites at the URR region (Fig. 1A,F) that was associated with a high level of expression of the oncoproteins E6 and E7 (Fig. 1F). Most importantly, we showed by chromatin immunoprecipitation (ChIP) that the E2 viral protein could not bind to the methylated E2-binding sites at the URR region (Fig. 1F), and thus led to E6 and E7 overexpression. Finally, the induction of hypomethylation events in the E2-binding sites at the URR region by a DNA demethylating agent (5-aza-2′-deoxycytidine) induced the recruitment of E2 to its URR-binding sites and a marked reduction of E6 and E7 expression (Fig. 1F). Most important, methylation of the E2-binding sites at the URR region was observed in six of eight (75%) primary cervical tumors that did not present E2 genomic deletion (Fig. 1G). Thus, our in vivo findings support the in vitro data about the inhibition of E2 binding to CpG methylated E2-binding sites at the URR (Thain et al. 1996).
We went one step further to confirm these results in a large collection (n = 99) of human primary samples from the different stages of cervical carcinogenesis. Once we have discarded those cervical tumors with L2 deletions (n = 12), we conducted in the remaining samples (n = 87), the technique of methylation-specific PCR to analyze the L2 and L1 DNA methylation status (Fig. 1G). We observed the progressive presence of hypermethylation at the L2 locus in tumorigenesis: 0% (0 of 10) in asymptomatic carriers, 29% (five of 17) in stage I of intraepithelial neoplasia (CIN I), 37% (16 of 43) in stages II and III of intraepithelial neoplasia (CIN II–III), and 94% (16 of 17) in primary cervical carcinoma. L1 methylation was analyzed in a subset of these cases (n = 14), and it was always concordant with L2 methylation. Overall, the DNA methylomes of HPV16 outlined are evidence of the existence of an adaptive DNA methylation pattern of the viral genome, whereby there was an increased methylated-CpG content during the progression of the disease arising from the cross-talk between the viral and the host genomes.
The DNA methylome of HPV18
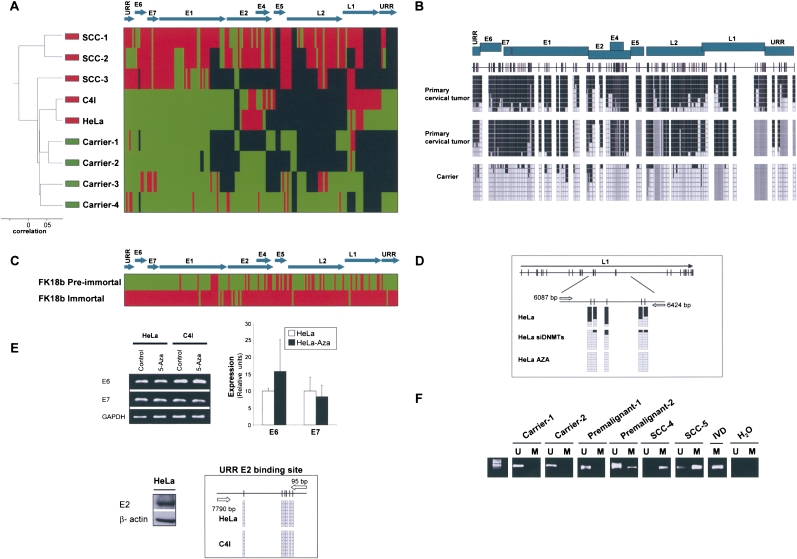
HPV16 is the most prevalent high-risk papilloma virus in the general population and is responsible for ∼50% of cervical cancers, but HPV18 occupies second place in both instances. HPV16 and HP18 are representative of two different HPV species (alpha9 and alpha7, respectively), but share a global genomic structure that codes for almost identical proteins (Butel 2000; zur Hausen 2002; Gatza et al. 2005; Wise-Draper and Wells 2008). Most importantly, a highly similar pattern was encountered when we obtained the complete DNA methylomes of the HPV18 virus. The bisulfite genomic sequencing of the entire HPV18 viral genome (168 CpGs in 7857 nt) obtained from asymptomatic carriers of the virus demonstrated a low level of methylation, while an increased methylated genome was observed in primary cervical carcinomas (Fig. 2A). The unsupervised clustering analysis was also able to distinguish infected tissues without clinical relevance from the full-blown tumors on the basis of the HPV18 DNA methylomes (Fig. 2A). Examples of the bisulfite genomic sequencing of multiple clones are shown in Figure 2B. The HPV18 DNA methylomes may be examined at http://ubio.bioinfo.cnio.es/Methylyzer/main/index.html. We not only analyzed the static nature of these DNA methylomes at given stages, but, as we did with HPV16, also obtained the HPV18 DNA methylomes present during the immortalization crisis that keratinocytes transfected with the entire HPV18 genome (integrated in the host genome) undergo (Steenbergen et al. 1996; Van Tine et al. 2004). The HPV18 DNA methylome from the pre-immortal keratinocytes was mostly unmethylated, whereas the immortal cells presented a densely methylated viral genome (Fig. 2C). Most importantly, no genetic or nucleotide changes in the HPV18 genome were observed. The HPV18 genome does not encode for any DNMTs, thus it is believed to “use” the host enzymes. Short interference RNA depletion of DNMT1 and DNMT3B did, indeed, induce hypomethylation events in the HPV18 positive cervical cancer cell line HeLa (Fig. 2D). Although HPV16 and HPV18 share many features in their DNA methylomes, one interesting distinction occurs for the viral oncoprotein expression standpoint. The levels of E6 and E7 expression in HPV18-infected cervical cancer cell lines, such as HeLa and C4I, were not modified by the treatment with a DNA demethylating agent (Fig. 2E). These findings fit perfectly well with the observation that the E2-binding sites at the URR region of HPV18 in these cells were not methylated (Fig. 2A,E), and thus the E2-mediated control of E6 and E7 expression could not be released by the DNA hypomethylating drug. The presence of DNA methylation at the E2-binding site of the URR region of HPV16, that drives the E6 and E7 expression, and its absence in HPV18, might be another factor that could explain the different prevalence and carcinogenicity of both strains.
Figure 2.
