Abstract
Structural variation is an important and abundant source of genetic and phenotypic variation. Here we describe the first systematic and genome-wide analysis of segmental duplications and associated copy number variants (CNVs) in the modern domesticated dog, Canis familiaris, which exhibits considerable morphological, physiological, and behavioral variation. Through computational analyses of the publicly available canine reference sequence, we estimate that segmental duplications comprise ∼4.21% of the canine genome. Segmental duplications overlap 841 genes and are significantly enriched for specific biological functions such as immunity and defense and KRAB box transcription factors. We designed high-density tiling arrays spanning all predicted segmental duplications and performed aCGH in a panel of 17 breeds and a gray wolf. In total, we identified 3583 CNVs, ∼68% of which were found in two or more samples that map to 678 unique regions. CNVs span 429 genes that are involved in a wide variety of biological processes such as olfaction, immunity, and gene regulation. Our results provide insight into mechanisms of canine genome evolution and generate a valuable resource for future evolutionary and phenotypic studies.
The unique evolutionary history of domesticated dogs (Canis familiaris), including strong artificial selection, population bottlenecks, and inbreeding, has resulted in over 400 genetically distinct breeds that make them well suited for addressing fundamental questions in population genetics, evolution, and the genetic architecture of phenotypic variation. In particular, domesticated dogs have engendered considerable interest because, since their domestication over 14,000 yr ago (Vila et al. 1997; Leonard et al. 2002; Savolainen et al. 2002), they have become one of the most phenotypically diverse mammalian species with an incredible assortment of shapes, sizes, and temperaments (Neff and Rine 2006). Beyond curiosity in outward appearances, canine genetics is also relevant to human health, as dogs are afflicted with over 350 inherited diseases (Patterson et al. 1988), many of which are similar to human diseases.
A number of enabling resources for canine genomics have recently become available including the development of an integrated canine linkage-radiation hybrid map (Mellersh et al. 2000), a 7.5× high-quality reference genome sequence (Lindblad-Toh et al. 2005), the construction of a dense map of over 2.5 million single nucleotide polymorphisms (SNPs) identified in a diverse panel of breeds (Lindblad-Toh et al. 2005), and the development of SNP genotyping arrays (Karlsson et al. 2007). These resources have provided important foundations for delimiting patterns of population structure among breeds (Irion et al. 2003; Parker et al. 2004; Karlsson et al. 2007; Quignon et al. 2007), inferring targets of artificial selection (Pollinger et al. 2005), and mapping traits such as Collie eye anomaly (Parker et al. 2007), body size (Sutter et al. 2007), and muscle mass (Mosher et al. 2007).
In contrast to SNPs and microsatellites, structural variation has received considerably less attention in dogs. Changes in DNA content are a significant source of genetic and phenotypic variation between individuals (Emanuel and Shaikh 2001; Bailey and Eichler 2006; Feuk et al. 2006; Beckmann et al. 2007; Conrad and Antonarakis 2007; Sebat 2007). Segmental duplications, in particular, are substrates of genome innovation, genomic rearrangements, and hotspots of CNV formation (Sharp et al. 2005; Graubert et al. 2007; She et al. 2008). Although segmental duplications and CNVs have been extensively studied in other organisms (Bailey et al. 2001, 2002, 2004; Iafrate et al. 2004; Tuzun et al. 2004; Cheng et al. 2005; Sharp et al. 2005; Tuzun et al. 2005; Conrad et al. 2006; Goidts et al. 2006; McCarroll et al. 2006; Perry et al. 2006, 2008; Redon et al. 2006; Graubert et al. 2007; Guryev et al. 2008; She et al. 2008), to date no such analyses have been performed in dogs. Recent studies demonstrate the potential contribution of CNVs to specific canine morphological phenotypes, such as dorsal hair ridge in Rhodesian and Thai Ridgebacks (Salmon Hillbertz et al. 2007). Thus, a more comprehensive understanding of the full spectrum of canine genomic variation is important for unraveling the genetic basis of variation in morphological, physiological, behavioral, and disease phenotypes segregating within and between breeds (Neff and Rine 2006).
Here we describe the first genome-wide and systematic analysis of segmental duplications and their associated CNVs in dogs. We find that similar to other mammalian genomes, recent segmental duplications comprise an appreciable fraction of the canine genome. Using high-density aCGH experiments specifically designed to interrogate putative segmental duplications, we identified 3583 CNVs in a panel of 17 genetically and phenotypically diverse breeds and a gray wolf.
Results and Discussion
Genome-wide identification and organization of segmental duplications
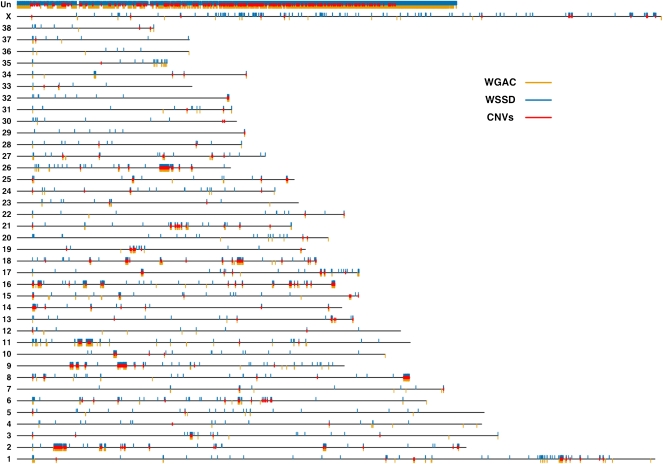
We applied two well-established computational approaches, whole-genome shotgun sequence detection (WSSD) (Bailey et al. 2002) and whole-genome assembly comparison (WGAC) (Bailey et al. 2001), to the publicly available canine genome sequence assembly (CanFam2.0) to detect putative segmental duplications. Briefly, WGAC identifies paralogous sequences ≥1 kb in length with ≥90% sequence identity, and WSSD identifies genomic regions that exhibit significant depth of coverage by aligning whole-genome shotgun sequencing reads to the reference genome sequence (see Methods). Using these computational algorithms, we predict 9137 segmental duplications, spanning ∼106.6 Mb of DNA sequence (Fig. 1; Supplemental Table 1). The average size of predicted segmental duplications is ∼11.7 kb (sd = 24.9 kb). We estimate that recent segmental duplications comprise ∼4.21% of the canine reference genome, which is consistent with similar observations in human and mouse (Bailey et al. 2001, 2002, 2004; She et al. 2008). As expected, the “uncharacterized chromosome” (chrUn), which consists of sequence that cannot be uniquely mapped to the genome, contains the majority of predicted duplication bases (65%).
