Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that control gene expression by inducing RNA cleavage or translational inhibition. Most human miRNAs are intragenic and are transcribed as part of their hosting transcription units. We hypothesized that the expression profiles of miRNA host genes and of their targets are inversely correlated and devised a novel procedure, HOCTAR (host gene oppositely correlated targets), which ranks predicted miRNA target genes based on their anti-correlated expression behavior relative to their respective miRNA host genes. HOCTAR is the first tool for systematic miRNA target prediction that utilizes the same set of microarray experiments to monitor the expression of both miRNAs (through their host genes) and candidate targets. We applied the procedure to 178 human intragenic miRNAs and found that it performs better than currently available prediction softwares in pinpointing previously validated miRNA targets. The high-scoring HOCTAR predicted targets were enriched in Gene Ontology categories, which were consistent with previously published data, as in the case of miR-106b and miR-93. By means of overexpression and loss-of-function assays, we also demonstrated that HOCTAR is efficient in predicting novel miRNA targets and we identified, by microarray and qRT-PCR procedures, 34 and 28 novel targets for miR-26b and miR-98, respectively. Overall, we believe that the use of HOCTAR significantly reduces the number of candidate miRNA targets to be tested compared to the procedures based solely on target sequence recognition. Finally, our data further confirm that miRNAs have a significant impact on the mRNA levels of most of their targets.
MicroRNAs (miRNAs) are a class of short noncoding RNAs controlling the expression levels of their target genes. They play a role in the differentiation of many tissues and organs and are involved in the pathogenesis of human diseases (Chang and Mendell 2007; Stefani and Slack 2008; Zhang 2008). At the molecular level, they exert their function in animal cells by binding, with imperfect base pairing, to target sites in the 3′ UTR of messenger RNAs. This binding either causes the inhibition of translational initiation or leads to mRNA degradation (Zamore and Haley 2005; Shyu et al. 2008). miRNA:mRNA base-pairing usually includes a “nucleus” (or “seed”), typically a perfect Watson-Crick−base-paired stretch of approximately seven nucleotides with a key role both in target site recognition and repression of the target transcript. The nucleus is located at the 5′ end of the miRNA, typically between nucleotides 2 and 8 (Lewis et al. 2005).
Currently, more than 600 miRNAs have been identified in the human and mouse genomes (miRBase database, http://microrna.sanger.ac.uk/sequences/; Griffiths-Jones 2004), but estimates suggest that their actual number may exceed 1000 (Bentwich et al. 2005). Taking into account the fact that each miRNA can regulate, on average, the expression of 100–200 target genes (Krek et al. 2005; Lim et al. 2005), the whole miRNA apparatus seems to participate in the control of gene expression for a significant proportion of the mammalian gene complement. To gain insight into the biological role of each miRNA, it is essential to identify the full repertoire of its mRNA targets. However, this is not an easy task as demonstrated by the limited number of bona fide miRNA targets that have been experimentally validated so far (see DIANA TarBase database; Sethupathy et al. 2006). In order to identify true miRNA targets, it is essential to improve the efficiency of their in silico prediction by means of computational techniques (Maziere and Enright 2007). Several computational approaches have recently been developed for the prediction of miRNA targets including, among the most popular ones, the miRanda, TargetScan, and PicTar softwares (Lewis et al. 2003; John et al. 2004; Krek et al. 2005; Rajewsky 2006; Kuhn et al. 2008), which mainly rely on the identification of the seed region between the miRNA and the corresponding target genes. Unfortunately, the presence of a seed region, although conserved across evolution, is not in itself a reliable way to identify functional miRNA targets. It has been shown that a significant proportion of predicted miRNA–mRNA target pairs, in spite of the presence of an appropriate seed region, are false positives (Lewis et al. 2005; Didiano and Hobert 2006), thus rendering the in silico preselection of miRNA targets very complex and laborious.
Recently, it has been suggested that the simultaneous expression profiling of miRNAs and mRNAs could be an effective strategy for miRNA target identification (Huang et al. 2007). This is because, contrary to the original idea that miRNAs mostly act at the translational level in animal cells, there is increasing evidence that many miRNAs cause degradation of their targets (Bagga et al. 2005; Lim et al. 2005; Wu and Belasco 2008). They can therefore determine on their targets significant effects at the transcriptional level that can be readily detectable by microarray and by quantitative (q)RT-PCR procedures. Based on this evidence, Huang et al. (2007) devised a strategy based on the evaluation of inverse expression relationships between miRNAs and mRNAs in large sets of transcriptome experiments to identify more reliably miRNA-target mRNA pairs. However, one limitation of this procedure is represented by the restricted number of global expression profiling experiments involving the direct analysis of the entire catalog of miRNAs, as compared to the amount of transcriptomic data available for mRNAs. This is mainly due to the relatively recent availability of suitable procedures to evaluate miRNA expression by microarrays (Yin et al. 2008).
We reasoned that it may be possible to overcome the latter problem by exploiting the fact that many miRNAs are intragenic, i.e., localized within the introns of transcriptional units (host genes) representing either protein-coding or noncoding mRNAs (Rodriguez et al. 2004). Several reports have shown that the expression profiles of intragenic miRNAs are highly correlated to those of their corresponding host genes at both the tissue and cellular levels (Baskerville and Bartel 2005; Karali et al. 2007; Kim and Kim 2007). Therefore, it is possible, in principle, to use the miRNA host gene as a proxy to monitor the expression of its embedded miRNA(s) (Tsang et al. 2007). This may provide the opportunity to analyze a larger set of transcriptome expression data for intragenic miRNAs, which will be comparable to that available for their putative mRNA targets.
