Abstract
Little is known about the telomere chromatin dynamics of embryonic stem (ES) cell. Here, we demonstrate localization of histone H3.3 at interphase telomeres and enrichment of Ser31-phosphorylated H3.3 at metaphase telomeres in pluripotent mouse ES cells. Upon differentiation, telomeric H3.3S31P signal decreases, accompanied by increased association of heterochromatin repressive marks and decreased micrococcal nuclease sensitivity at the telomeres. H3.3 is recruited to the telomeres at late S/G2 phase, coinciding with telomere replication and processing. RNAi-depletion of H3.3 induces telomere-dysfunction phenotype, providing evidence for a role of H3.3 in the regulation of telomere chromatin integrity in ES cells. The distinctive changes in H3.3 distribution suggests the existence of a unique and functionally essential telomere chromatin in ES cells that undergoes dynamic differentiation-dependent remodeling during the process of differentiation.
Pluripotent embryonic stem (ES) cells possess an unlimited capacity for self-renewal and the potential to differentiate into multiple lineages. Upon cellular differentiation, they undergo dramatic morphological and molecular changes through selective silencing and activation of specific genes. Recent studies have pointed to epigenetic phenomena as having fundamental roles in the differentiation process (Martens et al. 2005; Meshorer et al. 2006). Examples are the differentiation-dependent increase in the silenced chromatin marks, including H3K9me3, H3K20me2, H4K20me3, and H3K27me3, and a decrease in the level of acetylated H3 and H4. The hyperdynamic binding of structural chromatin proteins is also a hallmark of ES cells, but not of cells that are already lineage committed.
While epigenetic factors are essential for maintaining pluripotency, the capacity for continual telomere renewal is also critical. In vertebrates, telomeres consist of tandem arrays of TTAGGG repeats that are bound by a specialized multiprotein complex “shelterin.” Telomere length is maintained by telomerase, a reverse transcriptase that adds telomere repeats de novo after each cell division. High telomerase activity is a key factor that maintains telomere self-renewal and proliferative capacity in ES cells. Mammalian telomeres contain epigenetic markers such as H3K9me3 and H4K20me3 that are characteristic of silenced chromatin, and DNA hypermethylation at subtelomeric regions (Garcia-Cao et al. 2004; Gonzalo et al. 2006). Loss of these modifications results in defective telomere functions as shown by aberrantly increased telomere length and chromosomal instability, indicating that these repressive markers are essential for the formation of a compacted telomere chromatin and the regulation of telomere length.
The nucleosome consists of an octamer of four core histones, H2A, H2B, H3, and H4, wrapped inside 146 bp of dsDNA. Adjacent nucleosomes are connected via linker DNA bound by the linker histone H1. Higher-order organization of this basic subunit defines the nature of the chromatin structure and accessibility. Beside the core histones, several additional histone variants are present in mammalian cells. Histone H3.3 is highly evolutionarily conserved and is found in organisms ranging from yeasts to humans. It is a universal histone variant associated with active chromatin and is incorporated into sites of transcription independently of DNA replication (Ahmad and Henikoff 2002; McKittrick et al. 2004). Dynamic incorporation of H3.3 alters the chromatin state of the nucleosome by enriching it in post-translational covalent modifications that are associated with active chromatin and by depleting it in modifications associated with silent chromatin.
Although chromatin status is known to influence telomere structure and length, little is known about the epigenetic factors that drive continual telomere-length renewal in ES cells and changes in telomere chromatin during differentiation. In this study, we examine the epigenetic factors that regulate telomere chromatin in pluripotent mouse ES cells and changes in telomere chromatin properties during cellular differentiation, and perform our initial experiments using antibodies against histone variants and modifications. We report here the enrichment of Ser31-phosphorylated H3.3 at metaphase telomeres and localization of H3.3 at interphase telomeres in ES cells. We also show the dynamic changes in epigenetic modifications associated with telomeric chromatin during the cellular differentiation of ES cells, indicated by a decrease in the H3.3S31P signal and an increase in the association of repressive chromatin marks, including H3K9me3 and H4K20me3 at the telomeres in differentiated ES cells. In addition, we show that the recruitment of H3.3 to the telomeres coincides with telomere replication and processing at late S/G2 phase. Furthermore, RNAi-mediated inhibition of H3.3 induces an increased telomere-dysfunction phenotype, providing evidence for a functional role of H3.3 in the regulation of telomere structural integrity in ES cells. Our results indicate the presence of a distinctive telomere chromatin in ES cells that undergoes dynamic differentiation-dependent epigenetic remodeling and an essential function of H3.3 in modulating telomere chromatin dynamics.
Results
Dynamic changes in the chromatin properties of the telomeres during cellular differentiation of mouse ES cells
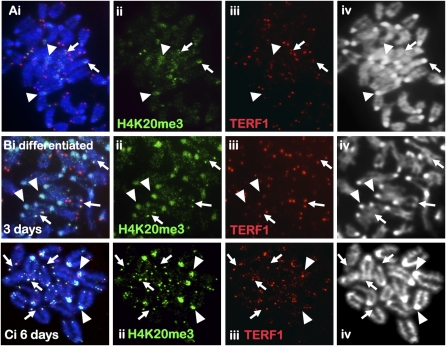
To study changes in telomere chromatin during mouse ES cell differentiation, we induced mouse ES cell differentiation by withdrawing leukemia inhibitory factor (LIF) and supplementing the culture with retinoic acid (RA). Prior to differentiation, >90% of the ES cells, ES129.1 and ES-W9.5, expressed stem cell surface marker POU5F1 (also known as OCT4). After 3 d, <50% expressed POU5F1. By 6 d, POU5F1 was barely detectable (data not shown). Induction of differentiation has no obvious effect on H3K4me2 (a euchromatin marker) distribution but resulted in increased H3K9me3 (a heterochromatin marker) level at the pericentric heterochromatin in ES129.1 (Supplemental Fig. 1A–D) and ES-W9.5 (data not shown) cells, as previously reported (Azuara et al. 2006; Meshorer et al. 2006). No significant increase in telomeric H3K9me3 signal was detected both before and after differentiation. The overall H4K20me3 (another marker of “repressed” chromatin) staining was weak in undifferentiated ES129.1 (Fig. 1A) and ES-W9.5 (data not shown) cells with some telomeric staining. Three days of induction of differentiation led to a significant increase in H4K20me3 level at both the telomeres and pericentric heterochromatin (Fig. 1B; Supplemental Fig. 2). Six days of differentiation led to an even greater increase in H4K20me3 level at both the telomeres and pericentric heterochromatin (Fig. 1C; Supplemental Fig. 2), indicating an increase in chromatin repression at pericentric heterochromatin and telomeric chromatin following ES cell differentiation.
Figure 1.
