Abstract
MicroRNA-based RNA interference is commonly used to produce loss-of-function phenotypes in mammalian systems, but is used only sparingly in invertebrates such as C. elegans and D. melanogaster. Here, we evaluate this method in transgenic strains of D. melanogaster and cultured S2 cells. High throughput-ready expression vectors were developed that permit rapid cloning of synthetic hairpin RNAs. As proof of concept, this method was used for the efficient silencing of dpp gene activity in the adult wing, and the analysis of the general RNA Polymerase II (Pol II) elongation factor, Nelf-E.
Introduction
Double-stranded RNAs (dsRNAs) are commonly used for gene silencing in Drosophila. However, these dsRNAs are typically quite long, >300-bp, and thereby produce dozens of unique ~21-nucleotide (nt) small interfering RNAs (siRNAs) (Elbashir et al., 2001). Sequence overlap of these RNAs with other regions of the genome, along with the unpredictable nature of the exact siRNAs derived from any given dsRNA, raises the likelihood of nonspecific, “off-target” phenotypes (Jackson and Linsley, 2004). Indeed, at least 40% of the dsRNAs in a widely used library might produce off-target effects in cultured Drosophila cell lines (Kulkarni et al., 2006). Nonetheless, considerable resources have been expended on this method for whole-genome assays (Dietzl et al., 2007).
siRNAs, defined as exogenous 21-nt silencing RNAs, can be expressed for targeted gene silencing assays through manipulation of the endogenous microRNA (miRNA) biogenesis pathway (Bartel, 2004). In this system, a customized siRNA sequence is swapped into a naturally occurring, imperfectly paired, ~70-90-base-pair (bp) stem-loop sequence (Chang et al., 2006). Cellular factors recognize the stem-loop as a miRNA precursor (pre-miRNA) and excise a specific ~21-nt sequence (the miRNA/siRNA) from one or both arms of the stem. Like exogenously delivered siRNAs, the resulting small RNA(s) is loaded into an effector complex (e.g. RISC) that can induce endonucleolytic cleavage of complementary RNAs (Hutvagner and Zamore, 2002; Miyoshi et al., 2005). The small RNA preferentially loaded into RISC from a given duplex RNA, e.g. a pre-miRNA or a transfected siRNA, is known as the mi/siRNA, while the less-incorporated strand is the star (mi/siRNA*) sequence. The incorporation of the mi/siRNA or mi/siRNA* into RISC depends, in part, on the local free energy of the duplex’s terminal base pairs (Khvorova et al., 2003; Schwarz et al., 2003). This property provides a means for controlling the exact sequence of an expressed siRNA, thereby diminishing nonspecific, off-target phenotypes. While several studies demonstrated the effectiveness of miRNA-based RNAi in mammalian systems, there have been just a few such studies in Drosophila (Chen et al., 2007; Huh et al., 2007).
Here, we present a versatile, miRNA-based system for the conditional silencing of gene activity in Drosophila. These newly developed expression vectors recapitulate the exact structure of the Drosophila pre-miRNA-1(miR-1) stem loop sequence, differing only by unique cloning sites within the stem itself. Synthetic oligonucleotides (oligos) are directly inserted into the pre-miR scaffold, thereby circumventing multiple PCR amplification and cloning steps. A subcloning vector was also developed for creating tandem hairpins, thereby permitting the expression of multiple siRNAs directed against different regions of a specific target mRNA. We have created separate one-step transgenesis vectors that are compatible with either Δ2,3 P-element transposase or phiC31 integrase (targeted integration)(Adams and Sekelsky, 2002; Groth et al., 2004), A web-based algorithm provides the exact design of the synthetic oligos for insertion into the pre-miR-1 scaffold. Both S2 tissue culture assays and targeted expression in transgenic larvae and adults were used to establish the effectiveness and specificity of the method. We discuss the results of targeted disruption of dpp and Nelf-E in wing imaginal disks, and compare these phenotypes with those produced by dsRNAs.
Material and Methods
Vector design
The original shmiR vector used in this study is based on the D. melanogaster pre-miR-1. A second-generation vector, pHB, includes ectopic cloning sites at the base of the pre-miR-1 sequence. Both types of vectors were constructed as mono or tri-cistronic synthetic genes by Celtek Bioscience (Nashville, TN), cloned into the KpnI and SacI restriction sites of pBluescript SK(+), and subsequently cloned into S2 cell expression or P-element transgenesis vectors as single or double hairpins by using appropriate restriction sites (see Supplemental File 1). Hairpins were inserted into pHB by directional ligation into the HindIII-BamHI cloning sites at the base of the pre-miR stem-loop.
The pNE2 vector was constructed in a modified pUAST backbone (Brand and Perrimon, 1993). EcoRI and XbaI cloning sites were removed by Klenow-mediated end fill. A mini-gene constructed as two annealed Ultramers (IDT) was inserted into this modified pUAST vector as a BglII-XhoI fragment. Hairpins were inserted into pNE2 by directional ligation into the NheI-EcoRI cloning sites at the base of the pre-miR stem-loop. pNE3 was constructed by end-filling a KpnI digested pNE2 vector, followed by linker-ligation of a BglII-KpnI-XbaI adaptor into the BglII-XbaI cloning site. This modification allows for directional insertion of a shmiR from pHB into pNE3, creating a tandem shmiR. pattB-NE3 is a variant of the phiC31 integrase-based pattB-UAST vector (Bischof et al., 2007). It was modified to include the same shmiR insertion and multi-cloning site as pNE3 (see Supplemental File 2 for complete restriction maps). A suggested method for creating two or more tandem shmirs in the pHB, pNE2, or pNE3 vectors is presented in Supplemental File 3.