In this report, we describe the design of a new procedure, HOCTAR (host gene oppositely correlated targets), based on the integration of expression profiling and sequence-based miRNA target recognition softwares. HOCTAR turned out to be very efficient in identifying a set of already validated miRNA targets, even those that had already been suggested to be translational targets. Furthermore, we demonstrate, by means of overexpression and down-regulation experiments performed on two miRNAs, i.e., miR-26b and miR-98, that the HOCTAR procedure is efficient also in predicting novel bona fide targets. A database collecting HOCTAR target predictions for 178 human miRNA is publicly available at http://hoctar.tigem.it.
Results
The HOCTAR procedure
Based on the evidence that it is possible to use a miRNA host gene as a proxy for the expression of the miRNA itself (Tsang et al. 2007), we hypothesized that the expression behavior of a miRNA host gene may be inversely correlated to that of the targets of the embedded miRNA. As a result, an increase in the expression levels of the host gene should correspond to a decrease in the expression levels of the targets of its embedded miRNA, at least in some tissues or cellular conditions. In this study, we tested whether such an inverse correlation can be exploited to improve the prediction of miRNA targets. To achieve this goal, we devised a novel strategy that we termed HOCTAR (see Fig. 1).
Figure 1.
Flowchart of the HOCTAR procedure (see text for further details).
As a first step, we extracted from miRBase (rel. 10.1) the lists of all human intragenic miRNAs and of their corresponding host genes (see Methods for further details). For each intragenic miRNA, we compiled a nonredundant list of predicted mRNA targets (hereafter referred to as PTs) by pooling all corresponding miRanda, TargetScan, and PicTar predictions. Expression correlation relationships between miRNA host genes and corresponding PTs were inferred by using the g:Sorter web tool (http://biit.cs.ut.ee/gprofiler/; Reimand et al. 2007). This resource allows the performance of gene expression similarity searches on the transcriptomic data available at the Gene Expression Omnibus (GEO) database (Barrett et al. 2007). g:Sorter can be queried with a gene of interest to retrieve the genes that have the most similar (correlated) or dissimilar (anti-correlated) expression profiles in a specific data set. To ensure data homogeneity, we focused our analysis on a single microarray platform type, namely the Affymetrix HG-U133A, for which 160 different experimental data sets (for a total of 3445 different microarray hybridization experiments) were available at the time of analysis (October 2007). The HOCTAR procedure consists in ranking the members of each PT list according to their cumulative occurrence as host gene anti-correlated genes across all investigated microarray experiments. In each of the examined 160 data sets, a PT was considered as anti-correlated to the tested host gene only when included within the top 3% of its reported anti-correlated probes.
We applied the HOCTAR procedure to all human miRNA host genes represented on the HG-U133A platform (n = 178). Results of this analysis are available on-line in the format of a searchable database (http://hoctar.tigem.it). The database contains the HOCTAR target predictions for all the analyzed intragenic miRNAs along with the annotation of the enriched Gene Ontology (GO) categories (see also below).
HOCTAR is able to predict efficiently already validated miRNA targets
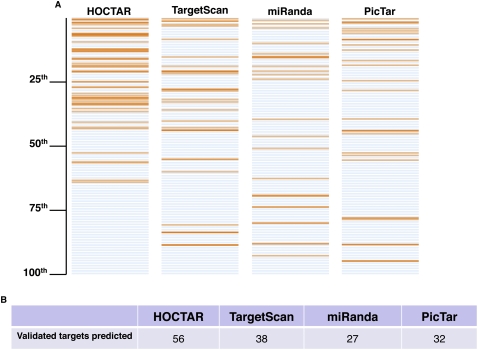
To test the efficacy HOCTAR in pinpointing the most likely miRNA targets, we decided to verify how the procedure performed with respect to already known, experimentally validated miRNA targets. We postulated that, if the HOCTAR tool is successful in improving miRNA target prediction, then the experimentally validated targets should display a tendency to be enriched at the top of the HOCTAR ranked prediction lists pertaining to the host genes of the corresponding embedded miRNAs. To test this hypothesis, we first selected from PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and DIANA TarBase (http://diana.cslab.ece.ntua.gr/tarbase/) 56 already known human miRNA target genes. These genes had previously been validated as targets of 20 different intragenic miRNAs either at the mRNA (n = 34) or at the translational (n = 22) level (Supplemental Table S1). We first mapped the selected validated targets onto the HOCTAR ranked lists of the corresponding miRNAs. We then compared the HOCTAR lists with the ranked lists of target predictions generated by the sequence-based prediction softwares miRanda, TargetScan, and PicTar for the corresponding miRNAs.
miRanda, TargetScan, and PicTar predicted 27, 38, and 32 out of the total number of 56 validated target genes, respectively. These predictions were uniformly distributed along the entire ranked lists pertaining to each of these softwares (Fig. 2). In contrast, the same genes were distributed preferentially at the top of the HOCTAR ranked lists (Fig. 2). In particular, we found that 51 out of the 56 (91%) validated targets fell within the first 50th percentile of the HOCTAR ranked lists, as compared to 15 out of 27 (56%), 27 out of 38 (71%), and 21 out of 32 (66%) in the miRanda, TargetScan, and PicTar prediction lists, respectively. It is important to point out that the average number of predicted targets present in each list is comparable, ranging from 450 to 700 (data not shown). Interestingly, we did not observe any significant difference in the distribution of “transcriptional” vs. “translational” targets within the HOCTAR ranked lists (data not shown).
Figure 2.
HOCTAR performance in recognizing previously validated miRNA targets. Comparison of HOCTAR with three sequence-based miRNA target prediction softwares (miRanda, TargetScan, and PicTar) in predicting 56 validated targets of 20 different miRNAs (see text for further details). (A) The panels display, for each of the algorithms analyzed, the ranking position, in a percentile format, of the validated targets selected for the analysis (orange horizontal lines). The distribution of the predictions by HOCTAR tends to be shifted toward the top 50th percentile in comparison to the other three softwares. (B) Summary of the number of validated targets predicted by each of the algorithms.