Figure 1.
The genomic architecture of canine segmental duplications and CNVs. Black lines represent all 38 canine autosomes, the X chromosome, and the uncharacterized chromosome (Un). Duplicated bases predicted by WGAC and WSSD are plotted as orange and blue rectangles below and above, respectively, each chromosome. Over 80% of chrUn contains duplicated bases. Of the autosomes, chromosomes 9, 16, and 26 possess the highest percentages of duplicated bases (over 4% of each chromosome), while chromosomes 12, 30, and 33 show the least amount of duplicated bases (<0.35% of each chromosome). Unique CNV regions (see text) are denoted by red rectangles.
Furthermore, similar to humans and mice, there is a greater proportion of intrachromosomal versus interchromosomal duplications, with ∼60% of predicted duplications being intrachromosomal. Pericentromeric regions represent 3.4% of genomic sequence, but show an enrichment of threefold for duplications (P-value <0.001) and contain 10.3% of all duplicated bases. Similarly, subtelomeric regions show an enrichment of 2.3-fold (P-value <0.001) and contain 7.9% of duplicated bases.
A total of 841 genes were located in predicted segmental duplications. In order to test the hypothesis that particular gene classes are over-represented in duplicated regions, we assigned PANTHER Molecular Function terms to all genes that overlapped duplications. Statistically significant enrichment was observed for seven categories (Supplemental Table 2). Consistent with similar analyses of duplications in other organisms (Bailey et al. 2002, 2004; Tuzun et al. 2004; Sharp et al. 2005), we observe significant enrichment in genes that participate in defense/immunity, receptors, and signaling (Supplemental Table 2). Interestingly, KRAB box transcription factors also were enriched significantly among our set of predicted duplications (Supplemental Table 2), which has not been observed previously in duplications from additional species. KRAB box transcriptional factors are a part of a large gene family that is believed to bind to DNA and exhibit transcriptional repression (Urrutia 2003). While the precise phenotypes that KRAB box transcription factors contribute to are largely unknown, recent work has shown that a particular KRAB box transcription factor influences mouse embryonic morphological development (Garcia-Garcia et al. 2008). No additional classes of transcription factors showed enrichment.
FISH characterization of predicted segmental duplication
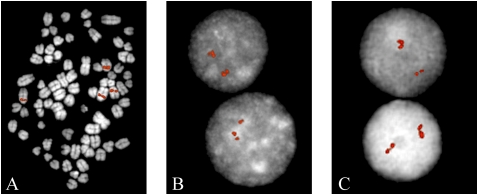
We experimentally validated a subset of the duplicated regions by fluorescent in situ hybridization (FISH). A total of 42 large-insert dog BAC clones corresponding to WGAC and WSSD duplicated regions (>20 kb in length) were used as probes and hybridized against a fibroblast C. familiaris cell line (Supplemental Table 3). We observed multiple signals either by examination of interphase or metaphase FISH for 20/42 of the probes, including 14 intrachromosomal, five interchromosomal, and a single probe that mapped to multiple centromeric regions. Only one of the interchromosomal probes showed more than three distinct signals, while the majority (10/14) of intrachromosomal duplication signals were clustered. Similar to the mouse genome (She et al. 2008), these data suggest that tandem intrachromosomal duplications predominate in the dog genome (Fig. 2). The basis for the remaining 22 BAC probes consistent with single copy sequence is unknown. We note, however, that the breed origin for the C. familiaris cell line used in the FISH experiments is not known, and structural polymorphism as well as limitations of BAC-FISH to detect duplications <40 kb (especially in the case of tandem duplications) may account for differences between the computational predictions and experimental data.
Figure 2.
Validation of duplicons by FISH analysis. (A) Example of an interchromosomal duplication detected with clone CH82-381N09. (B,C) Two representative examples of tandom intrachromosomal duplication detected with clones CH82-381N09 and CH82-331L01, respectively.
CNVs and segmental duplications
We designed a custom high-density tiling array covering all regions with significant WGAC or WSSD support to identify CNVs. Note, this includes both the 106.6 Mb of sequence predicted to be segmental duplications (both WGAC and WSSD support) as well as an additional 16.4 Mb of sequence with either WGAC or WSSD support. Obviously, our study design will not detect CNVs located outside of these regions, but we focused on them for two reasons. First, previous studies have demonstrated that segmental duplications are enriched four- to 20-fold for CNVs (Iafrate et al. 2004; Sebat et al. 2004; Sharp et al. 2005; Tuzun et al. 2005; Perry et al. 2006; Graubert et al. 2007; Guryev et al. 2008; She et al. 2008). Second, by restricting our aCGH experiments to regions most likely to harbor CNVs we were able to dramatically increase probe density (average probe spacing of 200 bp), resulting in a more exhaustive discovery of smaller CNVs as well as increasing the robustness of individual CNV calls.
