Abstract
Selective activity of a specific set of enhancers defines tissue-specific gene transcription. The pioneer factor FOXA1 has been shown to induce functional enhancer competency through chromatin openings. We have previously found that FOXA1 is recruited to thousands of regions across the genome of a given cell type. Here, we monitored the chromatin structure at FOXA1 binding sites on a chromosome-wide scale using formaldehyde assisted isolation of regulatory elements (FAIRE). Surprisingly, we find that a significant fraction of FOXA1-bound sites have a relatively closed chromatin conformation linked to a shift of the epigenetic signature toward repressive histone marks. Importantly, these sites are not correlated with gene expression in a given cell type suggesting that FOXA1 is required, but not sufficient, for the functional activity of bound enhancers. Interestingly, we find that a significant proportion of the inactive FOXA1-bound regulatory sites in one cell type are actually functional in another cellular context. We found that at least half of the FOXA1 binding sites from a given cell type are shared with another cell lineage. Mechanisms that restrict the activity of shared FOXA1-bound enhancers likely play a significant role in defining the cell-type-specific functions of FOXA1.
Organization of genomic DNA into chromatin and higher-order structures is at the center of gene expression regulation in eukaryotes (Ozsolak et al. 2007; Steinfeld et al. 2007; John et al. 2008; Schones et al. 2008). Accordingly, functional activity of cis-regulatory elements is linked to the recruitment of multiprotein complexes by transcription factors, including cofactors such as CBP/p300 and p160 family members, that can alter chromatin structure/function through control of histone post-translational modifications (Shang et al. 2000; Bauer et al. 2002; Metivier et al. 2003; Rosenfeld et al. 2006; Berger 2007; Torres-Padilla et al. 2007; Frietze et al. 2008; John et al. 2008; Schones et al. 2008). For instance, methylation of histone H3 on lysines 4 and 17 (H3K4me and H3R17me), as well as acetylation of H3K9 (H3K9ac), are associated with functionally competent genomic regions while methylation of H3K9 (H3K9me) is related to repression (Bauer et al. 2002; Barski et al. 2007; Heintzman et al. 2007; Kouzarides 2007; Mikkelsen et al. 2007; Frietze et al. 2008; Komashko et al. 2008; Krum et al. 2008). However, many transcription factors are unable to bind DNA when their target cis-regulatory elements are nucleosomal.
In this context, the discovery of specific proteins with crucial roles in development and differentiation, which act as pioneer factors binding condensed chromatin and allowing subsequent transcription factor recruitment, was an important breakthrough in our understanding of gene regulation (Gualdi et al. 1996; Cirillo et al. 1998; Bresnick et al. 2005; J. Xu et al. 2007; Escamilla-Del-Arenal and Recillas-Targa 2008). One such factor is the forkhead family member FOXA1, which is involved in differentiation of several endoderm-derived organs and whose pioneering activity was first described in the context of liver-specific induction of albumin expression (Gualdi et al. 1996; Cirillo et al. 2002; Lee et al. 2005; Friedman and Kaestner 2006). Indeed, FOXA1 was shown to establish competency of the albumin enhancer to which it binds before transcriptional induction of this gene in the endoderm (Zaret 1999). This was subsequently ascribed to FOXA1 ability to bind and relax condensed chromatin structures in vitro (Cirillo et al. 2002).
Recently, we found that FOXA1 binds to thousands of enhancers across the human genome (defined as the FOXA1 cistrome) (Lupien et al. 2008). In breast and prostate cancer cells, a fraction of these FOXA1-bound sites (20%–30%) is actively involved in steroid hormone receptor transcriptional regulatory functions (Gao et al. 2003; Carroll et al. 2005; Laganiere et al. 2005; Eeckhoute et al. 2006). However, a comprehensive knowledge of the activity of all FOXA1-bound enhancers in a given cell type is still lacking. Here, chromosome-wide analysis of chromatin structure, occurrence of histone marks and transcriptional regulatory factor recruitment, as well as correlation with gene expression were used to probe the functionality of FOXA1-bound cis-regulatory elements.