To evaluate further the reliability of this procedure, we determined the GO annotations of the genes that fell at least within the first 50th percentile of the HOCTAR prediction lists for each miRNA tested (Supplemental Table S2). This analysis revealed a significant enrichment in Gene Ontology Biological Process categories, which were consistent with previously published data. For example, miR-106b and miR-93 were already known to play a role in the expression control of genes involved in the negative regulation of cell cycle progression (Ivanovska et al. 2008). We found that the genes falling within the first 50th percentile of the HOCTAR list for miR-106b and miR-93 were enriched for the GO category “negative regulation of cell growth” as compared to a random subset of sequence-based target predictions for the same miRNA of equal size (http://hoctar.tigem.it).
These observations indicate that, by coupling miRNA target prediction softwares with the analysis of expression correlation using as proxies for intragenic miRNAs the corresponding host genes, it is possible to recognize known target genes with a high degree of efficiency. In addition, since we have verified that the vast majority of validated targets fall within the first 50th percentile of HOCTAR lists, we assume that the genes present at the top of the HOCTAR ranked lists have a higher probability of representing bona fide targets of miRNAs. This may significantly reduce the number of candidate targets to be tested for a given miRNA, at least in a first screening, compared to the procedure based solely on target sequence recognition.
miR-26b and miR-98 down-regulate HOCTAR predicted targets
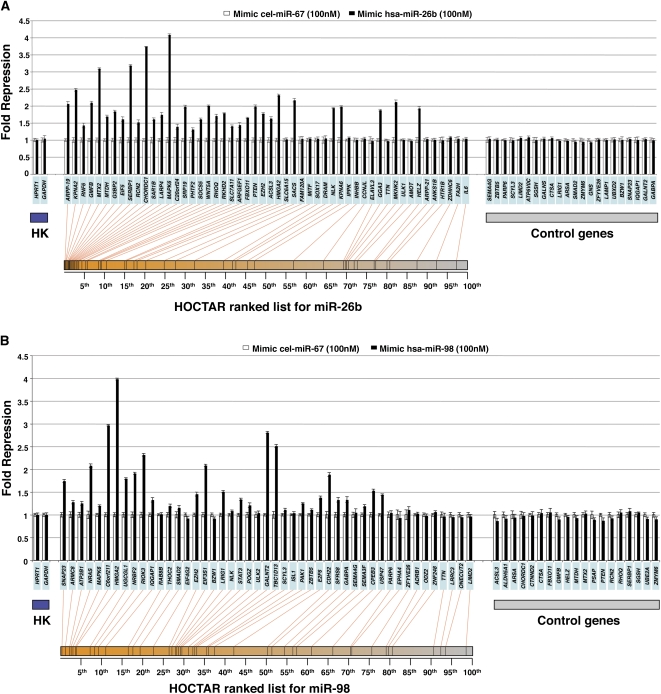
A possible limitation of the HOCTAR procedure is that a significant fraction of the genes with an anti-correlated expression behavior with respect to the miRNA host genes used as queries, although containing the specific target recognition sequence in their 3′ UTR, may not be direct targets of the corresponding embedded miRNA. They could represent targets of other independent regulatory processes controlled, directly or indirectly, by the protein product of the host gene and not by the embedded miRNA itself. To exclude this possibility as well as to test whether HOCTAR is effective also in predicting novel, not previously validated, miRNA target genes, we decided to apply the procedure to newly generated experimental data, namely those deriving from miRNA overexpression in human cells. For this purpose, we transiently transfected the synthetic RNA duplexes of the mature forms of human miR-26b and miR-98 (mimic-microRNA by Dharmacon) in HeLa cells. Transfection with the Caenorhabditis elegans miRNA cel-miR-67 duplex was used as control for each of the two overexpression experiments.
We tested by qRT-PCR the expression levels of 52 predicted target genes distributed along the entire HOCTAR ranked prediction list for miR-26b after transient transfection of mimic-miR-26b in HeLa cells. We observed that 34 out of 52 tested genes showed significant transcript down-regulation as compared to the cel-miR-67 transfection control (Fig. 3A). These genes were enriched within the first 50th percentile of the HOCTAR ranked list for miR-26b. The ratio between down-regulated and analyzed genes was 0.84 in the first 50th percentile of the ranked list (31 down-regulated out of 37 analyzed genes) and 0.2 in the second 50th percentile (three down-regulated out of 15 genes analyzed). As controls, we tested 22 genes not predicted to be miR-26b targets (without a seed for miR-26b) and none of these showed significant down-regulation (Fig. 3A).
Figure 3.
Experimental validation of a subset of HOCTAR predictions by overexpression of mimic-microRNAs in HeLa cells, as assessed by qRT-PCR. Histograms showing differences in the expression levels, between miRNA-overexpressed HeLa cells (black bars) and control (cel-miR-67 transfected) HeLa cells (white bars), of a subset of HOCTAR predicted target genes for miR-26b (A) and miR-98 (B) and of a subset of control genes by qRT-PCR assays. Y-axis: fold change repression (expressed as 2−ΔΔCt values). X-axis: predicted target genes tested (their symbols are indicated in the light blue boxes). The target genes tested were distributed along the entire HOCTAR ranked prediction lists for miR-26b (A) and miR-98 (B), and the red lines below the diagrams indicate the ranking of each target genes tested within the HOCTAR prediction ranked list for miR-26b (A) and miR-98 (B). The control genes shown on the right part of the two diagrams are not predicted to represent targets of the two analyzed miRNAs. The housekeeping (HK) genes used to normalize the expression of genes are HPRT1 and GAPDH. The analysis of each gene tested was performed in triplicate.