The remaining probes were placed in 5-kb flanking regions and putative single copy control regions with a mean spacing of 1 kb and 350 bp, respectively. Thus, in total, over 137 Mb of sequence was studied and 19 aCGH experiments were performed, each using a common reference sample derived from a female boxer. Specifically, hybridizations were performed on 17 diverse breeds (see Methods), a gray wolf, and a self–self hybridization.
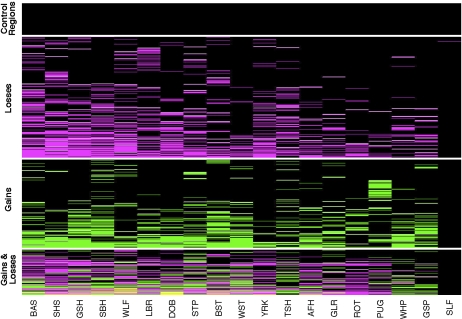
We used a previously described hidden Markov model method to identify changes in log2 signal intensity corresponding to gains and losses in copy number (Rueda and Diaz-Uriarte 2007). Using conservative criteria (see Methods), 3583 CNVs were identified (1578 gains and 2005 losses) that map to 678 distinct regions (Fig. 3; Supplemental Table 4). CNVs comprise 24 Mb of polymorphic sequence and ∼20% of the predicted segmental duplications exhibit CNVs (Fig. 1). Interestingly, ∼50% of CNVs exhibit both WGAC and WSSD support.
Figure 3.
Heatmap representation of CNVs. Each row represents one of the 678 unique CNV regions and columns correspond to dogs. For each CNV region, boxes are colored as black, magenta, and green, depending on whether the individual showed no copy number variation, a loss, or a gain, respectively. CNV regions that show both a loss and a gain within an individual dog (see text) are colored yellow. Horizontal white lines separate CNV calls from single copy control regions, and CNVs that exhibit only losses, gains, or both gains and losses. Within each class, CNV regions are sorted from low to high frequency and from left to right dogs are sorted by decreasing number of CNVs. Dog breeds are abbreviated as follows: Basenji (BAS), Shetland Sheepdog (SHS), German Shepherd (GSH), Siberian Husky (SBH), Wolf (WLF), Labrador Retriever (LBR), Doberman Pinscher (DOB), Standard Poodle (STP), Belgian Shepherd Tervuren (BST), West Highland White Terrier (WST), Yorkshire Terrier (YRK), Boxer (Tasha) (TSH), Afghan Hound (AFH), Golden Retriever (GLR), Rottweiler (ROT), Pug (PUG), Whippet (WHP), German Short Haired Pointer (GSP), and Self-Self hybridization (SLF).
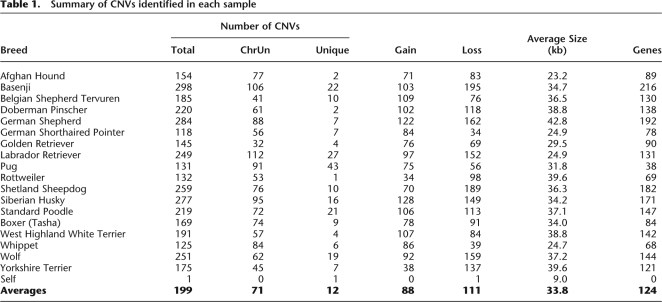
The average number of CNVs per breed was 199, ranging from 118 (German Shorthaired Pointer) to 298 (Basenji), and ∼68% of CNVs were found in two or more individuals (Table 1). In contrast, only one CNV was called in the self-self hybridization and no CNVs were called in the single copy control regions (Fig. 3). We also identified CNVs using an additional algorithm and found good overlap (Supplemental Note 1). Thus, these observations suggest that the false discovery rate (FDR) among the set of predicted CNVs is low (∼3%), although technical issues such as sequence divergence of individual dogs relative to the reference genome sequence or heterogeneity in DNA quality among samples makes it difficult to precisely quantify the FDR.
Table 1.
Summary of CNVs identified in each sample
In general, the number of CNVs identified in each sample is consistent with previous estimates (where available) of breed-specific founding and effective population sizes (Calboli et al. 2008) and levels of polymorphism based on 27,000 SNPs (Karlsson et al. 2007). Of interest, 169 CNVs were identified in the Boxer (Tasha), whose DNA was selected for the dog genome project (Lindblad-Toh et al. 2005) based on its relatively low level of genetic diversity. The somewhat lower level of polymorphism in this boxer, however, does not necessarily imply genetic homogeneity within boxers or other breeds (Parker et al. 2004; Sutter and Ostrander 2004; Wayne and Ostrander 2007). Indeed, a number of recent data sets and analyses suggest considerable genetic diversity exists within breeds (Quignon et al. 2007; Bjornerfeldt et al. 2008), which is consistent with our observation of segregating CNVs among boxers.
qPCR analysis of two CNV regions
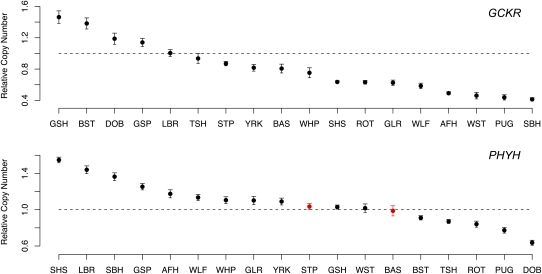
Quantitative PCR (qPCR) was performed using Taqman probes and primers on all dogs used in the aCGH experiments to further validate CNV regions and individual calls (see Methods). Probes were targeted to two regions that show homology with the human GCKR and PHYH genes. As shown in Figure 4, qPCR identified 16 and 15 individuals with copy number differences relative to the reference sample for GCKR and PHYH, respectively. Assuming the qPCR results represent the true copy number status of individual dogs, a total of two false positives occurred in the aCGH data (zero for GCKR and two for PHYH). Thus, the qPCR data confirms the designation of GCKR and PHYH as CNV regions, and suggests a low FDR of actual calls within such regions (2/31 = 6.5%). The qPCR data also confirm the conservativeness of thresholds for calling CNVs in the aCGH data, as 11 individuals were identified with copy number changes in qPCR but not in the aCGH data (false negative rate ∼30%).