Immunofluorescence analysis of H4K20me3 in mouse ES cells. (A) H4K20me3 (ii, green; Abcam) staining in ES129.1 cells was weak, with enrichment on pericentric heterochromatin (arrowheads) and on some telomeres (arrows) as indicated by TERF1 staining (iii, red). (B–C) Three days (B) to 6 d (C) of differentiation increased H4K20me3 staining (ii, green) at the pericentric heterochromatin (arrowheads) and telomeres (arrows) in ES129.1 cells.
H3.3S31P is enriched at the telomeres in mouse ES cells during mitosis
H3.3 is a universal histone variant associated with transcriptionally active, open chromatin (Ahmad and Henikoff 2002; McKittrick et al. 2004). There is currently no specific antiserum against unmodified H3.3. Immunofluorescence analysis was performed using anti-H3.3 phosphorylated at Serine 31 (H3.3S31P) antiserum. Specificity of the antiserum was confirmed by competitive immunofluorescence analysis with H3.3S31P peptide (Supplemental Fig. 1E–J). H3.3S31P is a mitotic-specific modification enriched in the pericentric heterochromatin (Hake et al. 2005). However, H3.3S31P mainly localized to the telomeres of metaphase chromosomes in undifferentiated ES129.1 (Fig. 2A) and ES-W9.5 cells (Supplemental Fig. 3A). Telomeric localization of H3.3S31P was confirmed by FISH analysis using a telomere-specific TTAGGG probe. Similar results were obtained using a different source of anti-H3.3S31P antibody (Upstate Biotechnology; data not shown). We also observed distinct H3.3S31P telomeric localization on a neocentromere-based human marker chromosome mardel(10) in ES129.1mar(10) cells (Supplemental Fig. 4A; Wong et al. 2005). In nonpluripotent mouse and human cells, including mouse NIH3T3 and L cells, and human primary fibroblast (telomere length of 7–8 kb), HT1080, and CCRF/CEM cells (telomere length of 15–20 kb; maintained via the Alternate Telomere Maintenance pathway) (Supplemental Fig. 4B–D), H3.3S31P mainly accumulated at the pericentric heterochromatin of metaphase chromosomes, with relatively little telomeric signal.
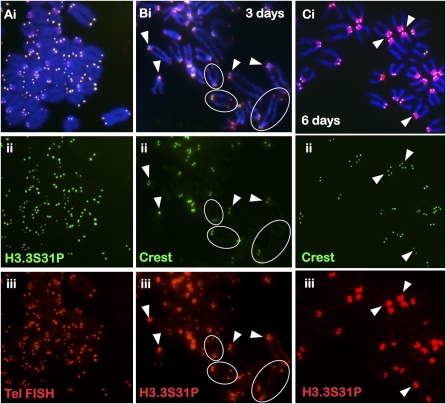
Figure 2.
Immunofluorescence analysis of H3.3S31P in mouse ES cells. (A) H3.3S31P (ii, green; Abcam) is enriched at the telomeres of all the chromosomes in ES129.1 cells, as confirmed by telomere FISH (iii, red). A second source of anti-H3.3S31P antiserum (Upstate; data not shown) gave a similar staining pattern. (B) Three days of differentiation resulted in decreased telomeric H3.3S31P signal (iii, red; some circled examples are shown) but enhanced staining at the pericentric DNA (arrowheads). (C) Six days of differentiation led to a near complete loss of telomeric H3.3S31P signals (iii, red) and a major increase in signals at the pericentric DNA. Centromeres were stained with CREST serum (B,C, ii, green).
Cellular differentiation of mouse ES cells leads to decreased H3.3S31P signals at the telomere ends but increased H3.3S31P signals at the pericentric heterochromatin
In ES129.1 (Fig. 2B,C; Supplemental Fig. 3D,E) and ES-W9.5 (Supplemental Fig. 3B,C) cells subjected to 3–6 d of differentiation, a decrease of H3.3S31P telomeric signal but a concomitant increase in pericentric H3.3S31P signal was detected. There was no change in telomere length in cells following differentiation (Supplemental Fig. 5), indicating that the post-differentiation reduction of H3.3S31P telomeric signal was not a consequence of telomere shortening but rather that of the existence of a distinctive telomeric chromatin in ES cells that undergoes dynamic differentiation-dependent remodeling.
Mouse embryonic germ cells also show H3.3S31P enrichment at the telomeres during mitosis
To further demonstrate that H3.3S31P incorporation at the telomere may be a feature associated with cellular pluripotency, we included embryonic germ (EG) cells in our analysis. We observed distinct H3.3S31P localization at the telomeres in two different mouse EG cell lines, EGRA2 (Fig. 3A) and EGRA3 (Supplemental Fig. 6A), although, interestingly, the telomeres on the short-arm ends of the chromosomes appeared to show, in general, more variable and weaker H3.3S31P staining compared to those seen on the long-arm telomeres. Furthermore, unlike the situation in ES cells, some H3.3S31P staining on chromosome arms was observed. It is interesting to note that H4K20me3 staining was also weak in EG cells, with slightly stronger signals at some of the telomeres (mostly at the long arms) (Fig. 3C; Supplemental Fig. 6B). Induction of differentiation led to a significant reduction in H3.3S31P signal at the telomeres and a concomitant substantial increase in H3.3S31P and H4K20me3 levels at the pericentric region (Fig. 3B,D). No significant change in H4K20me3 signals was seen at the telomeres in differentiated EG cells, suggesting that unlike the situation in differentiated mouse ES cells, delocalization of H3.3S31P in mouse EG cells is not directly correlated with an increase in the repressive chromatin marker H4K20me3.
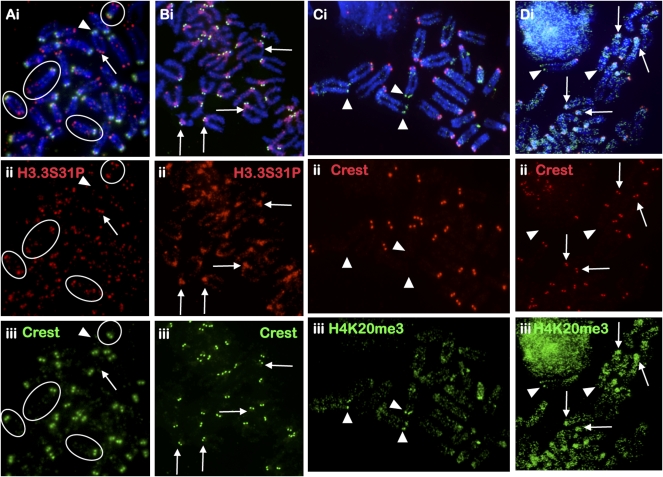
Figure 3.