Similar results were observed when we transiently transfected HeLa cells with a mimic-miR-98 (Fig. 3B). We found that 28 out of 46 miR-98 putative targets tested were significantly down-regulated in HeLa cells following miR-98 overexpression. Also in this case, down-regulated genes were preferentially localized at the top of the HOCTAR ranked list for miR-98 (first 50th percentile, n = 22). Nineteen control genes lacking a seed for miR-98 did not show any significant change in their expression levels following miR-98 overexpression (Fig. 3B). Among the control genes used in both overexpression experiments, we included a subset of genes that displayed an anti-correlated expression behavior (as assessed by the g:Sorter analysis) comparable to that of the real-time down-regulated targets falling in the top 50th percentile of the HOCTAR lists.
To assess further the validity of the above described results, we verified the effect of miR-26b and miR-98 overexpression on protein production for some of the HOCTAR predicted targets by dual luciferase reporter assays and we found a high correlation with the qRT-PCR results (Supplemental Fig. S1). Overall, these experiments show that the ranking provided by the HOCTAR tool is reliable and specific in predicting high-confidence transcriptional targets of miRNAs and is more efficient than the ranking provided by sequence-based target prediction softwares, as shown in Supplemental Figures S2 and S3.
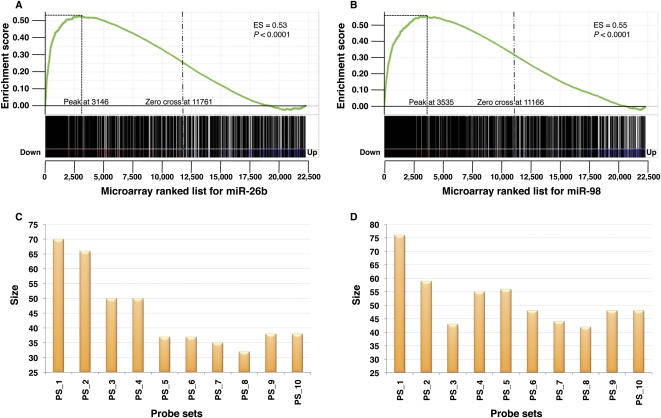
To get a more comprehensive assessment of the efficiency of HOCTAR, we also used global transcriptome analysis approaches. We used the microarray platform HG-U133A (Affymetrix) to profile the transcriptome changes in total RNA extracted from HeLa cells transfected with either the miR-26b or the miR-98 vs. the total RNA extracted from HeLa cells transfected with the cel-miR-67 control. After transfection with the miR-26b RNA duplex, 3603 probes (corresponding to 2645 genes) were significantly down-regulated and 3808 probes (corresponding to 2712 genes) were significantly up-regulated. Among the down-regulated ones, 359 probes (307 genes) were contained in the PT list (see above) for miR-26b. In order to verify whether the HOCTAR ranked list of miR-26b predicted targets was enriched in its top part for probes that were down-regulated following miR-26b transfection, we performed gene set enrichment analysis (GSEA). GSEA is a computational method for determining whether a defined subset of probes (in our case the set of probes present in the HOCTAR ranked list of miR-26b predicted targets) shows statistically significant enrichment at the top of a larger list of probes ranked according to their differential expression obtained from the analysis of two biological states (e.g., HeLa cells transfected with the miR-26b vs. HeLa cells transfected with the cel-miR-67 control). GSEA provides an enrichment score (ES) value that measures the degree of enrichment between the two analyzed lists: the ES value ranges from 0 to 1, where 1 indicates the highest enrichment.
We first evaluated the distribution of the entire HOCTAR ranked list for miR-26b (1024 probes) within the data set composed by the entire list of probes present in the microarray platform analyzed (n = 22,277). The latter data set was ranked by differential expression between cells transfected with miR-26b and cells transfected with the control RNA duplex in an ascending order. This analysis showed that the majority of the genes included in the HOCTAR list for miR-26b were preferentially distributed at the top of the analyzed microarray data set, where most down-regulated probes are localized (ES value = 0.53; P-value < 0.0001; Fig. 4A). This result is in line with previous observations that the overexpression of a given miRNA is responsible for the down-regulation of the transcript levels of many of its predicted targets. However, to test whether the ranking order provided by HOCTAR was effective in pinpointing the genes with the highest probability of representing bona fide miR-26b targets, we performed a more detailed GSEA analysis. We first divided the HOCTAR ranked prediction list (1024 probes) into 10 bins (probe sets), each of which containing 102 probes. We then repeated the GSEA analysis for each of these bins on the restricted data set of probes showing a differential expression in miR-26b- vs. control-transfected HeLa cells, i.e., all probes showing an FDR < 0.05 (n = 7410). Interestingly, we found that the number of probes with significant expression changes progressively decreased from bin1 to bin10 (Fig. 4C). In particular, 70 probes (corresponding to 60 genes) from bin1 and 66 probes (47 genes) from bin2 turned out to show significant changes in their expression levels in HeLa cells following miR-26b overexpression. The latter probes tend to be preferentially distributed at the top of the list of the most down-regulated probes of the miR-26b differentially expressed data set (Supplemental Fig. S4). In agreement, we found that the data set of probes differentially expressed in miR-26b-transfected cells vs. control was significantly enriched only for probes belonging to the two top-ranked bins of the HOCTAR prediction list for miR-26b (Supplemental Fig. S4). Finally, we did not observe any significant enrichment of up-regulated probes either in the entire HOCTAR miR-26b prediction list or in any of its bins (data not shown).
Figure 4.
Genes down-regulated after miR-26b and miR-98 overexpression are overrepresented in high-scoring HOCTAR predictions, as determined by microarray analysis. Enrichment plots generated by GSEA analysis of the HOCTAR predictions list for miR-26b and miR-98 are represented in A and B. The enrichment score is shown as a green line in each plot, and the vertical black bars below the plots indicate the position of probes belonging to the HOCTAR prediction lists for miR-26b within the rank ordering of the 22,277 probes present on the human U133A microarray from the probe most down-regulated (position 1 on the left) to the most up-regulated in HeLa cells after miR-26b (A) and miR-98 (B) transfection. (C,D) Histograms showing the number of probes with significant expression changes in HeLa cells after miR-26b (C) and miR-98 (D) overexpression (size, Y-axis). Each bar represents one of the 10 bins (probe sets, X-axis) in which the entire HOCTAR lists for miR-26b and miR-98 were subdivided (see the text for further details).