Figure 4.
qPCR of GCKR and PHYH regions. Each plot shows the relative copy number in comparison to the reference (y-axis) for each breed (x-axis). The reference sample was the same Boxer that was used as the reference in the aCGH experiments. Note that because the GCKR and PHYH regions are located in segmental duplications, a gain or loss is not expected to yield a relative copy change of 2 and 0.5, respectively. For example, if the reference sample contains three copies, a gain in the test sample would result in an expected relative copy number of 1.33. Vertical bars delimit 95% confidence intervals based on six independent replicates. False positives (CNVs predicted in the aCGH data but not confirmed by qPCR) are colored in red. Breed abbreviations are described in Figure 3.
Fine-scale architectural complexity of CNVs
Correctly delineating CNV boundaries is important for both understanding the molecular mechanisms governing CNV formation and correlating copy number changes with phenotypic variation (Perry et al. 2008). The high probe density of our tiling arrays allowed us to investigate patterns of breakpoint variation across individuals. Of the 460 distinct CNV regions where a gain or loss was observed in two or more samples, 235 exhibited relatively simple architectures with consistent patterns of breakpoints across individuals.
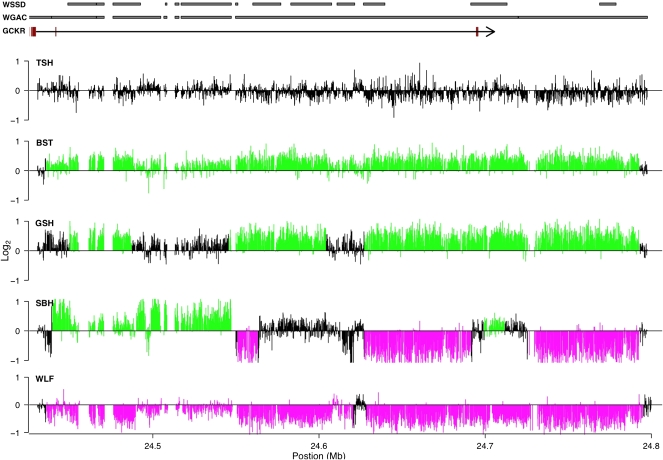
The remaining 225 CNV regions (∼50%) showed fine-scale architectural complexity in the form of substantial interindividual variation in breakpoints or spatial heterogeneity in copy number. Similarly complex CNV regions have been described (Redon et al. 2006; Perry et al. 2008; She et al. 2008), primarily in studies focusing on segmental duplications (Goidts et al. 2006). A particularly interesting pattern was observed for 20 CNV regions where alternating gains and losses occurred within individual dogs. An example of one such region spanning over 400 kb on chromosome 17 is shown in Figure 5. In this region we observed dogs that have gains or losses across the whole region, both gains and losses, and no copy number change relative to the reference sequence (Fig. 5). The precise mechanistic basis for such complex CNV patterns is unclear, and may be attributable to nonallelic homologous recombination or less-understood mechanisms such as the recently proposed replication-based fork stalling and template switching model (Lee et al. 2007).
Figure 5.
Example of a complex CNV region. Distribution of log2 probe intensities across a 400-kb region of chromosome 17 that shows substantial variation in breakpoints across individuals and spatial heterogeneity of copy number within individuals. Black, green, and magenta indicate regions called as no copy number variation, gains, and losses, respectively. The gene structure of GCKR is shown, with black lines and red boxes corresponding to introns and exons, respectively. For clarity, additional RefSeq genes are not shown. Regions with significant WGAC or WSSD scores are indicated by gray rectangles. Breeds are abbreviated as described in Figure 3.
Gene content of CNV regions
CNVs overlap 429 genes (Supplemental Table 5), 318 of which span the complete coding region. The set of copy number variable genes possess a wide spectrum of PANTHER molecular functions (Supplemental Tables 2,5), and provides a rich resource for testing hypotheses on the genetic basis of phenotypic variation within and among breeds. For example, in humans, copy number variation of cytochrome P450 genes, such as CYP2D6, contributes to interindividual variation in drug metabolism phenotypes (Daly 2004; Ledesma and Agundez 2005; Ouahchi et al. 2006). Similar to humans, adverse drug responses have been described in dogs, which often show marked variation in prevalence between breeds (Hickford et al. 2001; Mealey et al. 2001, 2003; Nelson et al. 2003; Neff et al. 2004; Trepanier 2004). Several CYP genes overlap CNVs, perhaps the most interesting of which is CYP3A12, the canine ortholog to human CYP3A4, which is the most abundant hepatic and intestinal cytochrome P450 isoform and is involved in metabolizing a substantial fraction of all drugs (Schuetz 2004). Specifically, nine dogs (Afghan Hound, Doberman Pinscher, German Shepherd, Labrador Retriever, Rottweiler, West Highland White Terrier, Yorkshire Terrier, Boxer, and Wolf) show partial loss of CYP3A12, and of these, adverse drug responses have been described in Doberman Pinschers (sulfonamide hypersensitivity) (Trepanier 2004), Labrador Retrievers (carprofen-induced hepatic toxicity) (Hickford et al. 2001), and Boxers (acepromazine sensitivity, although this result is controversial) (Wagner et al. 2003). Obviously, these observations, while interesting, require additional study to better delimit the relationship between CYP3A12 copy number and variation in drug metabolism phenotypes.