Cellular distribution of H3.3S31P and H4K20me3 in mouse EG cells. (A) H3.3S31P is enriched at the telomeres of EG cell lines EGRA2 using antiserum against H3.3S31P (ii, red; Abcam; some circled examples are indicated). Signals at the “short-arm” telomeres (arrowheads) of the telocentric chromosomes are in general weaker and more variable than those on the “long-arm” telomeres (arrows); telocentric ends are easily identifiable by their close proximity to the centromere, as marked by CREST staining (iii, green). (B) Some H3.3S31P signals are also detected on the long arms of the chromosomes. Six days of LIF depletion/RA treatment resulted in a significant decrease in telomeric H3.3S31P (ii, red) and increase in H3.3S31P signal at the pericentric heterochromatin (some examples are indicated by arrows). (C) Overall H4K20me3 (iii, green; Abcam) staining in EGRA2 cells was weak with some telomeres showing slightly stronger signals (arrowheads). (D) Six days of differentiation led to increased H4K20me3 signal (iii, green) at the pericentric heterochromatin (arrows). Red signals (C,D, ii) represent CREST at the centromeres. Immunofluorescence analysis of H3.3S31P and H4K20me3 in another mouse EG cell is shown in Supplemental Figure 6.
H3.3 also associates with the telomeres in mouse ES cells during interphase
To determine if H3.3 was present at the telomeres during interphase, ES129.1 cells were transfected with tetracycline-inducible vectors expressing H3.3 protein tagged with a MYC (Myc epitote tag) sequence at the N terminus, and examined for chromosomal localization 24 h post-transfection (Fig. 4A,B). In metaphase cells, MYC-H3.3 localized extensively to the chromosome arms and noticeably also to the telomeres. An enrichment of MYC-H3.3 at the telomeres was similarly seen in interphase cells, as indicated by colocalization with TERF1. In addition, MYC-H3.3 showed colocalization with H3.3S31P in metaphase but not in interphase cells (Fig. 4D,E). Similar results were obtained with the C-terminal constructs (data not shown). In parallel, we also transfected cells with N- and C-terminal MYC-tagged histone H3 expression-constructs into ES129.1 cells. In both metaphase (data not shown) and interphase (Fig. 4C) cells, MYC-H3 was incorporated into the entire chromosomes, with no observable enrichment at the telomeres.
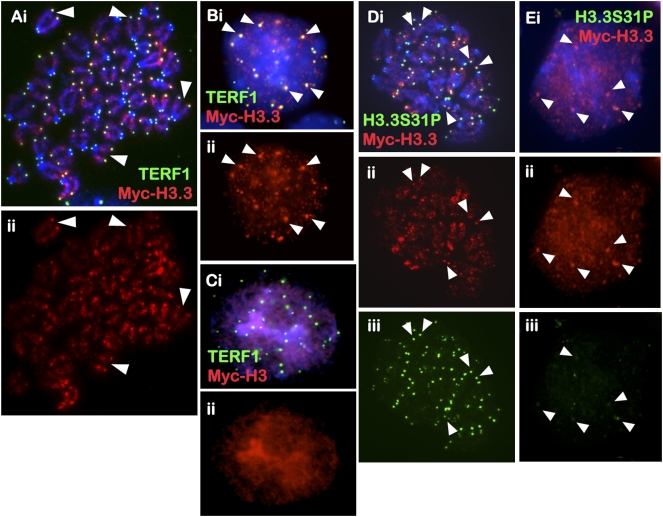
Figure 4.
Cellular distribution of MYC-H3.3 and MYC-H3 in mouse ES cells. (A–C) MYC-H3.3 and MYC-H3 constructs were transfected into ES129.1 cells and examined for chromosomal localization 24 h post-induction with 1 μM doxycycline. Both N-terminal and C-terminal (data not shown) MYC-H3.3 was enriched along the chromosome arms and at the telomeres in metaphase (A, ii, red) and interphase (B, ii, red) (arrowheads show some examples) ES129.1 cells, as indicated by costaining with anti-TERF1 antibody (A,B, i, green). N-terminal (C, ii, red) and C-terminal (data not shown) MYC-H3 localized to all regions of the chromosomes. (D,E) MYC-H3.3 (ii, red) also showed colocalization with H3.3S31P in metaphase (D, iii, green) but not interphase (E, iii, green) ES129.1 cells.
Cellular differentiation-mediated epigenetic modifications at the telomeres affect the underlying telomeric nucleosomal organization
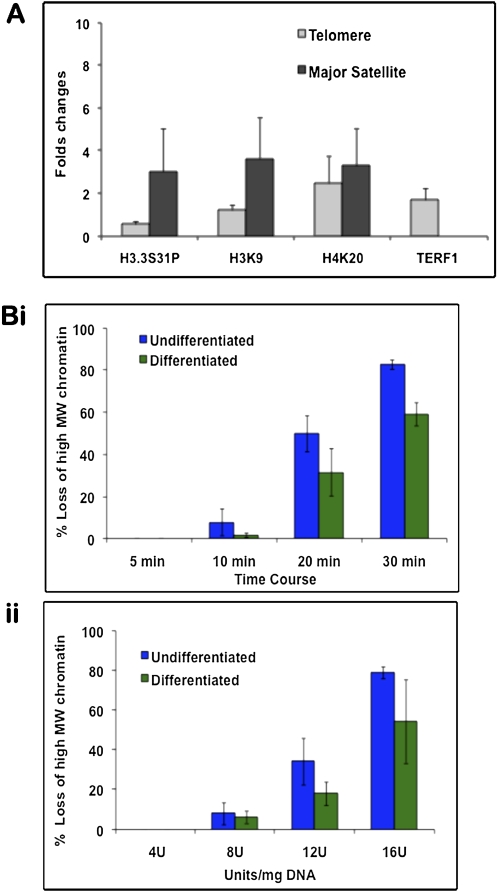
Combined cross-linked chromatin immunoprecipitation (ChIP) and quantitative telomere PCR assays (Cawthon 2002) were performed to assess changes in telomeric chromatin during ES129.1 differentiation (Supplemental Fig. 7). The telomere PCR assays method offers excellent sensitivity, accuracy and reproducibility compared with other protocols developed for telomere measurements, such as flow cytometry and cytometric assay. Data obtained by telomere PCR assays were also validated by dot blot analysis of the ChIP samples using a telomere DNA probe (Supplemental Fig. 8). Real-time PCR analysis of the ChIP samples showed decreased H3.3S31P but increased H3K9me3 and H4K20me3 levels at the telomeres in differentiated ES129.1 cells (Fig. 5A; Supplemental Table 1). ChIP using anti-TERF1 also showed an increased enrichment of TERF1 at the telomeres in differentiated ES129.1 cells. In parallel, we performed ChIP/real-time PCR using major satellite primers specific for mouse pericentric heterochromatin (Fig. 5A; Supplemental Fig. 7; Supplemental Table 1). Following differentiation, the levels of H3.3S31P, H3K9me3A, and H4K20me3 were increased at the pericentric region. Altogether these ChIP results indicate the dynamic changes in H3.3 localization and chromatin modifications at the telomeres and pericentric heterochromatin during ES cell differentiation.