We carried out the same experimental procedure for miR-98 and we obtained comparable results. We observed a more significant enrichment of miR-98 down-regulated probes in the top bin sets of the related HOCTAR list (Fig. 4B,D; Supplemental Fig. S5). In contrast, when the same analysis was carried out on the ranked lists of prediction by miRanda, TargetScan, and PicTar for both miR-26b and miR-98, we observed a homogeneous distribution of the differentially expressed genes across all 10 bins (Supplemental Fig. S6). Overall, these results indicate that the HOCTAR tool is able to provide a reliable ranking of miRNA target predictions.
Validation of the HOCTAR procedure by miRNA loss-of-function studies
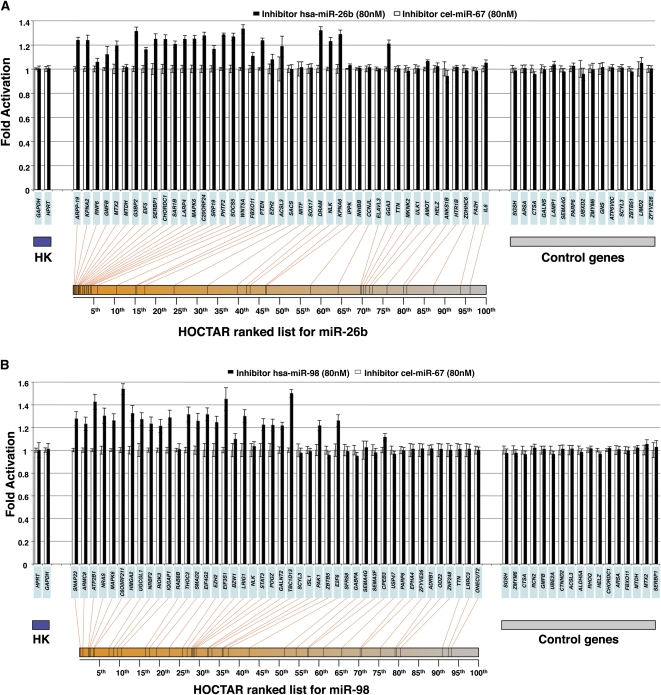
To assess further the validity of the HOCTAR procedure, we decided to test it on a miRNA-inactivation experimental model by down-regulating miR-26b and miR-98 expression in HeLa cells. We first assessed by qRT-PCR the expression levels of miR-26b and miR-98 in wild-type HeLa cells and we found that these miRNAs were expressed at significant levels in this cell line (data not shown). We then transfected HeLa cells with either an inhibitor-miR-26b or an inhibitor-miR-98 (Dharmacon). Transfection with the C. elegans miRNA cel-miR-67 duplex was used as control for each of the two inhibition experiments.
We tested by qRT-PCR the expression levels of 43 HOCTAR predicted target genes distributed along the entire HOCTAR ranked list for miR-26b (Fig. 5A). We found that 19 out of the 43 genes tested showed a statistically significant up-regulation of their expression levels following inactivation of miR-26b in HeLa cells, and the vast majority of these mapped in the top ranking position of the HOCTAR prediction list (first 50th percentile, n = 18). As negative controls, we also tested 15 genes not predicted to represent miR-26b targets and none of these showed a significant up-regulation.
Figure 5.
Experimental validation of the HOCTAR procedure by down-regulation of miR-26b and miR-98 in HeLa cells. Histogram showing differences in the expression levels between HeLa cells transfected with a miRNA-inhibitor (black bars) and control (cel-miR-67 transfected) HeLa cells (white bars), of a subset of HOCTAR predicted target genes for miR-26b (A) and miR-98 (B) and of a subset of control genes by qRT-PCR assays. Y-axis: fold change activation (expressed as 2−ΔΔCt values). X-axis: predicted target genes tested (their symbols are indicated in the light blue boxes). The target genes tested were distributed along the entire HOCTAR ranked prediction lists for miR-26b (A) and miR-98 (B) and the red lines below the diagrams indicate the ranking of each target genes tested within the HOCTAR prediction ranked list for miR-26b (A) and miR-98 (B). The control genes shown on the right part of the two diagrams are not predicted to represent targets of the two analyzed miRNAs. The housekeeping (HK) genes used to normalize the expression of genes are HPRT1 and GAPDH. The analysis of each gene tested was performed in triplicate.
Similar results were observed following the inactivation of miR-98 in HeLa cells. We tested by qRT-PCR the expression levels of 44 HOCTAR predicted target genes distributed along the entire HOCTAR ranked list for miR-98 (Fig. 5B). We found that 24 out of 44 genes were significantly up-regulated following miR-98 inactivation and that the majority of them were localized within the first 50th percentile of the HOCTAR prediction list (n = 23). As a control, none of the 17 control genes not predicted to be miR-98 targets displayed any significant change in their expression levels. Similar to the miRNA overexpression analyses (see previous section), the genes found to be up-regulated in HeLa cells following miR-26b and miR-98 inhibition genes were uniformly distributed within the ranked prediction lists of the miRanda, TargetScan, and PicTar softwares (Supplemental Figs. S7, S8). Since we analyzed by qRT-PCR largely overlapping subsets of genes in overexpression and loss-of-functions experiments (Figs. 3, 5), we could test the consistency of the results obtained in the two types of experiments. We found that 18/19 and 23/24 genes up-regulated after, respectively, miR-26b and miR-98 inhibition were significantly down-regulated in the corresponding overexpression experiments (data not shown). Taken together, these results further confirm the reliability of HOCTAR in pinpointing bona fide miRNA targets.