CNVs that span potential genes influencing disease susceptibility were also identified. For instance, the glucokinase regulatory protein gene (GCKR) is located in a complex CNV region (Fig. 5). Recent genome-wide association studies in humans have found that GCKR variation increases susceptibility to type 2 diabetes (Saxena et al. 2007). In our panel of dogs, there is a suggestive pattern of GCKR copy number status and risk of developing diabetes mellitus that warrants further study, with breeds considered at high risk preferentially showing deletions for varying parts of GCKR (Supplemental Table 6).
Conclusions
In summary, we have described the first genome-wide analysis of segmental duplications and associated CNVs in the modern domesticated dog. We found extensive copy number variation in segmental duplications across 17 phenotypically diverse breeds that affect 429 genes. Our study provides the foundation for correlating structural variation with phenotypic variation observed within and between breeds, which we suspect will be an important complement to SNP centric genome-wide association studies (Karlsson et al. 2007; Mosher et al. 2007; Parker et al. 2007; Quignon et al. 2007; Salmon Hillbertz et al. 2007; Sutter et al. 2007). However, in order to perform more principled phenotypic studies with structural variation, it will be necessary to better delimit the population genetic characteristics of CNVs including a broader sampling of breeds, extending CNV identification and characterization to regions of the genome outside of segmental duplications, assessing allele frequency distributions in unrelated individuals within breeds, and determining levels of linkage disequilibrium between CNVs and SNPs. Ultimately, understanding how the full spectrum of canine genomic variation influences phenotypic variation will provide insight into the genetic architecture of phenotypes and mechanisms of rapid short-term evolutionary change.
Methods
Computational analysis of segmental duplications
We downloaded CanFam2.0 genomic sequence data from the UCSC Genome Browser (http://genome.ucsc.edu/) and whole-genome shotgun sequence (WGS) reads were obtained through NCBI (http://www.ncbi.nlm.nih.gov/). WGAC and WSSD were performed as previously described (Bailey et al. 2001, 2002). Briefly, WGAC identifies paralogous stretches of sequence by fragmenting the genome into 400-kb fragments, which are masked for repeats with RepeatMasker and removed. Global alignments of these fragments are performed with alignments of >90% sequence identity and >1 kb in length deemed as paralogs. Masked regions are then reinserted and fragments are returned to their genomic locations.
WSSD identifies genomic regions of significant depth of coverage by aligning WGS reads to the reference genome sequence. We initially conducted WSSD on a set of 13 training BACs (also obtained from NCBI; AC090032.2, AC090890.3, AC091119.2, AC117937.4, AC147678.5, AC147681.8, AC147784.3, AC090889.3, AC090972.5, AC092249.3, AC147677.4, AC147680.4, AC147707.5). BACs were masked for repeats and MegaBLAST alignments of these BACs were performed against a database of WGS reads. We calculated duplication depth by counting the number of WGS reads aligning to 5-kb sliding windows. In addition, we calculated nucleotide divergence between the WGS reads and the BAC sequences for each 5-kb window. The distribution of alignment depth and divergence in this training set allows empirical thresholds to be determined. Consistent with previous studies (Bailey et al. 2002, 2004; Cheng et al. 2005; She et al. 2008), we define significant alignment depth and divergence scores as those that are greater than three standard deviations from the mean. After training, we masked the entire canine reference genome for common repeats with <10% divergence and performed MegaBLAST alignments of the WGS reads to the reference genome. Following previous studies (She et al. 2008), we defined segmental duplications based on the union of significant WGAC hits with <94% sequence identity and WSSD results (see Supplemental Table 1).
Bioinformatics analysis of segmental duplications
We investigated the genomic distribution of segmental duplications by testing the hypothesis that pericentromeric and subtelomeric regions were enriched for duplications (Bailey et al. 2001). Since the pericentromeric and subtelomeric regions are not well annotated, we defined pericentromeric and subtelomeric regions as 2 Mb from the most centromeric base and 2 Mb from the end(s) of chromosomes, respectively. Since all dog chromosomes are acrocentric, with the exception of the X chromosome, this results in a 2-Mb pericentromeric region at one end of the chromosome and a 2-Mb subtelomeric region at the other end of the chromosome. In the case of the X chromosome, the pericentromeric region was defined as two 1-Mb regions that flank the centromeric region and two 1-Mb subtelomeric ends on both ends of the chromosome. No sequence from chrUn was included. All predicted duplicated bases that overlap these regions were totaled and chi-square tests were used to test the null hypothesis of no enrichment as previously described (Bailey et al. 2001).
We obtained a catalog of all canine peptides from Ensembl (ftp://ftp.ensembl.org/pub/current_fasta/canis_familiaris/pep/). This yielded 25,546 peptides, 1078 of which overlap with predicted segmental duplications, and correspond to 841 unique Ensembl genes. PANTHER Molecular Function terms were assigned to all peptides using the PANTHER Hidden Markov Model scoring tools (http://www.pantherdb.org/downloads/). PANTHER Molecular Function terms with less than five observations among the duplicated genes were not analyzed further. Similar analyses were performed on the 547 peptides (corresponding to 429 Ensembl genes) that overlap with the 678 unique CNV regions. We tested the hypothesis that the remaining PANTHER Molecular Function terms were over-represented in segmental duplications and CNVs with the hypergeometric distribution. Bonferroni corrections were used to correct P-values for multiple hypothesis testing.