Figure 5.
Chromatin immunoprecipitation (ChIP)/PCR analysis and MNase digestion assays. (A) ChIP was performed on undifferentiated and differentiated ES129.1 cells using anti-sera against TERF1, H3.3S31P, H4K20me3, and H3K9me3, followed by real-time PCR analysis using primers specific for telomere or pericentric major satellite DNA. Fold-differences in the enrichment of the ChIP/PCR products (n = 3) were presented as histograms. Following differentiation, H3.3S31P levels at the telomeres was only 0.54-fold (i.e., reduced by 46%) of those in undifferentiated cells, whereas H3K9me3 and H4K20me3 levels at the telomeres were increased by 1.2- and 2.5-fold, respectively. At the pericentric heterochromatin, the levels of H3.3S31P, H3K9me3 and H4K20me3 were increased by 3.0-, 3.6-, and 3.3-fold, respectively. Data for the analysis are shown in Supplemental Table 1. (B) MNase assays (n = 3) were performed on both undifferentiated (i) and differentiated (ii, 6 d) ES129.1 cells by MNase digestion over a time-course (i, 4 units MNase/mg DNA for 5 to 30 min at 37°C) and with various concentrations of MNase (ii, 4–16 units/mg DNA for 5 min at 37°C). Southern blot analysis was performed using radioactive-labeled telomere probe. Change in MNase sensitivity was quantitated by measuring the distance between nucleosomes with the highest and lowest molecular sizes (Gilbert et al. 2007). Results were presented as percentage of loss of high-molecular-weight (MW) chromatin. Telomeric chromatin in differentiated cells was less sensitive to MNase digestion. Examples of individual MNase/Southern analysis are shown in Supplemental Figure 9.
To understand how the above-observed differentiation-dependent epigenetic modifications affected the underlying chromatin structure at the telomere, we assessed the micrococcal nuclease (MNase) sensitivity (Gilbert et al. 2007) of the telomeric chromatin over a time-course and at various concentrations (Fig. 5B; Supplemental Fig. 9). Both undifferentiated and differentiated ES129.1 cells showed identical digestion kinetics of bulk chromatin and nucleosome repeat length of the telomere. However, the telomere chromatin in ES129.1 cells became more resistant to MNase following differentiation, presumably as a result of a change in the telomere chromatin structure. As a control, we also compared the MNase sensitivity of centromeric and pericentric chromatin (as defined by the occupancy of the minor and major satellite DNA, respectively). There was no significant change in the MNase sensitivity of centromeric and pericentric chromatins (Supplemental Fig. 10), except for a slight increase in MNase resistance in the pericentric heterochromatin in differentiated ES cells during the early stage of nuclease treatment.
H3.3 telomeric incorporation synchronizes with telomeric replication and processivity activities
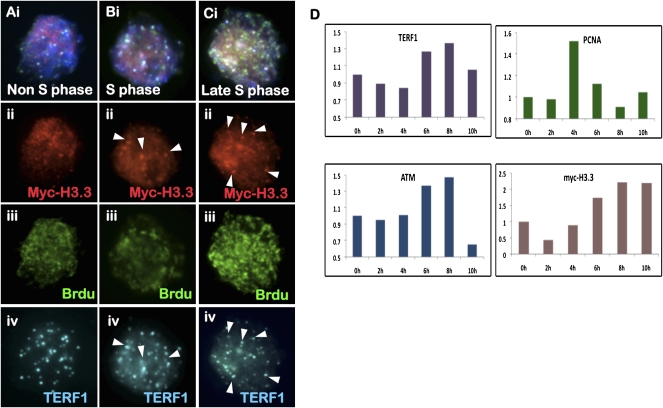
To determine the timing of H3.3 telomeric incorporation, combined 5-bromo-2′-deoxyuridine (BrdU) staining and immunofluorescence analysis was performed on ES129.1 cells expressing MYC-H3.3 (Fig. 6A–C). We also correlated the staining pattern of BrdU with that of DNA replication processivity marker PCNA (proliferative cell nuclear antigen—a DNA replication processivity marker; data not shown). In BrdU-negative non-S-phase cells, MYC-H3.3 staining pattern showed no observable colocalization with TERF1 at the telomeres. Whereas in BrdU-positive cells, telomeric colocalization of MYC-H3.3 and TERF1 increased in prominence from early to late S/G2 phase, suggesting that MYC-H3.3 telomeric loading is coupled with the timing of telomere replication and processing (Verdun and Karlseder 2006).
Figure 6.
Cell cycle–dependent telomeric incorporation of MYC-H3.3. (A–C) Immunofluorescence analysis of ES129.1 cells expressing MYC-H3.3. MYC-H3.3 is induced by doxycycline for 12 h. No MYC-H3.3 (A, ii, red) was detected at the telomere in non-S-phase cells (negative BrdU-staining; A, iii, green). MYC-H3.3 (B, ii, red) was enriched at the telomeres (as shown by costaining with TERF1 [A–C, iv, blue]; arrowheads) in replicating S-phase cells (positive BrdU-staining; B, iii, green), with even more prominent telomeric signals detected by late S/G2 phase (C). (D) ChIP/real-time PCR analysis (n = 3; using primers specific to telomere) of synchronized (thymidine-blocked) ES129.1 cells using antibodies against TERF1, MYC tag, ATMS1981P, and PCNA. MYC-H3.3 expression was induced by doxycycline for 12 h. Data and calculations for the experiments are shown in Supplemental Table 2. An example of the ChIP/PCR experiment is shown in Supplemental Figure 11. PCNA association indicated an increase in replication activity at the telomeres following 2–6 h of release from thymidine-induced G1/S block. TERF1 association at the telomeres increased after 6 h post-G1/S-release. ATMS1981P association with the telomeres also increased during S phase. MYC-H3.3 incorporation at the telomeres occurred predominantly after 6 h of release at late S/G2 phase, in synchrony with the timing of telomere replication and processing.
For further investigation, we performed ChIP/real-time PCR assays on synchronized ES129.1 cells (arrested at G1/S using a thymidine block protocol) expressing MYC-H3.3, using anti-sera against MYC tag, TERF1, phosphorylated ATM at Ser1981 (ATMS1981P; a DNA damage machinery marker) and PCNA (Fig. 6D; see Supplemental Fig. 11 for an example of ChIP/real-time PCR assays on synchronized ES129.1 cells; Supplemental Table 2). Following the release from G1/S thymidine-induced block, cell-cycle progression was determined by fluorescence-activated cell sorting (FACS) analysis. ES129.1 cells released from G1/S block progressed through S phase at 4–6 h post-release, entered M phase at 8–10 h, and re-entered G1 phase at 10–12 h. PCNA association indicated an increase in replication activity at the telomeres at 4–6 h post-G1/S-release. TERF1 association at the telomeres increased after 6 h post-G1/S-release. As previously reported (Verdun and Karlseder 2006) to be essential for telomere replication, ATMS1981P was also found associated with the telomeres during S phase. The real-time PCR analysis also showed increased loading of MYC-H3.3 at the telomeres during late S/G2 phase, in synchrony with the timing of telomere replication and processing.