Results
Chromosome-wide monitoring of chromatin structure reveals a high degree of heterogeneity at FOXA1-bound regulatory elements
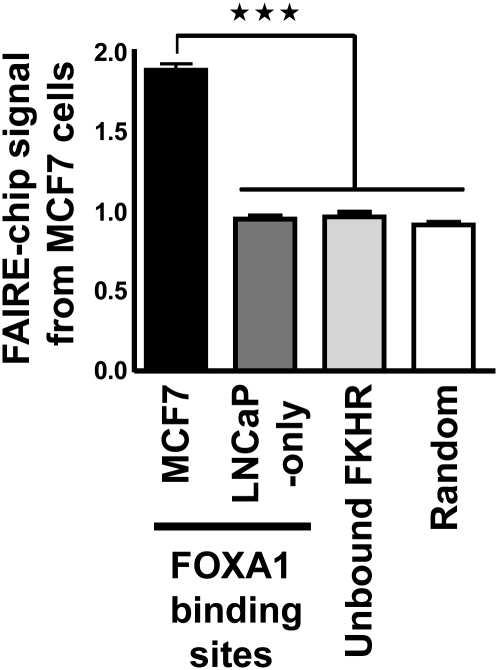
The degree of chromatin compaction is intimately related to its functionality and active cis-regulatory elements typically exist within open chromatin regions depleted in nucleosomes (Heintzman et al. 2007; Boyle et al. 2008). These domains can be identified using formaldehyde-assisted isolation of regulatory elements (FAIRE) that allows for enrichment of nucleosome-depleted genomic regions when cross-linked chromatin is subjected to phenol-chloroform extraction (Nagy et al. 2003; Hogan et al. 2006). Comparison of FAIRE and DNaseI sensitivity signals, as well as the occurrence of histone marks over large genomic regions has validated the use of FAIRE to identify active transcriptional regulatory sites (The ENCODE Project Consortium 2007; Giresi et al. 2007). Hence, FAIRE was performed to monitor on a genomic-scale chromatin structure at FOXA1 recruitment sites in MCF7 breast cancer cells (Lupien et al. 2008). DNA obtained from these cells after FAIRE was hybridized to tiled arrays covering the non-repetitive regions of chromosomes 8, 11, and 12 (hereafter FAIRE-chip). In a recent large-scale study of chromatin DNAseI sensitivity, it was reported that promoters generally harbor a more open conformation when compared with distal regulatory elements (Boyle et al. 2008). We observed a similar difference when we analyzed FAIRE-chip signal at RNA polymerase II (Pol II) and FOXA1-bound sites that were previously identified in MCF7 cells using chromatin immunoprecipitation coupled with hybridization on the same tiled arrays (ChIP-chip) (Carroll et al. 2006; Lupien et al. 2008). Indeed, the average FAIRE-chip enrichment was stronger at high confidence Pol II-bound sites compared to FOXA1-bound regions (Supplemental Fig. S1). This result is in agreement with Pol II binding primarily at promoters, while FOXA1 is recruited mainly to enhancers (Lupien et al. 2008). For subsequent analyses, we removed the small number of FOXA1 binding sites that map within 1 kilobase (kb) of proximal promoters (∼2% of all sites) (Lupien et al. 2008). This had a very modest effect on the average FAIRE-chip signal at FOXA1 binding sites (Supplemental Fig. S1). Importantly, FAIRE-chip signal at these enhancers is significantly higher than control sites specifically bound by FOXA1 in a distinct cell type, i.e., LNCaP prostate cancer cells (Fig. 1; Lupien et al. 2008). Similarly, significantly lower FAIRE signals were found at randomly selected regions or at regions containing a Forkhead motif that are not bound by FOXA1 (Fig. 1). Analysis of the FAIRE signal at recognition motifs for two other transcription factors in MCF7 cells, ESR1 and E2F1, also revealed greater FAIRE enrichment at sites that were bound versus unbound (Bieda et al. 2006; Carroll et al. 2006; Rabinovich et al. 2008; Supplemental Fig. S2).
Figure 1.
FAIRE-chip signal from MCF7 cells at FOXA1 binding sites. FAIRE-chip signal from MCF7 cells within high confidence FOXA1 recruitment sites from MCF7 cells (lying outside of 1 kb promoters) or specific to LNCaP cells is indicated. A set of randomly selected Forkhead motif-containing regions not bound by FOXA1 in MCF7 cells (Unbound FKHR) as well as randomly selected regions were also used. Data represent mean ± SEM of signals derived from MAT analysis of FAIRE-chip data. *** Indicates a statistically significant difference (P < 0.001).
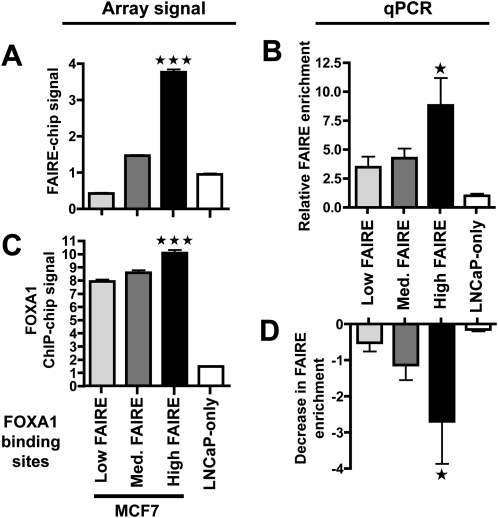
In order to further characterize the chromatin structure at FOXA1 binding sites, we divided them into tertiles based on their FAIRE-chip signal. Hence, three subsets of FOXA1-bound regions were defined corresponding to low, medium, or high FAIRE. Interestingly, there was a very pronounced difference in the average FAIRE enrichment between low and high FAIRE sites (Fig. 2A). This difference was validated on randomly selected sites using real-time PCR (FAIRE-qPCR) (Fig. 2B). Importantly, low FAIRE signals at a subset of FOXA1 recruitment regions did not stem from a lower confidence in the authenticity of these binding sites since (1) only high confidence FOXA1 binding sites with an extremely high rate of validation by directed ChIP were used (Lupien et al. 2008); (2) sites with low FAIRE enrichments showed an average evolutionary conservation and enrichment for the Forkhead motif as significant as sites with high FAIRE signals (Supplemental Figs. S3 and S4); (3) the fraction of FOXA1 binding sites independently identified as also recruiting FOXA1 in another cell type (LNcaP cells) was similar between sites with low and high FAIRE enrichments (Supplemental Fig. S5); and (4) FOXA1 binds strongly to low FAIRE enrichment sites though FOXA1 recruitment is slightly greater at high FAIRE sites (Fig. 2C). Next, we asked whether FOXA1 was actually involved in defining FAIRE enrichments at bound regulatory elements. To answer this question, we transfected MCF7 cells with a siRNA directed against FOXA1 or luciferase, as a control (Eeckhoute et al. 2006). These cells were then used in FAIRE-qPCR experiments to monitor effects on FAIRE enrichments at regions of FOXA1 recruitment. As shown in Figure 2D, FOXA1 silencing was able to reduce FAIRE enrichment at FOXA1 binding sites, with the most significant effect on sites with high FAIRE signals. Hence, FOXA1 is required, but not sufficient to trigger high FAIRE enrichment at bound cis-regulatory regions.
Figure 2.
