Abstract
Dominant-negative mutations in the homopentameric extracellular matrix glycoprotein cartilage oligomeric matrix protein (COMP) result in inappropriate intracellular retention of misfolded COMP in the rough endoplasmic reticulum of chondrocytes, causing chondrocyte cell death, which leads to two skeletal dysplasias: pseudoachondroplasia (PSACH) and multiple epiphyseal dysplasia (EDM1). COMP null mice show no adverse effects on normal bone development and growth, suggesting a possible therapy involving removal of COMP mRNA. The goal of this study was to assess the ability of a hammerhead ribozyme (Ribo56, designed against the D469del mutation) to reduce COMP mRNA expression. In COS7 cells transfected with plasmids that overexpress wild-type or mutant COMP mRNA and Ribo56, the ribozyme reduced overexpressed normal COMP mRNA by 46% and mutant COMP mRNA by 56% in a dose-dependent manner. Surprisingly, the use of recombinant adenoviruses to deliver wild-type or mutant COMP mRNA and Ribo56 simultaneously into COS7 cells proved problematic for the activity of the ribozyme to reduce COMP expression. However, in normal human costochondral cells (hCCCs) infected only with adenoviruses expressing Ribo56, expression of endogenous wild-type COMP mRNA was reduced in a dose-dependent manner by 50%. In chondrocytes that contain heterozygous COMP mutations (D469del, G427E and D511Y) that cause PSACH, Ribo56 was more effective at reducing COMP mRNA (up to 70%). These results indicate that Ribo56 is effective at reducing mutant and wild-type COMP levels in cells and suggests a possible mode of therapy to reduce the mutant protein load.
Keywords: PSACH, ribozyme, gene therapy, adenovirus, chondrocyte
INTRODUCTION
Cartilage oligomeric matrix protein (COMP/TSP-5) is one of the proteins that comprise the complex extracellular matrix (ECM) networks of cartilage, tendon, and ligament that cushion and stabilize skeletal elements (Kempson et al. 1968; Urban et al. 1979; Schmidt et al. 1990). COMP, the fifth member of the thrombospondin (TSP) gene family, is a multidomain homopentameric protein comprised of an N-terminal pentamerization domain, an epidermal growth factor (EGF)-like domain, a highly conserved type 3 (calcium-binding) repeat domain, and a C-terminal globular region (Fife and Brandt 1984; DiCesare et al. 1994; Adams et al. 1995; Smith et al. 1997; Hecht et al. 1998a; Carlson et al. 2008). Recent work suggests that COMP is multifunctional and is involved in (1) regulation of collagen fiber assembly/diameter (Rosenberg et al. 1998); (2) interaction with other ECM molecules (Holden et al. 2001; Thur et al. 2001; Mann et al. 2004); (3) regulation of chondrocyte proliferation (Kipnes et al. 2003; Xu et al. 2007); (4) regulation of apoptosis (Duke et al. 2003; Hashimoto et al. 2003; Hecht et al. 2004); and (5) enhancement of cellular attachment (Chen et al. 2005).
Mutations in COMP cause two skeletal dysplasias: a severe disproportionate dwarfing condition, pseudoachondroplasia (PSACH), and a milder condition, multiple epiphyseal dysplasia (MED/EDM1 subtype) (Briggs et al. 1995; Hecht et al. 1995; Thur et al. 2001; Briggs and Chapman 2002). PSACH is a dwarfing condition in which infants have a normal birth length and weight and are first diagnosed when their linear growth decelerates between 1 and 2 years of age (Unger and Hecht 2001; Posey et al. 2004). Characteristic PSACH clinical findings include waddling gait beginning in early childhood; widened and lax joints; brachydactyly; windswept, knock knee, or bowing deformities of the lower extremities; exaggerated lumbar lordosis; and early onset osteoarthritis (Unger and Hecht 2001; Posey et al. 2004). Replacement of the weight-bearing joints, particularly knees and hips, often is required in early adulthood (Unger and Hecht 2001). In contrast to PSACH, MED is diagnosed around five years of age when the children present with abnormal gait and/or stiff painful hips (Unger and Hecht 2001). Mild short stature and osteonecrosis of the hips (Legg-Perthes disease) are also frequent findings in MED (Unger and Hecht 2001). Hip replacement often is needed in early adulthood (Unger and Hecht 2001).