The DNA methylome of HPV18. (A) Unsupervised clustering analysis of the HPV18 DNA methylome in asymptomatic carriers of the virus, primary tumors (SCCs), and cancer cell lines (HeLa and C4I). (Red) Methylated, (green) unmethylated, and (black) deleted sequences. (Top) The HPV18 genome. (B) Example of bisulfite genomic sequencing analysis of multiple clones for the HPV18 genome. (Black squares) Methylated and (white squares) unmethylated CpG dinucleotides; (gray squares) deleted genome sequences. (C) Bisulfite sequencing showing how the mostly unmethylated HPV18 DNA methylome from pre-immortal keratinocytes undergo hypermethylation in immortalized cells. (Red) Methylated and (green) unmethylated sequences. (D) Depletion of DNMT1 and DNMT3B by short interference RNA or upon treatment with the DNA demethylating agent causes a DNA hypomethylation of the HPV18 genome in HeLa cells. (E) The use of a DNA demethylating agent (5-aza-2′-deoxycytidine) does not change the expression of the HPV18 E6 and E7 oncoproteins in HeLa cells in association with an unmethylated URR E2-binding site. (Top) Conventional RT-PCRs and q-RT-PCRs; (bottom) expression of E2 and bisulfite sequencing that demonstrates an unmethylated URR E2-binding site. (F) Methylation-specific PCR analysis of HPV16 E2 sequence in cervical tumorigenesis. The presence of a band under the U or M lanes indicates unmethylated or methylated sequences. In vitro methylated DNA (IVD) is shown as a positive control.
Finally, to extend our analysis to a larger collection (n = 32) of human primary samples from the different stages of HPV18-mediated cervical carcinogenesis, we used the technique of methylation-specific PCR for the E2 region of the virus (Fig. 2F). We confirmed the progressive presence of hypermethylation at the E2 locus in tumorigenesis: 0% (0 of 6) in asymptomatic carriers, 14% (one of seven) in stage I of cervical intraepithelial neoplasia (CIN I), 67% (six of nine) in stages II and III of cervical intraepithelial neoplasia (CIN II–III), and 90% (nine of 10) in primary cervical carcinoma.
The DNA methylome of HBV
The third genome whose complete DNA methylome we obtained is the Hepatitis B Virus (HBV). The HBV infectious virion has an outer envelope, formed by the hepatitis B surface antigen (HBsAg) in a lipid bilayer. This encloses the nucleocapsid core, within which lies the viral genome (Gatza et al. 2005; Lupberger and Hildt 2007). The latter is a relaxed circular, partially double-stranded DNA molecule of 3215 bp in length and contains four partially overlapping open reading frames (ORFs). These code for the envelope glycoproteins (L, M, and S HBsAgs), the pre-core/core (precursor of the soluble hepatitis B e antigen, HBeAg, and the core protein), the X protein (HBx), and the polymerase, which acts as reverse transcriptase and also has DNA polymerase activity (Gatza et al. 2005; Lupberger and Hildt 2007). It is interesting that while HBV is a DNA virus, it replicates through an RNA intermediate and thus requires the described active viral reverse transcriptase polymerase enzyme. HBV infects more than 2 billion people worldwide, 400 million of which are chronically infected and are at high risk for the development of active hepatitis, cirrhosis, and hepatocarcinoma (HCC) (Gatza et al. 2005; Lupberger and Hildt 2007). The HBV carriers with chronic liver disease have 100-fold greater risk of developing HCC, which is the third leading cause of cancer death in the world (Gatza et al. 2005; Lupberger and Hildt 2007). Ninety percent of HBV-associated liver cancers show integration of the HBV genome within the human genome (Gatza et al. 2005; Lupberger and Hildt 2007). Two major HBV-specific mechanisms contribute to the development of HCC: The viral genome integration in the human genome causes cis-effects that inactivate tumor-suppressor genes and activate oncogenes; and the expression of trans-activating factors derived from the HBV genome, such as the HBx protein and the PreS2 activators (Schluter et al. 1994), which disrupt the signal transduction pathways and alter the gene expression of the host cell (Gatza et al. 2005; Lupberger and Hildt 2007).
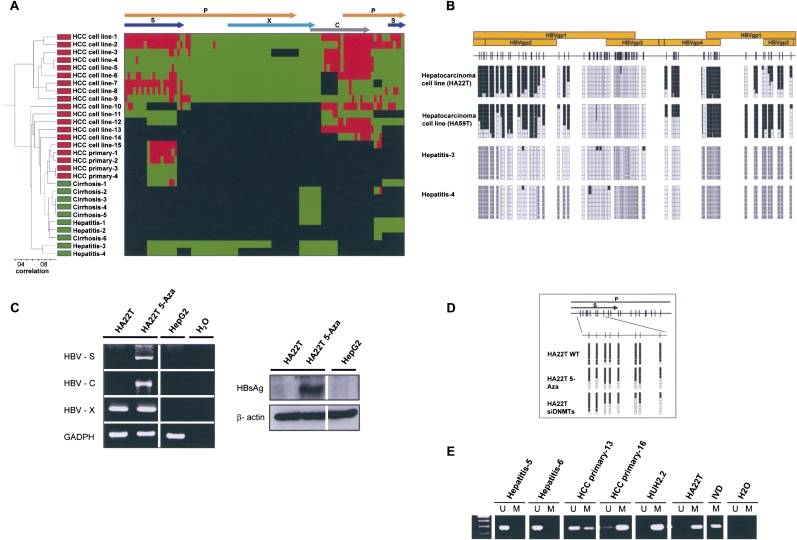
We completed the HBV DNA methylomes (n = 32) from different stages of the liver tumorigenesis, from chronic active hepatitis (CAH) and hepatic cirrhosis (HC) to primary hepatocarcinomas (HCCs) and a well-characterized panel of hepatic cancer cell lines. The samples included genotypes D and A of HBV (Supplemental Table S3). The results are in concordance with those observed for HPV16 and HPV18: The HBV genome is almost completely unmethylated in the early stages of carcinogenesis, such as hepatitis and cirrhosis, while it becomes more methylated in the established liver tumors, both in patients and in cultured cancer cell lines (Fig. 3A). Unsupervised clustering analysis was also able to classify these liver samples as pre-malignant or malignant stages on the basis of their HBV DNA methylomes (Fig. 3A). The DNA methylation patterns and deletions of the HBV genomes are both responsible in the establishment of the clustering. The HBV DNA methylomes may be examined at http://ubio.bioinfo.cnio.es/Methylyzer/main/index.html. Examples of the bisulfite genomic sequencing of multiple clones are shown in Figure 3B. Most importantly, the presence of DNA methylation at the HBVgp4 and HBVgp2 genes, which, respectively, code for the C and S viral proteins, is associated with their lack of expression (Fig. 3C), while release of transcriptional silencing can be achieved by the use of a DNA demethylating agent (Fig. 3C).
Figure 3.