Synthetic 71-base-pair oligonucleotides
71-nt oligos (Elim Biopharmaceuticals or IDT) were annealed at a final concentration of 50 µM in 1X annealing buffer (75 mM KCl, 20 mM Tris [pH 8.0]), boiled for ~2 minutes, and then cooled to room temperature for ~30 minutes. Annealed oligos were diluted 1:100 in 1X annealing buffer, and then ligated to ~500 ng of the appropriate linearized vector for ~15 minutes at room temperature using T4 DNA ligase (New England Biolabs). All inserts were sequenced and assessed for proper folding of the pre-miR stem loop via mFold (Mathews et al., 1999; Zuker, 2003).
siRNA sequence design
siRNAs directed against dpp recognize all dpp mRNA isoforms. siRNAs targeting mRNAs encoding pGL3-based Luciferase (Promega) and the mCherry red fluorescent protein were designed using the Dharmacon “siDesign” center (Shaner et al., 2004). Nelf-E siRNAs were designed using the Ambion “siRNA target finder” algorithm. The target-specificity of each 21-nt sequence was determined using parameters described by Khvorova and colleagues (Birmingham et al., 2007). Further, miR-1, like most miRNAs, begins with a 5′ uracil (Du and Zamore, 2005). In accordance with this generality, all siRNAs designed for use with these vectors contain a 5′-U. The complete sequences of all of the siRNAs used in this study are presented in Supplemental File 1.
Online Hairpin Design Tool
A Perl-CGI script was written to automate the construction of 21-mer siRNAs. This interactive web page provides the shmiR sequence required to produce a given 21-nt siRNA, using either the pHB or pNE expression vector. Information is also provided for the predicted secondary structure of the top (sense) strand hairpin produced with UNAfold (formerly mFold) (Dimitrov and Zuker, 2004). The structure of the resultant shmiR incorporates specific mismatched bases (at nts 2 and 11, as measured from the siRNA’s 5′-most base) to mimic the native D. melanogaster pre-miR-1 structure.
It has recently been found that siRNAs containing the same seed residues (nts 2–8) as those present within known miRNAs can induce systematic off-target phenotypes in cultured cell lines (Birmingham et al., 2006; Jackson et al., 2006). Therefore, each input 21-mer is cross-referenced against a database with all annotated D. melanogaster miRNA sequences, as of June 2008 (Griffiths-Jones, 2004). An alert will be displayed if the user-defined 21-mer contains a seed sequence overlapping a known miRNA.
S2 cell culture and transient transfection assays
S2 cells were plated at a density of 5×105 cells/well in 24-well plates. Synthetic shmiRs were inserted into a pAc5.1-B-based expression vector (Invitrogen), and used to transfect plated cells the following day with Effectene (Qiagen), as suggested by the manufacturer. Cell lysates were collected 48 hours after transfection, and then analyzed using a dual-Luciferase assay kit (Promega). For mCherry RNAi, the shmiR vector was transfected at a ~4-fold excess over the target vector, and fluorescence was assayed at 48 and 72 hrs following transfection. shmiRs directed against mCherry, as well as the mCherry ORF were cloned into pAc5.1, as above.
An ~500-bp dsRNA directed against the firefly Luciferase coding sequence was amplified by PCR using primers that contain a T7 RNA polymerase promoter sequence—primer sequences are published in Supplemental File 1. RNA was transcribed using MEGAscript T7 (Ambion) and processed for transfection as described (Forstemann et al., 2005).
AGO Immunoprecipitation
Cells were transfected with 450 ng shmiR-Luc 1HB and 50 ng mCherry, as described above. After 48 hrs, cells were collected by centrifugation, washed 3X with PBS [pH 7.4] (Gibco), and resuspended in 15 µl of lysis buffer per mL of cell culture media, as described (Forstemann et al., 2005). Cell debris was removed following centrifugation, producing lysate with a final protein concentration of ~2µg/µl). Monoclonal antibodies against Drosophila AGO1 or AGO2 were bound to sheep anti-mouse-Ig conjugated Dynabeads (Dynal) by rocking at 4°C overnight (Miyoshi et al., 2005). Antibody-conjugated beads were collected on a magnetized stage and washed 3X in PBS. 80 µl of lysate was added to the beads per IP, and this mixture was incubated on ice for 4 hours, with occasional shaking. Following IP, beads were collected on a magnetized stage, the supernatants were removed, and then washed 3X with cold lysis buffer. Beads were resuspended in 100 µl of lysis buffer and bound proteins were removed by boiling for 5 min. Both the supernatant and recovered solutions were added directly to 900 µl of Trizol (Invitrogen) and 200 µl of chloroform. RNA was extracted and precipitated with 1 V/V isopropanol and 20 µg glycogen (Roche).
Fly stocks
P-element-mediated transformations of yw strains with the pUAST-based shmiR vectors were done using traditional methods, either in-house or by BestGene Inc. (Chino Hills, CA). At least 3 independent lines were recovered and assayed for each UAS-shmiR construct, except for UAS-shmiR-dpp1. The following Bloomington Stock Center lines were used in this study: A9-GAL4 (stock #8716), sd-GAL4 (stock #8609), daughterless-GAL4 (stock #5460), dppshv, also known as dpps1 (stock #397), dppd5 (stock #2071), and UAS-Nelf-e-IR/dsRNA (stock #6788) (Enerly et al., 2002). UAS-dpp-dsRNA lines “47A.1” and “48A.1”—also identified as TR00047A.1 and TR00048A.1, respectively--were kindly provided by Norbert Perrimon (Ni et al., 2008),
Wing mounting
Adult flies were recovered 1–3 days after eclosion, and stored for 24 hours or more in absolute ethanol. Wings were dissected in ethanol, mounted immediately in CMCP-10 (Polysciences Inc.), and baked overnight in a desktop convection oven.
Larval RNA and Northern blot assays
GAL4 and UAS transgenic strains were mated as follows. Three males carrying a specific UAS shmiR construct were mated with three virgin females carrying a particular GAL4 expression vector. RNA was extracted from 10–20 wandering larvae or 0–1 day pupae in ~700 µl Trizol and 140 µl choloroform, followed by precipitation of the aqueous phase with 3V/V absolute ethanol and 20 µg glycogen. The final RNA concentration was determined using a Nanodrop ND-1000 spectrophotometer. ~15µg of total RNA per lane RNA was fractionated on 20% PAGE (SequaGel System, National Diagnostics) using a Hoefer Scientific SE250 Mighty Small Gel Electrophoresis unit in 0.5X TBE. RNA size markers (IDT) were designed to minimize 2° structures, the oligonucleotides were pooled, end labeled with γ-32P-ATP (MP Biochemicals) end-labeled using T4 Polynucleotide kinase (NEB), and unincorporated label was removed using an Illustra Microspin G-25 spin column (GE Healthcare). ~2 fmoles of the size markers were used for each Northern blot. RNA was transferred onto a Hybond N+ membrane (Amersham Biosciences), pre-wetted with 0.5X TBE in a TransBlot-SD semi-dry apparatus (Bio-Rad) at constant 20V for one hour. Following transfer, the membrane was UV crosslinked in a Stratalinker (Stratagene), pre-hybridized in ULTRAhyb Oligo (Ambion), and treated according to manufacturer's recommendations. The membrane and ~50 pmoles of γ-32P-ATP end-labeled deoxyoligonucleotides complementary to the appropriate small RNA were incubated at ~40°C in rolling 50-ml conical tubes overnight. Membranes were washed 3X 20 minutes in 2X SSC supplemented with 0.2% SDS, exposed on a Fuji phoshorimagery plate and analyzed on a Typhoon TRIO Phosphorimager (Amersham Biosciences). Probes were stripped by boiling in RNAse-free water supplemented with 0.1% SDS for 5 minutes, followed by immediate pre-hybridization. Probe and marker sequences are published in Supplemental File 1.