Discussion
We have demonstrated that HOCTAR, a procedure based on the analysis of expression correlation between host genes and the candidate targets of the corresponding intragenic miRNAs, can be a valuable resource to improve the efficacy of miRNA target prediction. HOCTAR takes advantage of the following observations: (1) miRNAs can down-regulate some of their targets not only at the translational but also at the transcript level (Shyu et al. 2008); (2) it is therefore possible to use the paired expression analysis of miRNAs and mRNAs to identify mRNA targets of miRNAs; and (3) the expression profiles of intragenic miRNAs and of their corresponding host genes are very similar both at the tissue and cellular level (Baskerville and Bartel 2005; Kim and Kim 2007), which makes it possible to use the expression data pertaining to host genes to infer the expression data of the corresponding embedded miRNAs. For the target prediction, HOCTAR relies on the use of three established miRNA target prediction softwares, miRanda, TargetScan, and PicTar, which have already been proved to be very effective. We evaluated the efficacy of the HOCTAR procedure by analyzing a set of 56 already validated miRNA:mRNA target pairs. The vast majority of these miRNA:mRNA pairs (91%, 51 out of 56) were localized within the first 50th percentile of the HOCTAR ranked lists for these miRNAs, thus demonstrating the efficacy of the procedure when compared to the sequence-based target prediction softwares (Fig. 2).
It was generally believed, until recently, that miRNAs exerted their repressive action on their targets via translation down-regulation. However, it is now widely accepted that miRNAs can determine in animal cells a down-regulation of their targets also at the transcriptional levels via mRNA degradation (Bagga et al. 2005; Lim et al. 2005; Wu and Belasco 2008) and that this action does not require the presence of a perfect sequence complementarity between miRNA and mRNA targets. More recently, high-throughput methods have been used to determine on a large scale the amount of protein repression mediated by miRNAs (Baek et al. 2008; Selbach et al. 2008). These studies have shown that the translational repression operated by miRNAs is very significant and that a subset of miRNA targets are modestly derepressed by miRNAs at the protein level with little or no change at the mRNA level (Baek et al. 2008; Selbach et al. 2008). Obviously, the HOCTAR procedure cannot be used at present to identify miRNA targets exclusively regulated at the translational level, However, by testing the HOCTAR procedure on a set of experimentally validated miRNA targets (Fig. 2), we did not observe any significant differences in the behavior of targets that had previously been ascertained only at the translational level vs. targets that had been shown to be affected at the transcript level (data not shown). This observation further confirms that the transcriptional effects of miRNA action on the expression levels of their targets represent a widespread phenomenon (Lim et al. 2005; Baek et al. 2008; Selbach et al. 2008), which is not limited to a restricted subset of targets, thus increasing the usefulness of HOCTAR in miRNA target identification.
We have also demonstrated, by means of experimental miRNA overexpression and loss-of-function procedures in HeLa cells, that HOCTAR can be reliably used to predict effectively novel transcriptional targets, not previously experimentally validated. In particular, we validated, by both microarray and qRT-PCR procedures, 34 and 28 novel targets for miR-26b and miR-98, respectively. The sequence-predicted mRNA targets of these two miRNAs, that proved to undergo the most significant extent of down-regulation following miRNA overexpression, were preferentially distributed at the top of the HOCTAR ranked lists of miR-26b and miR-98. These validation experiments further support the idea that the candidates identified by HOCTAR are bona fide miRNA targets and not targets of other independent regulatory processes controlled, directly or indirectly, by the protein product of the host gene itself. Therefore, we conclude that the use of HOCTAR can facilitate the preselection of targets to be tested for a given miRNA.
Obviously, we cannot completely rule out the possibility that some of the targets predicted by HOCTAR, although containing the specific miRNA recognition sequence in their 3′ UTR, are not direct targets of the analyzed miRNAs and that the anti-correlated expression behavior they exhibit is due to other indirect molecular mechanisms. However, both the high performance obtained by HOCTAR in predicting a set of already validated targets (Fig. 2) and the fact that HOCTAR high-scoring targets showed a significant enrichment in Gene Ontology Biological Process categories, which were consistent with already published data, make this hypothesis less likely. On the other hand, the transcriptional changes mediated by miRNAs on the expression levels of their targets may not be entirely explained by the direct repression operated by miRNAs but may also reflect the activation of feedback and feedforward transcriptional loops within gene regulatory networks of which miRNAs represent important players (Tsang et al. 2007; Marson et al. 2008). In particular, miRNA-mediated “coherent” and “incoherent” feedforward loops (Marson et al. 2008) are now recognized as important components of cellular gene regulatory networks. The relative role of these transcriptional circuits in the overall picture of miRNA function remains to be further established through additional experimental work.
It has already been suggested that high-throughput expression data analysis could be exploited to improve miRNA target prediction procedures (Huang et al. 2007). However, to the best of our knowledge, HOCTAR is the first tool for systematic miRNA target prediction that utilizes the same set of microarray experiments to monitor the expression of both miRNAs (through their host genes) and candidate targets. The expression data set used by HOCTAR is much larger than any currently available miRNA-specific microarray data set. Moreover, this data set is representative not only of static evaluations of wild-type or disease cells/tissues, but also of a large variety of dynamic states following different types of stimulations and perturbations that can be of physical, biological, and genetic nature (Supplemental Table S3). It is also important to point out that HOCTAR is the first tool for miRNA target recognition that is able to provide different prediction target lists for identical miRNAs present in multiple copies in the genome and localized within different host genes. These lists differ in the ranking of the predictions, which is based on the expression data pertaining to the corresponding host gene. On the other hand, a drawback of HOCTAR is represented by the fact that, by virtue of its design, it is expected to have a lower performance in the particular case in which a miRNA targets its own host gene. Overall, we believe that the specific features of HOCTAR strongly contribute to its capability to pinpoint targets of a large number of miRNAs, which have a different expression specificity and diverse biological roles.
