Abstract
We applied genome-wide profiling to successive salt-extracted fractions of micrococcal nuclease-treated Drosophila chromatin. Chromatin fractions extracted with 80 mM or 150 mM NaCl after digestion contain predominantly mononucleosomes and represent classical “active” chromatin. Profiles of these low-salt soluble fractions display phased nucleosomes over transcriptionally active genes that are locally depleted of histone H3.3 and correspond closely to profiles of histone H2Av (H2A.Z) and RNA polymerase II. This correspondence suggests that transcription can result in loss of H3.3+H2Av nucleosomes and generate low-salt soluble nucleosomes. Nearly quantitative recovery of chromatin is obtained with 600 mM NaCl; however, the remaining insoluble chromatin is enriched in actively transcribed regions. Salt-insoluble chromatin likely represents oligonucleosomes that are attached to large protein complexes. Both low-salt extracted and insoluble chromatin are rich in sequences that correspond to epigenetic regulatory elements genome-wide. The presence of active chromatin at both extremes of salt solubility suggests that these salt fractions capture bound and unbound intermediates in active processes, thus providing a simple, powerful strategy for mapping epigenome dynamics.
Recent technological advances in microarray fabrication and massively parallel sequencing have led to the general dissemination of powerful tools for genome-wide profiling of chromatin proteins. As a result, the coverage and resolution that can be obtained in a single experiment has increased by orders of magnitude in the past few years. However, methods that are used for the preparation of chromatin for affinity capture, such as chromatin immunoprecipitation with hybridization to microarrays (ChIP-chip) and ChIP with sequencing (ChIP-seq), are not substantially different from those that were developed in the 1990s to study single loci. This raises questions as to whether the full potential of current genomic technologies is being realized. Moreover, the much greater resolution and coverage obtained with the collection of millions of data points per sample will magnify any inhomogeneities in chromatin preparations that went undetected in the past. Of particular concern is that methods for chromatin preparation might not result in quantitative recovery of starting material, and this leads to uncertainty as to whether the DNA fragments that are recovered are a representative sample of chromatin in vivo.
Chromatin isolation methods used for ChIP and other affinity capture strategies fall into two general classes. X-ChIP typically involves cross-linking of cells or nuclei followed by chromatin extraction and sonication prior to affinity capture (Orlando 2000). X-ChIP is applicable to any epitope that can be cross-linked to DNA, and so is well-suited for profiling of DNA-binding proteins. N-ChIP typically involves nuclear isolation, fragmentation with micrococcal nuclease (MNase), and release of the resulting mono- and oligo-nucleosomes by lysis and moderate salt extraction (O'Neill and Turner 2003). N-ChIP is well-suited for profiling of histone epitopes but is inappropriate for profiling proteins that are not tightly bound to DNA. Other methods that have been applied to chromatin profiling employ cleavage reagents such as Deoxyribonuclease I (DNase I) and MNase to map sites of differential accessibility (Yuan et al. 2005; Crawford et al. 2006; Sabo et al. 2006; Mito et al. 2007) and tethered Dam methylase to map binding sites of chromatin proteins in vivo (van Steensel et al. 2001). An advantage of these latter methods over those based on affinity capture is that they do not require preservation of the protein component of the genome, and so, the DNA to be profiled can be extracted with virtually 100% efficiency. Quantitative recovery of DNA eliminates the concern that what is being profiled is not representative of the chromatin state in vivo, and is a desirable goal for affinity capture methods.
Here we show that nearly quantitative recovery of chromatin for affinity capture can be achieved by using a salt extraction method that was first described 30 years ago (Sanders 1978). Successive washing of intact MNase-treated nuclei with increasing salt results in the isolation of chromatin fractions with dramatically different genome-wide profiles, including a low-salt soluble fraction of highly accessible chromatin, a higher-salt soluble fraction representing the bulk of chromatin, and an insoluble fraction that is largely derived from transcribed regions of the genome. Using affinity capture of biotin-tagged histones, we show that the low-salt soluble fraction is enriched in nucleosomes that are replaced independent of replication, are enriched in histone H2Av, and are preferentially located upstream of active promoters and within epigenetic regulatory elements. The radically different landscapes obtained from salt fractions of native chromatin suggest that our method maps intermediates in active processes, providing an explanation for the generation of classical active chromatin and introducing a novel strategy for profiling chromatin dynamics.
Results
Genome-wide profiling of salt-fractionated chromatin
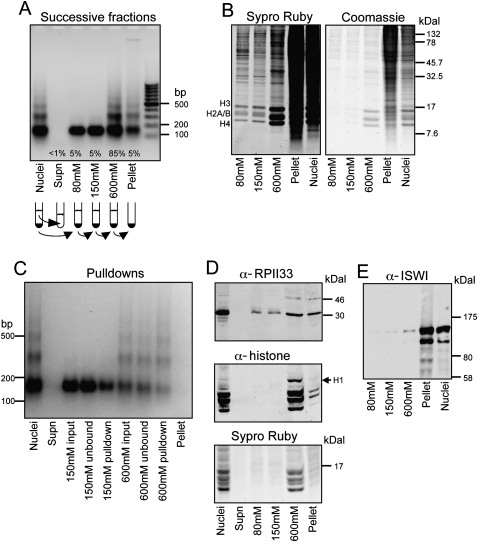
In the course of profiling histone variants in the Drosophila melanogaster genome, we wondered if we could improve methods by completely solubilizing chromatin from nuclear preparations. Because methods of chromatin purification might affect the types of nucleosomes recovered, we first set out to characterize the products recovered in successive solubilization steps. We treated Drosophila S2 cell nuclei with micrococcal nuclease (MNase) in 10 mM Tris-HCl (pH 7.4), 2 mM MgCl2 and washed the nuclei successively with increasing concentrations of NaCl in solubilizing buffer. Successive chromatin fractions were collected and analyzed on agarose gels. We used EGTA instead of EDTA to preferentially chelate calcium and stop the MNase digestion, while maintaining Mg2+-dependent chromatin compaction and nuclear integrity (Sanders 1978). As has been reported by others, chromatin that is soluble in up to 150 mM NaCl accounts for a small proportion of the total chromatin and is mostly mononucleosomal (Fig. 1A; Davie and Saunders 1981; Rocha et al. 1984; Hebbes and Allen 2000). The fraction of chromatin recovered increases with higher salt washes, and a 600 mM NaCl wash solubilizes ∼90% of the chromatin. Protein analysis of salt fractions indicates that successive 80 mM, 150 mM, and 600 mM salt fractions release levels of core histones consistent with the amounts of DNA recovered, and the pellet is devoid of histones relative to high-molecular-weight protein (Fig. 1B).