FOXA1 is required but not sufficient for high FAIRE enrichment at bound regulatory elements. (A) FOXA1 binding sites were divided into tertiles and the average FAIRE-chip signal (based on MAT scores, see Methods) for each subset of sites (low, medium, and high FAIRE) was calculated. Each subset was comprised of 645 sites. FAIRE-chip enrichments at FOXA1 LNCaP-specific sites were also analyzed. Data represent mean ± SEM of signals derived from MAT analysis of FAIRE-chip data. (B) FAIRE-qPCR experiments were performed in MCF7 cells to monitor FAIRE enrichment at the indicated categories of FOXA1 recruitment regions (at least 8 different sites were analyzed for each subset). Relative enrichment compared to negative control regions is shown. Data are mean ± SD from three independent experiments. (C) Signals from FOXA1 ChIP-chip in MCF7 cells within FOXA1 binding sites with low, medium or high FAIRE-chip enrichments. LNCaP-specific sites were also analyzed. Data represent means ± SEM of signals derived from MAT analysis of the FOXA1 ChIP-chip data from Lupien et al. (2008). (D) FAIRE-qPCR experiments performed as in B. Decrease in FAIRE enrichments triggered by FOXA1 silencing is shown. Data are mean ± SD from three independent experiments. *** and * Indicate a statistically significant difference between FOXA1 recruitment sites with high and low FAIRE enrichments (P < 0.001 and P < 0.05, respectively).
Levels of FAIRE enrichment correlate with the transcriptional regulatory activity at FOXA1-bound enhancers
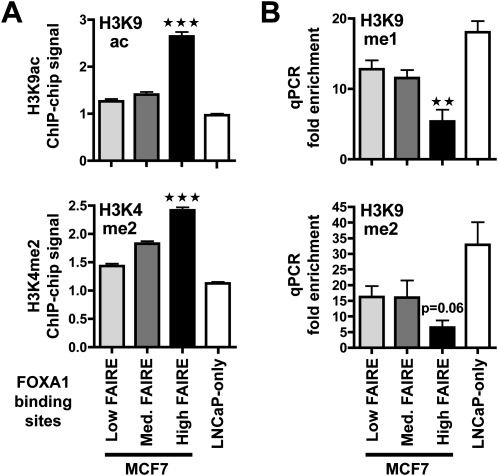
We next sought to determine the functional role of FOXA1 binding sites associated with low, medium, or high FAIRE signals in MCF7 cells. Elevated ESR1 recruitment and CARM1 activity at FOXA1 binding sites supports the identification of these sites as functional enhancers (Carroll et al. 2006; Lin et al. 2007; Gao et al. 2008; Lupien et al., unpubl.). Interestingly, both ESR1 binding and CARM1 activity (including factor and H3R17 dimethylation) were predominantly associated with high FAIRE FOXA1 binding sites (Supplemental Fig. S6). This supports the conclusion that high FAIRE FOXA1 sites are more likely to be functional. To substantiate this finding, we monitored histone marks levels typically associated with active (H3K9ac and H3K4me2) or repressed (H3K9me1 and 2) regulatory elements at the FOXA1-bound regions according to FAIRE class. We found that FOXA1-bound sites with high FAIRE enrichment exhibited significantly higher levels of H3K9ac and H3K4me2 (Fig. 3A), but lower levels of H3K9me1 and 2 (Fig. 3B) when compared to sites with low FAIRE-chip signals.
Figure 3.
High FAIRE enrichment at FOXA1 binding sites correlate with a shift toward active histone marks. (A) H3K9ac and H3K4me2 ChIP-chip signals at FOXA1 binding sites from MCF7 or specific to LNCaP cells were analyzed. Data represent mean ± SEM of signals derived from MAT analysis of the ChIP-chip data. *** Indicates a statistically significant difference (P < 0.001) between FOXA1 sites from MCF7 with high FAIRE versus low FAIRE enrichment. (B) ChIP-qPCR experiments were performed to monitor H3K9me1 and me2 levels in MCF7 cells at the indicated categories of FOXA1 recruitment regions from MCF7 cells or specific to LNCaP cells (LNCaP only). Data show relative enrichments compared to negative control regions. Data are mean ± SEM from at least three independent experiments. Statistical significance of the difference between FOXA1 recruitment sites with high versus low FAIRE enrichments is shown (** indicates P < 0.01).
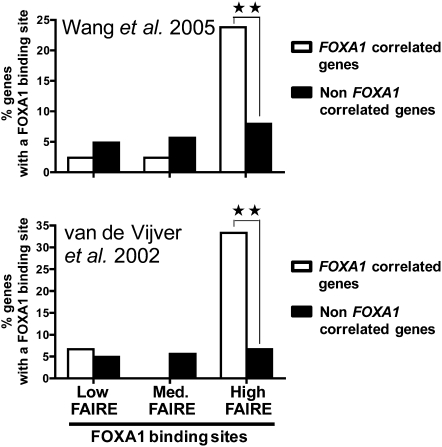
To determine whether these differences in chromatin structure at high and low FAIRE enrichment FOXA1 binding sites were indeed linked to their regulatory activity we next investigated their correlation with gene transcription. First, we analyzed the distribution of FOXA1 binding sites from the three different subsets of FAIRE enrichment relative to genes whose transcriptional start site (TSS) is bound by Pol II in MCF7 cells. Interestingly, sites of FOXA1 recruitment with high FAIRE signals were more associated with Pol II promoter-bound genes than FOXA1 binding sites with low FAIRE signals (Supplemental Fig. S7). We previously showed that FOXA1 binding sites from MCF7 cells were also enriched nearby genes co-expressed with FOXA1 in primary breast tumors (Lupien et al. 2008). Hence, we analyzed the distribution of the different FAIRE subsets of FOXA1 recruitment regions relative to genes co-expressed with FOXA1 in breast cancer. Importantly, using both the Wang and van de Vijver expression data sets (van de Vijver et al. 2002; Wang et al. 2005), we found that only FOXA1 binding sites with high FAIRE signals were significantly enriched in the vicinity of genes co-expressed in breast cancer compared to non-co-expressed genes used as control (Fig. 4). Hence, altogether our data indicate that only a fraction of FOXA1 bound cis-elements in MCF7 cells with an open chromatin structure are functionally connected to gene regulation in breast cancer.