The HOCTAR procedure could so far be applied to 178 intragenic miRNAs whose host genes were represented in the microarray data set selected for the analysis (see Results). The increasing availability of transcriptomic data generated by both microarray and high-throughput sequencing procedures (Gresham et al. 2008; Mortazavi et al. 2008; Sultan et al. 2008) is expected to further improve the efficacy of the HOCTAR procedure and to extend its use to a higher number of intragenic miRNAs, particularly those embedded within poorly characterized transcriptional units such as EST clusters representing noncoding RNAs (Kim and Nam 2006).
In conclusion, based on both bioinformatic and experimental analyses, we have demonstrated that the HOCTAR integrated procedure represents a valuable tool to identify bona fide miRNA targets. Furthermore, thanks to a systematic application of this procedure, we have also been able to assess that the action that miRNAs exert on the transcript levels of their targets is more common than previously recognized. Overall, these results are expected to lead to a deeper insight into the biological role of this class of noncoding RNAs in both physiological and pathological conditions.
Methods
The HOCTAR procedure
The list of human intragenic miRNAs and corresponding host genes was retrieved from miRBase (release 10.1) (http://microrna.sanger.ac.uk/sequences/; Griffiths-Jones 2004). We considered host genes only those whose RefSeq sequences overlapped the miRNA either in introns, exons, or UTR and that were transcribed on the same strand as the miRNA. All the above-mentioned features were manually verified using the UCSC Human Genome Browser database (release 2006/March; http://genome.ucsc.edu/).
Expression correlation analyses of miRNA host genes and putative targets were performed by using the g:Sorter tool (http://biit.cs.ut.ee/gprofiler/gsorter.cgi), which is part of the g:Profiler package (Reimand et al. 2007). g:Sorter is a tool for gene expression similarity search. For a selected gene, protein, or probe ID, g:Sorter retrieves a number of most similar coexpressed (correlated) or dissimilar reversely expressed (anti-correlated) profiles in a specified GEO data set. For the analysis, we focused on the HG-U133A GeneChip array (GPL96, Feb 19, 2002), for which a total of 160 microarray data sets were available at g:Sorter at the time of the analysis. These experiments are widely heterogeneous, including analyses of tissue differentiation, comparisons of cancerous and healthy cells/tissues, responses to biological or physical stimuli, etc. (for more details, see Supplemental Table S3). As input to the HOCTAR procedure we used all the probes corresponding to the selected miRNA host genes and represented in the HG-U133A array, as assessed through the analysis of the Affymetrix website (http://www.affymetrix.com/index.affx). The probes were mapped to the human genome sequence by using the UCSC Human Genome Browser to verify their actual correspondence to the miRNA host genes. As a result, we selected 220 probes covering 130 miRNA host genes encompassing 178 distinct miRNAs.
The lists of putative target genes (PT lists) were built by retrieving, using default parameters, target predictions from PicTar (release 2007/March, http://pictar.mdc-berlin.de/), miRanda (release 2005/July, http://cbio.mskcc.org/mirnaviewer/), and TargetScan (version 4.1, release 2008/January, http://www.targetscan.org/vert_40/). In the case of PicTar, we selected the targets which were found to be conserved in mammalian genomes (Krek et al. 2005).
We queried the selected experimental data set of g:Sorter with all the individual probes covering the 178 selected host genes. For each analyzed probe, we retrieved at g:Sorter the first 3% of most anti-correlated genes for each microarray data set. Then, we ranked all relative putative target genes based on their occurrence in the 160 different lists of most anti-correlated genes. Genes with an equal number of occurrences were ranked according to their average ranking within the subset of experiments in which they were found to be anti-correlated. The entire procedure resulted in the building of ranked lists of putative target genes for each miRNA, ordered by their anti-correlated expression with respect to the corresponding miRNA host gene.
Cell transfection assays
The Human cervical cancer-derived cells (HeLa) were grown in Dulbecco's Modified Eagle's Medium (DMEM, Invitrogen), supplemented with 10% heat-inactivated Fetal Bovine Serum (FBS, Euroclone). All cells were incubated at 37°C in a humidified chamber supplemented with 5% CO2. Cells were seeded in six-well plates at 10% confluence (1 × 105 cells) before transfection. Transfection of HeLa cells was performed using DharmaFECT 1 Transfection Reagent (Dharmacon Research) according to the manufacturer's protocol. Cells were transfected with either miRIDIAN™ Dharmacon microRNA Mimics (miR-26b, miR-98, or negative control cel-miR-67), at a final concentration of 100 nM or with miRIDIAN™ Dharmacon microRNA Inhibitor (miR-26b, miR-98, or negative control cel-miR-67) at a final concentration of 80 nM. Cells were harvested after 48 h for total RNA extraction. Total RNA was obtained using the miRNeasy kit (Quiagen) according to the manufacturer's instructions. RNA was quantified using the NanoDrop 1000 (Thermo Fischer). Quality of RNA was assessed by gel electrophoresis.
Quantitative real-time PCR
Quantitative (q) Reverse Transcriptase (RT-)PCR-based detection of mature miR-26b and miR-98 was performed using the TaqMan microRNA assays (Applied Biosystems). The qRT-PCR results, recorded as threshold cycle numbers (Ct), were normalized against an internal control (RNU48), and then expressed as fold changes (Chen et al. 2005).
We used samples with high-quality RNA to prepare cDNA synthesis using Quantitect Reverse Transcription kit (Quiagen, Inc.) starting from 1 μg of DNase-treated RNA.
In order to unambiguously distinguish spliced cDNA from genomic DNA contamination, specific exons primers were designed to amplify across introns of the genes tested. All primers were previously tested by reverse transcription (RT)-PCR and -RT controls reactions were performed. The primers for all target genes tested were designed with PrimerDesigner 2.0 software (Applied Biosystems). Primer sequences are available in supplemental methods (Supplemental Tables S4, S5).