DNA samples
Breeds used in this study include an Afghan Hound, Basenji, Belgian Shepherd (Tervuren), Boxer, Doberman Pinscher, German Shepherd, German Shorthaired Pointer, Golden Retriever, Labrador Retriever, Pug, Rottweiler, Shetland Sheepdog, Siberian Husky, Standard Poodle, West Highland White Terrier, Whippet, and Yorkshire Terrier. In addition, genomic DNA was obtained from Tasha, a boxer whose DNA was used in the canine genome project, and also a gray wolf. Sample collection was approved by the Animal Care and Use Committees from Washington State University. We isolated genomic DNA from whole blood samples using Qiagen's QIAamp DNA Blood Maxi Kit. All dogs were screened for correct AKC status. We assessed DNA quality and purity by OD260/280 and OD260/230 readings and by digesting genomic DNA with a salt sensitive restriction enzyme (NlaIII).
FISH and image analysis
C. familiaris metaphase spreads were prepared from PHA-stimulated peripheral lymphocytes of a normal donor of unknown breed identity by standard procedures. DNA extraction from BACs has already been reported. FISH experiments were performed essentially as previously described (Ventura et al. 2003). Briefly, DNA probes were directly labeled with Cy3-dUTP (Perkin-Elmer) by nick-translation. Two hundred nanograms of labeled probe was used for the FISH experiments. Hybridization was performed at 37°C in 2× SSC, 50% (v/v) formamide, 10% (w/v) dextran sulfate, and 3 mg of sonicated salmon sperm DNA in a volume of 10 μL. Post-hybridization washing was at 60°C in 0.1× SSC (three times, high stringency).
Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3 (red) and DAPI (blue) fluorescence signals, detected with specific filters, were recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
aCGH chip design and hybridizations
The production of the aCGH chip was carried out by NimbleGen Systems, Inc. (www.NimbleGen.com), with final probe design and locations being approved after individual inspection by eye. We targeted probes to regions designated as segmental duplications (significant WGAC and WSSD support; 106.6 Mb), as well as all regions not formally designated as segmental duplications but possessing significant WGAC or WSSD support (16.4 Mb). In total, 368,360 probes were placed into these regions (spanning 123 Mb) with an average probe spacing of 200 bp. Each segmental duplication region was flanked with 5 kb of putative single copy sequence. In cases where the added flanks resulted in overlaps between separate segmental duplications, the segmental duplications were merged together with new flanking regions added to the merged segmental duplication. A total of 9278 probes were placed in flanking regions (10.6 Mb) with a mean spacing of 1 kb. Finally, we placed 8790 probes with a mean spacing of 350 bp into single copy control regions (3.5 Mb). Single copy control regions were defined as being at least 5 Mb away from any predicted segmental duplications (with the exception of three control regions on the X chromosome that were at least 2.5 Mb away from any predicted segmental duplication). All genomic DNA samples were sent to NimbleGen for the hybridizations to be performed. A female Boxer distinct from Tasha was used as a reference sample in each hybridization.
aCGH data analysis and CNV calling
For each hybridization, normalized log2 ratios were first averaged across 2-kb windows. Due to the unique assembly of chrUn sequences, each chrUn assembly contig was analyzed as if it were an individual chromosome, and separately from the assembled chromosomes. All averaged 2-kb windows were analyzed using the R package RJaCGH (Rueda and Diaz-Uriarte 2007). We used the self-self hybridization and control regions to define suitable thresholds to apply to the RJaCGH calls in order to minimize false positives. Specifically, we retained predicted CNVs if it had at least five datapoints supporting it, thus limiting the minimum CNV size to ∼10 kb. The chrUn results were further filtered by requiring the average log2 value of the CNV to be >0.25 or <−0.25. We merged overlapping CNV coordinates across hybridizations to form unique CNV regions. All unique CNV regions on the X chromosome that were only supported by male dogs were removed to avoid potential complications, since the reference sample was a female. We repeated the entire RJaCGH analyses for a subset of the arrays to ensure consistency in calls. We assessed breakpoint variation by analyzing each individual CNV in a defined unique region and determining whether its length varied by more than 5 kb from the entire CNV region length.
The false discovery rate (FDR) was estimated based on the observation of a single false positive in the self-self hybridization, and thus a rough estimate of the FDR is the expected number of false positives per array (1) times the number of total arrays (18) divided by the total number of unique CNV regions (678), resulting in an estimated FDR of 2.65%. Note that this calculation is only approximate because it assumes that each false positive results in a unique CNV region and does not take into account the potential for varying false positive rates across arrays.
qPCR
We performed qPCR on all dogs from two CNV regions (GCKR and PHYH). Specifically, Taqman probes and primers (Applied Biosystems) were designed for three regions; a single copy control region (labeled with FAM dye) and two test regions (each labeled with VIC dye). Primer and probe sequences are available upon request. Assays were performed on an ABI 7900HT (Applied Biosystems) using 20-μL reactions containing 10 μL of Taqman Universal PCR Master Mix, 250 nM of FAM probe, 900 nM of forward and reverse primers for FAM probe, 250 nM of VIC probe, 900 nM of forward and reverse primers for VIC probe, and 30 ng of genomic DNA. Amplification was done under the following conditions: one cycle at 50°C for 2 min, one cycle at 95°C for 10 min, 40 cycles at 95°C for 15 sec, and 60°C for 1 min. Serial dilutions were performed for each assay to estimate the PCR efficiency (E). Using the ΔCT method, relative copy number was determined with respect to the same reference Boxer sample used in the aCGH experiments. CT-values were adjusted for PCR efficiency (E) as log2(ECT). The CT-values then were normalized by subtracting the VIC CT-value from the FAM CT-value (FAM CT – VIC CT). The relative copy number was determined as 2-(normalized CTfor test strain–normalized CT for reference strain). In total, six independent replicates were performed for each individual. Statistical significance was determined by a one-sample t-test. Similar results were obtained with a one-sample Wilcoxon test (data not shown).