More than 100 novel COMP mutations have been identified, with the majority found in the highly conserved type 3 calcium-binding repeat domain (Briggs et al. 1995; Hecht et al. 1995; Unger and Hecht 2001; Kennedy et al. 2005). The most common mutation, D469del, accounts for ∼30% of all PSACH cases (Unger and Hecht 2001; Kennedy et al. 2005). These mutations produce a COMP molecule consisting of a mixture of mutant and wild-type subunits that causes protein stalling in the rER and intracellular COMP accumulation in large rER cisternae (Cooper et al. 1973; Hecht et al. 1998b, 2005; Chen et al. 2004). This massive intracellular protein retention eventually leads to chondrocyte death; the loss of chondrocytes translates into markedly diminished linear growth in affected individuals (Unger and Hecht 2001; Briggs and Chapman 2002).
Transgenic mouse models expressing mutant COMP demonstrate a mild PSACH phenotype (Pirog-Garcia et al. 2007; Schmitz et al. 2007; Posey et al. 2008). In contrast, COMP knockout mice develop normally, suggesting that COMP may not be necessary or has a redundant function in cartilage formation, bone growth, and extracellular cartilage matrix (ECM) development (Svensson et al. 2002). These observations led to the hypothesis that elimination of a majority of the mutant COMP from the secretory pathway would prevent the cellular pathology associated with the PSACH phenotype without side effects owed to partial reduction of its normal counterpart. RNAi (interference) technology is one approach that could be used to reduce in vitro and in vivo gene expression.
Ribozymes are short catalytic RNA oligonucleotides that either splice or cleave RNA molecules and were hailed in the early years of gene therapy research for their potential to control gene expression (Cameron and Jennings 1989; Haseloff and Gerlach 1992; McCall et al. 1992; James and Turner 1997). However, interest in ribozymes faded when they did not reliably knock down targeted genes and with the advent of siRNA technologies. Recently, there has been a renewed interest in the use of ribozymes as a means to knock down expression of specific genes because of recent advances in design and delivery, better stabilization, and their ability to remain active over long periods in vivo. Hammerhead ribozymes are small ribozymes consisting of 24–35 nucleotides and three stem–loop structures that induce a site-specific cleavage of RNA based on Watson–Crick base pairing with their target sequences (Haseloff and Gerlach 1988; Ruffner et al. 1990; Shimayama et al. 1995). Because the hammerhead ribozyme functional sequence is short and conserved, novel designs can quickly be synthesized and tested in cell culture for knockdown of mRNA expression of genes that cause disease (Hendry et al. 1997; Tabler and Sczakiel 1997; Blount and Uhlenbeck 2002; Liu et al. 2002; Khan 2006). In addition, ribozymes, unlike siRNA, are not thought to activate interferon responses (Shiota et al. 2004), so they may have longer-term utility in whole animal/humans. These studies were undertaken to test the efficacy and potential utility in gene therapy applications of a novel ribozyme targeting COMP.