qPCR
~10 transgenic males carrying UAS-shmiR-dpp2HB or UAS-NLS-GFP were mated with ~10 transgenic females carrying daughterless-GAL4 at 25°C. 2nd and 3rd instar larvae were collected as they emerged from the fly food and total RNA was extracted from 4–8 individuals per genotype, as described earlier. cDNA was prepared using the Quantitect RT-PCR kit (Qiagen) from 1 µg of total RNA. qPCR was performed using Applied Biosystems SYBR Green PCR Master Mix and analyzed on an Applied Biosystems 7300 Real-Time PCR system. Sequences for the RP49 and dpp primers are published in Supplemental File 1.
Results
Design and rationale of miRNA-based vectors
Previous attempts to use miRNA-based siRNA expression vectors {termed short hairpin RNAs or (sh)miRs} relied on the manual design of an appropriate hairpin sequence, synthesis of ~80-bp oligos, followed by one or two rounds of PCR amplification, and digestion with a restriction enzyme to produce the final expression vector (Fig. 1A, left) (Chen et al., 2007). We sought to streamline this process for high-throughput applications by creating a one-step cloning system (Fig. 1A, right).
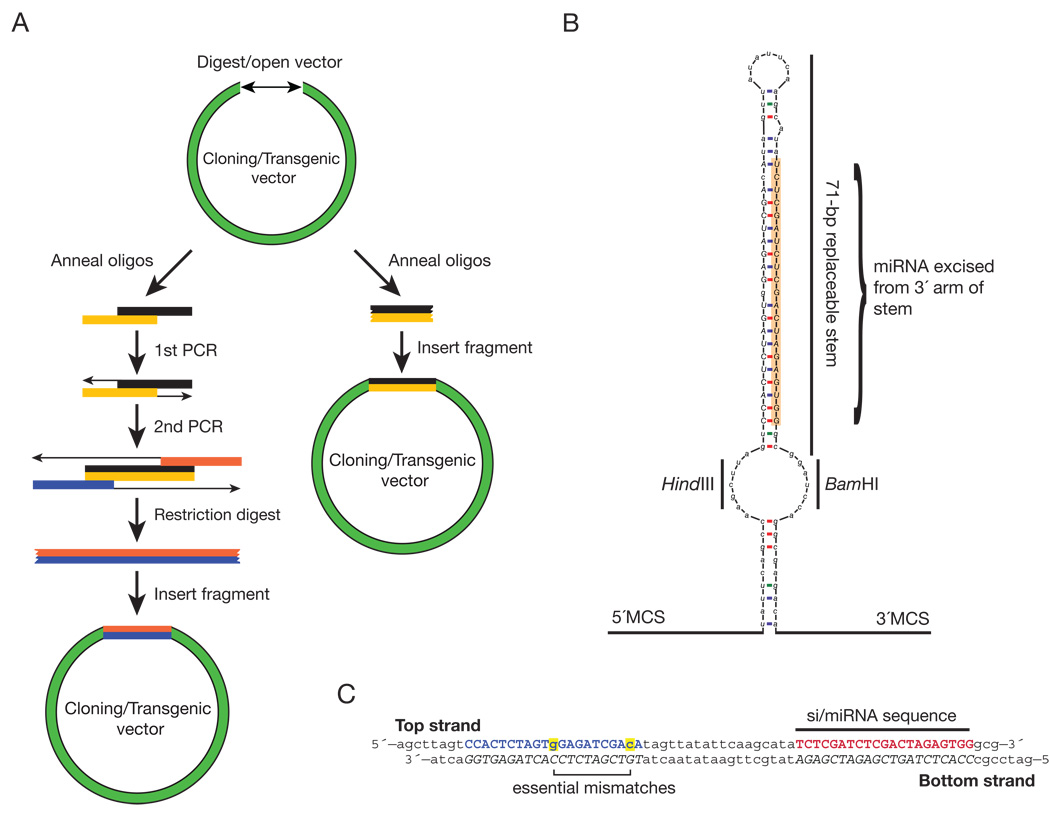
Fig. 1. Schematic of miRNA-based RNAi vector—shmiRs.
(A) Standard method for cloning miRNA-based RNAi vectors, left, is compared to miRNA-based vectors described in this study, right. (B) Structure of shmiR-dpp2 in pHB scaffold. Annealed 71-nt DNA oligos are inserted into the open-loop region with the specified restriction sequences, HindIII-BamHI in this example. Resultant siRNAs will be excised, like miR-1, from the 3′ arm of the stem-loop (orange highlighted region). This construct is flanked by 5′ and 3′ cloning sites (M.C.S.), enabling the creation of polycistronic hairpins. (C) shmiR-dpp2 knockdown oligos inserted into the HindIII-BamHI site of pHB. The siRNA sequence is highlighted in red, while the blue nucleotides represent the presumptive star sequence of the dpp2 siRNA. Highlighted in yellow are two specific mismatched bases necessary to maintain the endogenous pre-miR-1 stem-loop structure.