The DNA methylome of HBV. (A) Unsupervised clustering analysis of the HBV DNA methylome in carriers of the virus with pre-malignant lesions such as cirrhosis and hepatitis, primary liver tumors (HCC primary), and liver cancer cell lines (HCC cell line). (Red) Methylated, (green) unmethylated, and (black) deleted sequences. (Top) The HBV genome. (B) Example of bisulfite genomic sequencing analysis of multiple clones for the HBV genome. (Black squares) Methylated and (white squares) unmethylated CpG dinucleotides; (gray squares) deleted genome sequences. (C) DNA methylation-associated silencing of the HBVgp4 (HBV-C) and HBVgp2 (HBV-S) genes in the hepatic carcinoma cell line HA22T and reactivation upon the use of a DNA demethylating agent (5-aza-2′-deoxycytidine). The HBVgp3 remains unmethylated and expressed. (Left) RT-PCRs; (right) Western blots. The hepatoblastoma cell line HepG2, negative for the HBV virus, is used as a negative control. (D) Depletion of DNMT1 and DNMT3B by short interference RNA or upon treatment with the DNA demethylating agent (5-Aza) causes a DNA hypomethylation of the HBV genome in HA22T cells. (E) Methylation-specific PCR analysis of the HBVgp2 sequence in liver tumorigenesis. The presence of a band under the U or M lanes indicates unmethylated or methylated sequences. In vitro methylated DNA (IVD) is shown as a positive control.
Our sequencing data also show that most of the HBV genomes, although more methylated than the pre-malignant lesions, retained the HBVgp3 gene that codes for the X protein (HB-X) in an unmethylated state (Fig. 3C). HB-X was transcribed in all the studied liver cancer cell lines (Fig. 3C). Interestingly, the HB-X protein might regulate DNMT activity (Park et al. 2007). In this regard, double depletion of DNMT1 and DNMT3B by short interference RNA in HA22T cells did, indeed, cause DNA hypomethylation of the HBV genome (Fig. 3D). Our extensive bisulfite genomic sequencing effort also established that the integrated HBV genome undergoes significant deletions, as previously reported (Gatza et al. 2005; Lupberger and Hildt 2007), which are more common in CAH and HC than in liver tumors. We developed a FISH analysis for the HBV virus to demonstrate further its presence in the liver tumors (Supplemental Fig. S2). Finally, to expand our study of the HBV DNA methylome and to confirm its dynamic nature, we tested a larger collection (n = 35) of human primary samples from the different stages of HBV-mediated liver carcinogenesis, using the technique of methylation-specific PCR for the HBVgp2 gene of the virus (Fig. 3E). We have confirmed the progressive presence of hypermethylation at the HBVgp2 locus in the hepatic tumorigenesis: 0% (zero of five) in HC, 11% (one of nine) in CAH, and 52% (11 of 21) in primary liver tumors.
Thus, these data suggest that DNA methylation of the HBV genome may “mark” those cells that progress toward tumorigenesis, as was implied by the analysis of the HPV16 and HPV18 DNA methylomes.
The DNA methylome of EBV
The final extensive DNA methylation of a viral genome we analyzed in the present study is that of the Epstein-Barr virus (EBV), which is several orders of magnitude larger than those considered above (171,823 bp). EBV was discovered more than 40 yr ago examining cells cultured from Burkitt's lymphoma, a childhood tumor that is common in sub-Saharan Africa. However, instead of showing a restricted distribution, EBV has been found to be widespread in all human populations. Ninety percent of the world's adult population is infected by the virus, and it persists in the vast majority of individuals as a lifelong, asymptomatic infection of the B-lymphocyte cells (Young and Rickinson 2004; Gatza et al. 2005; Williams and Crawford 2006; Klein et al. 2007), but can occasionally evolve to an infectious mononucleosis (IM). In immunocompromised humans, such as AIDS and post-transplant patients, EBV infection is implicated in the etiology of several different lymphoid and epithelial malignancies, such as nasopharyngeal carcinoma (Young and Rickinson 2004; Gatza et al. 2005; Williams and Crawford 2006; Klein et al. 2007). There are two alternative states of EBV infection: lytic and latent. Virions are only produced in lytic infection, and its genome remains as an episome in the host cell (Young and Rickinson 2004; Gatza et al. 2005; Williams and Crawford 2006; Klein et al. 2007). EBV has the ability to transform resting B-cells into permanently infected lymphoblastoid cell lines that constitutively express a limited set of viral products, which are called “latent proteins.” These oncogenic genes are the nuclear antigens EBNAs 1, 2, 3A, 3B, 3C, and LP; and the latent membrane proteins LMPs 1, 2A, and 2B. Other gene products are the immediate early genes (which are considered the switch between latent and lytic cycle), the early genes (e.g., enzymes influencing the host cell nucleotide metabolism and DNA synthesis), and the late gene products (e.g., the virion structural proteins) (Young and Rickinson 2004; Gatza et al. 2005; Williams and Crawford 2006; Klein et al. 2007).
The large EBV genome, of almost 172 kb, prevented us from deriving the entire EBV DNA methylome using the same technology we had used for the HPV16, HPV18, and HBV genomes. Instead, we undertook the most extensive DNA methylation analysis of the EBV genome to date using bisulfite genomic sequencing analysis of multiple clones for 77 amplicons that contains the transcription start sites that code for the 94 EBV proteins and the two structural RNAs, EBER1 and EBER2. The complete set of EBV transcription start sites studied included 95% (72 of 76) CpG islands (Takai and Jones 2003) and covered 1344 CpGs of the EBV genome. Overall, 22 EBV DNA methylomes were obtained from a collection of lymphoid samples that included benign proliferating cells, such as reactive lymphadenitis and infectious mononucleosis, non-tumorigenic lymphoblastoid cell lines, post-transplant lymphoproliferative disorder, and EBV-associated lymphomas from primary tissues and cell lines, such as non-Hodgkin lymphoma (diffuse large B-cell lymphoma and peripheral T-cell lymphoma), Hodgkin lymphoma, and Burkitt's lymphoma. All the samples corresponded to EBV type A (Supplemental Table S3). In addition, EBV-positive nasopharyngeal tumors were also included.