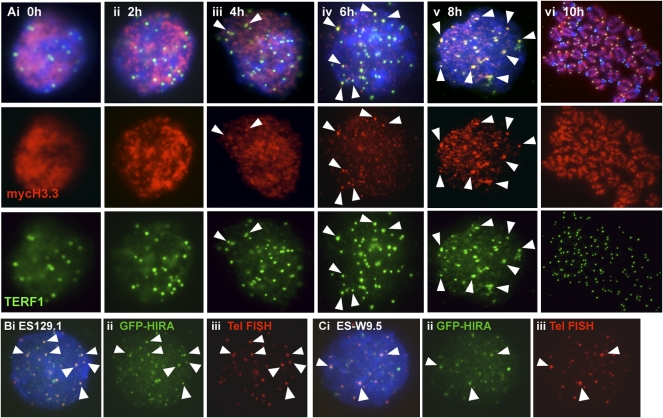
We also performed immunofluorescence analysis on synchronized ES129.1 cells (Fig. 7A; Supplemental Table 3). During the initial 4 h post-G1/S release, only 0%–20% of cells showed positive telomeric localization of MYC-H3.3. H3.3 recruitment predominantly occurred at late S/G2 phase, with 48%, 64%, and 80% of cells showing MYC-H3.3 incorporation following 6, 8, and 10 h post-release, respectively, providing further support that H3.3 telomeric loading coupled with telomere replication and processivity. Furthermore, H3.3 loading is likely to be facilitated by nucleosome assembly factor HIRA (a chaperon that promotes H3.3 nucleosome assembly) (Tagami et al. 2004), given that GFP-HIRA also localized to the telomeres in both ES129.1 and ES-W9.5 cells (Fig. 7B,C). Interestingly, the level of GFP-HIRA at the telomeres in metaphase cells showed with an increase in signals at the pericentric heterochromatin (Supplemental Fig. 4E–G). Following 3 d of differentiation, the increase of GFP-HIRA signals at the pericentric heterochromatin also became more prominent.
Figure 7.
Immunofluorescence analysis of MYC-H3.3 telomeric loading and GFP-HIRA. (A) Immunofluorescence analysis of synchronized ES129.1 cells. MYC-H3.3 was induced by doxycycline. MYC-H3.3 (red) distribution was homogenous in cells 0–4 h post-release from thymidine-G1/S block. MYC-H3.3 was mostly loaded onto telomeres (indicated by colocalization with TERF1, green; indicated by arrowheads) after 6 h of release, and became even more apparent 8 h of release. MYC-H3.3 remained associated with the telomeres as cells entered metaphase after 10 h of release. (B,C) Immunofluorescence analysis of ES129.1 (B) and ES-W9.5 (C) cells 24 h post-transfection with N-terminal GFP-HIRA construct. Telomeric localization of GFP-HIRA (B,C, ii, green) was indicated by colocalization with telomere FISH signals (B,C, iii, red). Cellular localization of GFP-HIRA in differentiated cells is shown in Supplemental Figure 4E–G.
RNAi-depletion of H3.3 expression in mouse ES cells induces the formation of TIFs
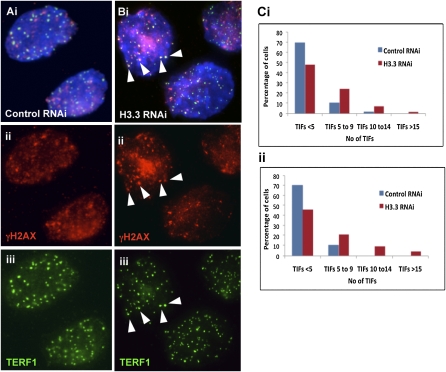
To study the functional role of H3.3 at the telomeres, we transfected ES129.1 cells with RNAi-duplex oligonucleotides corresponding to H3.3A and H3.3B genes. Specificity and efficiency of three sets of oligonucleotides were tested, with one giving up to 66.64% depletion of H3.3 expression (Supplemental Table 4). H3.3 RNAi-depletion had no significant effect on cell growth during the initial 24 h, although it led to a slight reduction in cell growth 48 h post-RNAi knockdown (data not shown). It also did not induce any formation of telomeric replication intermediate as shown by the two-dimensional gel electrophoresis analysis (Cesare and Griffith 2004) (Supplemental Fig. 12). Next, we quantitated telomere dysfunctional-induced foci (TIFs, cytological telomeric foci containing DNA damage response factors, such as gamma-H2AX) to assess the integrity of telomeric structure (Hockemeyer et al. 2005). ES129.1 cells containing five or more TIFs increased by 13.53% and 26.37%, following 24 and 48 h of H3.3 RNAi knockdown, respectively (Fig. 8; Supplemental Fig. 13A). Whereas, H3.3 RNAi knockdown in NIH3T3 did not lead to a significant increase in the number of cells containing five or more TIFs (Supplemental Fig. 13B). These results suggest that H3.3 contributes to the functional integrity of telomere chromatin in ES cells.
Figure 8.
Telomere damage response upon H3.3-depletion. (A,B) Immunofluorescence analysis of ES129.1 cells subjected to 48-h knockdown with either control or H3.3-specific RNAi-duplex oligonuclotides using anti-gamma-H2AX (A,B, ii, red) antiserum. Increased number of TIFs was detected at the telomeres (indicated by colocalization with TERF1; A,B, iii, green) in cells depleted of endogenous H3.3 (B) (arrowheads show some examples of TIFs). (C) Induction of TIFs by H3.3 inhibition for 48 h. Data are presented in histograms by subgrouping the cells according to the number of TIFs per cell (<5 TIFs, 5–9 TIFs, 10–14 TIFs, and ≥15 TIFS). A normal cell can contain 1–2 TIFs on average, thus, a threshold of 5 TIFs was used, as described in other studies (Hockemeyer et al. 2005). When transfected with H3.3-RNAi oligonuclotides, the number of cells with ≥5 TIFs increased from 12.94% to 37.65% (n = 85; 24.71% increase; P = 3.80 × 10−5) and 11.76% to 40.0% (n = 85; 28.24% increase; P = 6.26 × 10−5) for the two experiments (i and ii), respectively, with an average increase of 26.47%, compared with the population transfected with control scrambled RNAi-oligonucleotide. Data for 24-h H3.3 depletion in ES129.1 and NIH3T3 cells are shown in Supplemental Figure 13.