RESULTS
COMP-specific ribozyme (Ribo56) reduces steady-state levels of wild-type COMP mRNA or mutant COMP (D469del) mRNA and protein in transfected COS7 cells
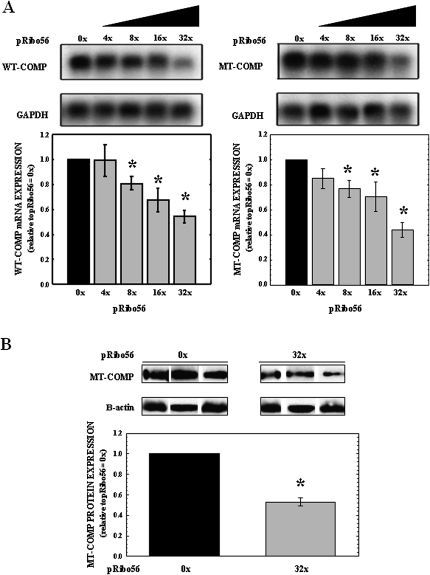
To determine whether Ribo56 can reduce steady-state wild-type (WT) and mutant (MT) COMP mRNA in cells, COS7 cells were transfected with plasmids that express WT-COMP (pWT-COMP) or MT-COMP (pMT-COMP). Since these cells do not express COMP or other cartilage-specific ECM proteins (Hashimoto et al. 2003), all of the WT and MT-COMP mRNA assessed in these studies was expressed from the plasmids. Cells were cotransfected with increasing amounts (0×–32×) of a plasmid that express Ribo56 (pRibo56). Transfection efficiency in COS7 cells was consistently found to be 30%–35% (data not shown). RNA and protein were isolated 72-h post-transfection and subjected to Northern and Western analysis. Northern analysis of WT and MT-COMP mRNA (shown in Fig. 1A) indicates that Ribo56 decreased steady-state levels of WT- and MT-COMP mRNA in a dose-dependent manner. The reduction in mRNA expression is significant (P < 0.05) for Ribo56 at concentrations of 8, 16, and 32× compared with RNA levels without Ribo56 (Fig. 1A). The most efficient knockdown of COMP mRNA occurred at 32× Ribo56, with 46% reduction of WT-COMP mRNA and 56% reduction of MT-COMP mRNA levels. Importantly, Ribo56 had comparable activity against WT- and MT-COMP in this assay. Surprisingly, Ribo56 appeared to be more effective against MT-COMP (D469del) mRNA than WT-COMP mRNA, although the difference was not significant (P = 0.16) in this assay.
FIGURE 1.
WT- and MT-COMP expression is decreased in a dose-dependent manner in COS7 cells cotransfected with increasing amounts of Ribo56 plasmid. COS7 cells were cotransfected with 250 ng of either pWT-COMP or pMT-COMP and increasing amounts of pRibo56. RNA and cellular protein were harvested 72 h after transfection. (A) Total RNA (7 μg) was subjected to Northern analysis using radiolabeled probes specific for COMP and human GAPDH (loading control). Representative autoradiographs (N = 6) are shown above, while normalization of COMP mRNA knockdown is displayed in the bar graph. COS7 cells expressing only COMP (0×) were normalized to 1. Error bars represent the standard error of sample means (SEM), and P < 0.05 was considered significant (*). (B) Protein (30 μg) was subjected to Western analysis of three independent experiments with pMT-COMP using antibodies specific for COMP and β-actin (loading control). Shown are the resulting autoradiographs and bar graph normalization of the results.
Western analysis was performed to assess MT-COMP protein knockdown by Ribo56 in the COS7 cells that were transfected with pMT-COMP and pRibo56 at concentrations 0 and 32×. Protein was isolated 72-h post-transfection. The results of three independent transfections are shown in Figure 1B. Quantification of intracellular MT-COMP protein shows that, in the presence of Ribo56 at a concentration of 32×, COMP was significantly reduced by 45%. These findings demonstrate that Ribo56 has the ability to reduce COMP protein levels in COS7 cells by reducing the steady-state levels of MT-COMP mRNA.