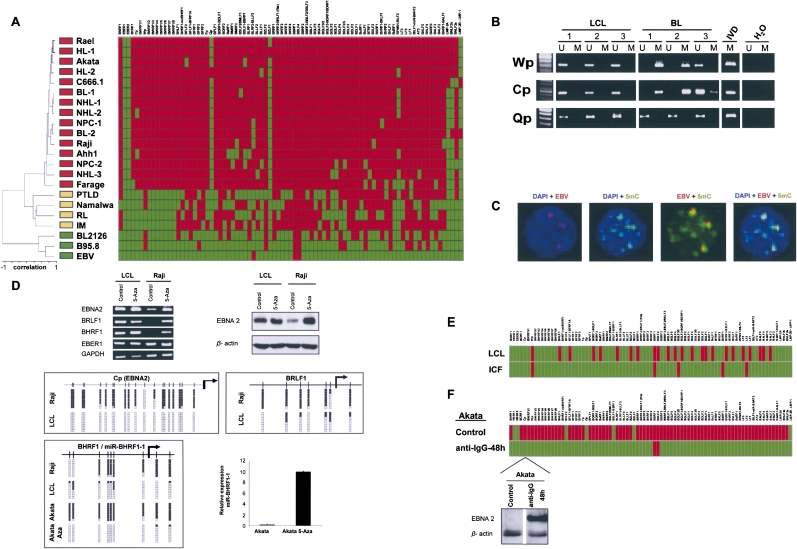
We observed a similar EBV DNA methylation profile to that observed for HPV16, HPV18, and HBV. The EBV DNA obtained from free viral particles is devoid of DNA methylation (Fig. 4A); however, the EBV genome present in human cells corresponding to benign diseases, such as reactive lymphadenitis in tonsils and infectious mononucleosis, showed the incipient presence of methylated EBV transcription start sites (Fig. 4A). The same occasional hypermethylation was observed in non-tumorigenic human B-cell-derived lymphoblastoid cell lines (Fig. 4A). Remarkably, this pattern was significantly changed in cancer cells infected with EBV, where the majority of lymphoma and nasopharyngeal samples from primary tumors and cell lines had a large number of hypermethylated EBV transcription start sites (Fig. 4A). Unsupervised clustering analysis was able to distinguish free virus DNA, EBV-related benign conditions, and EBV-associated human lymphomas and epithelial carcinomas on the basis of the EBV DNA methylome (Fig. 4A). These results were confirmed by methylation-specific PCR in a second set of EBV-positive non-tumorigenic (n = 16) versus EBV-lymphoma (n = 21) cell lines: Lymphoblastoid cell lines did not present promoter hypermethylation of Wp and Cp genes, while Burkitt's lymphoma cell lines had 43% and 23% hypermethylation frequencies, respectively (Fig. 4B). Confocal studies demonstrated the colocalization of the EBV viral genome determined by FISH (Supplemental Fig. S3) and the 5-methylcytosine DNA staining in Burkitt's lymphoma cells (Fig. 4C). The EBV DNA methylomes may be examined at http://ubio.bioinfo.cnio.es/Methylyzer/main/index.html.
Figure 4.
The DNA methylome of EBV. (A) Unsupervised clustering analysis of the EBV DNA methylome in the wild EBV, the B95-8 type EBV, lymphoblastoid cell line (BL2126), benign lesions ([RL] reactive lymphadenitis in tonsil and [IM] infectious mononucleosis), post-transplant lymphoproliferative disorder (PTLD), primary lymphomas ([NHL] non-Hodgkin lymphoma, [HL] Hodgkin lymphoma, [BL] Burkitt's lymphoma), lymphoma cell lines (Rael, Akata, Raji, Ahh1, Farage, Namalwa), nasopharyngeal primary carcinoma (NPC), and cancer cell line (C666.1). (Red) Methylated and (green) unmethylated CpG dinucleotides encompassing the corresponding 5′-end transcription start sites. (Top) The EBV genome. (B) Methylation-specific PCR analysis of the EBV FP, WP, and CP 5′-end CpG islands in lymphoblastic and Burkitt's lymphoma cell lines. The presence of a band under the U or M lanes indicates unmethylated or methylated sequences. In vitro methylated DNA (IVD) is shown as a positive control. (C) Colocalization of the EBV viral genome determined by FISH and the 5-methylcytosine DNA staining in Akata cells. (D) 5′-CpG island methylation-associated silencing of the EBNA2, BRLF1, and BHRF1-H2H3 genes and the microRNA miR-BHRF1-1 and release of silencing upon the use of a DNA demethylating agent (5-aza-2′-deoxycytidine). (Top) RT-PCRs and Western blots; (middle) bisulfite genomic sequencing; (bottom) q-RT-PCR for miR-BHRF1-1. (E) EBV-immortalized ICF lymphocytes (DNMT3B defective) demonstrate a hypomethylated EBV genome in comparison to the one observed in DNMT3B-proficient EBV-transformed lymphocytes (LCL). (Red) Methylated and (green) unmethylated CpG dinucleotides encompassing the corresponding 5′-end transcription start sites. (F) The induction of the EBV lytic cycle in a lymphoma cell line (Akata) upon addition to the media of anti-IgG caused a massive DNA hypomethylating event in the EBV-transcription start sites. (Red) Methylated and (green) unmethylated CpG dinucleotides encompassing the corresponding 5′-end transcription start sites. (Bottom) Example of Western blot for the EBNA2 protein showing the restoration of expression upon the demethylation of the corresponding CpG island.
Interestingly, of the 77 transcription start site regions that originate the entire EBV mRNA transcriptome, five amplicons “escaped” the DNA methylation mark: the two structural RNAs, EBER1 and EBER2, and the Qp, BZLF1, and LMP-2B/LMP-1 sequences (Fig. 4A). Methylation-specific PCR analyses for the Qp sequence confirmed the absence of DNA methylation from the additional set of both lymphoblastoid (n = 16) and Burkitt's lymphoma (n = 21) cell lines (Fig. 4B). RT-PCR expression confirmed that the always unmethylated EBER1 was expressed in all the analyzed samples (Fig. 4D). Most importantly, the presence of a hypermethylated EBV transcription start site in the cancer cells was associated with the corresponding transcriptional silencing of the neighboring gene. We demonstrated this close association between a methylated transcription start site and its loss of transcription at the single gene level, using RT-PCR and Western blotting (Fig. 4D) for the EBNA2, BRLF1, and BHRF1 genes, and at the global level, matching the obtained EBV methylomes with our previous study of the EBV transcriptome by expression microarrays (Supplemental Fig. S4; Yuan et al. 2006). Notably, as it has also been shown in human cells (Saito et al. 2006; Lujambio et al. 2007), the presence of DNA methylation in a microRNA locus was also associated with its loss of expression. This was the case for miR-BHRF1-1, located in the 5′-untranslated region of the BHFR1 gene (Pfeffer et al. 2004), which was hypermethylated in Akata cells (Fig. 4D). The link between DNA methylation and silencing was further reinforced by the use of the DNA demethylating agent 5-aza-2′-deoxycytidine, which was able to restore the expression of the EBV methylation-inactivated genes and of miR-BHRF1-1 (Fig. 4D).