The newly developed expression vectors are based on the Drosophila melanogaster pre-miR-1 stem-loop sequence since it contains all of the features predicted to be required for efficient processing of a mature miRNA (Fig. 1B). This includes an open secondary structure at the penultimate 5′ position of the mature miR-1 sequence, which ensures efficient loading into RISC (Khvorova et al., 2003; Schwarz et al., 2003). A similar mismatch at position 11 is thought to direct the small RNA into the miRNA, not RNAi, biogenesis pathway (Tomari et al., 2007). In order to facilitate the rapid cloning of siRNAs into the miR-1 scaffold, specific mutations were introduced in the pre-miR-1 stem loop sequence that maintain the structure of the predicted pre-miR, including a fixed 7-nt unpaired region implicated in optimal miRNA processing (Han et al., 2006). For example, these mutations allow directional cloning of 71-bp synthetic oligos into HindIII-BamHI restriction sites (vector “pHB”, Fig. 1B and C). The stem-loop sequence is flanked by multiple cloning sites (M.C.S.), thereby facilitating the construction of polycistronic shmiRs, which are particularly potent in disrupting gene activity in mammalian systems (restriction maps of each vector are published as Supplemental File 2) (Sun et al., 2006). We have also prepared derivatives of this vector--pNE2, pNE3, and pattB-NE3—that also contain the pre-miR-1 scaffold for direct cloning of 71-nt synthetic oligos harboring unique NheI-EcoRI cloning sites into derivatives of the previously described pUAST or pattB-UAST P-element transformation vectors (Bischof et al., 2007; Brand and Perrimon, 1993). This approach facilitates the creation of transgenic strains for targeted expression assays using the GAL4-UAS expression system (Phelps and Brand, 1998). An accompanying web-tool provides optimal hairpin design for either the pHB or pNE-based vectors (http://flybuzz.berkeley.edu/cgi-bin/constructhairpin.cgi).
Gene silencing assays in cultured S2 cells
In order to assess the effectiveness of the miRNA-based RNAi system, shmiRs were designed that target the firefly Luciferase RNA (Fig. 2A). We placed shmiRs in a mono-or bi-cistronic configuration in either a “naked” vector or in the 3′ UTR of EGFP (hereafter referred to as GFP-shmiRX, with X identifying the target gene). The shmiR sequences were placed under the control of a strong Pol II promoter in order to mediate constitutive expression in transfected S2 cells. The GFP-shmiR expression vectors allow simple monitoring of transfection efficiency, and also permit FACS-based sorting of cells expressing shmiR constructs. Separate siRNAs derived from the pHB and pNE vectors, individually or linked as part of a bi-cistronic construct (connecting one pHB-shmiR to a separate pNE-shmiR), provided >95% knockdown of firefly Luciferase (Fig. 2A).
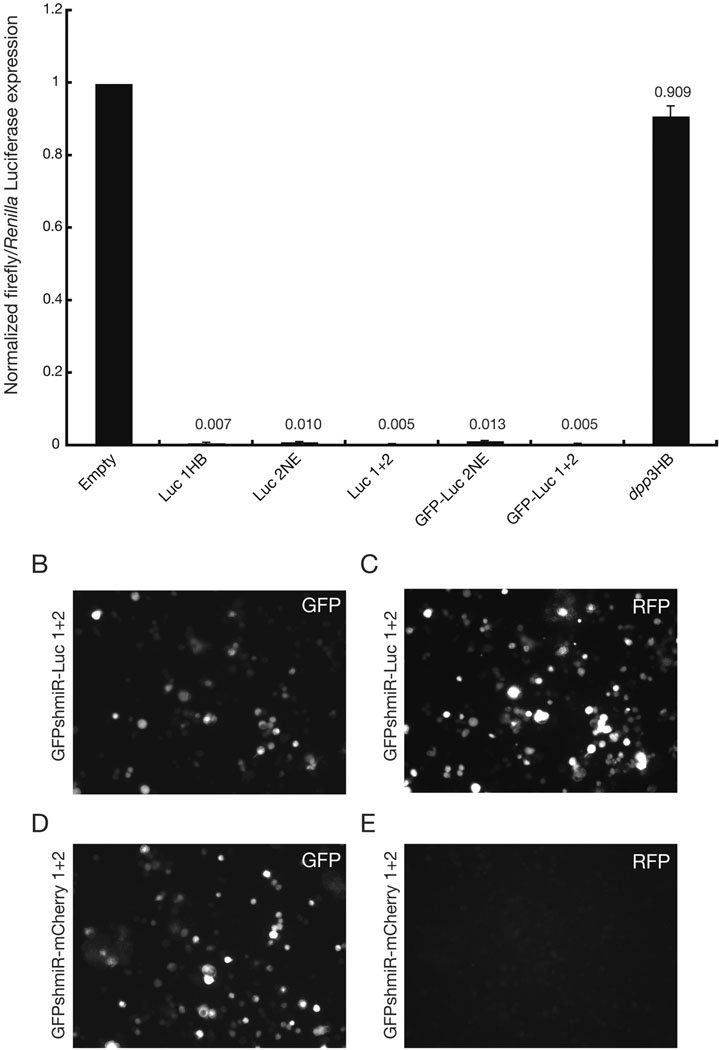
Fig. 2. pre-miR-1-based shmiRs effectively silence a reporter gene in vitro.
(A) Normalized ratios of firefly (target) to Renilla (control) Luciferase co-transfected with indicated constructs are presented, values are displayed above appropriate bars. Empty refers to an empty expression vector, and Luc 1HB and Luc 2NE refer to shmiRs targeting separate regions of the pGL3-derived firefly Luciferase gene, cloned using the pHB or pNE scaffold, respectively. Luc 1+2 is a bicistronic construct with shmiRs-Luc 1HB and Luc 2NE in tandem. GFP-shmiRs contain aforementioned shmiRs in the 3′ UTR of EGFP, and dpp2HB expresses a dpp-targeting shmiR not expected to interact with the firefly Luciferase transcript. (B–C) RNAi control transfection using GFP-shmiR-Luc 1+2 from (A): EGFP expression (B) and mCherry/RFP expression (C). (D–E) targeted RNAi transfection using GFP-shmiR-mCherry 1+2 (pHB and pNE shmiR tandem): EGFP expression (D) and mCherry/RFP expression (E).
Overall, we observed peak silencing efficiency when shmiRs were expressed in tandem, consistent with previous reports from mammalian cell culture experiments (Sun et al., 2006). Moreover, shmiRs expressed individually or in tandem retained high efficiency when placed in the 3′ UTR of EGFP. Overall efficiency of the shmiRs were comparable to analogous knockdown conditions using an ~500-bp dsRNA direct against Luciferase—published as Supplemental File 4.