Figure 4.
FOXA1 binding sites with high FAIRE-chip signals correlate with gene expression in breast cancer. Distribution of FOXA1 binding sites with different FAIRE enrichments relative (within 20 kb of the TSS) to genes co-expressed or non-co-expressed with FOXA1 in primary breast tumors (van de Vijver et al. 2002; Wang et al. 2005). ** Indicates a statistically significant difference between the distribution of FOXA1 binding sites near correlated and non-correlated genes (P < 0.01).
Cell-lineage-specific activities of conserved FOXA1-bound enhancers
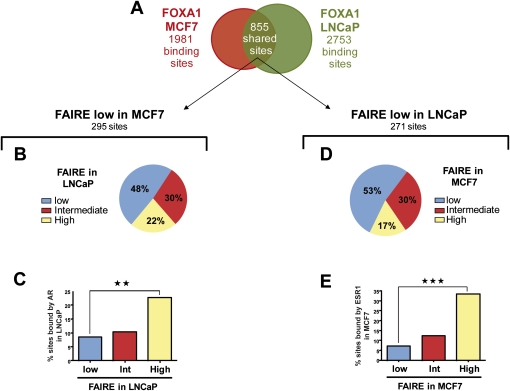
In light of these results, we sought to determine whether low FAIRE FOXA1 binding regions from MCF7 cells could be active in another cellular context. Indeed, in addition to significant cell-type-specific recruitment, comparison of the FOXA1 cistromes from MCF7 and LNCaP cells revealed a substantial number of shared binding regions (Fig. 5A; Lupien et al. 2008). We performed FAIRE-chip in LNCaP cells and, as for MCF7 cells we divided FOXA1 binding sites in tertiles according to their FAIRE-chip enrichment (Supplemental Fig. S8). In keeping with the results in MCF7 cells, there was a significant difference in the average FAIRE-chip signals between the low and high FAIRE subsets of sites in LNCaP (Supplemental Fig. S8B). While high FAIRE signals at FOXA1 binding sites were correlated with the presence of estrogen response elements (ERE) and strong ESR1 recruitment in MCF7 cells (Supplemental Fig. S6A), FOXA1 sites harboring high FAIRE enrichments in LNCaP cells showed greater levels of enrichment for androgen response elements (ARE) and AR recruitment (Supplemental Fig. S9). To determine whether FOXA1 recruitment regions found in a closed structure in MCF7 cells could be active and in an open conformation in LNCaP cells, we analyzed the FAIRE-chip signals obtained in LNCaP cells for shared sites harboring low FAIRE enrichment in MCF7 (Fig. 5B). Interestingly, 22% of these sites showed high FAIRE signals in LNCaP cells (Fig. 5B). Differential FAIRE signals between the two cell types at common FOXA1 recruitment regions were also apparent when sites with high FAIRE in MCF7 cells were considered (Supplemental Fig. S10). The differential FAIRE enrichments did not stem from false negative signals from the FAIRE-chip data as analysis of ESR1 and histone marks at sites with selectively low FAIRE in MCF7 verified they were inactive in this cell line (Supplemental Fig. S11). In addition FAIRE-qPCR experiments performed in MCF7 and LNCaP cells on several shared FOXA1 binding regions validated their differential FAIRE enrichments between the two cell types (Supplemental Fig. S12). Interestingly, the subset of sites with low FAIRE in MCF7, but high FAIRE in LNCaP, had a significantly greater likelihood of recruiting the androgen receptor (AR), a central transcriptional regulator in prostate cancer cells (Fig. 5C). On the other hand, shared FOXA1 sites with low FAIRE enrichment in LNCaP cells, but high FAIRE signals in MCF7 cells had a significantly greater overlap with ESR1 recruitment regions than sites with low FAIRE signals in both cell-lines (Fig. 5D,E). Moreover, these sites showed higher levels of histone marks associated with active regulatory elements (Supplemental Fig. S13). Clustering of shared FOXA1 binding sites according to their FAIRE-chip enrichment, ESR1, and AR binding, also revealed sites with different FAIRE-chip signals within the two cell types that correlate with alternate nuclear receptor recruitment (Supplemental Fig. S14). Altogether, these results strongly suggest that FOXA1 binding sites found within two different cell lineages can act as cell-type-specific enhancers that are distinguished by their chromatin structure. Specific examples of the correlation between the cell-type-specific activities of common FOXA1 binding sites and differential gene regulation between MCF7 and LNCaP cells are shown in Figure 6 and Supplemental Figure S15.
Figure 5.
Cell-specific activity of conserved FOXA1 recruitment sites. (A) Comparison of FOXA1 binding sites identified in MCF7 and LNCaP cells by ChIP-chip (Lupien et al. 2008). (B) FOXA1 binding sites shared between MCF7 and LNCaP and harboring low FAIRE enrichment in MCF7 cells were selected. These sites were then classified relative to their FAIRE enrichment levels (low, medium, or high) in LNCaP cells. (C) Histogram indicating the percentage of sites defined in (B) that also recruits AR in LNCaP cells. ** Indicates a statistically significant difference (P < 0.01) between FOXA1 binding sites with high and low FAIRE enrichment in LNCaP cells. (D) FOXA1 binding sites shared between MCF7 and LNCaP and harboring low FAIRE enrichment in LNCaP cells were selected. These sites were then classified relative to their FAIRE enrichment levels (low, medium, or high) in MCF7 cells. (E) Histogram indicating the percentage of sites defined in (B) that also recruits ESR1 in MCF7 cells. *** Indicates a statistically significant difference (P < 0.001) between FOXA1 binding sites with high and low FAIRE enrichment in MCF7 cells.