The EBV genome could be a target of the human DNMTs. In the case of EBV, we have a research avenue that has been provided by nature itself. Some individuals are born with a germline defect in DNMT3B (Immunodeficiency, Centromere Instability, Facial Anomalies syndrome, ICF, OMIM #242860) (Okano et al. 1999; Xu et al. 1999), and lymphocytes immortalized with EBV from these patients have been widely used to understand the DNA methylation machinery (Matarazzo et al. 2007; Jin et al. 2008). The analysis of DNA methylation of EBV-immortalized ICF lymphocytes revealed a more hypomethylated EBV genome than that observed in DNMT3B-proficient EBV-transformed lymphocytes (Fig. 4E), which is evidence of the role of DNMT3B in establishing the EBV DNA methylome. Interestingly, in a similar fashion to what occurs with HPV16, HPV18, and EBV, there may be a cross-talk between the EBV-viral proteins and the DNA methyltransferases of the host human cell. The viral LMP1 protein induces the expression and activity of human DNMTs (Tsai et al. 2002; Tsai et al. 2006), but it is not known whether it induces hypermethylation of the EBV genome. We have addressed this issue by the depletion of LMP1 using the RNA interference approach in the RAJI lymphoma cell line. Knockdown of LMP1 induced DNMT1 and DNMT3B depletion and hypomethylation of particular CpG sites in the EBV genome (Supplemental Fig. S5).
Finally, as we showed for the other three viral double-strand DNA methylomes, the EBV DNA methylome was highly adaptable to the biological and environmental circumstances surrounding the host cell. The induction of the EBV lytic cycle in a lymphoma cell line (Akata) upon addition of anti-IgG to the media (Yuan et al. 2006) caused a major change in the EBV DNA methylome, in just 48 h, characterized by the presence of unmethylated transcription start sites, most likely associated with the newly replicated viral DNA (Fig. 4F). The viral hypomethylated DNA was accompanied by the restoration of gene expression of the previously methylated genes, which matched our expression microarray data (Supplemental Fig. S4; Yuan et al. 2006), and as is confirmed here by Western blot (Fig. 4F). Thus, the dynamic nature of the epigenome, exemplified here by the changing EBV DNA methylome, was further highlighted.
Discussion
Our analysis provides for the first time the complete DNA methylomes of organisms associated with human disease, the three double-stranded DNA viruses HPV16, HPV18, and HBV, in addition to the DNA methylation status of all the transcription start sites of another double-stranded DNA virus, EBV. These DNA methylomes have been studied in more than 80 human samples infected with these viruses at different stages of the disease, and this large-scale sequencing effort has revealed the dynamic nature of the epigenome, at least at the level of DNA methylation. One of the main findings of our study is the progressive increase in the DNA methylation content in these viruses from asymptomatic carriers, through benign lesions and pre-malignant disease, to full-blown human tumors. The DNA methylation of “foreign” (including viral) DNA integrated into the human genome has been studied for decades (Doerfler 2008), but one particularly interesting possibility is that DNA methylation might be a device to camouflage the virus from our immune system (Tao and Robertson 2003; Hilleman 2004). A DNA methylation-associated blockade of viral antigen presentation could be used to evade immune control. Most interestingly, the use of clinically approved DNA demethylating agents for the treatment of hematological malignancies, in addition to restoring the expression of epigenetically silenced tumor-suppressor genes of the host cell (Bernstein et al. 2007; Esteller 2007; Feinberg 2007; Jirtle and Skinner 2007; Jones and Baylin 2007), might also reactivate the immune response, thereby enhancing its therapeutic benefits. The DNA methylomes presented here could be a good starting point for biomedical researchers who wish to address these questions or to understand how the viral proteins themselves are able to use our own DNMTs to favor the establishment of persistent infection. These issues go beyond basic research and might have a great impact on public health, since many millions of people in the world are carriers of these viruses. The potential clinical applications of these findings include the non-invasive detection of methylated viral genomes in biological fluids, serum, and blood. From a geographical location standpoint, we did not observe different DNA methylation profiles, for example, between the NPC cell line from Southern China and the primary NPCs from Southern Europe, but larger studies are necessary to address this issue and other environmental factors. Higher-sequence resolution might help to discriminate between the scenario wherein a small number of viral genomes are already methylated among the large population of unmethylated viral genomes and expand into the cancerous cells or if there is a methylation change in the viral genome that drives the carcinogenic change. Above all, the DNA methylomes obtained here, although owned by very small organisms, could be an excellent starting point for launching ultra-deep sequencing projects aimed at the complete description of the human DNA methylome, in a similar way to which microbial genomes stimulated the race to derive the human genome. Our viral DNA methylomes, in this regard, could be an excellent proof of principle for the successful completion of ongoing human epigenome projects.
Methods
Cell lines and primary tissues
The human cell lines used in this study were obtained from the American Type Culture Collection (Rockland, MD), the German Collection of Microorganisms and Cell Cultures (DMSZ, Braunschweig, Germany), and the Coriell Institute for Medical Research (Camden, NJ). Cell lines were maintained in the appropriate medium and treated with 5-aza-2-deoxycytidine at a concentration of 1 μmol/L for 3 d to achieve DNA demethylation. Cell lines of primary human foreskin keratinocytes transfected with HPV16 or HPV18 were obtained as previously described (Steenbergen et al. 1996). The EBV lytic cycle was induced in Akata cells upon addition of anti-IgG to the media as previously described (Yuan et al. 2006). Primary tissues from normal, pre-malignant, and tumoral samples were all obtained at the time of the clinically indicated procedures.
Western blot analysis
To study the levels of viral proteins in the cell lines used in the study, cells were lysed with RSB buffer (10 mmol/L Tris at pH 7.5, 10 mmol/L NaCl, 3 mmol/L MgCl2) in the presence of 1% NP-40 (17) and resuspended in SDS-PAGE loading buffer. Equal amounts of proteins were separated on 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membranes were then hybridized with antibodies against HPV16 E6 (N-17, sc-1584 1:200), HPV16 E7 (ED17, sc-6981 1:200), HPV16 E2 (TVG261, ab17185: 1:250), Hepatitis B Virus X antigen antibody (3F6-G10, ab 235, 1:250), Hepatitis B Virus Surface Antigen (Ad/Ay) antibody (86C, ab20758, 1:200; Abcam), HBV HBcAg (C1-5, sc-23945 1:200), and EBNA2 (DakoCytomation M7004 1:100). The proteins were detected with the Enhanced Chemiluminescence Plus Western Blotting Detection System (Amersham Biosciences).