Discussion
In this study, we examine the epigenetic factors that regulate telomere chromatin dynamic in ES cells. Specifically, we showed localization of histone H3.3 at interphase telomeres and enrichment of H3.3S31P enrichment at the telomeres in ES cells (and EG cells) during mitosis, instead of localizing at pericentric heterochromatin as reported for non-ES cell types (Hake et al. 2005). The telomeric enrichment of H3.3S31P is also observed in pluripotent mouse EG cells but not in other nonpluripotent mouse and human cell types, including NIH3T3, L cells, HT1080, 293T, and CCRF/CEM cells (with exceptional telomere length of 15–20 kb), suggesting a unique function of H3.3 in the control of chromatin properties at the telomeres that is distinct from its proposed role in transcription activation. The observed presence of H3.3 at the telomeres of EG cell chromosomes and its delocalization from the telomeres following cellular differentiation also suggest the possibility of a more general role of H3.3 in modulating telomere chromatin dynamics in a wide range of other pluripotent cells.
In this study, we also show that the telomere chromatin in pluripotent mouse ES cells undergoes dynamic epigenetic remodeling during cellular differentiation. Upon differentiation, H3.3S31P signals are greatly reduced at the telomeres without any observable telomere-length shortening. In parallel, the telomere chromatin concomitantly becomes more “repressed/closed,” as indicated by increased association of H3K9me3 and H4K20me3 and decreased MNase sensitivity following differentiation. Furthermore, we show that the underlying changes in epigenetic modifications and chromatin properties at the telomeres are not a consequence of the shortening of telomere lengths following the process of cellular differentiation.
Telomere-length homeostasis is achieved via a binary switch between telomerase-extendible and nonextendible state; however, it is unclear how the equilibrium is achieved. Blasco (2007) has recently suggested that the assembly of heterochromatin at the telomeres forms a “closed” conformation that results in less accessibility to telomere-elongating machinery/telomerase activity. A relatively more “open” telomeric chromatin structure in ES cells, as shown in our study, compared with their differentiating counterparts should favor a greater accessibility to telomere-elongating activities. However, whether the unique composition of telomere chromatin contributes directly to the equilibrium of telomere-length homeostasis and the capacity for continual telomere-renewal in pluripotent ES cells awaits further investigation.
We also show that telomeric MYC-H3.3 incorporation in ES cells during late S/G2 phase is coupled with the events of telomere replication and processing. Although mammalian telomeres replicate throughout S phase, telomere replication is completed at late S/G2, facilitated by the DNA damage machinery to ensure optimal end-replication and acquisition of a functionally essential telomere structure (Verdun and Karlseder 2006). Despite that H3.3 RNAi knockdown over 24 to 48 h does not induce extremely severe telomere dysfunction phenotype, including the formation of replication intermediates (also indicating no severe compromise in telomere DNA replication) and telomere fusions, it noticeably induces an increase in DNA damage/TIF phentotype, suggesting that H3.3 loss compromises the proper formation of telomere chromatin structure.
In summary, our results provide evidence for the existence of a distinctive telomere chromatin in ES cells that undergoes dynamic differentiation-dependent epigenetic remodeling, and suggest a unique function of H3.3 in modulating telomere chromatin dynamics. Consistent with recent studies that have shown the ability of H3.3 in chromatin remodeling during mammalian sex chromosome inactivation (van der Heijden et al. 2007) and Drosophila sperm nucleus decondensation upon fertilization (Loppin et al. 2005), the loading and unloading of the chromatin-remodeling histone H3.3 variant at the telomeres may potentially serve as a primary epigenetic reprogramming cue operating at the nucleosomal level that controls telomere-length homeostatic equilibrium and structural organization in pluripotent ES cells.
Methods
Cell cultures
Cell lines including human HT1080 (fibrosarcoma), primary human fibroblast (derived from a 5-yr-old child), CCRF/CEM (acute lymphoblastic leukemic cells; ECACC), and mouse NIH3T3 and L cells (transformed fibroblasts) were cultured in DMEM with 10% FCS. Mouse ES129.1, ES-W9.5, and ES129.1mar(10) (ES129.1 cell hybrid carrying a neocentromeric chromosome mardel(10) that was tagged with zeocin-resistance gene; 100 μg/mL zeocin was added into the culture) (Wong et al. 2005) in DMEM with 15% heat-inactivated FCS and 103 units/mL of leukemic inhibiting factor (LIF) and 0.1 mM β-mercaptoethanol. EG cell lines EGRA2 and EGRA3 were derived from primordial germ cells of strain 129 mice (J. McWhir, Roslin Institute, Edinburg, UK). They were cultured on LIF expressing STO fibroblast feeder cells.
Antibodies
Primary antibodies used were as follows: rabbit polyclonal anti-sera against TERF1, H3K4me2 (Upstate Biotechnology), H3K9me3 (Upstate Biotechnology), H4K20me3 (Abcam and Upstate Biotechnology), H3.3S31P (Abcam and Upstate Biotechnology), and GFP (Abcam); mouse monoclonal antiserum against MYC tag (Invitrogen), POU5F1 (Santa Cruz); gamma-H2AX phosphorylated at Ser139 (Upstate Biotechnology), ATM kinase phophorylated at Ser1981 (Upstate Biotechnology), PCNA (Oncogene), and BrdU (Roche; conjugated with FITC); human autoimmune anti-centromere anti-serum CREST6 (predominantly anti-CENPA and CENPB). Secondary anti-sera were donkey anti-human Texas red (Jackson Laboratory), and donkey anti-mouse or anti-rabbit Alexa Fluoro 488/594/647 (Molecular Probes). AlexaFluoro rabbit polyclonal 488 and 594 conjugation kits were used for the conjugation of anti-sera (Invitrogen).
Immunofluorescence analysis, FISH, and BrdU incorporation
Cells were treated with microtubule-depolymerizing agent Colcemid for 1 h at 37°C, harvested for hypotonic treatment in 0.075 M KCl, cytospun on slides, and incubated in KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris.HCl at pH 7.2, 0.5 mM EDTA, 0.1% [v/v] Triton X-100 and protease inhibitor) (Uren et al. 2000; Wong et al. 2005). Slides were blocked in KCM buffer containing 1% BSA and incubated with the relevant primary and secondary antibodies for 1 h at 37°C. After each round of antibody incubation, slides were washed three times in KCM buffer. Slides were then fixed in KCM with 4% formaldehyde and mounted in mounting medium (Vetashield). Images were collected using a fluorescence microscope linked to a CCD camera system.
For peptide blocking experiments, the peptide (see below) was incubated together with the primary antibody for 1 h at 37°C. Specificity of the antiserum against H3.3S31P (Abcam) was validated by successful competition with a synthetic H3.3S31P peptide derived from within residues 1–100 of human H3.3S31P protein (Abcam).