Figure 1.
DNA and protein characterization of S2 cell chromatin fractions. (A) Ethidium bromide–stained agarose gel showing a typical MNase ladder of DNA purified from MNase-treated nuclei (Nuclei); followed by ladders of DNA purified from the nuclear supernatant (Supn); successive 80 mM, 150 mM, and 600 mM extractions; and the remaining pellet, as indicated in the cartoon below. The average percentage of total chromatin isolated at each step is indicated. (B) SDS–polyacrylamide gel analysis (SDS-PAGE) of proteins in salt fractions. Equal aliquots of fractions from successive extraction steps in a typical experiment were loaded onto a 16% polyacrylamide gel, which was electrophoresed and stained with Sypro ruby for sensitive detection of histones (left). Restaining with Coomassie blue (right) shows that the pellet includes mostly high-molecular-weight proteins. (C) MNase ladders of 150 mM and 600 mM fractions from a typical pulldown experiment. (D) Western blot analysis of the RPII33 subunit of RNA polymerase II (Pol II) and histones in salt fractions. Equal aliquots of samples were loaded onto a 16% SDS-PAGE gel, electrophoresed, blotted, probed with rabbit RPII33 (Muse et al. 2007) and mouse pan-histone (Roche MAB052) antibodies, followed by IRD700 anti-rabbit and IRD800 anti-mouse secondary antibodies (LI-COR). After blotting, the gel was stained with Sypro ruby. RPII33 is enriched in the insoluble fraction relative to histones. Histone H1 is mostly extracted with the 600 mM fraction as expected (Davie and Saunders 1981). (E) Western blot analysis of the ISWI nucleosome remodeling protein in salt fractions. Equal aliquots of samples were loaded onto a 4%–12% NuPAGE (Invitrogen), gel electrophoresed and blotted to nitrocellulose, and probed with an ISWI antibody (Tsukiyama et al. 1995). Successive extractions release increasing amounts of ISWI, although most of the ISWI (MW 119) that is recovered resides in the insoluble fraction.
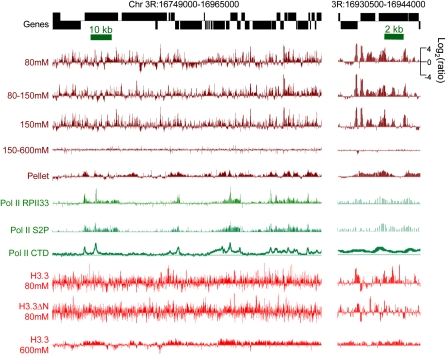
In order to characterize chromatin extracted from salt fractions at high resolution, we profiled salt fractions on single high-density NimbleGen isothermal arrays (with 2.1 million 50–75-base pair [bp] probes) that tile the entire annotated fly genome at ∼65-bp resolution. Each sample was profiled versus whole MNase-treated chromatin to control for the extent of nuclease digestion in individual experiments. When salt-fraction profiles were directly compared for the same chromosomal regions, we saw dramatic differences. Extraction with 600 mM NaCl gave essentially equal representation relative to whole MNase-treated chromatin, confirming that this concentration of salt solubilizes virtually all nucleosomes. In contrast, 80 mM, 80–150 mM, and 150 mM salt extracts displayed numerous highly reproducible sharp peaks and broader features throughout the genome (Fig. 2, brown tracks; Supplemental Figs. S1, S2A). Low-salt soluble chromatin features are not restricted to particular genic features and are found in genes, promoters and intergenic regions. These peaks must arise from selective extraction of particular sequences by low-salt treatment.
Figure 2.
Salt-extracted chromatin fractions display different chromatin landscapes. Tracks show profiles for a typical gene-rich region of the fly genome. Salt-extracted chromatin fractions (brown), Pol II (green), and H3.3 (red) recovered from these fractions are all displayed on the same log2-ratio scale. Pol II tracks are calculated from published data for different Pol II epitopes: the RPII33 subunit, serine-2-phosphate on the C-terminal domain (CTD), and the unphosphorylated CTD (Muse et al. 2007; Misulovin et al. 2008). An expansion of a subregion is shown on the right. The 80–150 mM fraction derives from a 150 mM extraction following an 80 mM extraction, and the 150–600 mM fraction derives from a 600 mM extraction following a 150 mM extraction. All tracks represent single data sets; examples of tracks representing biological replicates of this region are shown in Supplemental Figure S1.
Transcribed genes are enriched for low-salt soluble chromatin genome-wide
Many of the peaks seen in the low-salt soluble chromatin landscape correspond to annotated transcriptional start sites (TSSs). To test if low-salt soluble chromatin is enriched in transcriptionally active genes, we compared chromatin profiles for genes with different expression levels. We measured transcript levels using oligo(dT)-primed cDNA made to total RNA extracted from our S2 cells, labeled the cDNA with Cy3, and hybridized to tiling microarrays together with Cy5-labeled fragmented genomic DNA. Cy3/Cy5 ratios from algorithmically selected probes were used to assign expression levels and to rank-order all 9247 genes for which both the TSS and poly(A) addition sites are known from experimental evidence (Supplemental Table S1). Rank-ordered genes were divided into five 20th-percentile classes according to expression level. By lining up genes at their 5′ ends and averaging log-ratio intensities in 25-bp intervals ±1.5 kb on either side of the TSS, we generated average chromatin profiles for the each of the salt fractions, each divided into five expression classes. We excluded values from regions within neighboring, overlapping, or nested transcription units, although approximately similar profiles were obtained when these regions were included (data not shown).
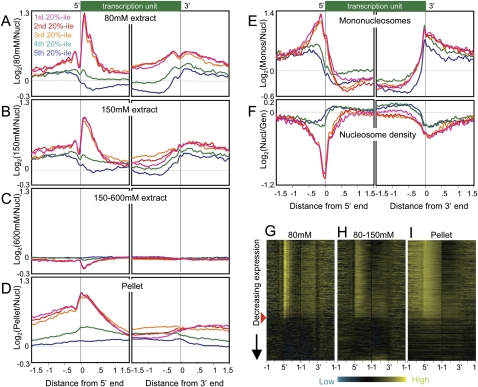
For all three low-salt fractions, profiles were similar for genes in the first, second, and third 20th-percentile classes based on expression level. In the 80 mM fraction, 5′ end profiles showed a strong peak 100 bp downstream from the TSS, followed by a sharply attenuated series of peaks spaced ∼200 bp apart over gene bodies (Fig. 3A; Supplemental Fig. S2B). This fraction is also characterized by a dip ∼50 bp upstream of the TSS that is preceded by a peak 150 bp upstream. The presence of conspicuous peaks indicates that, on average, nucleosomes in this fraction are phased relative to the TSS. These are active gene patterns, because profiles of genes in the fourth quintile based on expression show almost no features and those of genes in the fifth quintile show genic depletion for the 80 mM fraction relative to flanking regions. Similar profiles were seen for the 150 mM fraction except that downstream phasing is less distinct (Fig. 3B; Supplemental Fig. S2C). Notably, the profiles for genes in the top three quintiles essentially superimpose, suggesting that salt solubility of chromatin around 5′ ends is an all-or-none feature of expressed genes. Indeed, heat map analysis showed almost no trend with decreasing expression down to an approximate threshold of expression above which the first several hundred base pairs of transcriptionally engaged chromatin are saturated for low-salt soluble nucleosomes (Fig. 3G,H).