Quantitative RT-polymerase chain reaction (qRT-PCR) experiments were performed using the ABI Prism 7900HT Fast Sequence Detection System with ABI Power SYBR Green reagents (Applied Biosystems). Real-time PCR results were analyzed using the comparative Ct method normalized against the housekeeping genes HPRT1 and GAPDH (Vandesompele et al. 2002). The range of expression levels was determined by calculating the standard deviation of the ΔCT (Pfaffl 2001). We considered as down-regulated and up-regulated the genes showing a change in their expression with a P-value < 0.01.
Luciferase assays
HeLa cells were transfected with firefly luciferase reporter plasmids containing the 3′ UTR of the genes analyzed and with psiUx plasmid (Denti et al. 2004) constructs containing the precursor sequences of hsa-miR-26b and hsa-miR-98 (see Supplemental Table S6 for a list of olignucleotides used to amplify both the 3′ UTR and the pre-miR sequences). Twenty-four hours before transfection, HeLa cells were plated in a six-well plate. Luciferase assays were performed 48 h after transfection using Dual Luciferase Reporter Assay System (Promega), normalized for transfection efficiency by cotransfected Renilla luciferase.
Microarray experiments
Total RNA from HeLA-transcfected cells was used to prepare cRNA for hybridization to the Affymetrix HG-U133A array platform. Microarray hybridizations were performed in triplicates at the Coriell Genotyping and Microarray Center, Coriell Institute for Medical Research, Camden, NJ, USA. Microarray results are available from the GEO database with the accession number GSE12091 (miR-26b overexpression) and GSE12092 (miR-98 overexpression). A false discovery rate (FDR) <0.05 was used to assess significant gene differential expressions.
Gene set enrichment analysis
GSEA was performed as previously described (Subramanian et al. 2005). The cumulative distribution function was constructed by performing 1000 random gene set membership assignments. A nominal P-value < 0.01 and an FDR < 0.25 are used to assess the significance of the enrichment score (ES).
Gene Ontology analysis
Gene Ontology (GO) analyses were performed with the web tool DAVID at http://david.abcc.ncifcrf.gov/home.jsp using default parameters (Sherman et al. 2007). GO analyses were performed on the 30th, 50th, and 100th percentile of HOCTAR ranked lists for all 178 miRNA analyzed. After performing the analysis, only Biological Process (BP) categories with a P-value ≤ 0.001, FDR ≤ 5, and fold enrichment ≥ 2 in the analysis of the 50th percentile lists were retained. Redundant terms and noninformative terms (e.g., multigene family) were eliminated. Results are collected in the HOCTAR database at http://hoctar.tigem.it.
HOCTAR database
The data set is stored in a relational form using MySQL 4.1.14 database and it is freely accessible through a web interface written in php and supported by all the common browsers. The database can be accessed at http://hoctar.tigem.it.
Acknowledgments
We thank Graciana Diez-Roux, Vincenzo Nigro, and Diego di Bernardo for critical reading of the manuscript. We are grateful to Irene Bozzoni for kindly providing us with the psiUx plasmid, Giulia Cuccato and Michela Palmieri for technical assistance, and Giampiero Lago for the database website design. This work was supported by the Italian Telethon Foundation.
Footnotes
[Supplemental material is available online at www.genome.org. The microarray expression data from this study have been submitted to GEO under accession nos. GSE12091 and GSE12092.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.084129.108.
References
- Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., Kim I.F., Soboleva A., Tomashevsky M., Edgar R. NCBI GEO: Mining tens of millions of expression profiles—Database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Chang T.C., Mendell J.T. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denti M.A., Rosa A., Sthandier O., De Angelis F.G., Bozzoni I. A new vector, based on the PolII promoter of the U1 snRNA gene, for the expression of siRNAs in mammalian cells. Mol. Ther. 2004;10:191–199. doi: 10.1016/j.ymthe.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Didiano D., Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Gresham D., Dunham M.J., Botstein D. Comparing whole genomes using DNA microarrays. Nature reviews. 2008;9:291–302. doi: 10.1038/nrg2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.C., Babak T., Corson T.W., Chua G., Khan S., Gallie B.L., Hughes T.R., Blencowe B.J., Frey B.J., Morris Q.D. Using expression profiling data to identify human microRNA targets. Nat. Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- Ivanovska I., Ball A.S., Diaz R.L., Magnus J.F., Kibukawa M., Schelter J.M., Kobayashi S.V., Lim L., Burchard J., Jackson A.L., et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karali M., Peluso I., Marigo V., Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest. Ophthalmol. Vis. Sci. 2007;48:509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Kim V.N. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Nam J.W. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kuhn D.E., Martin M.M., Feldman D.S., Terry A.V., Jr, Nuovo G.J., Elton T.S. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J., et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziere P., Enright A.J. Prediction of microRNA targets. Drug Discov. Today. 2007;12:452–458. doi: 10.1016/j.drudis.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl.):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Reimand J., Kull M., Peterson H., Hansen J., Vilo J. g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(Web Server issue):W193–200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sethupathy P., Corda B., Hatzigeorgiou A.G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman B.T., Huang da W., Tan Q., Guo Y., Bour S., Liu D., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID Knowledgebase: A gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A.B., Wilkinson M.F., van Hoof A. Messenger RNA regulation: To translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G., Slack F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M., Schulz M.H., Richard H., Magen A., Klingenhoff A., Scherf M., Seifert M., Borodina T., Soldatov A., Parkhomchuk D., et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Tsang J., Zhu J., van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Belasco J.G. Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Yin J.Q., Zhao R.C., Morris K.V. Profiling microRNA expression with microarrays. Trends Biotechnol. 2008;26:70–76. doi: 10.1016/j.tibtech.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang C. MicroRNomics: A newly emerging approach for disease biology. Physiol. Genomics. 2008;33:139–147. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]