For telomere-FISH, slides were fixed in cold methanol followed by dehydration in an ethanol series prior to denaturation and hybridization with Cy3-conjugated telomere DNA probe (Dako). For BrdU staining, cells were incubated with 10 μM BrdU (Sigma) for 1 h. BrdU incorporation was assessed by a denaturation protocol using 0.5 N hydrochloric acid; followed by immunofluorescence analysis with anti-BrdU FITC antibody (Roche).
Cell-cycle synchronization and FACS analysis
Cells were treated with 2 mM thymidine for 12 h, released from cell cycle blockage by washing with warm medium, and incubated in medium at 37°C for various time points. For FACS analysis, cells were harvested, washed in 1× PBS, fixed with ice-cold ethanol, stained with propidium iodide, and analyzed on a LSR II Becton Dickinson Flow Cytometry Analyzer.
Generation and transfection of MYC-tagged H3.3 and H3, and GFP-HIRA expression vector constructs
Cells were transfected with either pcDNA-TR (expression vector encoding tetracycline repressor); pcDNA4/TO/MYC-HisA (tetracycline-inducible expression vector) carrying either N- or C-terminal tagged MYC-H3.3, H3.3-MYC, MYC-H3, or H3-MYC; or GFP-HIRA (Green Fluorescent Protein mammalian expression vector) using Lipofectamine LTX (Invitrogen). Overexpression of MYC-H3.3, H3.3-MYC, MYC-H3, or H3-MYC was induced by addition of 1 μM doxycycline (a tetracycline analog) for either 12 or 24 h.
H3.3 cDNA was generated using H3.3cDNAforward and H3.3cDNAreverse. To generate C-terminal MYC-tagged H3.3 construct, H3.3 cDNA was amplified using H3.3mycForward and H3.3cmycReverse and was cloned into a tetracycline-inducible expression vector, pcDNA4/TO/myc-His A (Invitrogen; containing MYC sequence and 6× histidines epitopes), at BamHI and ApaI. The sequences of the primers were as follows:
H3.3cDNAforward (5′-GTAAGTAAGGAGGTCTCTGTACCATGGCT-3′) and H3.3cDNAreverse (5′-CCATCATAGTGGATTCTTAAGCACGTTCTC-3′); and
H3.3mycForward (5′-CGGGATCCACCATGGCTCGTACAAAGCAG-3′ [carrying a BamHI site, underlined, and a start codon, italic]) and H3.3cmycReverse (5′-GAAGGGCCCAGCACGTTCTCCACGTATGC-3′ [carrying an ApaI site, underlined]).
To generate N-terminal MYC-tagged H3.3 and H3 constructs, three rounds of PCR extensions were performed using forward and reverse primer corresponding to H3.3 and H3 cDNA sequences, respectively. The resulting H3.3 and H3 fragments were cloned into pcDNA4/TO/myc-His A at BamHI and ApaI. The sequences of H3.3 primers for serial PCR extension were as follows:
First PCR, Forward H3.3a primer 5′-CAGAAGAGGATCTGGCTCGTACAAAGCAGA-3′;
Second PCR, Forward H3.3b primer 5′-ACAAAAACTCATCTCAGAAGAGGATCTGGC-3′;
Third PCR, Forward H3.3c primer 5′-CGGGATCCACCATGGAACAAAAACTCATCT-3′ (carrying a BamH1 site, underlined, and a start codon, italic); and
Reverse H3.3 primer 5′-GAAGGGCCCTTAAGCACGTTCTCCACGTAT-3′ (carrying a ApaI site, underlined, and a stop codon, italic).
The sequences of H3 primers for serial PCR extension were as follows:
First PCR, Forward H3a primer 5′-CAGAAGAGGATCTGGCCCGGACGAAGCAGA-3′;
Second PCR, Forward H3b primer 5′-ACAAAAACTCATCTCAGAAGAGGATCTGGC-3′; and
Third PCR, Forward H3c primer 5′-CGGGATCCACCATGGAACAAAAACTCATCT-3′ (carrying a BamHI site, underlined, and a start codon, italic); and
Reverse H3 primer 5′-GAAGGGCCCTTATGCTCGCTCGCCACGGAT-3′ (carrying a ApaI site, underlined, and a stop codon, italic).
To generate C-terminal GFP-tagged HIRA construct, mouse HIRA cDNA was PCR amplified using XhoIHIRAforward and SacIIHIRAreverse and was cloned into pEGFP-C1 mammalian expression vector (GIBCO) at XhoI and SacII. The sequences of the primers were:
XhoIHIRAforward 5′-CCGCTCGAGCTATGAAGCTCTTGAAGCCAACC-3′ (carrying a XhoI site, underlined) and
SacIIHIRAreverse 5′-TCCCCGCGGTCTTGCCCCACAGGATGTCCAG-3′ (carrying a SacII site, underlined).
RNAi-depletion of H3.3 and real-time PCR analysis
Transfections of H3.3-RNAi-duplex oligonucleotides were performed using Lipofectamine LTX (Invitrogen). The RNAi-duplex oligonucleotides (three sets of oligonucleotides corresponding to both H3.3A and H3.3B genes) were purchased from Invitrogen. These oligonucleotides have been prevalidated to confirm their targeting specificity to H3.3A and H3.3B mRNAs. A set of control scrambled RNAi-duplex oligonucleotides was also used. Cells were harvested 24–48 h post-RNAi knockdown for the assessment of the levels of endogenous H3.3 by real-time PCR analysis using the SYBR system (Applied Biosystem). As internal controls, specific primers corresponding to house-keeping gene Actin were used in real-time PCR analysis. Changes in expression levels were calculated according to the manufacturer's instruction . The sequences of the H3.3 RNAi-duplexes were as follows:
H3.3A2 (AUGUACUUGCAUGUAGGACAACUCA and UGAGUUGUCCUACAUGCAAGUACAU);
H3.3A3 (UUACAGGAACAGCACAGAAGACAGA and UCUGUCUUCUGUGCUGUUCCUGUAA);
H3.3B2 (UUAGCGUGGAUGGCACACAGGUUGG and CCAACCUGUGUGCCAUCCACGCUAA); and
H3.3B3 (AACUGGAUGUCUUUGGGCAUGAUGG and CCAUCAUGCCCAAAGACAUCCAGUU).
The H3.3 and Actin primer sets used for real-time PCR analysis were as follows:
H3.3AutrF1 (ATTTCTTATTGACGGCAACTAAATG) and H3.3AutrR1 (AATAGCAAACTTACAGGAACAGCAC);
H3.3ButrF1 (TTGCTTGGTTAGAAAAGGCTAAAGTA) and H3.3ButrR1 (CCTATTAGAAAGATCCAAAACAGCA); and
Actin 5′- TCCCTGGAGAAGAGCTACGA and Actin 3′- AGCACTGTGTTGGCGTACAG.