Figure 3.
Active and inactive genes display contrasting profiles in salt-extracted chromatin fractions. (A) Ends analysis of chromatin extracted with 80 mM salt, divided into quintile groupings based on gene expression levels in Supplemental Table S1. Averages from eight independent experiments are shown. The 9247 genes for which both ends are known were aligned at their 5′ and their 3′ ends and averaged for each 25-bp interval on either side. Contributions from neighboring transcription units were omitted. (B) Same as A except that 150 mM fractions from three independent experiments were profiled, averaged, and plotted on the same scale. (C) Same as A except that the 150–600 mM fractions from three independent experiments were profiled, averaged, and plotted on the same scale. (D) Same as A except that pellet fractions from three independent experiments were profiled, averaged, and plotted on the same scale. (E) Ends analysis of mononucleosomes isolated by agarose gel purification from DNA obtained from MNased nucleosomes extracted using EDTA and shearing. An average from two experiments is shown. (F) Ends analysis profile of nucleosome density averaged from three independent experiments. (G) A representative heat map profile for 80 mM–extracted chromatin. The red arrowhead indicates the cutoff used to separate active from inactive genes for the analyses shown in Fig. 4. 5′ (left) and 3′ (right) end profiles for ±1 kb from single representative experiments are shown, ordered by decreasing expression, based on cDNA levels measured as a log-ratio over genomic DNA using the same array design as for chromatin profiling, with probes for each gene chosen algorithmically. Contributions from neighboring transcription units were omitted, which resulted in some horizontal gray streaks toward the upstream and downstream sides of each map. Contrast levels are equal for all stacks (Java TreeView setting of 2.0). (H) Same as G for a representative chromatin fraction extracted with 150 mM NaCl following an 80 mM extraction (80–150 mM). (I) Same as H for the corresponding pellet fraction.
In contrast to the prominent peaks seen for profiles of active genes in the low-salt fractions, the 150–600 mM fraction shows active gene profiles with slight depressions upstream and downstream of TSSs, approximately the inverse of the patterns seen for low-salt fractions (Fig. 3C). Considering that this fraction represents the bulk (∼80%) of the genome after low-salt extraction, such mild overall depletion mirroring low-salt extraction is as expected.
We considered the possibility that low-salt soluble chromatin contains mononucleosomes simply because mononucleosomes are smaller than oligonucleosomes and so are more easily extracted from MNase-treated nuclei. To test this, we solubilized MNase-treated nuclei by EDTA extraction and cavitation shearing under minimal salt conditions (<15 mM) (Jin and Felsenfeld 2007), gel-electrophoresed purified DNA (Supplemental Fig. S3), and excised the bulk mononucleosomal DNA. Mononucleosomal DNA was then profiled versus whole MNase-treated DNA. Strikingly, we found that mononucleosome particles are not uniformly distributed across the genome; instead, the mononucleosome fraction shows a very distinctive profile when aligned across genes (Fig. 3E). A broad peak of ∼600 bp is found upstream of the TSS, and a sharp peak is centered just upstream of the 3′ end. Notably, gene bodies themselves are nuclease-resistant relative to intergenic DNA. As mononucleosomes result from two nearby cleavages by MNase, the mononucleosomal fraction must originate from nuclease-sensitive regions of chromatin. Low-salt solubilization does yield mononucleosomal sized DNA but must extract a distinct subset of nucleosomal particles as this displays a very different profile from bulk mononucleosomal DNA (Fig. 3, cf. A and E).
TSSs of active genes are slightly depleted in low-salt soluble chromatin compared with immediately upstream and downstream regions, but are enriched relative to the genome-wide average and relative to TSSs of inactive genes (Fig. 3A,B). This was unexpected, because of the general perception that active promoters are “nucleosome-free regions” (Lee et al. 2004; Yuan et al. 2005). We tested whether active promoters in whole S2 cell nuclei show the expected nucleosome-free regions by profiling nucleosome density from the same batches of nuclei used for isolation of salt-soluble chromatin. As expected, nucleosome density reaches a minimum at promoters of active genes, whereas inactive genes display profiles that are close to the genome-wide average but with a slight increase in density over gene bodies (Fig. 3F). Therefore, low-salt treatment preferentially recovers nucleosomes from active gene promoters, despite the fact that there are conspicuously few nucleosomes in these regions.
Low-salt soluble nucleosomes are enriched in histone H3.3 at promoters
Previous studies of low-salt soluble nucleosomes showed them to be hyper-acetylated, hyperphosphorylated on H3Ser28, and enriched in the H3.3 histone variant (Perry and Chalkley 1981; Hebbes and Allen 2000; Sun et al. 2007). H3.3 is deposited independent of replication and marks sites of active chromatin (Ahmad and Henikoff 2002). Tagged H3.3 has been used for quantitative recovery of chromatin (Mito et al. 2005). To profile H3.3 in salt fractions, we used S2 cells expressing biotin-tagged H3.3 induced under control of the Drosophila metallothionein promoter (Mito et al. 2005) and used Streptavidin–Sepharose to pull down nucleosomes from successive 80 mM and 600 mM salt-extracted fractions. We also included an H3.3 construct that is missing the N-terminal tail (H3.3ΔN), which our previous work has shown cannot be deposited into chromatin by replication-coupled assembly (Ahmad and Henikoff 2002; Schwartz and Ahmad 2005).
Examination of a representative gene-rich region for its H3.3 distribution in 80 mM salt fractions reveals numerous peaks of H3.3 (Fig. 2, red tracks), only some of which correspond to peaks of low-salt-extracted nucleosomes from which they derive. Approximately similar results were obtained for H3.3ΔN (r = 0.71), indicating that the low-salt fraction contains in part H3.3 nucleosomes that are assembled by a replication-independent pathway. In contrast, the 600 mM fraction of H3.3 shows broad regions of enrichment that closely correspond to active genes (Fig. 2, bottom track; Fig. 4, A,I), consistent with our previous findings using lower density tiling arrays (Mito et al. 2005).