Dual adenovirus infection increases steady-state levels of COMP mRNA and protein in COS7 cells
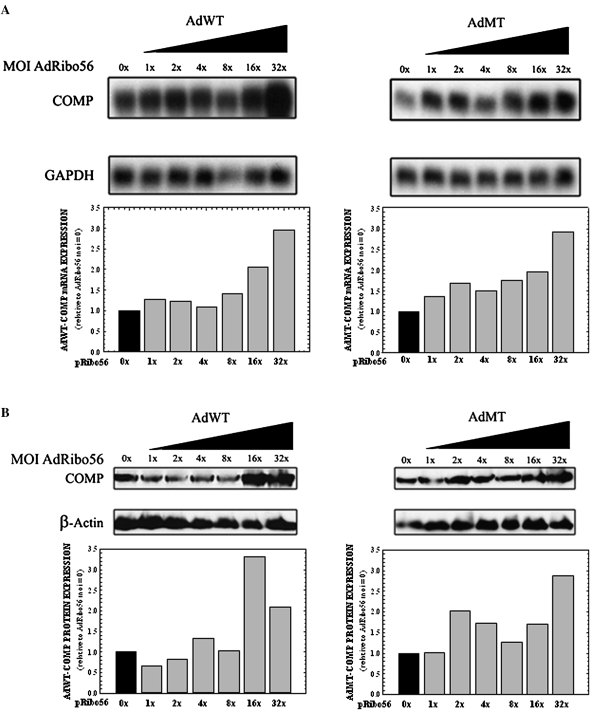
The feasibility of a recombinant adenovirus system to deliver Ribo56 to cells was assessed in COS7 cells. COS7 cells were coinfected at a constant multiplicity of infection (MOI) of a recombinant adenovirus that expressed either WT-COMP (AdWT) or MT-COMP (AdMT) with an increasing MOI (0–32×) of a recombinant adenovirus expressing Ribo56 (AdRibo56). Infected cells were incubated for 96 h, and the RNA was isolated. Northern analysis showed that neither WT- nor MT-COMP mRNAs decreased in the presence of increasing Ribo56 in these cells (Fig. 2A). In fact, steady-state COMP mRNA levels increased with increasing MOI of AdRibo56 (Fig. 2A). This increase also was reflected in the steady-state levels of WT- and MT-COMP protein (Fig. 2B). Surprisingly, the coinfection unexpectantly appeared to increase steady-state levels of COMP mRNA and protein.
FIGURE 2.
Adenovirus-based COMP expression is increased in a dose-dependent manner in COS7 cells when coinfected with adenovirus expressing Ribo56. COS7 cells were coinfected with either AdWT- or AdMT-COMP with an increasing MOI of AdRibo56. RNA and cellular proteins were collected after 96 h. (A) Total RNA (7 μg) was subjected to Northern analysis using radiolabeled probes specific for COMP and human GAPDH (loading control). Resulting autoradiographs are shown above, while normalization of COMP mRNA knockdown is displayed in the bar graph. COS7 cells expressing only COMP (0×) were normalized to 1. (B) Protein (30 μg) was subjected to Western analysis using antibodies specific for COMP and β-actin (loading control). Shown are the resulting autoradiographs and normalized data.
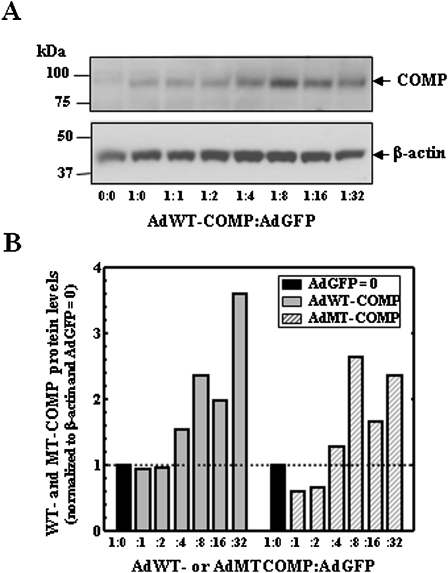
To assess whether the increase of COMP mRNA was due to increased Ribo56 or adenovirus, the experiment was repeated using increasing AdGFP with constant AdWT- or AdMT-COMP. The results in Figure 3 show that the expression of both WT- and MT-COMP mRNA increases in the presence of an increasing amount of AdGFP. These data suggest that increasing adenovirus infection leads to activation of the CMV promoter driving COMP expression. Therefore, the dual infection approach for disease modeling was abandoned.
FIGURE 3.