Transfected cells were examined for the production of active siRNAs from the GFP-shmiR fusion RNAs (Fig. 2B–E). Here, tandem shmiRs targeting mCherry RNA were inserted in the 3′UTR of EGFP, and then co-expressed with mCherry. Indeed, all cells co-expressing GFP-shmiR-mCherry and the mCherry target RNA had nearly undetectable levels of red fluorescence (Fig. 2E), while those cells co-expressing the mCherry target RNA and a control expression vector (GFP-shmiR-Luciferase) exhibited strong red fluorescence (Fig. 2C). Thus, the siRNAs directed against mCherry RNA produce specific and quantitative loss of mCherry protein synthesis.
In order to assess whether the shmiR-derived siRNAs were entering into the miRNA or RNAi pathways, antibodies directed against AGO1 and AGO2 were used to immunoprecipitate (IP) RNA/protein complexes from S2 cells transfected with the shmiR-Luc 1HB-siRNA expression vector (Fig. 3). As expected, the control miRNA, miR-184, was enriched in the AGO1-IP sample, but not in the AGO2-IP (Fig. 3A). In contrast, shmiR-derived products are partitioned almost equally between AGO1 and AGO2. Thus, it would appear that the shmiR-derived RNAs are loaded into both the AGO1-RISC and AGO2-RISC complexes, suggesting engagement of both the siRNA and miRNA pathways of gene silencing.
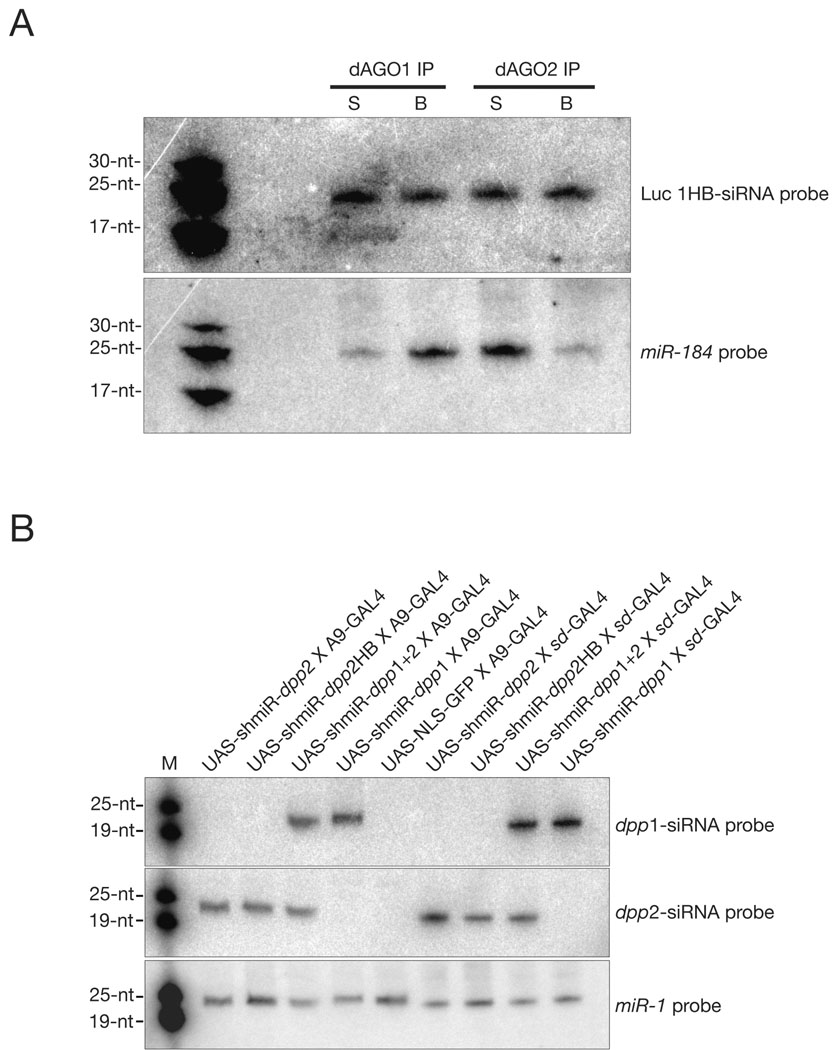
Fig. 3. Processing of small RNAs from endogenous miR-1 or pHB-based vectors in vitro and in vivo.
(A) Northern blot for a shmiR-derived siRNA (Luc 1HB) or endogenous miRNA (miR-184) following AGO1 or AGO2 immunpreciptation of S2 cell cytoplasmic extract. Unbound fraction is marked as “S” and antibody bound fraction is marked as “B.” 30, 25, and 17-nt RNAs were used as size markers. (B) Northern blot for siRNAs derived from transgenic strains of Drosophila upon UAS-GAL4 mediated expression of indicated dpp shmiRs or NLS-GFP transgenes, top. Each lane contains ~15 µg of total RNA extracted from climbing third instar larvae and 0–1 day-old pupae. 19 and 25-nt RNAs were used as markers.
Gene silencing assays in transgenic imaginal disks
Both the pHB version of the pre-miR-1 scaffold (shmiR-dppXHB, where X identifies a unique siRNA directed against the dpp mRNAs in the pHB pre-miR), as well as an unmodified version lacking the cloning sites (shmiR-dppX), was used to express dpp-specific siRNAs in vivo in transgenic larvae and adults. dpp was selected as a target since it functions in a highly dosage-sensitive manner to produce a spectrum of well-defined mutant phenotypes in adult tissues, thereby providing an immediate measure of the effectiveness of siRNAs in suppressing dpp activity (Segal and Gelbart, 1985; Spencer et al., 1982). All synthetic hairpins were introduced into a modified version of pUAST, a Drosophila P-element expression vector containing five tandem UAS-response elements for the yeast GAL4 activator (Brand and Perrimon, 1993). We also compared the dpp mutant phenotypes obtained with mono- or bi-cistronic shmiR-dpp expression vectors. shmiRs were expressed in the Drosophila wing disc using A9-GAL4, which produces moderate levels of expression along the dorsal surface of the wing imaginal disc, or sd-GAL4, which directs strong expression in the developing wing pouch (Klein et al., 1997; Marquez et al., 2001). A9-GAL4 is located on the Drosophila X-chromosome, and consequently, stronger mutant phenotypes are observed in males as compared with females.