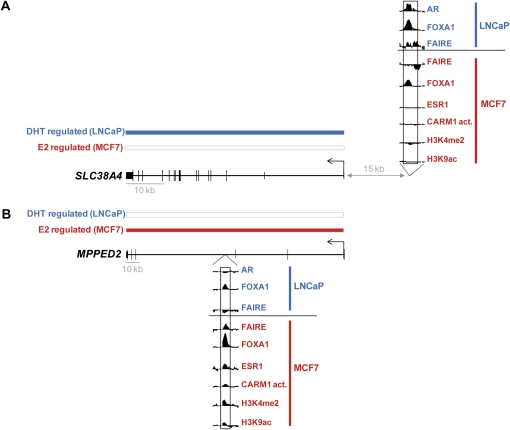
Figure 6.
Correlation between selective activity of common FOXA1 binding sites and cell-type-specific gene regulation. Examples of genes associated with FOXA1 binding sites common to MCF7 and LNCaP cells, but regulated in a cell-type-specific manner. Individual probe level signal for FAIRE-chip and ChIP-chip of the indicated factors or histone marks within the FOXA1 binding sites is shown. Transcriptional regulation by dihydrotestosterone (DHT) in LNCaP cells (Wang et al. 2007) or by estradiol (E2) in MCF7 cells (Carroll et al. 2006) was determined using a t-test (P < 5 × 10−3) and is indicated by blue and red bars, respectively. (A) Example of a gene regulated by DHT in LNCaP, but not by E2 in MCF7. (B) Example of a gene regulated by E2 in MCF7, but not by DHT in LNCaP. Note that shared FOXA1 binding sites with selective activities are often associated with cell-type-specific ones in the vicinity of target genes. This is consistent with the clustering of transcription factor recruitment regions nearby regulated genes (Chan and Song 2008; Krum et al. 2008).
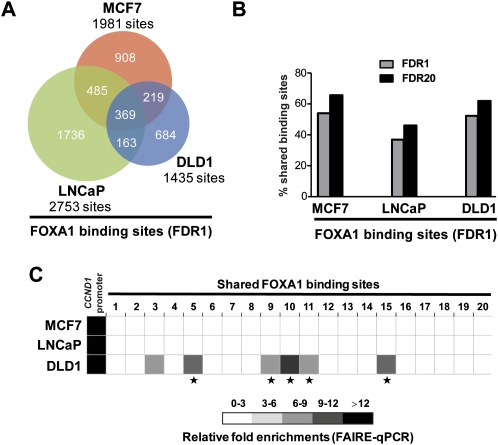
To further determine the importance of cell-lineage selective activity of conserved FOXA1 recruitment regions, we decided to determine by ChIP-chip the FOXA1 cistrome in another cell line of different lineage, i.e., the DLD1 colon carcinoma cell line. Interestingly, comparison of the MCF7, LNCaP, and DLD1 FOXA1 cistromes revealed cell-type-specific sites, as well as all possible combination of overlapping sites (Fig. 7A). Hence, subsets of the sites that we previously defined as MCF7 or LNCaP specific (Lupien et al. 2008) are actually shared with DLD1. Indeed, these data now reveal that at least half of FOXA1 binding sites from a given cell type are also bound in at least another cellular background when three FOXA1 expressing cell lineages are considered (Fig. 7B). This suggests that the majority of FOXA1 binding sites might actually be bound in at least two different cell types and mechanisms that differentially regulate chromatin structure and activity of a given binding site must exist. In support of this notion, FAIRE-qPCR at FOXA1 binding sites with low FAIRE enrichment in both MCF7 and LNCaP, and shared among all three cell types, revealed a significant proportion with high FAIRE enrichment only in DLD1 cells (Fig. 7C). FOXA1 silencing verified that FAIRE enrichments at most of those sites were dependent upon FOXA1 expression in DLD1 cells as well (Supplemental Fig. S16).
Figure 7.
Comparison of FOXA1 recruitment sites within MCF7, LNCaP, and DLD1 cells. (A) Venn diagram showing the overlap between FOXA1 recruitment sites identified in three different cell lineages, i.e., breast (MCF7), prostate (LNCaP), and colon (DLD1) cells. Only high confidence binding sites were used here (FDR1). (B) Histogram showing the percentage of high confidence FOXA1 binding sites (FDR1) from each cell-line that is also found in at least one of the other two cells at FDR1 or 20. (C) FAIRE-qPCR experiments were performed in MCF7, LNCaP, and DLD1 cells. CCND1 proximal promoter, used as a positive control (Eeckhoute et al. 2006), and 20 FOXA1 binding sites shared between the three cell-lines were analyzed. Relative enrichments compared to negative control regions are shown. Data are mean ± SD from two or three independent experiments. * Indicates statistically significant differences (P < 0.05) between FAIRE enrichments in DLD1 and both MCF7 and LNCaP cells.