ChIP assays
Cells were fixed in 0.8% paraformaldehyde in 1× PBS, washed extensively in 1× PBS, lysed in ice-cold Lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl at pH 8.0 and protease inhibitors; sonicated chromatin products of ∼100–300 bp), and diluted 10× in dilution buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 2 mM EDTA, protease inhibitors, and 1 mg/mL bovine serum albumin). ChIP was performed with the relevant antibody and captured with Protein A/G–Sepharose. DNA–protein complex was washed with 1× wash buffer I (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, and 2 mM EDTA), 2× wash buffer II (20 mM Tris-HCl at pH 8.0, 250 mM NaCl, 0.1% SDS, 1% Triton X-100, and 2 mM EDTA) and eluted in 1% SDS and 100 mM NaHCO3. Eluate fraction was de-cross-linked by high-salt treatment (200 mM NaCl) at 60°C followed by proteinase K treatment at 50°C. DNA extracted was subjected to either PCR or dot blot analysis (see below).
Standard and telomere PCR analysis
PCR analysis was performed using the primer sets specific for telomere (for details on Telomere PCR assays, see below) (Cawthon 2002) and major satellite DNA, followed by gel electrophoresis. Signal intensities of the products were quantitated using Syngene Gene Tools software (Synoptics Ltd). Telomere PCR assay (Cawthon 2002) has been widely used for quantitative measurement of telomere length for a variety of cell types. It offers excellent accuracy, sensitivity, and reproducibility compared with other protocols developed for telomere measurements such as flow cytometry and cytometric assay. PCR amplification of telomere DNA was performed using primers tel1 and tel2 according to a modified protocol (Cawthon 2002) with PCR cycling parameter as described—10 min at 95°C, followed by 30 cycles of 15 sec at 95°C, and 2 min at 54°C. Real-time PCR was performed using Gold AmpliTaq (Applied Biosystem). PCR amplification of pericentric major satellite DNA was carried out by standard PCR procedure using primers major satellite1 and major satellite2. The sequences of the primers were as follows:
tel1 (5′-GGTTTTTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGT-3′) and tel2 (5′-TCCCGACTATCCCTATCCCTATCCCTATCCCTATCCCTA-3′);
Major satellite1 (5′-TGGCGAGAAAACTGAAAATCACGGA-3′) and
Major satellite2 (5′-TCTTGCCATATTCCACGTCCTACAG-3′).
Dot blot analysis
DNA (50 ng) was diluted with 0.6 M NaCl, denatured (by heating DNA at 100°C for 5 min and cooling on ice for 5 min), and transferred onto Hybond N+ nitrocellulose membrane. The membrane was washed in 0.4 N NaOH at RT for 5 min, followed by neutralization with 2× SSC twice. Subsequently, the membrane was subjected to Southern blot analysis with [γ-32P]ATP-end-labeled (TTAGGG)4 telomere probe. Signal intensities were analyzed with Typhoon PhosphoImager System and ImageQuant software.
Pulse field gel electrophoresis and Southern blot analysis
Total genomic DNA was digested with RsaI and HinfI and subjected to either standard or pulse field (6 V/cm for 20 h at 14°C; switch times ramped linearly from 0.5 to 6 sec using CHEF PFGE Biorad system) gel electrophoresis, followed by Southern blot analysis with a [γ-32P]ATP-end-labeled (TTAGGG)4 telomere repeat probe. Signal intensities were analyzed with Typhoon PhosphoImager System and ImageQuant software.
MNase digestion assays
Cells were lyzed in ice-cold TBS buffer (10 mM Tris at pH 7.5, 3 mM CaCl2, 2 mM MgCl2, and protease inhibitor) containing 0.5% Tween40 for 1 h and homogenized using a Dounce Homogeniser Tight/A pestle. Nuclei were pelleted and incubated in digestion buffer (50 mM Tris at pH 7.5, 320 mM sucrose, 1 mM CaCl2, 4 mM MgCl2, and protease inhibitor) with MNase (4–16 units/μg DNA) at 37°C for various time course. Reaction was stopped with the addition of 1/100 of 0.5 M EDTA, followed by gel electrophoresis and Southern blot analysis with either [γ-32P]ATP-end-labeled (TTAGGG)4, α-32P-labeled centromeric minor, or pericentric major satellite probe. Signal intensities were analyzed with Typhoon PhosphoImager System and ImageQuant software.
Two-dimensional gel electrophoresis
Cells were resuspended in 1× PBS with 1.4% agarose (1:1 v/v), digested with 1mg/mL Proteinase K in TENs buffer (100 mM EDTA, 0.2% sodium deoxycholate, and 1% sodium lauryl sarcosine) and washed in 1× TE buffer (10 mM Tris at pH 7.5 and 1 mM EDTA). For the first dimension, DNA plugs digested with HinfI and HaeIII were fractionated on a 0.4% agarose gel at 1 V/cm for 25.5 h, followed by second-dimensional electrophoresis on a 1% agraose gel at 6 V/cm for 8 h and Southern blot analysis with a [γ-32P]ATP-end-labeled (TTAGGG)4 .probe. Signal intensities were analyzed with Typhoon PhosphoImager System and ImageQuant software.
Acknowledgments
We thank Roger Reddel and Anthony Caeser for discussion, and Jim McWhir for EG cell lines. This work was supported by the National Health and Medical Research Council of Australia.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.084947.108.
References
- Ahmad K., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Blasco M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- Cawthon R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare A.J., Griffith J.D. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M., O'Sullivan R., Peters A.H., Jenuwein T., Blasco M.A. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- Gilbert N., Thomson I., Boyle S., Allan J., Ramsahoye B., Bickmore W.A. DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J. Cell Biol. 2007;177:401–411. doi: 10.1083/jcb.200607133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S., Jaco I., Fraga M.F., Chen T., Li E., Esteller M., Blasco M.A. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Hake S.B., Garcia B.A., Kauer M., Baker S.P., Shabanowitz J., Hunt D.F., Allis C.D. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc. Natl. Acad. Sci. 2005;102:6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Sfeir A.J., Shay J.W., Wright W.E., de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B., Bonnefoy E., Anselme C., Laurencon A., Karr T.L., Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- Martens J.H., O'Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E., Gafken P.R., Ahmad K., Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Uren A.G., Wong L., Pakusch M., Fowler K.J., Burrows F.J., Vaux D.L., Choo K.H. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 2000;10:1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- van der Heijden G.W., Derijck A.A., Posfai E., Giele M., Pelczar P., Ramos L., Wansink D.G., van der Vlag J., Peters A.H., de Boer P. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat. Genet. 2007;39:251–258. doi: 10.1038/ng1949. [DOI] [PubMed] [Google Scholar]
- Verdun R.E., Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Wong L.H., Saffery R., Anderson M.A., Earle E., Quach J.M., Stafford A.J., Fowler K.J., Choo K.H. Analysis of mitotic and expression properties of human neocentromere-based transchromosomes in mice. J. Biol. Chem. 2005;280:3954–3962. doi: 10.1074/jbc.M410047200. [DOI] [PubMed] [Google Scholar]