Adenovirus-based COMP expression is increased in COS7 cells when coinfected with adenovirus expressing only GFP. COS7 cells were coinfected with either AdWT- or AdMT-COMP with an increasing MOI of AdGFP. Cellular proteins were collected after 96 h. (A) Protein (30 μg) from AdWT-COMP:AdGFP was subjected to Western analysis using antibodies specific for FLAG and β-actin (loading control) and visualized by ECL. Resulting autoradiographs are shown. (B) Normalized COMP protein from AdWT- or MT-COMP:AdGFP is shown in the bar graph. COS7 cells expressing only COMP (0×) were normalized to 1.
Expression of endogenous COMP mRNA is decreased in a dose-dependent manner in primary human chondrocytes (hCCCs) infected with increasing MOIs of AdRibo56
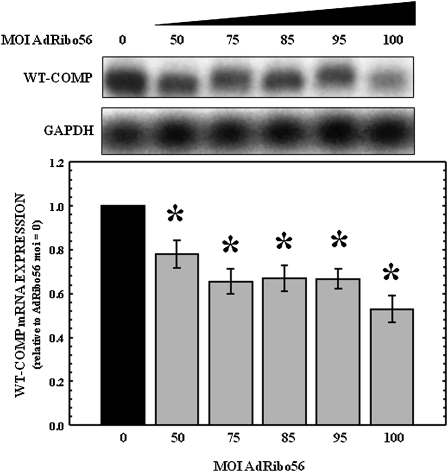
Previous experiments suggested that adenoviral delivery of genes to hCCCs can be efficiently undertaken (Merritt et al. 2006). However, the coinfection results described above suggest that in the presence of two different adenoviruses, one encoding the target and one encoding the ribozyme, there are unexpected interactions that lead to an overall increase in the levels of COMP (Fig. 2). To determine whether this was an artifact of cotransfection of two viral expression vectors, studies were undertaken to investigate the knockdown of endogenous COMP in hCCCs. The hCCCs were infected with increasing MOIs of AdRibo56 ranging from 0 to 200. RNA was isolated 96-h post-infection and subjected to Northern analysis. As shown in Figure 4, the presence of AdRibo56 significantly (P < 0.05) decreased endogenous steady-state normal COMP mRNA levels in the hCCCs at the highest MOIs. As the AdRibo56 MOI increased to 100, dose-dependent knockdown was observed; endogenous COMP mRNA expression was reduced by 47% for AdRibo56 at an MOI of 100.
FIGURE 4.
Expression of MT-COMP mRNA is decreased in a dose-dependent manner in primary hCCCs infected with increasing MOIs of adenovirus expressing Ribo56. Normal costochondral chondrocytes were isolated as previously described and were transfected in monolayer with an increasing MOI of AdRibo56 (shown above the gel). RNA was collected after 96 h. Total RNA (7 μg) was subjected to Northern analysis using radiolabeled probes specific for COMP and human GAPDH (loading control). Representative autoradiographs (N = 5) are shown above, and COMP expression in uninfected cells was used to normalize to 1. Error bars represent the SEM, and P < 0.05 was considered significant (*).