Analysis of CpG methylation status
DNA methylation patterns in the CpG islands were determined by bisulfite-mediated conversion of unmethylated, but not methylated, cytosines to uracil as previously described (Clark et al. 1994; Herman et al. 1996). Bisulfite genomic sequencing of multiple clones was performed following the detailed protocol described at http://www.epigenome-noe.net/researchtools/protocol.php?protid=34. In brief, we first modified the DNA with sodium bisulfite as described (Herman et al. 1996) and then used Methyl Primer Express software (Applied Biosystems) to design bisulfite-modified DNA-specific oligonucleotides spanning the whole genomes of the viruses HPV16, HPV18, and HBV viruses, and all the transcription start sites of the EBV virus. We next amplified the promoter regions under standard PCR conditions, cloned the PCR products into pGEM-TEasy Vector (Promega), randomly selected at least eight colonies per sample, and performed sequencing using an ABI Prism 3130XL Applied Biosystems DNA sequencer. Finally, we analyzed the methylation of every CpG using Sequencing Analysis 5.2 Software from Applied Biosystems. DNA methylation at each CpG site was determined in at least eight clones by bisulfite sequencing of the forward or reverse strands. For primary samples, we also developed a second analysis using methylation-specific PCR (MSP) (Herman et al. 1996). A complete table with all the primers used is provided in Supplemental Tables S4–S7. To view all the viral DNA methylomes, please go to http://ubio.bioinfo.cnio.es/Methylyzer/main/index.html.
Quantitative chromatin immunoprecipitation (qChIP) assays
To investigate the presence of E2 protein at the E2-binding sites of the URR region of the HPV16 genome, chromatin immunoprecipitation (ChIP) assays were done as described previously (Ballestar et al. 2003) using the E2 antibody (TVG261, ab17185). Cell lysates were sonicated for 20 min with 30 sec on-and-off cycles at the high setting of a Bioruptor (Diagenode) to produce chromatin fragments of 0.5 kb on average. At least three independent ChIP experiments were done for each cell line. PCR amplification was done in 20 μL with specific primers encompassing the URR region. The sensitivity of PCR amplification was evaluated in serial dilutions of total DNA collected after sonication (input fraction). PCR conditions for the ChIP assay are available upon request. Negative controls (no antibody) and input were included for each PCR experiment, and each experiment was carried out in triplicate. For the real-time PCR reactions, we used the SYBR Green PCR master mix (Applied Biosystems) and a 7900HT Fast Real-Time PCR system (Applied Biosystems). Standard curves were calculated from serial dilutions (100–0.1 ng) of input genomic DNA. To evaluate the relative enrichment of target sequences after ChIP, we calculated the ratios of the signals in the immunoprecipitated DNA versus input DNA (relative enrichment over input).
Semiquantitative reverse-transcription PCR expression analysis
We reverse-transcribed total RNA (2 μg) treated with DNase I (Ambion) using oligo(dT) primer with SuperScript II reverse transcriptase (Life Technologies). We used 100 ng of cDNA for PCR amplification and amplified all of the genes with multiple (20–35) cycles to determine the appropriate conditions for obtaining semiquantitative differences in their expression levels. Reverse transcription-PCR (RT-PCR) primers were designed between different exons to avoid any amplification of DNA. PCRs were done simultaneously with two sets of primers, with glyceraldehyde-3-phosphate dehydrogenase as an internal control to ensure cDNA quality and loading accuracy.
Viral gene expression by quantitative RT-PCR
Total RNA was isolated from CaSki, SiHa, and HeLa cells by TRIzol (Invitrogen) extraction according to the manufacturer's instructions. Then, we transcribed total RNA (2 μg) treated with TURBO DNA-free (Ambion), using oligo(dT) primer with SuperScript II reverse transcriptase (Life Technologies). The gene-specific primer sequences are available in the Primer Tables. The real-time PCR reactions typically contained sense and antisense gene-specific primers (350 nM), 7 μL of SYBR Green PCR Master Mix (Applied Biosystems), and 2 μL of diluted cDNAs (10 ng/μL) in a total volume of 14 μL. The reaction was done in an ABI 7900 HT. Glyceraldehyde-3-phosphate dehydrogenase expression was used as an internal control to ensure cDNA quality and loading accuracy.
miR-BHRF1-1 detection by quantitative RT-PCR
Total RNA was isolated from Akata cells and Akata cells treated with 5-aza-2′-deoxycytidine (1 μmol/L for 72 h) by TRIzol (Invitrogen), with extraction according to the manufacturer's instructions. Trace amounts of unwanted DNA were eliminated using TURBO DNA-free, from Ambion. We then measured miRNA expression using quantitative-real-time reverse transcription PCR (qRT-PCR) using the Ncode SYBR Green ER miRNA qRTPCR Kit from Invitrogen. Briefly, 1 μg of RNA was polyadenylated following the manufacturer's instructions, and the reverse transcription was then performed to obtain first-strand cDNA. The gene-specific primer sequence was identical to the entire mature miRNA sequence (see Supplemental Table). The real-time PCR reactions typically contained gene-specific primer (200 nM), universal qPCR primer (200 nM), 0.24 μL of ROX Reference Dye, 6 μL of Platinum SYBR Green qPCR SuperMix-UDG, and 1.2 μL of diluted cDNAs (1:5) in a total volume of 12 μL. The reaction was done in an ABI 7900 HT. hsa-miR-16 expression was used to normalize ebv-miR-BHRF1-1 expression levels.
Short interference RNA (siRNA) experiments
Short interference RNA experiments were undertaken as previously described (Ropero et al. 2006). DNMT1- and DNMT3B-specific siRNAs were designed and synthesized by QIAGEN. siRNA for LMP1 (EBV) was provided by Genelink. Scramble siRNA was also purchased by QIAGEN and used as a control. For adherent cells, transfection was carried out using oligofectamine (Invitrogen) according to the manufacturer's specifications. Suspension cells were transfected by electroporating 107 cells in 0.8 mL of PBS with 10 μg of siRNA at 250 V and 975 μF. The cells were harvested at specific times for total RNA extraction, and the target genes were assessed by RT-PCR (primer sequences available upon request).