Discussion
FOXA1 has been defined as a pioneer factor that induces competency of enhancers allowing for subsequent recruitment of collaborating transcription factors. Here, we performed a genome-scale analysis of chromatin structure using FAIRE at FOXA1 binding sites. We found that a substantial fraction of FOXA1 recruitment sites within a given cell type harbor a relatively closed chromatin structure and lack apparent functional activity in positive gene regulation. This indicates that numerous FOXA1-bound enhancers might become active only under particular stimuli. However, we found that FOXA1 binding sites with low FAIRE in MCF7 cells did not correlate with gene expression in primary breast tumors suggesting they are not of functional importance in this cell type in vivo. Moreover, we showed that a subset of these FOXA1-bound cis-regulatory regions with low FAIRE signals is actually active in a different cellular context. Since this differential activity is correlated with the co-recruitment of a secondary cell-specific factor (ESR1 or AR, respectively), this suggests that these sites might remain non-functional in the cell type that lacks the required factor. Inactive sites in a differentiated cell could represent spurious elements that were active at some point during the differentiation of the cell and/or that are within a chromatin environment allowing for FOXA1 binding even though they are not meant to be functional in this particular cellular context. Of note, several other regulatory factors have binding sites that are conserved between different cell types (Odom et al. 2004; Bieda et al. 2006; Acosta-Alvear et al. 2007; Kim et al. 2007; X. Xu et al. 2007). Moreover, recent genome-wide studies of transcription factor binding to chromatin identified numerous recruitment sites of which only a fraction seemed associated with target gene regulation (Yang et al. 2006; Bolton et al. 2007; Zheng et al. 2007; Chan and Song 2008; Chen et al. 2008; Hua et al. 2008; Kim et al. 2008). In the light of our findings, we propose that restricted activities of cis-regulatory elements bound by a given factor in a given cellular environment might represent an important means to ensure cell-type-specific functions of this transcription factor. Indeed, we show here that FOXA1 cell-specific functions are linked to selective activities of a subset of the cis-regulatory elements bound by this pioneer factor.
Our study also reveals chromatin-related properties of active versus inactive FOXA1-bound enhancers. Indeed, FOXA1 binding regions with low FAIRE enrichment harbor a shift toward histone marks typical of “repression,” compared with sites with high FAIRE signals (higher H3K9me1 and 2, but lower H3K9ac, H3K4me2, and potentially H3R17me2 levels). Importantly, this difference was not as pronounced as the one observed at FKHR motifs not able to recruit FOXA1 in a specific cell type (Fig. 3 and Supplemental Fig. S5). Hence, our study shows that the balance between repressive and active histone marks at potential FOXA1-dependent enhancers is linked not only to the regulation of FOXA1 recruitment (Lupien et al. 2008), but also to subsequent levels of chromatin decompaction. The state of this balance could dictate the ability of FOXA1 (and/or collaborating factors) to promote chromatin openings. Alternatively, our data could point to additional levels of regulation subsequent to pioneer factor recruitment that could be involved in shifting this balance toward positive histone marks to trigger enhancer activity. Even if FOXA1 binding is not sufficient to establish a chromatin structure characteristic of active enhancers, silencing experiments confirmed that high FAIRE enrichment depends on FOXA1. Hence, our work shows on a broad scale that pioneer factor recruitment is required, but not sufficient to trigger full openings and functional activities of cis-regulatory regions. The ability of FOXA1 to selectively cooperate with specific transcriptional regulatory factors might play a crucial role in defining active versus inactive FOXA1-bound enhancers.
Cell-specific activities of enhancers play a crucial role in defining differential gene expression (Crawford et al. 2006; Pennacchio et al. 2006, 2007; Xi et al. 2007). Our determination of the FOXA1 cistrome in three distinct cell lineages reveals that most FOXA1 binding sites in a given cell lineage are actually shared with at least another cell type. Extrapolation of these findings would suggest that there might actually be few, if any, FOXA1 recruitment sites absolutely specific to a single cell type when all FOXA1 expressing tissues are considered. Our work suggests that FOXA1 cell-lineage-specific functions are determined by a combination between its specific pattern of recruitment to chromatin within a given cell type (Lupien et al. 2008) and selective activity of a subset of the bound regulatory elements. Hence, there is an unexpectedly high degree of flexibility regarding the activities of cis-regulatory elements recruiting FOXA1. This study therefore reveals, on a genomic scale, that FOXA1 pioneering activity is actually integrated within a multilayer regulatory process that ultimately defines selective activities of bound enhancers. Indeed, FOXA1 binding is not sufficient to establish functional enhancers, which is highly dependent upon the cellular and physiologic context.
Methods
Formaldehyde-assisted isolation of regulatory elements (FAIRE)
FAIRE was performed as described by Giresi et al. (2007) with slight modifications. Asynchronously growing cells (60%–70% confluence) were cross linked with 1% formaldehyde for 10 min at room temperature. Glycine was added to a final concentration of 125 mM and the cells were rinsed with cold PBS and harvested. Cells were then lysed with 1% SDS, 10 mM EDTA and 50 mM Tris-HCl (pH 8.1) containing a protease inhibitor cocktail (Roche) and sonicated for 14 min (30 sec on/off cycles) using a Bioruptor (Diagenode) set up at the highest intensity. After centrifugation at 13,000g for 10 min at 4°C to precipitate cellular debris, the soluble chromatin was isolated and subjected to three consecutive phenol-chloroform extractions (Sigma, P3803). Each time, the aqueous phase was recovered and mixed with an equal volume of phenol-chloroform. After vortexing, the mixture was centrifuged at 13,000g for 1 min in phase lock gels (Eppendorf) to help separate the phenol and aqueous phases. Samples were then incubated overnight at 65°C to reverse cross linking. DNA was finally purified using the MinElute PCR purification kit (Qiagen).
FAIRE-qPCR
Real-time PCR were performed on purified DNA as in Eeckhoute et al. (2006, 2007). All primer sequences are available upon request. Statistical significance was determined using Student's t-test comparison for unpaired data.