Two distinct regions of the dpp mRNA were targeted by the dpp1- and dpp2-siRNAs, respectively. Northern blot assays were performed with RNA extracted from transgenic stains expressing a unique shmiR construct to determine the size and expression levels of the predicted siRNAs (Fig. 3B). For each construct, we observe the expected ~21-nt mature siRNA. All shmiR-dpp1 and dpp2 variants were efficiently and specifically processed, since larger precursor products (pre-miRNAs) were not detected. Moreover, insertion of an exogenous siRNA into the pre-miR-1 scaffold does not affect the inherent miR-1/miR-1* duplex asymmetry. This is shown by the different levels of dpp2 and dpp2* siRNAs produced by the shmiR-dpp2 expression vector (see Supplemental File 5).
To assess potential non-specifc effects, we created a mutant form of the dpp2HB shmiR (shmiR-dpp2HBmutant) that contains <15 bp with any region of the D. melanogaster genome (Fig. 4A; see Supplemental File 1). The shmiR-dpp2HBmutant produced no obvious mutant phenotypes in adult wings over a range of experimental conditions. Efficient expression of the mature dpp2HBmutant siRNA is shown in Supplemental File 4. The directed expression of a nuclear-localized GFP (NLS-GFP) transgene also produced no wing phenotype (see Figure 5B).
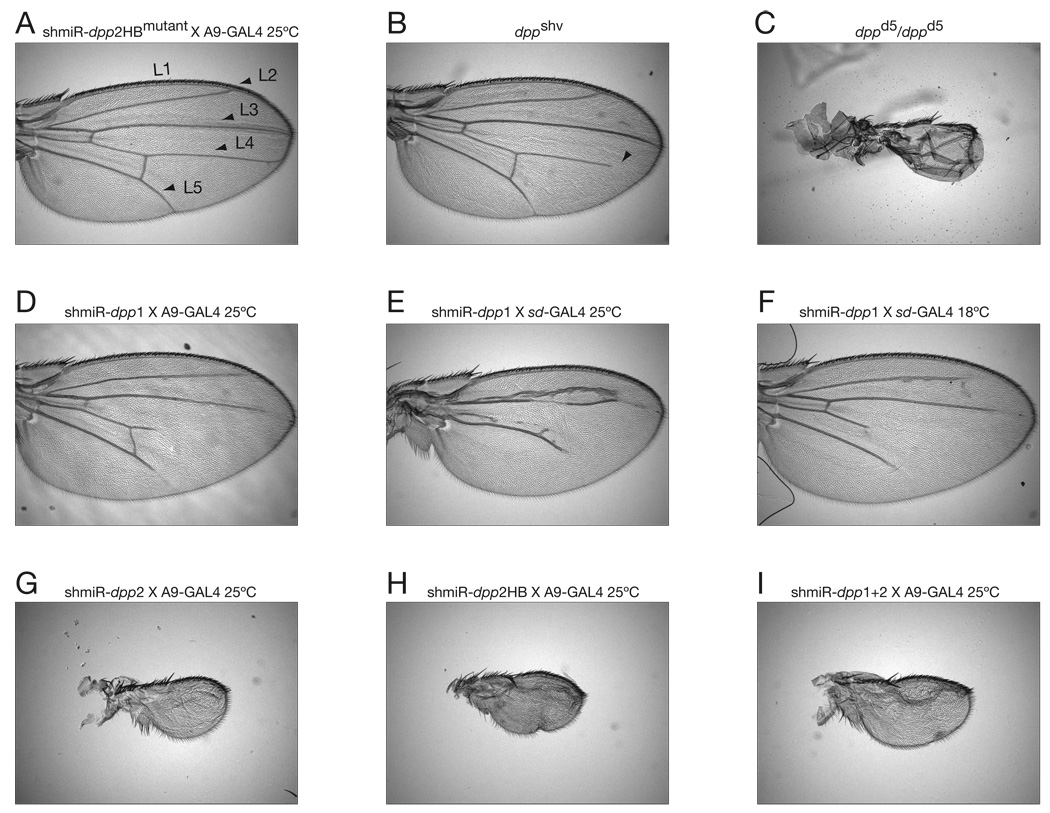
Fig. 4. Phenocopy of classical dpp wing mutations via GAL4-mediated expression of miRNA-based RNAi vectors.
Only wings from male UAS/GAL4 flies are displayed in appropriate panels. (A) Ectopic expression of a control shmiR—dpp2HBmutant—in the wing by A9-GAL4. Flies were reared at 29°C to induce any potential GAL4 or nonspecific shmiR effects at the highest level of expression. Arrowheads identify L1–L5 wing veins. (B) Classical, weak dppshv phenotype, with arrowhead indicating shortened L4 wing vein. (C) Severe dppd5/dppd5 phenotype, showing small wings with no vein structures. (D) Weak dpp knockdown phenotype with dpp siRNA 1 in endogenous miR-1-based vector. (E) dpp knockdown phenotype obtained with the stronger sd-GAL4 driver. (F) Same as (E) except that a weaker phenotype is obtained at the lower temperature of 18°C. (G) Phenocopy of strong dpp mutation with dpp siRNA 2, derived from an umodified miR-1 vector. (H) dpp knockdown phenotype with dpp siRNA 2 in a pHB-based vector. (I) dpp knockdown phenotype with bi-cistronic endogenous mir-1-based hairpins containing dpp siRNAs 1 and 2. See Supplemental Figure 6 for additional characterization of dpp2-siRNA-induced phenotypes.
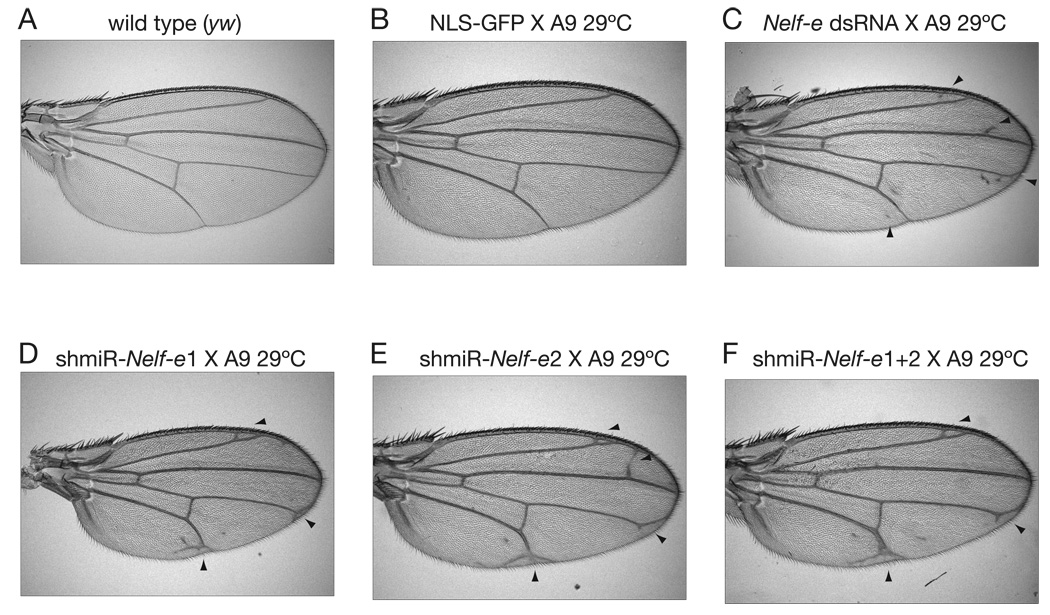
Fig. 5. Nelf-e knockdown phenotypes in the adult Drosophila wing.