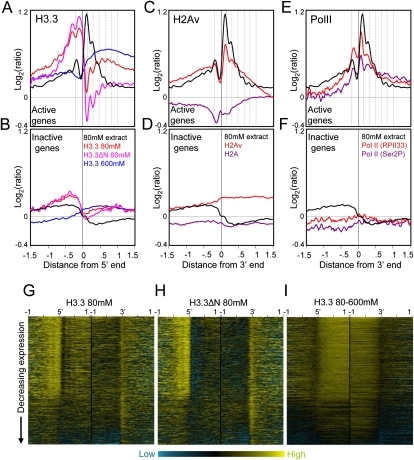
Figure 4.
H3.3-enriched low-salt soluble nucleosomes occupy active promoters. (A) Ends analyses of H3.3 obtained from salt fractions as indicated for active genes. H3.3 peaks are out-of-phase with low-salt soluble chromatin peaks (dotted vertical lines). (B) same as A except for inactive genes. To separate active and inactive genes, loci were rank-ordered based on expression levels in Supplemental Table S1. For simplicity, genes were divided into active and inactive fractions based on the visible discontinuity in salt solubility and H3.3 heat maps after rank-ordering by expression (Fig. 3G). For comparison, profiles are shown for 80 mM salt-extraction averaged from eight experiments (black curve). (C) Ends analyses of H2Av and H2A obtained by pulldown of biotin-tagged variants after a low-salt/EDTA/shearing extraction procedure (Jin and Felsenfeld 2007), showing active gene profiles. (D) Same as C except for inactive genes. (E) Ends analyses of Pol II ChIP-chip of S2 cell chromatin using RPII33 and Ser2P antibodies (Muse et al. 2007). Profiles for active genes are shown together with low-salt chromatin peaks (dotted lines), which align with but are slightly offset from Pol II peaks. (F) Same as E except for inactive genes. (G) Heat map display of H3.3/input for chromatin from an 80 mM extract. Contrast levels are equal for all stacks (Java TreeView setting of 2.0). (H) Same as G but for H3.3ΔN 80 mM. (I) Same as G but for the corresponding 80–600 mM extract.
Averaged chromatin profiles for these samples reveal that H3.3 in low-salt soluble chromatin is strongly enriched at promoters of active genes (Fig. 4A). Similar peaks are seen for H3.3 and H3.3ΔN profiles in promoters, but the overall abundance of H3.3ΔN is lower than that of H3.3 in active gene bodies (e.g., Fig. 4, cf. G and H). This difference appears to be linked to transcription, because inactive genes show no difference in H3.3 and H3.3ΔN (Fig. 4B), suggesting that removal of the N-terminal tail impedes histone replacement during transcriptional elongation.
Interestingly, these profiles are not simply subsets of the 80 mM fraction peaks. Instead, H3.3 and H3.3ΔN from 80 mM–extracted chromatin display average peaks that are out of phase with average 80 mM–extract peaks (Fig. 4A). Out-of-phase profiles are most conspicuous downstream of the TSS, where the sharp peak of low-salt soluble nucleosomes centered at +100 bp corresponds to a sharp dip in H3.3 occupancy, following by detectably out-of-phase profiles downstream (Fig. 4A). In addition, low-salt soluble H3.3 and H3.3ΔN profiles show a small dip at −150 bp that aligns with the upstream peak in the low-salt soluble fraction itself. At genic 3′ ends of both active and inactive genes, there is also a sharp peak in both H3.3 and H3.3ΔN profiles that aligns with a dip in low-salt-extracted chromatin (Supplemental Fig. S4). Thus it appears that the process that results in phased low-salt soluble nucleosomes causes partial depletion of H3.3.
Low-salt soluble chromatin is enriched in H2Av
One possible explanation for local depletion of H3.3 in low-salt soluble chromatin is suggested by the recent report that bulk H3.3 nucleosomes lose dimers, especially those containing H2A.Z (Jin and Felsenfeld 2007). Drosophila encodes a single H2A variant, H2Av, which is both an H2A.Z and an H2A.X (van Daal and Elgin 1992). In yeast, H2A.Z is enriched in unstable nucleosomes at promoters of active genes (Dion et al. 2007), and so we would expect it to be enriched in low-salt soluble chromatin. H2A.Z appears to be a component of active chromatin, and in Arabidopsis, H2A.Z excludes DNA methylation (Santisteban et al. 2000; Farris et al. 2005; Zilberman et al. 2008). Genome-wide profiling of human, yeast, Drosophila, and Arabidopsis H2A.Z yields average patterns downstream from TSSs that resemble patterns of low-salt soluble nucleosomes (Raisner et al. 2005; Barski et al. 2007; Mavrich et al. 2008; Zilberman et al. 2008). To confirm that H2Av is similarly enriched in S2 cell chromatin, we profiled biotin-tagged H2Av and H2A genome-wide using the low-salt/EDTA/shearing extraction method (Jin and Felsenfeld 2007). Indeed, the averaged H2Av profile closely resembles that for 80 mM salt extraction over promoters, with H2A showing partial depletion where H2Av is strongly enriched (Fig. 4C,D). The +1 nucleosome is especially striking, in displaying both strong H2Av enrichment in bulk nucleosomes and strong depletion of H3.3 from the 80 mM salt fraction. This correspondence suggests that nucleosomes containing both H3.3 and H2Av in the +1 position are preferentially lost, consistent with evidence that bulk H3.3+H2Av nucleosomes are relatively unstable (Jin and Felsenfeld 2007).
Correspondences between salt fractions and RNA polymerase II
We wondered whether the out-of-phase peaks seen in comparisons of H3.3/input from 80 mM–extracted nuclei versus 80 mM–extract/whole nuclei result from nucleosome loss associated with transcription. Perhaps chromatin that transiently lacks nucleosomes is instead occupied by nonhistone protein complexes, such as RNA polymerase II (Pol II). To test this possibility, we used recently published ChIP-chip data for Pol II from S2 cells (Muse et al. 2007). When we computed average Pol II profiles from these data, we observed a striking similarity with the low-salt soluble chromatin profile (Fig. 4E,F). Relative abundances are similar across genes, and peaks of low-salt soluble chromatin correspond to peaks of Pol II, slightly offset such that Pol II peaks are closer to the TSS than phased nucleosome peaks. These close correspondences suggest that low-salt soluble nucleosomes in genes are generated by active transcription or by transcription-coupled remodeling.