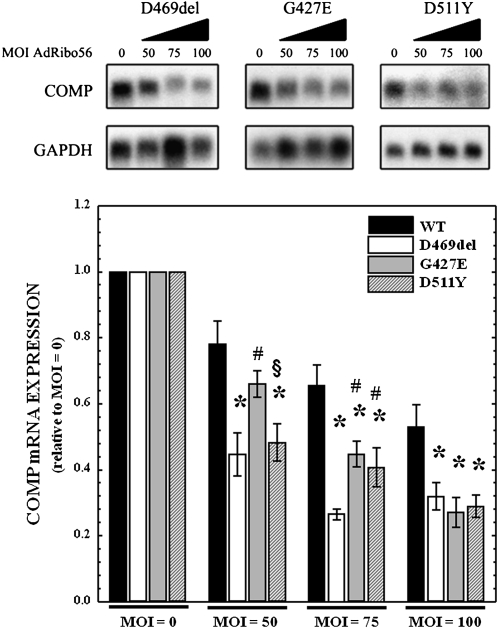
AdRibo56 reduces COMP mRNA expression in primary mutant PSACH chondrocytes with three different COMP mutations more efficiently than in normal chondrocytes
Results shown in Figure 4 indicated that the adenovirus delivery system can be used to knock down endogenous WT-COMP in primary hCCCs. The next series of experiments were performed to determine whether AdRibo56 reduces COMP mRNA levels in three primary PSACH chondrocytes with G427E, D469del, or D511Y COMP mutations and to compare the extent of knockdown to that of the normal COMP. Mutant chondrocytes were infected with AdRibo56 at MOIs of 0, 50, 75, and 100. As seen in Figure 5, Northern analysis of mRNA from these cells shows a reduction in the steady-state COMP with increasing MOIs of AdRibo56 for all PSACH mutations. Quantification shows that at all MOIs greater than 50, AdRibo56 significantly reduced the WT- and to a greater extent the MT-COMP mRNA levels. The D469del mRNA was maximally reduced by 73% at an MOI of 75, the G427 and D511Y mRNAs were maximally reduced by 70% and 77%, respectively, at an MOI of 100. These results indicate that Ribo56 is very effective at reducing steady-state levels of MT-COMP for all three mutations but specifically appears more effective against D469del MT-COMP at lower MOIs. These data indicate that Ribo56 is more effective against MT-COMP compared with endogenous WT-COMP when expressed in chondrocytes (Fig. 5).
FIGURE 5.
COMP mRNA expression is decreased in a dose-dependent manner in primary PSACH chondrocytes with three different COMP mutations infected with increasing MOIs of adenovirus expressing Ribo56. PSACH chondrocytes heterozygous for WT COMP and one of three COMP mutations (D469del, G427E, or D511Y) were isolated as previously described and were infected with increasing MOIs of AdRibo56. RNA was collected after 96 h. Total RNA (7 μg) was subjected to Northern analysis using radiolabeled probes specific for COMP and human GAPDH (loading control). Representative autoradiographs (N = 3) are shown above, while normalization of COMP mRNA knockdown is displayed in the bar graph. COMP expression in uninfected cells (0×) was normalized to 1. Error bars represent the SEM, and P < 0.05 was considered significant. (*) P ≤ 0.01 versus wild type; (#) P ≤ 0.01 versus D469del; (§) P ≤ 0.01 versus G427E.
DISCUSSION
This is the first report to show reduction of COMP using a ribozyme that is amenable to use in future gene therapy applications for patients suffering from COMP-related skeletal dysplasias. Ribo56, a hammerhead ribozyme directed against the COMP sequence, targets the GUC triplet near nucleotide 56 of the mature mRNA and successfully reduces protein levels of both WT- and MT-COMP in chondrocytes. Most importantly, the ribozyme is more active in knockdown of three well-described MT-COMP species in chondrocytes when compared with normal WT-COMP (Fig. 4). Such preference presumably occurs because of differences in the secondary structure (Andronescu et al. 2005) or accessibility of target sequences in ribonucleoprotein complexes (Welting et al. 2008) of the transcripts encoding wild-type versus mutant COMP. Selective targeting of mutant COMP over WT-COMP by a ribozyme has never been reported.
In the initial phases of this study, COS7 cells were used as an in vitro model because the cells are easily transfected and, importantly, they do not synthesize COMP or any other ECM protein that might potentially interfere with the outcome (Hashimoto et al. 2003). Therefore, these studies directly tested the ability of Ribo56 to knock down either WT- or MT-COMP mRNA and protein without competition from endogenous COMP. Transfected cells can take up more than one type of plasmid efficiently, eliminating the possibility of singly transfected cells and allowing dose-dependent assessment of the ribozyme. The results clearly demonstrate that Ribo56 significantly reduced COMP mRNA at most concentrations (8×–32×) but was most effective at the highest concentration (32×). Under these conditions, when both target and ribozyme were expressed from a plasmid, Ribo56 reduced WT- or MT-COMP equally. In these studies, MT-COMP mRNA was reduced by at least 50% and protein levels by 45%. These results indicate that COMP mRNA knockdown is reflected in the reduction of COMP protein levels in this model system.