(A) Wild type adult wing from yw stock. (B) Ectopic expression of a NLS-GFP under the control of A9-GAL4. Flies were reared at 29°C to induce any non-specific phenotypes due to general transgene overexpression. (C) Ectopic expression of a GAL4-responsive, ~800-bp dsRNA containing an inverted repeat that overlaps the Nelf-e ORF. Aberrant wing vein formation is marked by black arrowheads in panels (C–F). (D) shmiR expression targeting region 1 of the Nelf-e transcript. (E) shmiR expression targeting region 2 of the Nelf-e transcript. (F) bicistronic shmiR expression targeting both regions 1 and 2 of the Nelf-e transcript.
Both the dpp1 and dpp2 shmiRs recapitulated an allelic series of dpp mutants in adult wings, including progressive loss of wing vein tissue and reduction of tissue size (Fig. 4). shmiR-dpp1 expressing flies induced a moderate dpp phenotype, resembling the shortened wing veins of the classical dppshv mutant (Fig. 4B, D–F) (Segal and Gelbart, 1985). Overall stronger phenotypes were observed for dpp2 siRNAs derived from either pHB or the unmodified pre-miR-1 expression vectors (Fig. 4G,H; compare with C).
The dual-hairpin scaffold containing tandem dpp1 and dpp2 shmiRs triggered consistently more severe phenotypes among independent transgenic strains (Fig. 4I), although similar phenotypes were observed in strains with the strongest expression of dpp2 alone (Fig. 4G,H). The transgenic lines expressing the highest levels of either single or bi-cistronic hairpins produced adult flies with a virtual loss of the wings and halteres, similar to the complete loss of dpp activity, exemplified by dppd5 homozygotes (Fig. 4C) (Spencer et al., 1982). The specificity of these phenotypes suggests efficient and specific disruptions in dpp activity. Position effects are observed, as predicted for randomly integrated transgenes (see Supplemental File 6).
To assess whether the dpp2 siRNAs cause degradation of dpp mRNA, larvae were produced from the mating of transgenic strains carrying UAS-shmiR-dpp2HB and the ubiquitous daughterless-GAL4 (da-GAL4) driver. Larvae are translucent and sluggish, similar to mutations in glass bottom boat, which encodes a TGFb signaling molecule related to dpp (data not shown, and Khalsa et al., 1998). RNA was extracted from these larvae and used for qPCR assays. There is a ≥75% reduction in the normal levels of dpp mRNA as compared with larvae expressing GFP—published as Supplemental File 7.
The efficiency of shmiR and dsRNAs were compared using UAS-dpp-dsRNA transgenic lines (Ni et al., 2008). Two different dsRNAs directed against the dpp mRNA were expressed in wing imaginal disks using the A9-GAL4 driver. One of them, line 48A.1, overlaps the same region of the dpp mRNA as the shmiR-dpp1. Neither dsRNA produced a very strong phenotype. Females completely lacked a phenotype, while males displayed a weak dppshv-like phenotype (evidenced by a thinning L4 wing vein), even at the highest levels of expression (29°C) (see Supplemental File 8). This result suggests that, shmiRs promote much stronger attenuation of dpp+ gene activity as compared with dsRNAs.
Wing phenotypes were also observed upon expression of mono- or bi-cistronic shmiRs directed against Nelf-e (Fig. 5). The NELF-E protein is part of a complex that binds to nascent transcripts associated with paused Pol II (Narita et al., 2003; Yamaguchi et al., 1999). Recent studies have shown that a large number of developmental control genes are paused in the early Drosophila embryo (Wang et al., 2007; Zeitlinger et al., 2007). Reduced Nelf-e activity might cause ectopic expression or hyperactivation of genes that are normally paused in the wing imaginal disk (Aida et al., 2006). Indeed, shmiR-Nelf-e expression vectors cause the formation of ectopic wing veins, quite distinct from the loss of veins caused by reductions in dpp activity (Fig. 5D–F). This phenotype might reflect ectopic activation of dpp and/or EGF signaling components (Marquez et al., 2001). As seen for dpp shmiRs, consistently more severe phenotypes were observed with tandem Nelf-e shmiRs (Fig. 5F).
Discussion
We have described the development and use of a series of novel expression vectors that permit targeted silencing of defined genes. The method depends on the expression of specific siRNAs via synthetic 71-nt stem-loop sequences within a modified pre-miR-1 scaffold. The resulting ~21-nt silencing RNAs are pre-defined and work through known molecular mechanisms. In contrast, the use of long dsRNAs creates the potential for unexpected siRNAs with unknown targeting activities. The direct comparison of different shmiRs and dsRNAs directed against dpp mRNAs suggests that the defined siRNAs produce much more efficient disruptions in dpp+ gene activity. Moreover, tandem shmiRs producing two different siRNAs yield consistently stronger disruptions in gene activity, consistent with recent studies in mammalian systems (Sun et al., 2006).
We have also described P-element-based shmiR expression vectors that can be used with either the conventional Δ2,3 transposase or the recently developed phiC31 integrase system (Brand and Perrimon, 1993; Groth et al., 2004). The phiC31 system utilizes site-specific insertion of transgenes, effectively eliminating line-to-line variation between constructs. However, it is sometimes of interest to generate an allelic series for a particular gene (Hemann et al., 2003). Such a series is presented for dpp in Supplemental File 6. Less phenotypic variation is observed among independent lines using the tandem shmiR expression vectors, indicating that the single shmiR vectors should be used for the creation of an allelic series.