We also characterized insoluble chromatin that had been extensively extracted with 600 mM NaCl, and were surprised to find that this fraction displays a distinct profile (Fig. 2, pellet track). When these data were subjected to 5′ and 3′ ends analysis, we observed a peak over the TSS and just downstream of active genes followed by a gradual decrease over gene bodies (Fig. 3D,I). Upstream of TSSs there is a broad shoulder. The insoluble chromatin profile approximately resembles profiles for low-salt soluble nucleosomes and Pol II but is broader (Fig. 2, green tracks; Supplemental Fig. S5). Engaged Pol II has been reported to be highly insoluble when extracted under standard conditions (Kimura et al. 1999; Otero et al. 1999). Thus, the profile of our pellet fraction is consistent with the idea that it is insoluble because of its association with Pol II. To confirm that Pol II is enriched in the pellet fraction, we performed Western blot analysis using an antibody to the RPII33 (also known as RPB3) Pol II subunit. As expected, the pellet fraction contains levels of RPII33 that are high relative to the levels of histones (Fig. 1D).
We also observed enrichment of pellet chromatin at regions that do not appear to correspond to Pol II, suggesting that other chromatin-associated complexes have rendered regions of chromatin insoluble. ATP-dependent chromatin remodelers are abundant in cells, and to test the possibility that these render chromatin insoluble, we probed Western blots of salt fractions with an antibody to the ISWI remodeler (Tsukiyama et al. 1995). Very little ISWI was recovered in the successive salt fractions, and a substantial fraction remained behind in the insoluble pellet (Fig. 1E). Therefore, salt extraction removes nearly all of the histones but leaves behind high-molecular-weight chromatin proteins that likely render the remaining DNA insoluble.
Low-salt extracted and insoluble nucleosomes are enriched regionally within transposons
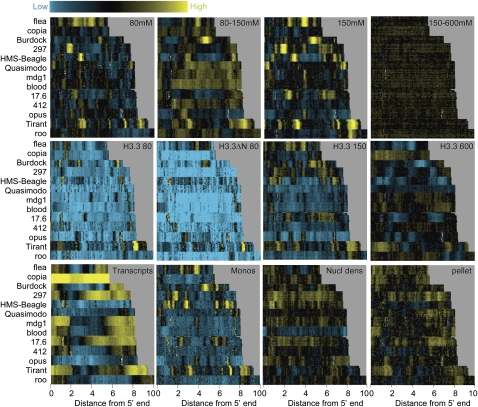
Our tiling array design covered all regions of the euchromatic genome, including repeated sequences. This allowed us to ask whether repeat families are packaged by nucleosomes that differ in solubility. Because there are hundreds of known repeat families in the Drosophila genome, we limited our survey to the most regular and abundant class of large dispersed repeats: LTR retrotransposon families with 10 or more annotated copies. For each family, we chose the longest representatives within a 5–10 kb range and generated heat maps with transposons aligned at their 5′ ends. We created a “stack” by displaying up to 10 representatives for each of the 13 families ordered by length (Fig. 5).
Figure 5.
Enrichment of low-salt soluble chromatin locally within transposons. Heat map stacks of 13 transposon families for salt fractions, H3.3, mononucleosomes, nucleosome density, and cDNAs, showing single representative samples. (Top row) Successive salt-extraction fractions (versus MNased nuclei) for 80 mM, 80–150 mM, 150 mM, and 150–600 mM extracts. (Second row) H3.3/input levels for 80 mM, 80 mM H3.3ΔN, 150 mM, and 600 mM pulldowns. (Third row) cDNA/genomic DNA from poly(A)+ RNAs, gel-purified mononucleosomes/input bulk DNA, MNased nuclei/genomic DNA, and pellet/nuclei after a 600 mM wash. Vertical gray streaks, most noticeable in the 600 mM stack, result from alignment gaps. Contrast levels are equal for all stacks (Java TreeView setting of 3.0).
The transposon stack for the 80 mM and 150 mM fractions showed sharp patterns of high and low intensities, whereas the 150–600 mM fraction was essentially featureless. Pellet chromatin displayed distinct, but less prominent, patterns over transposons. The sharply defined features seen for active genes and different classes of LTR retrotransposons indicate that locally enhanced nucleosome solubility is a general feature of Drosophila chromosomes. However, we observed no relationship between low-salt solubility and the overall level of transcription of a transposon family. H3.3 and H3.3ΔN from 80 mM salt extracts are strongly depleted in transposon stacks relative to the genome as a whole, consistent with the idea that most LTR transposons are silent copies. Even copia, which is expressed at a very high level in S2 cells, appears almost completely devoid of H3.3 in the 80 mM fraction. The exceptional lack of correspondence between transcription of these transposon families and their average chromatin state suggests that transcripts are produced by a small number of representatives among a large number of copies in the genome.
Low-salt extracted and insoluble nucleosomes are enriched at epigenetic regulatory sites
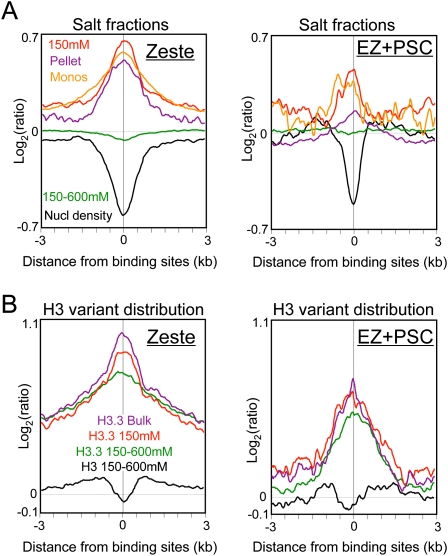
We have previously shown that nucleosome occupancy is decreased and H3.3 is enriched at binding sites for both Zeste, a trithorax-group (trxG) protein, and two Polycomb-group (PcG) proteins, EZ and PSC, genome-wide (Mito et al. 2007). To test whether nucleosome depletion at these sites also corresponds to particular chromatin fractions, we aligned averaged profiles around sites of Zeste and EZ+PSC binding. We used factor-binding data from early fly embryos (Moses et al. 2006; Li et al. 2008) and averaged intervals in either direction out to 3 kb. As previously observed, Zeste and EZ+PSC sites are depleted of nucleosomes overall (Fig. 6A, black curves) and are enriched in H3.3 relative to H3 (Fig. 6B). For both Zeste and EZ+PSC, we observed marked enrichment of 150 mM salt-extracted and insoluble nucleosomes, and slight depletion of the 600 mM fraction (Fig. 6A). Therefore, like promoters of active genes, binding sites for both trxG and PcG proteins are enriched in both the low-salt soluble and insoluble subsets of nucleosomes. The association of large Polycomb-containing complexes at these sites presumably accounts for the insolubility of these binding sites (Schwartz et al. 2005).