Recombinant adenovirus has been used to successfully transfect a wide variety of primary and transformed cells and hCCCs, in particular (Wickham et al. 1996; Merritt et al. 2006). Therefore, recombinant adenovirus technology was used to deliver AdRibo56 in the next series of studies. Coinfection of WT- and MT-COMP into COS7 cells in the presence of Ribo56 first was performed to assess the effectiveness of adenovirally delivered ribozyme against overexpressed COMP (James and Turner 1997; Tabler and Sczakiel 1997; Merritt et al. 2006) in these cells. Surprisingly, coinfection resulted in a net increase of both WT- and MT-COMP mRNA and protein. This increase in steady-state levels of COMP could be the result of interactions of gene products of the two independent adenoviruses that alter transcript processing, export, or stability or increase gene expression in unanticipated ways (Timpe et al. 2006). Alternatively, Ribo56 is not able to recognize or cleave the target sequence when both are expressed from an adenovirus. Interestingly, when the COMP adenovirus MOI was reduced even further, the increase in steady-state levels of COMP still occurred in the presence of the ribozyme but only when it was delivered using an adenoviral vector (data not shown). This finding may suggest that cellular stress from the presence of two separate adenoviral vectors may stimulate COMP expression.
The disease pathology in PSACH is specific for the growth plate chondrocytes (Cooper et al. 1973; Hecht et al. 1998b; Hecht et al. 2004, 2005). Therefore, a system with the capacity to deliver the ribozyme sequence into these cells must be obtained. To eliminate the complications of synergistic effects on ribozyme activity from two different adenoviruses and to test the function of Ribo56 in a more native system, hCCCs were infected with different MOIs of adRibo56 to determine the effect of the ribozyme on endogenous COMP. As seen in Figure 3, steady-state WT-COMP mRNA levels were reduced by Ribo56 in a dose-dependent manner up to 50% at the highest concentrations. The same approach was used to test the effectiveness of Ribo56 against MT-COMP in three different PSACH patient hCCCs. The results shown in Figure 4 clearly show that COMP mRNA expression was reduced by ∼70% for D468del, G427E, and D511Y mutations. The magnitude of the reduction suggests that this ribozyme may be more effective against MT-COMP than WT-COMP in native chondrocytes. The difference in efficiency of mRNA cleavage may be related to the difference in folding patterns of the WT-COMP mRNA compared with the MT-COMP (Fedor and Uhlenbeck 1990; Liu et al. 2000; Liu et al. 2002). Even though the site of cleavage of Ribo56 is near the 5′ end of the COMP transcript, the changes in the secondary structures of MT-COMP compared with WT-COMP assessed by S-fold extend to the region complementary to the annealing arms of the ribozyme. Of interest, the preference of the ribozyme for the mutant transcript over the native one may selectively reduce the levels of the mutant rather than the native form of COMP when both coexist in PSACH chondrocytes (Merritt et al. 2006).
Mutations in COMP cause the PSACH and MED (EMD1 type) skeletal dysplasias. The disease pathology of these conditions results from the massive retention of misfolded COMP and inappropriate formation of intracellular matrix in the rER (Merritt et al. 2007). In contrast, COMP null mice are normal, suggesting that loss of COMP does not compromise normal growth, development, or longevity. Thus, a treatment strategy removing or reducing steady-state levels of mutant COMP in chondrocytes of affected individuals is needed. Here, we show that Ribo56 can reduce COMP mRNA and protein with a preference for knocking down the mutant form. This opens the door for the use of mutant-recognizing ribozymes as an effective gene therapy for these skeletal dysplasias.