FAIRE-chip
Input DNA or DNA from FAIRE (pooled DNA from cells cross linked for 6 or 10 min) was amplified, fragmented, labeled, and hybridized on Affymetrix array F covering human chromosomes 8, 11, and 12 as previously described (Carroll et al. 2005, 2006). Three independent experiments were performed and processed together using MAT algorithm (Johnson et al. 2006).
Chromatin immunoprecipitation (ChIP) and ChIP-chip
H3K9me1 and H3K9me2 levels were determined by ChIP-qPCR using the following antibodies from Abcam: Ab9045 and Ab1220, respectively. Statistical significance was determined using Student's t-test comparison for unpaired data. Chromatin fragments bound by FOXA1 in DLD1 cells were immunoprecipitated as previously described using antibody Ab5089 (Abcam) (Lupien et al. 2008). For ChIP-chip, ChIPed DNA was processed and hybridized on Affymetrix array F. Two independent experiments were performed and processed together using MAT algorithm (Johnson et al. 2006).
FOXA1 silencing
FOXA1 was silenced using siRNA as described previously (Eeckhoute et al. 2006).
Array signal intensity analysis
Intensity of the signal from FAIRE-chip or FOXA1, ESR1, CARM1 activity, H3K9ac and H3K4me2 ChIP-chip at different binding sites data sets was determined using the intensity of the strongest probe within each site after probe signal normalization by MAT algorithm (MAT score) (Johnson et al. 2006). Statistical significance was determined using Student's t-test comparison for unpaired data. The following ChIP-chip data were used in these analyses: ESR1 (Carroll et al. 2006), E2F1 (Bieda et al. 2006), FOXA1, and H3K4me2 (Lupien et al. 2008), as well as H3K9ac (Nagashima et al. 2008), from MCF7 cells and AR and FOXA1 from LNCaP cells (Lupien et al. 2008). CARM1 activity data were obtained using antibody 07-214 (Upstate biotechnology) directed against H3R17me2. Since the signal provided by this antibody could also stem from recognition of CARM1-dependent NCOA3 (also known as AIB1) methylation it was therefore referred to as CARM1 activity (CARM1 act.) (Naeem et al. 2007). As controls, 1 kb long regions containing or not containing a FOXA1 recognition motif in their center [using FOXA1 recognition sequence defined in (Lupien et al. 2008)] were randomly selected within the non-repetitive sequences of chromosomes 8, 11, and 12 covered by the DNA chips. Statistical significance was determined using Student's t-test comparison for unpaired data.
Binding site overlap
Two binding sites were considered to overlap as long as they had one base pair in common. Taking into account the average size of enriched regions, this means that the center of the two enriched sites had to be, on average, within 1 kilobase of each other to be considered overlapping.
Association of trends in gene expression with FOXA1 binding sites
Correlation between subsets of FOXA1 binding sites with different FAIRE enrichments and genes co-expressed in primary breast tumors was performed as in Lupien et al. (2008). The correlation coefficient used to define co-expressed genes was 0.5. Fisher's exact test was used to assess the statistical significance of the difference in the distribution of the different subsets of FOXA1 binding sites relative to co-expressed genes.
Cluster analysis
We generated a set of genomic intervals derived from the union of all high confidence sites associated with FOXA1 binding in MCF7 and LNCaP cells. Each interval was assigned a score for FAIRE enrichment, ESR1 and AR binding corresponding to the intensity of the strongest probe within each site after probe signal normalization by MAT algorithm (MAT score) (Johnson et al. 2006). For each data set, MAT scores were trimmed at the 2 and 98 percentiles and scaled to lie between zero and one. Genomic regions were clustered using k-means clustering.
Transcription factor recognition motif enrichment analysis
Analyses of transcription factor recognition motif enrichment were performed as in Lupien et al. (2008).
Acknowledgments
The authors thank members of the SPARTE team for helpful discussions during the preparation of this manuscript. This work was supported by funds from the Centre National de la Recherche Scientifique (CNRS) and the University of Rennes 1 and by grants from the NIDDK (R01DK074967 to M.B.), the NCI (P01 CA8011105 and the DF/HCC Breast Cancer SPORE Grant to M.B.), and the DFCI Women's Cancers Program.
Footnotes
[Supplemental material is available online at www.genome.org. ChIP-chip and FAIRE-chip data have been submitted to the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession nos. GSE12801 and GSE11579.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.084582.108.