Figure 6.
Enrichment of low-salt soluble chromatin at epigenetic regulatory sites. (A) Genome-wide profiles of salt-fractionated soluble and insoluble nucleosomes for 1428 aligned Zeste (trxG) binding sites (left) and for 197 aligned Enhancer-of-Zeste and Primordial-sex-combs (EZ + PSC = PcG) binding sites (right). (B) Biotin-H3.3 and biotin-H3 variant distributions from bulk-extracted and salt-extracted samples.
Discussion
We have shown that a simple salt-fractionation procedure can separate chromatin into fractions that display radically different genome-wide landscapes. Extraction of chromatin from intact nuclei under low ionic conditions yields soluble nucleosomes that are conspicuously enriched around promoters of active genes, 3′ ends, and trxG and PcG protein binding sites genome-wide. Many low-salt soluble chromatin peaks are also found within and between genes, suggesting that some of them represent previously unannotated features of the epigenome, perhaps involved in nuclear architecture. These more soluble nucleosomes are mostly mononucleosomal but nevertheless comprise a different subset of chromatin than the subset that is purified by gel-extraction of mononucleosome-sized fragments, suggesting that solubility, and not just preferential cleavage by MNase, underlies fractionation. Low-salt soluble chromatin likely represents transcriptionally disrupted H3.3+H2Av nucleosomes (Jin and Felsenfeld 2007), because H3.3 is most strongly depleted at the +1 position, where H2A.Z and Pol II are most strongly enriched. By recovering the remaining soluble chromatin using 600 mM salt, we were able to profile the large majority of chromatin in cells. Our findings confirm on a genome-wide scale classical salt-fractionation studies in which the most soluble nucleosomes are “active” chromatin (Annunziato et al. 1981; Davie and Saunders 1981; Rocha et al. 1984; Rose and Garrard 1984). But in addition, we demonstrate that the most insoluble chromatin is enriched in a different active fraction, extending previous work showing that components of active chromatin are enriched in the insoluble residue after EDTA and salt extraction (Rose and Garrard 1984; Delcuve and Davie 1989; Hendzel et al. 1994). Our findings are likely to be applicable generally to chromatin studies, insofar as salt fractionation has been applied to a variety of cell types in a range of eukaryotes (Cooper et al. 1981; Hebbes and Allen 2000; Sun et al. 2007).
Salt fractionation for profiling chromatin landscapes provides a novel tool for identification of regulatory sites genome-wide. Although salt fractionation was first described 30 years ago (Sanders 1978) and has been used to characterize global chromatin accessibility (Weil et al. 2004), it had not been previously applied to regulatory site identification. In this regard, our adaptation of a classical method for chromatin characterization parallels the introduction of genome-scale methods for DNase I hypersensitive site (HS) mapping (Crawford et al. 2006; Sabo et al. 2006). The simplicity of salt fractionation—combined with characterization of nearly all chromatin in a parallel step—is especially attractive for epigenome mapping and regulatory site identification in general. Unlike methods for mapping of chromatin features that yield only DNA, such as DNase I HS mapping and formaldehyde-assisted isolation of regulatory elements (FAIRE), the recovery of native chromatin by salt fractionation allows histone epitopes to be readily profiled (e.g., Fig. 4A). Salt fractionation also provides single-nucleosome mapping resolution for active chromatin, and results can be interpreted directly in terms of nucleosome properties, rather than via a surrogate measure such as nuclease accessibility (HS mapping) or solubility after cross-linking and sonication (FAIRE). Accordingly, we have adopted our chromatin purification protocol for histone variant profiling in the D. melanogaster and Caenorhabditis elegans modENCODE project (The ENCODE Project Consortium 2007).
Quantitative recovery of chromatin using our procedure stands in contrast to current widely used ChIP-based methods (Orlando 2000; Hanlon and Lieb 2004), where extraction and processing can lead to incomplete recovery of chromatin, resulting in profiles that might not accurately represent a protein's in vivo distribution. The fact that we could obtain very different profiles from the same nuclear preparation raises questions as to whether current methods extract only the most soluble chromatin. For example, the FAIRE fractionation method for profiling active chromatin uses an extraction method that is similar to extractions used for X-ChIP and is based on the partitioning of soluble formaldehyde cross-linked material into the aqueous phase during phenol extraction, separating it from bulk chromatin (Giresi et al. 2007). Whether or not current ChIP-based analyses are complicated by incomplete recovery and fractionation is not clear; however, a simple control is to profile the DNA recovered from chromatin used for ChIP input versus DNA from the whole starting nuclear material. If the dynamic range of the profile is low relative to that of the ChIPed chromatin (e.g., Fig. 2, 150–600 mM track), then it seems fair to assume that the chromatin being profiled represents the bulk of the genome.
It is intriguing that transcriptionally active chromatin was isolated at both extremes of solubility, which indicates that these two fractions are distinct, despite being derived from the same or overlapping regions of the genome. These distinct fractions are unlikely to originate from genetically or epigenetically distinct subpopulations of cells maintained over large numbers of cell generations, because we have obtained very similar results using different S2-derived cell lines (Supplemental Fig. S6). Rather, changes in proteins complexed with DNA during transcription can account for this partitioning. Polymerases are thought to transiently uncoil nucleosomes and reassemble them as they pass through (Studitsky et al. 1994). Disrupted nucleosomes have been detected in the wake of DNA polymerase during replication and are more accessible to cleavage on both sides by MNase to release mononucleosomes (Annunziato et al. 1981). Transient disruption by Pol II might also increase the solubility of these nucleosomes, perhaps because of loss of nonhistone chromatin proteins. The close match of profiles for low-salt soluble chromatin with those for engaged Pol II over active genes (Fig. 4E) suggests that this fraction captures transient intermediates generated during polymerase transit. This model could explain the relative depletion of H3.3 at phased peaks of low-salt soluble chromatin around active gene promoters (Fig. 4A) as partially disrupted nucleosomal intermediates that have not completed transcription-coupled histone replacement.