All of the shmiR expression vectors incorporate a single-step cloning method. Moreover, these vectors do not require the specialized strains of bacteria often used for the cloning of inverted repeats (Bao and Cagan, 2006). Instead, synthetic 71-nt shmiRs are directly inserted into the vectors, which could be done at a high throughput scale. In contrast, dsRNAs require template DNA preparation, primer design and synthesis, PCR amplification of the target region, and at least two additional cloning steps in order to create the final expression plasmid (Bao and Cagan, 2006).
At present, predictions of off-target effects by potential siRNAs produced from long-dsRNAs invoke a ≥19-bp complementarity cut-off (Dietzl et al., 2007; Kulkarni et al., 2006). This criterion may be rather stringent and significantly underestimate off-target effects. Indeed, only 11 of 21-contiguous nucleotides were needed for detectable target RNA cleavage in Drosophila extracts, and only 15 total bps were necessary for off-target effects in human tissue culture experiments (Haley et al., 2003; Jackson et al., 2003; Martinez and Tuschl, 2004). The present method employs specific siRNAs that are designed to minimize off-target activities. Moreover, an integrated database containing all annotated Drosophila miRNAs was incorporated into the shmiR design webtool. All input siRNAs are tested for the presence of a seed sequence matching that of a known miRNA. Eliminating such seed sequence overlap has been shown to reduce off-target effects for a given siRNA (Birmingham et al., 2006; Jackson et al., 2006).
In summary, we have demonstrated the effectiveness of the miRNA-based RNAi system for silencing defined genes in S2 cells and transgenic flies. Since this method employs preciese 21-nt siRNAs, it is less likely to produce non-specific, off-target effects than more commonly used long dsRNAs. The newly developed expression vectors permit one-step cloning through the use of inexpensive synthetic oligos, facilitating creation of whole-genome libraries for systematic gene disruption assays. The present system is based on current knowledge of miRNA processing and siRNA design. Since this field is advancing at a rapid pace, it is likely that simple modifications in the current method will yield even more efficient gene silencing in future years.
Supplementary Material
Acknowledgements
We thank Heather Melichar, Frederic Biemar, Matt Ronshaugen, and members of the Levine lab for support and advice. We also thank Johannes Bischof and Konrad Basler for supplying the pattB-UAST vector, Norbert Perrimon for the dpp-dsRNA fly stocks, and Mikkiko Siomi for the gift of AGO1 and AGO2 antibodies. B.H. is supported by an American Cancer Society Postdoctoral Fellowship. This work was funded by a grant from the NIH to M.L. (GM34431).
Abbreviations
- RNAi
RNA interference
- siRNA
small interfering RNA
- miRNA
microRNA
- shmiR
short hairpin/miRNA
- RISC
RNA induced silencing complex
- dpp
decapentaplegic
- Nelf-e
negative elongation factor e
- GFP
green fluorescent protein
- RFP
red fluorescent protein
- M.C.S.
multiple cloning site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note: Supplemental information accompanies this manuscript.
Competing Interest Statement
The authors declare that they have no competing financial interests.
References
- Adams MD, Sekelsky JJ. From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nat Rev Genet. 2002;3:189–198. doi: 10.1038/nrg752. [DOI] [PubMed] [Google Scholar]
- Aida M, et al. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Cagan R. Fast cloning inverted repeats for RNA interference. Rna. 2006;12:2020–2024. doi: 10.1261/rna.258406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Birmingham A, et al. A protocol for designing siRNAs with high functionality and specificity. Nat Protoc. 2007;2:2068–2078. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- Birmingham A, et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Bischof J, et al. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chang K, et al. Lessons from Nature: microRNA-based shRNA libraries. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- Chen CH, et al. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dimitrov RA, Zuker M. Prediction of hybridization and melting for double-stranded nucleic acids. Biophys J. 2004;87:215–226. doi: 10.1529/biophysj.103.020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, et al. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerly E, et al. Reverse genetics in Drosophila: from sequence to phenotype using UAS-RNAi transgenic flies. Genesis. 2002;34:152–155. doi: 10.1002/gene.10111. [DOI] [PubMed] [Google Scholar]
- Forstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, et al. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, et al. In vitro analysis of RNA interference in Drosophila melanogaster. Methods. 2003;30:330–336. doi: 10.1016/s1046-2023(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Han J, et al. Molecular basis for the recognition of primary microRNAs by theDrosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hemann MT, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- Huh JR, et al. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J Biol Chem. 2007;282:2056–2068. doi: 10.1074/jbc.M608051200. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson AL, et al. Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. Rna. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Khalsa O, et al. TGF-beta/BMP superfamily members, Gbb-60A and Dpp, cooperate to provide pattern information and establish cell identity in the Drosophila wing. Development. 1998;125:2723–2734. doi: 10.1242/dev.125.14.2723. [DOI] [PubMed] [Google Scholar]
- Khvorova A, et al. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Klein T, et al. An intrinsic dominant negative activity of serrate that is modulated during wing development in Drosophila. Dev Biol. 1997;189:123–134. doi: 10.1006/dbio.1997.8564. [DOI] [PubMed] [Google Scholar]
- Kulkarni MM, et al. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- Marquez RM, et al. Transgenic analysis of the Smad family of TGF-beta signal transducers in Drosophila melanogaster suggests new roles and new interactions between family members. Genetics. 2001;157:1639–1648. doi: 10.1093/genetics/157.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T. RISC is a 5' phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, et al. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, et al. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps CB, Brand AH. Ectopic gene expression in Drosophila using GAL4 system. Methods. 1998;14:367–379. doi: 10.1006/meth.1998.0592. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Segal D, Gelbart WM. Shortvein, a new component of the decapentaplegic gene complex in Drosophila melanogaster. Genetics. 1985;109:119–143. doi: 10.1093/genetics/109.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Spencer FA, et al. Decapentaplegic: a gene complex affecting morphogenesis in Drosophila melanogaster. Cell. 1982;28:451–461. doi: 10.1016/0092-8674(82)90199-4. [DOI] [PubMed] [Google Scholar]
- Sun D, et al. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- Tomari Y, et al. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 2007;21:1031–1036. doi: 10.1101/gad.1521207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.