Fluorescent in situ hybridization (FISH) and 5mC immunodetection
Cell cultures were exposed to colchicine (0.5 μg/mL) for 4 h at 37°C and harvested routinely. Metaphases were prepared from the immortalized cell lines following a conventional cytogenetic protocol: Cells were pelleted, gently resuspended in 2–3 mL of 75 mM KCl pre-warmed to 37°C, and incubated for 15 min at 37°C. Cells were pelleted and initially resuspended in a residual volume of 100 μL of freshly made fixation solution (3:1 [v/v] methanol:acetic acid). Up to 3 mL of fresh fix was added dropwise while the tubes were vortexed at low speed. Two more fixation washes in the same fixation solution were done. To prepare spreads, 100 μL of cells was dropped onto a pre-cleaned microscope slide, which was dried slowly in a fume hood and left overnight to age the chromosomes naturally. Bacterial artificial chromosome (BAC) clones containing the entire genomic region of the viruses were used to generate the FISH probes. BAC for HBV16 was kindly provided by Pulivarthi H. Rao (Texas Children's Cancer Center, Baylor College of Medicine, Houston, TX). BAC for HBV was kindly provided by Tian-Hua Huang (Research Center of Reproductive Medicine, Shantou University Medical College, Shantou 515,041, Guangdong Province, China). Teru Kanda (Center for Virus Vector Development, Institute for Genetic Medicine, Hokkaido University, Sapporo 060-0815, Japan) kindly provided us with BAC for EBV virus. In brief, all BACs were labeled directly by nick translation according to the manufacturer's specifications using the CGH Nick Translation Reagent Kit with Spectrum Red dUTP (Abbot Molecular, Inc.). The probes were blocked with Cot-1 Human DNA (Roche Diagnostic Corp.) to suppress repetitive sequences. The probe was denatured for 5 min at 96°C and hybridized overnight at 37°C in a humid chamber. After post-hybridization washes, the cell metaphases were counterstained with 4′,6-diamidino-2-phenylindole (DAPI II) and included in a VectaShield mounting medium kit (Vector Laboratories, Inc.) for chromatin counterstaining and fluorescent protection before microscopy. Cell images were captured using a CCD camera (Photometrics SenSys) installed on an Olympus BX microscope connected to a computer running the analysis system CytoVision v. 3.1 Software package (Applied Imaging Corp.). For 5mC immunodetection after FISH, cells were treated with 0.5N ClH for 30 min at 37°C and neutralized in Tris-Borate-EDTA buffer for 5 min at room temperature. Treated samples were blocked in PBS–0.5% BSA for 15 min at room temperature and incubated with mouse monoclonal antibodies recognizing 5-methylcytidine (a gift from A. Nivelau). Primary antibodies were detected using Cy2-labeled secondary antibodies. Confocal optical sections were obtained using a Leica TCS SP2 microscope (Leica Microsystems) equipped with krypton and argon lasers. Images were acquired with Leica LCS Lite software and processed with the publicly available GNU Image Manipulation Program.
Computational analysis
We developed a new Java software package, in the form of a web server, called “Methylyzer.” It is able to analyze DNA methylation data obtained from sequences previously modified with bisulfite and to publish them on the Internet. Data obtained from the experiments (multiple sequence alignment files) were uploaded to and analyzed by Methylyzer. Original, new, and lost CpG sites are located and a methylation value is assigned. Mutations (replacements, insertions, and deletions) are also identified. To navigate through the genomes of these viruses, we set up a genome browser (GBrowse) (Stein et al. 2002) that lets the user jump directly to Methylyzer and visualize the annotations. A set of tracks, which are very easy to activate, shows the different types of information, such as “DNA methylation sequence,” “Genetic sequence,” “Transcription start site,” “Gene,” “Exon,” and so on. By clicking, for example, over an annotated “DNA methylation sequence,” one can view the methylation data for the available samples of a particular amplicon. The samples are represented by a band colored according to the average level of DNA methylation. It is possible to click on these bands and the accompanying description, and this will take the user to another web page with detailed information for each CpG site of that sample. The information can be seen in the form of two types of image: those of the overall bisulfite genomic sequence and those of the bisulfite genomic sequence of multiple clones. By selecting all or a subset of the samples, Methylyzer builds images of methylation profiles that allow the direct comparison of the methylation over the different samples. The data matrix containing the CpGs methylation profiles for HPV16, HPV18, HBV, and EBV samples were obtained from Methylyzer. Unsupervised hierarchical clustering (UPGMA) analyses were carried out applying the City Block Metric (Manhattan) and Single Linkage methods.
Acknowledgments
This work was supported by grants I+D+I MCYT08-03, FU2004-02073/BMC, Consolider MEC09-05, SAF2007-65134, SAF2006-03681, SAF2005-04340, FIS-PI061244, Transfog LSHC-CT-2004-503438, FP7-CANCERDIP-200620, Fundació Marató TV3 052310, grants of the National Institute for Bioinformatics (http://www.inab.org), a platform of “Genoma España,” Spanish Association Against Cancer (AECC), Fundación Mutua Madrileña, and the Swiss Bridge Award. M.E. is an ICREA (Institucio Catalana de Recerca i Estudis Avançats) Research Professor.
Footnotes
[Supplemental material is available online at www.genome.org. Viral DNA methylomes are freely available for viewing at http://ubio.bioinfo.cnio.es/Methylyzer/main/index.html.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083550.108.
References
- Ambinder R.F., Robertson K.D., Tao Q. DNA methylation and the Epstein-Barr virus. Semin. Cancer Biol. 1999;9:369–375. doi: 10.1006/scbi.1999.0137. [DOI] [PubMed] [Google Scholar]
- Ballestar E., Paz M.F., Valle L., Wei S., Fraga M.F., Espada J., Cigudosa J.C., Huang T.H., Esteller M. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 2003;22:6335–6345. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Meissner A., Lander E.S. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Brena R.M., Huang T.H., Plass C. Toward a human epigenome. Nat. Genet. 2006;38:1359–1360. doi: 10.1038/ng1206-1359. [DOI] [PubMed] [Google Scholar]
- Burgers W.A., Blanchon L., Pradhan S., de Launoit Y., Kouzarides T., Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–1655. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butel J.S. Viral carcinogenesis: Revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405–426. doi: 10.1093/carcin/21.3.405. [DOI] [PubMed] [Google Scholar]
- Clark S.J., Harrison J., Paul C.L., Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sanjose S., Diaz M., Castellsague X., Clifford G., Bruni L., Munoz N., Bosch F.X. Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: A meta-analysis. Lancet Infect. Dis. 2007;7:453–459. doi: 10.1016/S1473-3099(07)70158-5. [DOI] [PubMed] [Google Scholar]
- Doerfler W. In pursuit of the first recognized epigenetic signal-DNA methylation: A 1976 to 2008 synopsis. Epigenetics. 2008;3:125–133. doi: 10.4161/epi.3.3.6249. [DOI] [PubMed] [Google Scholar]
- Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Fraga M.F., Ballestar E., Paz M.F., Ropero S., Setien F., Ballestar M.L., Heine-Suner D., Cigudosa J.C., Urioste M., Benitez J., et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini M.G., Hurst S.D., Lim H.B., Swaminathan S., Laimins L.A. Abrogation of a mitotic checkpoint by E2 proteins from oncogenic human papillomaviruses correlates with increased turnover of the p53 tumor suppressor protein. EMBO J. 1997;16:318–331. doi: 10.1093/emboj/16.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J., Song J., Stirzaker C., Hinshelwood R.A., Peinado M.A., Clark S.J. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat. Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- Gatza M.L., Chandhasin C., Ducu R.I., Marriott S.J. Impact of transforming viruses on cellular mutagenesis, genome stability, and cellular transformation. Environ. Mol. Mutagen. 2005;45:304–325. doi: 10.1002/em.20088. [DOI] [PubMed] [Google Scholar]
- Herman J.G., Graff J.R., Myohanen S., Nelkin B.D., Baylin S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleman M.R. Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections. Proc. Natl. Acad. Sci. 2004;101(Suppl 2):14560–14566. doi: 10.1073/pnas.0404758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpherys D., Eggan K., Akutsu H., Hochedlinger K., Rideout W.M., III, Biniszkiewicz D., Yanagimachi R., Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- Jin B., Tao Q., Peng J., Soo H.M., Wu W., Ying J., Fields C.R., Delmas A.L., Liu X., Qiu J., et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum. Mol. Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- Jirtle R.L., Skinner M.K. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A., Martienssen R. A blueprint for a Human Epigenome Project: The AACR Human Epigenome Workshop. Cancer Res. 2005;65:11241–11246. doi: 10.1158/0008-5472.CAN-05-3865. [DOI] [PubMed] [Google Scholar]
- Keshet I., Schlesinger Y., Farkash S., Rand E., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Kim K., Garner-Hamrick P.A., Fisher C., Lee D., Lambert P.F. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J. Virol. 2003;77:12450–12459. doi: 10.1128/JVI.77.23.12450-12459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E., Kis L.L., Klein G. Epstein-Barr virus infection in humans: From harmless to life endangering virus–lymphocyte interactions. Oncogene. 2007;26:1297–1305. doi: 10.1038/sj.onc.1210240. [DOI] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A.V., Colot V., Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat. Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- Lister R., O'Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A., Ropero S., Ballestar E., Fraga M.F., Cerrato C., Setien F., Casado S., Suarez-Gauthier A., Sanchez-Cespedes M., Git A., et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Lupberger J., Hildt E. Hepatitis B virus–induced oncogenesis. World J. Gastroenterol. 2007;13:74–81. doi: 10.3748/wjg.v13.i1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo M.R., Boyle S., D'Esposito M., Bickmore W.A. Chromosome territory reorganization in a human disease with altered DNA methylation. Proc. Natl. Acad. Sci. 2007;104:16546–16551. doi: 10.1073/pnas.0702924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud E.J., van Vugt M.J., Bultman S.J., Sweet H.O., Davisson M.T., Woychik R.P. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes & Dev. 1994;8:1463–1472. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- Minarovits J. Epigenotypes of latent herpesvirus genomes. Curr. Top. Microbiol. Immunol. 2006;310:61–80. doi: 10.1007/3-540-31181-5_5. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Park I.Y., Sohn B.H., Yu E., Suh D.J., Chung Y.H., Lee J.H., Surzycki S.J., Lee Y.I. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476–1494. doi: 10.1053/j.gastro.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Pfeffer S., Zavolan M., Grasser F.A., Chien M., Russo J.J., Ju J., John B., Enright A.J., Marks D., Sander C., et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Rakyan V.K., Hildmann T., Novik K.L., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., et al. DNA methylation profiling of the human major histocompatibility complex: A pilot study for the Human Epigenome Project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropero S., Fraga M.F., Ballestar E., Hamelin R., Yamamoto H., Boix-Chornet M., Caballero R., Alaminos M., Setien F., Paz M.F., et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat. Genet. 2006;38:566–569. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- Saito Y., Liang G., Egger G., Friedman J.M., Chuang J.C., Coetzee G.A., Jones P.A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Schluter V., Meyer M., Hofschneider P.H., Koshy R., Caselmann W.H. Integrated hepatitis B virus X and 3′ truncated preS/S sequences derived from human hepatomas encode functionally active transactivators. Oncogene. 1994;9:3335–3344. [PubMed] [Google Scholar]
- Steenbergen R.D., Walboomers J.M., Meijer C.J., van der Raaij-Helmer E.M., Parker J.N., Chow L.T., Broker T.R., Snijders P.J. Transition of human papillomavirus type 16 and 18 transfected human foreskin keratinocytes towards immortality: Activation of telomerase and allele losses at 3p, 10p, 11q and/or 18q. Oncogene. 1996;13:1249–1257. [PubMed] [Google Scholar]
- Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The Generic Genome Browser: A building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D., Jones P.A. The CpG Island Searcher: A new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- Tao Q., Robertson K.D. Stealth technology: How Epstein-Barr virus utilizes DNA methylation to cloak itself from immune detection. Clin. Immunol. 2003;109:53–63. doi: 10.1016/s1521-6616(03)00198-0. [DOI] [PubMed] [Google Scholar]
- Thain A., Jenkins O., Clarke A.R., Gaston K. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J. Virol. 1996;70:7233–7235. doi: 10.1128/jvi.70.10.7233-7235.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.N., Tsai C.L., Tse K.P., Chang H.Y., Chang Y.S. The Epstein-Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyltransferases. Proc. Natl. Acad. Sci. 2002;99:10084–10089. doi: 10.1073/pnas.152059399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.L., Li H.P., Lu Y.J., Hsueh C., Liang Y., Chen C.L., Tsao S.W., Tse K.P., Yu J.S., Chang Y.S. Activation of DNA methyltransferase 1 by EBV LMP1 involves c-Jun NH2-terminal kinase signaling. Cancer Res. 2006;66:11668–11676. doi: 10.1158/0008-5472.CAN-06-2194. [DOI] [PubMed] [Google Scholar]
- Van Tine B.A., Kappes J.C., Banerjee N.S., Knops J., Lai L., Steenbergen R.D., Meijer C.L., Snijders P.J., Chatis P., Broker T.R., et al. Clonal selection for transcriptionally active viral oncogenes during progression to cancer. J. Virol. 2004;78:11172–11186. doi: 10.1128/JVI.78.20.11172-11186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn M.W., Tanurd Ic M., Lippman Z., Jiang H., Carrasquillo R., Rabinowicz P.D., Dedhia N., McCombie W.R., Agier N., Bulski A., et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5:e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Williams H., Crawford D.H. Epstein-Barr virus: The impact of scientific advances on clinical practice. Blood. 2006;107:862–869. doi: 10.1182/blood-2005-07-2702. [DOI] [PubMed] [Google Scholar]
- Wise-Draper T.M., Wells S.I. Papillomavirus E6 and E7 proteins and their cellular targets. Front. Biosci. 2008;13:1003–1017. doi: 10.2741/2739. [DOI] [PubMed] [Google Scholar]
- Xu G.L., Bestor T.H., Bourc'his D., Hsieh C.L., Tommerup N., Bugge M., Hulten M., Qu X., Russo J.J., Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Young L.S., Rickinson A.B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Yuan J., Cahir-McFarland E., Zhao B., Kieff E. Virus and cell RNAs expressed during Epstein-Barr virus replication. J. Virol. 2006;80:2548–2565. doi: 10.1128/JVI.80.5.2548-2565.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nat. Rev. Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]