Where Pol II engages the template it renders chromatin almost completely insoluble (Kimura et al. 1999; Otero et al. 1999), so that oligonucleosomes attached to engaged RNA polymerases would end up in the pellet. Not all low-salt soluble and insoluble nucleosomes appear to correspond to Pol II–engaged regions, but it is likely that engagement with the many abundant ATP-dependent nucleosome remodelers, such as ISWI (Fig. 1E), will also reduce solubility, resulting in preferential release of mononucleosomes and short oligonucleosomes by MNase digestion. In this way, active processes of transcription and remodeling at a site in the genome will produce nucleosomes that (1) have recently been disrupted and released and so are low-salt soluble; (2) are closely linked to engaged RNA polymerases, and so are insoluble upon extraction; and (3) are unaffected and so have intermediate solubility. Thus, successive chromatin salt fractions derived from a single cell population would provide snapshots of dynamic processes that act on the epigenome (Henikoff 2008).
Methods
Cell culture
Histones H3.3 and H3.3ΔN were tagged at their N termini with biotin ligase recognition peptide and expressed individually with Escherichia coli biotin ligase during log-phase growth in D. melanogaster S2 cells as previously described (Mito et al. 2005). Western analysis and measurements of yields of Streptavidin-bound chromatin confirmed that the levels of biotinylated histones were low compared with those of endogenous histones.
Chromatin preparation
Nuclei were prepared as previously described from D. melanogaster S2 cells (McKittrick et al. 2004), and chromatin was extracted by modification of our previous procedure (Mito et al. 2005). Briefly, cells were suspended in TM2 (10 mM Tris at pH 7.4, 2 mM MgCl2, 0.5 mM phenylmethanesulphonylfluoride [PMSF]) with gentle vortexing to a concentration of 1 × 108/mL to 2 × 108/mL, held 1 min in ice water, and 10% NP-40 was added with gentle vortexing to 1.5%. After 2–5 min on ice, crude nuclei were pelleted at 800 rpm in a Sorvall SS-34 rotor at 4°C, the supernatant was removed carefully, and the pellet was transferred with cold TM2 to a 2 mL microfuge tube. Nuclei were spun in an Eppendorf microcentrifuge at 1000 rpm, and the pellet was resuspended in TM2 to an estimated concentration of 0.5 × 109/mL to 1 × 109/mL for MNase digestion. CaCl2 was added to 1 mM; the tubes were warmed to 37°C for 1 min and then digested by addition of MNase (Sigma 1.6 U/mL) for 10 min. Tubes were transferred to ice, and reactions were stopped by addition of 0.2 M EGTA (Na+) to 2 mM. Nuclei were pelleted for 10 min at 2000 rpm 4°C, washed in TM2, and resuspended in 0.7 mL cold 80 mM or (150 mM) buffer: 70 mM (or 140 mM) NaCl, 10 mM Tris (pH 7.4), 2 mM MgCl2, 2 mM EGTA, 0.1% Triton X-100, 0.5 mM PMSF (pH 7.5). After 1–2 h rotation on a nutator at 4°C, nuclei were pelleted for 10 min at 2000 rpm, and the pellet was resuspended gently in 0.7 mL cold 600 mM NaCl, 10 mM Tris (pH 7.4), 2 mM MgCl2, 2 mM EGTA, 0.1% Triton X-100, and 0.5 mM PMSF (pH 7.5). Supernatants from each extraction were clarified by spinning for 2 min at 13,200 rpm and held on ice to be used for pulldowns with aliquots taken for DNA and protein analyses (80 mM, 150 mM, and 80–150 mM extractions). The 600 mM resuspended pellet was rotated overnight at 4°C, pelleted for 10 min at 2000 rpm, and the supernatant was clarified and held on ice (80–600 mM and 150–600 mM extractions).
For some experiments, MNase treatment was stopped by addition of EDTA (Na+ at pH 8) to 10 mM, and after pelleting and saving the supernatant, nuclei were suspended in 0.6 mL 10 mM Tris, 0.25 mM EDTA (pH7.4). The suspension was passed five times through a 26-gauge needle and combined for profiling with the supernatant (EDTA extraction) (Jin and Felsenfeld 2007). For experiments described in Figure 3E, DNAs extracted from samples prepared in this way were electrophoresed, the mononucleosome band was excised, and the DNA was extracted using the Qiagen Qiaquick kit.
Affinity purification was performed by addition of Streptavidin–Sepharose and rotating for 2 h at 4°C as described (Mito et al. 2005), except that the same buffer (without Triton) was used for washing the resin, which was brought up in 0.2 mL 150 mM buffer (without Triton) and (usually) frozen at −20°C prior to extraction. Aliquots of MNase-digested nuclei and clarified soluble nucleosomes were also frozen prior to extraction.
Prior to DNA isolation, aliquots of chromatin samples were removed and mixed with SDS-PAGE load buffer for electrophoresis and Western blot analyses using standard protocols (Ausubel et al. 1994). ISWI and RPII33 antibodies were kindly provided by John Tamkun (University of California, Santa Cruz, CA) and Karen Adelman (National Institute of Environmental Health Sciences, Research Triangle Park, NC), respectively. DNA was purified from chromatin fractions and pulldown samples as described (Mito et al. 2005). Briefly, samples were RNase-treated, deproteinized with SDS and proteinase K, and extracted using a mixture of phenol/chloroform/isoamyl alcohol (25:24:1) followed by chloroform extraction, and DNA was precipitated with 2 vol ethanol. Overall yields were ∼10%–15% for 150 mM, ∼5% for 80 mM, and ∼70%–85% for 600 mM salt extractions, and the pellet contained ∼3%–15% of the total.
Microarray-based profiling
DNA samples were labeled with Cy3 or Cy5 by random priming following the NimbleGen protocol as described (Mito et al. 2005) with the following modifications: 50 μL strand-displacement reactions were performed at 37°C overnight and stopped by addition of 5 μL 0.5M EDTA, transferred to a 1.5-mL tube, mixed with 5.7 μL 5M NaCl, and precipitated by addition of 60 μL isopropanol. After dissolving in water and measuring A260-levels, equal amounts of Cy3- and Cy5-labeled samples were mixed, and volumes were dried or reduced to ∼12 μL and delivered to Hutchinson Center Genomics Shared Resource for hybridization to NimbleGen microarrays. Starting with 0.2–3 μg of DNA, this procedure generally yielded >30 μg of Cy-labeled DNA, except it was very inefficient for mononucleosomal DNA. Therefore, we relabeled 1–3 μg aliquots of the labeled DNA in a second 50 μL reaction, and this consistently yielded >60 μg.
Profiling was performed on single custom designed 2.1 million feature isothermal microarrays (GEO GPL6888) purchased from NimbleGen, Inc. and was hybridized and scanned by the Fred Hutchinson Center Genomics Facility using NimbleGen protocols. Data are available from GEO (GSE13217).