Data Release
All aCGH data has been submitted to the gene expression omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE13266.
Acknowledgments
We thank the pet owners, and their dogs, for participating in this research; Dayna Akey for helpful discussions; and Kerstin Lindblad-Toh for providing DNA for the sample Boxer (Tasha) used to construct the reference dog genome sequence. This work was supported in part by grants from the University of Washington Royalty Research Fund (J.M.A.), the American Kennel Club (J.M.A. and K.M.), Howard Hughes Medical Institute (E.E.E.), and a Sloan Fellowship in Computational Biology (J.M.A.).
Footnotes
[Supplemental material is available online at www.genome.org. All aCGH data from this study have been submitted to Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE13266.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.084715.108.
References
- Bailey J.A., Eichler E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Church D.M., Ventura M., Rocchi M., Eichler E.E. Analysis of segmental duplications and genome assembly in the mouse. Genome Res. 2004;14:789–801. doi: 10.1101/gr.2238404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J.S., Estivill X., Antonarakis S.E. Copy number variants and genetic traits: Closer to the resolution of phenotypic to genotypic variability. Nat. Rev. Genet. 2007;8:639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- Bjornerfeldt S., Hailer F., Nord M., Vila C. Assortative mating and fragmentation within dog breeds. BMC Evol. Biol. 2008;8:28. doi: 10.186/1471-2148-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calboli F.C., Sampson J., Fretwell N., Balding D.J. Population structure and inbreeding from pedigree analysis of purebred dogs. Genetics. 2008;179:593–601. doi: 10.1534/genetics.107.084954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Conrad B., Antonarakis S.E. Gene duplication: A drive for phenotypic diversity and cause of human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:17–35. doi: 10.1146/annurev.genom.8.021307.110233. [DOI] [PubMed] [Google Scholar]
- Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Daly A.K. Pharmacogenetics of the cytochromes P450. Curr. Top. Med. Chem. 2004;4:1733–1744. doi: 10.2174/1568026043387070. [DOI] [PubMed] [Google Scholar]
- Emanuel B.S., Shaikh T.H. Segmental duplications: An “expanding” role in genomic instability and disease. Nat. Rev. Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- Feuk L., Carson A.R., Scherer S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia M.J., Shibata M., Anderson K.V. Chato, a KRAB zinc-finger protein, regulates convergent extension in the mouse embryo. Development. 2008;135:3053–3062. doi: 10.1242/dev.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goidts V., Cooper D.N., Armengol L., Schempp W., Conroy J., Estivill X., Nowak N., Hameister H., Kehrer-Sawatzki H. Complex patterns of copy number variation at sites of segmental duplications: An important category of structural variation in the human genome. Hum. Genet. 2006;120:270–284. doi: 10.1007/s00439-006-0217-y. [DOI] [PubMed] [Google Scholar]
- Graubert T.A., Cahan P., Edwin D., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V., Saar K., Adamovic T., Verheul M., van Heesch S.A., Cook S., Pravenec M., Aitman T., Jacob H., Shull J.D., et al. Distribution and functional impact of DNA copy number variation in the rat. Nat. Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- Hickford F.H., Barr S.C., Erb H.N. Effect of carprofen on hemostatic variables in dogs. Am. J. Vet. Res. 2001;62:1642–1646. doi: 10.2460/ajvr.2001.62.1642. [DOI] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Irion D.N., Schaffer A.L., Famula T.R., Eggleston M.L., Hughes S.S., Pedersen N.C. Analysis of genetic variation in 28 dog breed populations with 100 microsatellite markers. J. Hered. 2003;94:81–87. doi: 10.1093/jhered/esg004. [DOI] [PubMed] [Google Scholar]
- Karlsson E.K., Baranowska I., Wade C.M., Salmon Hillbertz N.H., Zody M.C., Anderson N., Biagi T.M., Patterson N., Pielberg G.R., Kulbokas E.J., 3rd, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Ledesma M.C., Agundez J.A. Identification of subtypes of CYP2D gene rearrangements among carriers of CYP2D6 gene deletion and duplication. Clin. Chem. 2005;51:939–943. doi: 10.1373/clinchem.2004.046326. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Leonard J.A., Wayne R.K., Wheeler J., Valadez R., Guillen S., Vila C. Ancient DNA evidence for Old World origin of New World dogs. Science. 2002;298:1613–1616. doi: 10.1126/science.1076980. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade C.M., Mikkelsen T.S., Karlsson E.K, Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., rd, Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- McCarroll S.A., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Mealey K.L., Bentjen S.A., Gay J.M., Cantor G.H. Ivermectin sensitivity in collies is associated with a deletion mutation of the mdr1 gene. Pharmacogenetics. 2001;11:727–733. doi: 10.1097/00008571-200111000-00012. [DOI] [PubMed] [Google Scholar]
- Mealey K.L., Northrup N.C., Bentjen S.A. Increased toxicity of P-glycoprotein-substrate chemotherapeutic agents in a dog with the MDR1 deletion mutation associated with ivermectin sensitivity. J. Am. Vet. Med. Assoc. 2003;223:1453–1455. 1434. doi: 10.2460/javma.2003.223.1453. [DOI] [PubMed] [Google Scholar]
- Mellersh C.S., Hitte C., Richman M., Vignaux F., Priat C., Jouquand S., Werner P., Andre C., DeRose S., Patterson D.F., et al. An integrated linkage-radiation hybrid map of the canine genome. Mamm. Genome. 2000;11:120–130. doi: 10.1007/s003350010024. [DOI] [PubMed] [Google Scholar]