References
- Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N.H., Arias C., Lennon C.J., Kluger Y., Dynlacht B.D. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bauer U.M., Daujat S., Nielsen S.J., Nightingale K., Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bieda M., Xu X., Singer M.A., Green R., Farnham P.J. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton E.C., So A.Y., Chaivorapol C., Haqq C.M., Li H., Yamamoto K.R. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes & Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick E.H., Martowicz M.L., Pal S., Johnson K.D. Developmental control via GATA factor interplay at chromatin domains. J. Cell. Physiol. 2005;205:1–9. doi: 10.1002/jcp.20393. [DOI] [PubMed] [Google Scholar]
- Carroll J.S., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F., et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chan C.S., Song J.S. CCCTC-binding factor confines the distal action of estrogen receptor. Cancer Res. 2008;68:9041–9049. doi: 10.1158/0008-5472.CAN-08-2632. [DOI] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J., et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cirillo L.A., McPherson C.E., Bossard P., Stevens K., Cherian S., Shim E.Y., Clark K.L., Burley S.K., Zaret K.S. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., Zaret K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Crawford G.E., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J., Carroll J.S., Geistlinger T.R., Torres-Arzayus M.I., Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes & Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J., Keeton E.K., Lupien M., Krum S.A., Carroll J.S., Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor αexpression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla-Del-Arenal M., Recillas-Targa F. GATA-1 modulates the chromatin structure and activity of the chicken α-globin 3′ enhancer. Mol. Cell. Biol. 2008;28:575–586. doi: 10.1128/MCB.00943-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Kaestner K.H. The Foxa family of transcription factors in development and metabolism. Cell. Mol. Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietze S., Lupien M., Silver P.A., Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- Gao H., Falt S., Sandelin A., Gustafsson J.A., Dahlman-Wright K. Genome-wide identification of estrogen receptor α-binding sites in mouse liver. Mol. Endocrinol. 2008;22:10–22. doi: 10.1210/me.2007-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N., Zhang J., Rao M.A., Case T.C., Mirosevich J., Wang Y., Jin R., Gupta A., Rennie P.S., Matusik R.J. The role of hepatocyte nuclear factor-3 α(Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol. Endocrinol. 2003;17:1484–1507. doi: 10.1210/me.2003-0020. [DOI] [PubMed] [Google Scholar]
- Giresi P.G., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdi R., Bossard P., Zheng M., Hamada Y., Coleman J.R., Zaret K.S. Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes & Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hogan G.J., Lee C.K., Lieb J.D. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S., Kallen C.B., Dhar R., Baquero M.T., Mason C.E., Russell B.A., Shah P.K., Liu J., Khramtsov A., Tretiakova M.S., et al. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol. Syst. Biol. 2008;4:188. doi: 10.1038/msb.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S., Sabo P.J., Johnson T.A., Sung M.H., Biddie S.C., Lightman S.L., Voss T.C., Davis S.R., Meltzer P.S., Stamatoyannopoulos J.A., et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Johnson W.E., Li W., Meyer C.A., Gottardo R., Carroll J.S., Brown M., Liu X.S. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl. Acad. Sci. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., Orkin S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komashko V.M., Acevedo L.G., Squazzo S.L., Iyengar S.S., Rabinovich A., O'Geen H., Green R., Farnham P.J. Using ChIP-chip technology to reveal common principles of transcriptional repression in normal and cancer cells. Genome Res. 2008;18:521–532. doi: 10.1101/gr.074609.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krum S.A., Miranda-Carboni G.A., Lupien M., Eeckhoute J., Carroll J.S., Brown M. Unique ERα cistromes control cell type-specific gene regulation. Mol. Endocrinol. 2008;22:2393–2406. doi: 10.1210/me.2008-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganiere J., Deblois G., Lefebvre C., Bataille A.R., Robert F., Giguere V. Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.S., Friedman J.R., Fulmer J.T., Kaestner K.H. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Lin C.Y., Vega V.B., Thomsen J.S., Zhang T., Kong S.L., Xie M., Chiu K.P., Lipovich L., Barnett D.H., Stossi F., et al. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet. 2007;3:e87. doi: 10.13711/journal/pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M., Eeckhoute J., Meyer C.A., Wang Q., Zhang Y., Li W., Carroll J.S., Liu X.S., Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R., Penot G., Hubner M.R., Reid G., Brand H., Kos M., Gannon F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem H., Cheng D., Zhao Q., Underhill C., Tini M., Bedford M.T., Torchia J. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell. Biol. 2007;27:120–134. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima T., Suzuki T., Kondo S., Kuroki Y., Takahashi K., Ide K., Yumoto N., Hasegawa A., Toyoda T., Kojima T., et al. Integrative genome-wide expression analysis bears evidence of estrogen receptor-independent transcription in heregulin-stimulated MCF-7 cells. PLoS One. 2008;3:e1803. doi: 10.1371/journal.pone.0001803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.L., Cleary M.L., Brown P.O., Lieb J.D. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc. Natl. Acad. Sci. 2003;100:6364–6369. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom D.T., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F., Song J.S., Liu X.S., Fisher D.E. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Loots G.G., Nobrega M.A., Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich A., Jin V.X., Rabinovich R., Xu X., Farnham P.J. E2F in vivo binding specificity: Comparison of consensus versus nonconsensus binding sites. Genome Res. 2008;18:1763–1777. doi: 10.1101/gr.080622.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld M.G., Lunyak V.V., Glass C.K. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Hu X., DiRenzo J., Lazar M.A., Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Steinfeld I., Shamir R., Kupiec M. A genome-wide analysis in Saccharomyces cerevisiae demonstrates the influence of chromatin modifiers on transcription. Nat. Genet. 2007;39:303–309. doi: 10.1038/ng1965. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla M.E., Parfitt D.E., Kouzarides T., Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver M.J., He Y.D., van't Veer L.J., Dai H., Hart A.A., Voskuil D.W., Schreiber G.J., Peterse J.L., Roberts C., Marton M.J., et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Wang Y., Klijn J.G., Zhang Y., Sieuwerts A.M., Look M.P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M.E., Yu J., et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Wang Q., Li W., Liu X.S., Carroll J.S., Janne O.A., Keeton E.K., Chinnaiyan A.M., Pienta K.J., Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H., Shulha H.P., Lin J.M., Vales T.R., Fu Y., Bodine D.M., McKay R.D., Chenoweth J.G., Tesar P.J., Furey T.S., et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Pope S.D., Jazirehi A.R., Attema J.L., Papathanasiou P., Watts J.A., Zaret K.S., Weissman I.L., Smale S.T. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc. Natl. Acad. Sci. 2007;104:12377–12382. doi: 10.1073/pnas.0704579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Bieda M., Jin V.X., Rabinovich A., Oberley M.J., Green R., Farnham P.J. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Zhu Z., Kapranov P., McKeon F., Church G.M., Gingeras T.R., Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Zaret K. Developmental competence of the gut endoderm: Genetic potentiation by GATA and HNF3/fork head proteins. Dev. Biol. 1999;209:1–10. doi: 10.1006/dbio.1999.9228. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Josefowicz S.Z., Kas A., Chu T.T., Gavin M.A., Rudensky A.Y. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]