Data analysis
Ends analysis was performed essentially as previously described (Mito et al. 2007) except that we used 25-bp intervals for averaging to take advantage of the denser tiling (averaging ∼65 bp). K-means clustering was performed using Cluster 3.0 for OSX (Eisen et al. 1998), and heat maps were created using Java TreeView v. 1.1.0 using the default contrast level of 3.0 for all maps. Tracks were displayed using SignalMap (NimbleGen, Inc.).
Acknowledgments
We thank Yoshiko Mito, Takehito Furuyama, Roger Deal, Siew-Loon Ooi, and Steve Hahn for helpful discussions; Andy Marty and Jeff Delrow of the FHCRC Genomics Shared Resource for microarray processing and advice; Christine Codomo, Martin Riedel, and Terri Bryson for technical assistance; and John Tamkun and Karen Adelman for antibodies. This work was supported by the NIH-NHGRI modENCODE project (U01 HG004274).
Footnotes
[Supplemental material is available online at www.genome.org. The microarray data sets from this study have been submitted to GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE13217.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.087619.108.
References
- Ahmad K., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Annunziato A.T., Schindler R.K., Thomas C.A., Jr, Seale R.L. Dual nature of newly replicated chromatin. Evidence for nucleosomal and non-nucleosomal DNA at the site of native replication forks. J. Biol. Chem. 1981;256:11880–11886. [PubMed] [Google Scholar]
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Current protocols in molecular biology. Wiley; New York: 1994. [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Cooper E., Palmer R.J., Spaulding S.W. Effect of thyrotropin on 32P-labelled histones H1 and H3 in specific populations of nucleosomes in the thyroid. Nucleic Acids Res. 1981;9:3389–3401. doi: 10.1093/nar/9.14.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford G.E., Davis S., Scacheri P.C., Renaud G., Halawi M.J., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S. DNase-chip: A high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie J.R., Saunders C.A. Chemical composition of nucleosomes among domains of calf thymus chromatin differing in micrococcal nuclease accessibility and solubility properties. J. Biol. Chem. 1981;256:12574–12580. [PubMed] [Google Scholar]
- Delcuve G.P., Davie J.R. Chromatin structure of erythroid-specific genes of immature and mature chicken erythrocytes. Biochem. J. 1989;263:179–186. doi: 10.1042/bj2630179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion M., Kaplan T., Friedman N., Rando O.J. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris S.D., Rubio E.D., Moon J.J., Gombert W.M., Nelson B.H., Krumm A. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J. Biol. Chem. 2005;280:25298–25303. doi: 10.1074/jbc.M501784200. [DOI] [PubMed] [Google Scholar]
- Giresi P.G., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon S.E., Lieb J.D. Progress and challenges in profiling the dynamics of chromatin and transcription factor binding with DNA microarrays. Curr. Opin. Genet. Dev. 2004;14:697–705. doi: 10.1016/j.gde.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Allen S.C. Multiple histone acetyltransferases are associated with a chicken erythrocyte chromatin fraction enriched in active genes. J. Biol. Chem. 2000;275:31347–31352. doi: 10.1074/jbc.M004830200. [DOI] [PubMed] [Google Scholar]
- Hendzel M.J., Sun J.M., Chen H.Y., Rattner J.B., Davie J.R. Histone acetyltransferase is associated with the nuclear matrix. J. Biol. Chem. 1994;269:22894–22901. [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Jin C., Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes & Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Tao Y., Roeder R.G., Cook P.R. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: Little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol. Cell. Biol. 1999;19:5383–5392. doi: 10.1128/mcb.19.8.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Li X.Y., MacArthur S., Bourgon R., Nix D., Pollard D.A., Iyer V.N., Hechmer A., Simirenko L., Stapleton M., Luengo Hendriks C.L., et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich T.N., Jiang C., Ioshikhes I.P., Li X., Venters B.J., Zanton S.J., Tomsho L.P., Qi J., Glaser R.L., Schuster S.C., et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E., Gafken P.R., Ahmad K., Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z., Schwartz Y.B., Li X.Y., Kahn T.G., Gause M., MacArthur S., Fay J.C., Eisen M.B., Pirrotta V., Biggin M.D., et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y., Henikoff J., Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Mito Y., Henikoff J., Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Moses A.M., Pollard D.A., Nix D.A., Iyer V.N., Li X.Y., Biggin M.D., Eisen M.B. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput. Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill L.P., Turner B.M. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31:76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- Otero G., Fellows J., Li Y., de Bizemont T., Dirac A.M., Gustafsson C.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Perry M., Chalkley R. The effect of histone hyperacetylation on the nuclease sensitivity and the solubility of chromatin. J. Biol. Chem. 1981;256:3313–3318. [PubMed] [Google Scholar]
- Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E., Davie J.R., van Holde K.E., Weintraub H. Differential salt fractionation of active and inactive genomic domains in chicken erythrocyte. J. Biol. Chem. 1984;259:8558–8563. [PubMed] [Google Scholar]
- Rose S.M., Garrard W.T. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J. Biol. Chem. 1984;259:8534–8544. [PubMed] [Google Scholar]
- Sabo P.J., Kuehn M.S., Thurman R., Johnson B.E., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- Sanders M.M. Fractionation of nucleosomes by salt elution from micrococcal nuclease-digested nuclei. J. Cell Biol. 1978;79:97–109. doi: 10.1083/jcb.79.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban M.S., Kalashnikova T., Smith M.M. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- Schwartz B.E., Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes & Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y.B., Kahn T.G., Pirrotta V. Characteristic low density and shear sensitivity of cross-linked chromatin containing polycomb complexes. Mol. Cell. Biol. 2005;25:432–439. doi: 10.1128/MCB.25.1.432-439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studitsky V.M., Clark D.J., Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- Sun J.M., Chen H.Y., Espino P.S., Davie J.R. Phosphorylated serine 28 of histone H3 is associated with destabilized nucleosomes in transcribed chromatin. Nucleic Acids Res. 2007;35:6640–6647. doi: 10.1093/nar/gkm737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T., Daniel C., Tamkun J., Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- van Daal A., Elgin S.C. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell. 1992;3:593–602. doi: 10.1091/mbc.3.6.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., Delrow J., Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- Weil M.R., Widlak P., Minna J.D., Garner H.R. Global survey of chromatin accessibility using DNA microarrays. Genome Res. 2004;14:1374–1381. doi: 10.1101/gr.1396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Coleman-Derr D., Ballinger T., Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]