- Mosher D.S., Quignon P., Bustamante C.D., Sutter N.B., Mellersh C.S., Parker H.G., Ostrander E.A. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007;3:e79. doi: 10.1371/journal.pgen.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.W., Rine J. A fetching model organism. Cell. 2006;124:229–231. doi: 10.1016/j.cell.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Neff M.W., Robertson K.R., Wong A.K., Safra N., Broman K.W., Slatkin M., Mealey K.L., Pedersen N.C. Breed distribution and history of canine mdr1-1Delta, a pharmacogenetic mutation that marks the emergence of breeds from the collie lineage. Proc. Natl. Acad. Sci. 2004;101:11725–11730. doi: 10.1073/pnas.0402374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson O.L., Carsten E., Bentjen S.A., Mealey K.L. Ivermectin toxicity in an Australian Shepherd dog with the MDR1 mutation associated with ivermectin sensitivity in Collies. J. Vet. Intern. Med. 2003;17:354–356. [PubMed] [Google Scholar]
- Ouahchi K., Lindeman N., Lee C. Copy number variants and pharmacogenomics. Pharmacogenomics. 2006;7:25–29. doi: 10.2217/14622416.7.1.25. [DOI] [PubMed] [Google Scholar]
- Parker H.G., Kim L.V., Sutter N.B., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Parker H.G., Kukekova A.V., Akey D.T., Goldstein O., Kirkness E.F., Baysac K.C., Mosher D.S., Aguirre G.D., Acland G.M., Ostrander E.A. Breed relationships facilitate fine-mapping studies: A 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Res. 2007;17:1562–1571. doi: 10.1101/gr.6772807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson D.F., Haskins M.E., Jezyk P.F., Giger U., Meyers-Wallen V.N., Aguirre G., Fyfe J.C., Wolfe J.H. Research on genetic diseases: Reciprocal benefits to animals and man. J. Am. Vet. Med. Assoc. 1988;193:1131–1144. [PubMed] [Google Scholar]
- Perry G.H., Tchinda J., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S.W., Eichler E.E., et al. Hotspots for copy number variation in chimpanzees and humans. Proc. Natl. Acad. Sci. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G.H., Ben-Dor A., Tsalenko A., Sampas N., Rodriguez-Revenga L., Tran C.W., Scheffer A., Steinfeld I., Tsang P., Yamada N.A., et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollinger J.P., Bustamante C.D., Fledel-Alon A., Schmutz S., Gray M.M., Wayne R.K. Selective sweep mapping of genes with large phenotypic effects. Genome Res. 2005;15:1809–1819. doi: 10.1101/gr.4374505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignon P., Herbin L., Cadieu E., Kirkness E.F., Hedan B., Mosher D.S., Galibert F., Andre C., Ostrander E.A., Hitte C. Canine population structure: Assessment and impact of intra-breed stratification on SNP-based association studies. PLoS One. 2007;2:e1324. doi: 10.1371/journal.pone.0001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda O.M., Diaz-Uriarte R. Flexible and accurate detection of genomic copy-number changes from aCGH. PLoS Comput. Biol. 2007;3:e122. doi: 10.1371/journal.pcbi.0030122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon Hillbertz N.H., Isaksson M., Karlsson E.K., Hellmen E., Pielberg G.R., Savolainen P., Wade C.M., von Euler H., Gustafson U., Hedhammar A., et al. Duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat. Genet. 2007;39:1318–1320. doi: 10.1038/ng.2007.4. [DOI] [PubMed] [Google Scholar]
- Savolainen P., Zhang Y.P., Luo J., Lundeberg J., Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Schuetz E.G. Lessons from the CYP3A4 promoter. Mol. Pharmacol. 2004;65:279–281. doi: 10.1124/mol.65.2.279. [DOI] [PubMed] [Google Scholar]
- Sebat J. Major changes in our DNA lead to major changes in our thinking. Nat. Genet. 2007;39:S3–S5. doi: 10.1038/ng2095. [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Cheng Z., Zollner S., Church D.M., Eichler E.E. Mouse segmental duplication and copy number variation. Nat. Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter N.B., Ostrander E.A. Dog star rising: The canine genetic system. Nat. Rev. Genet. 2004;5:900–910. doi: 10.1038/nrg1492. [DOI] [PubMed] [Google Scholar]
- Sutter N.B., Bustamante C.D., Chase K., Gray M.M., Zhao K., Zhu L., Padhukasahasram B., Karlins E., Davis S., Jones P.G., et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepanier L.A. Idiosyncratic toxicity associated with potentiated sulfonamides in the dog. J. Vet. Pharmacol. Ther. 2004;27:129–138. doi: 10.1111/j.1365-2885.2004.00576.x. [DOI] [PubMed] [Google Scholar]
- Tuzun E., Bailey J.A., Eichler E.E. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res. 2004;14:493–506. doi: 10.1101/gr.1907504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., Mudge J.M., Palumbo V., Burn S., Blennow E., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S., et al. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 2003;13:2059–2068. doi: 10.1101/gr.1155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila C., Savolainen P., Maldonado J.E., Amorim I.R., Rice J.E., Honeycutt R.L., Crandall K.A., Lundeberg J., Wayne R.K. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- Wagner A.E., Wright B.D., Hellyer P.W. Myths and misconceptions in small animal anesthesia. J. Am. Vet. Med. Assoc. 2003;223:1426–1432. doi: 10.2460/javma.2003.223.1426. [DOI] [PubMed] [Google Scholar]
- Wayne R.K., Ostrander E.A. Lessons learned from the dog genome. Trends Genet. 2007;23:557–567. doi: 10.1016/j.tig.2007.08.013. [DOI] [PubMed] [Google Scholar]