MATERIALS AND METHODS
Cell culture
Cells (hCCCs) were obtained from patients undergoing pectus excavatum surgery after informed consent at Shriners Hospital for Children-Houston (Hecht et al. 1998b, 2004). hCCCs were cultured as described above in 20% fetal bovine serum (FBS). Two PSACH patient chondrocyte lines, D469del (deletion of aspartic acid 469) and G427E, were obtained from the Iowa skeletal dysplasia cell bank (Hecht et al. 1998b). The third PSACH patient cell line, D511Y, was obtained from a growth plate biopsy (Hecht et al. 2005). All of the chondrocyte samples were from prepubertal children. These three PSACH cell lines were heterozygous for the COMP mutations. Normal growth plate chondrocytes were isolated as previously described (Hecht et al. 2004).
Hammerhead ribozyme vectors
The hammerhead ribozyme (Ribo56) and annealing arms were designed using the methods previously described (Liu et al. 2002) and its target sequence validated using S-ribo (http://sfold.wadsworth.org/sribo.pl). S-fold was used to compare the predicted secondary structures for normal and three mutant COMP sequences chosen for this study, all of which affected the predicted folding pattern of the target transcripts in a manner that could impact interactions with targeted ribozymes. Ribozyme 56 cuts at nucleotide 56 of the COMP mRNA: 5′-CUGCCCUCGGCGCGUCCGGACAGGGCCAG-3′. The cut site (underlined) lies in the region of the transcript encoding the C-terminal half of the signal sequence extending just into the coding sequence for the mature protein. Hence, blockade or cleavage of this region will reduce transcript levels when cut and interfere with translation of the key portion of the molecule governing entry into the secretory pathway when functioning as an antisense molecule. DNA encoding the hammerhead ribozyme first was ligated into the BLOCK-iT U6 RNAi entry vector of the ViraPower Lentiviral Gateway expression system (Invitrogen). The Ribo56 construct includes a U6 promoter sequence, the small nuclear RNA (snRNA) U1 sequence for stabilization of the ribozyme and a Pol III termination sequence. To generate the AdRibo56, the expression cassette was excised from the original vector and ligated into the multiple cloning site of the pAdTrack-GFP shuttle vector (Qbiogene). AdRibo56 was expanded to working titers by Vector Biolabs.
Two COMP expression plasmid vectors were provided by Dr. Jeff W. Stevens (University of Iowa) (Chen et al. 2004). These pc-DNA3.1+ vectors contain either a WT-COMP (GenBank accession number: L32137) or MT-COMP (D469del) that have a FLAG tag amino acid sequence (DYKDDDDK) added to the carboxyl terminus. The recombinant adenoviruses (AdWT and AdMT) were constructed as previously described (Merritt et al. 2006) and expanded to working titers by Vector Biolabs.
Plasmid transfection
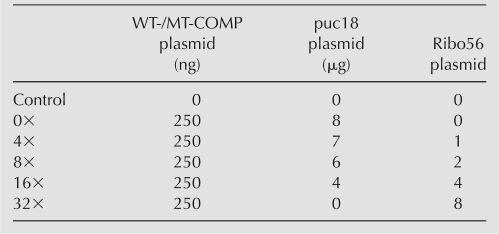
COS7 cells were cultured in 6-well tissue culture dishes and grown to 80%–90% confluency. Cells were transfected per manufacturer's instructions with plasmid DNA using a 3:2 ratio of FuGene6 (Roche) to plasmid DNA. All transfections contained 250 ng of WT- or MT-COMP plasmid and were cotransfected with varying amounts of Ribo56 and pUC18 plasmids for a total of 8.25 μg DNA per transfection to obtain steady-state COMP plasmid with an increasing concentration of Ribo56 plasmid (Table 1). The COS7 cells were transfected and incubated for 96 h. The cells were collected at 96 h using either Trizol (Invitrogen) for RNA or, for protein, RIPA buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% [v/v] Triton X-100, Mini complete protease inhibitors [Roche]; 1% [v/v] Na-deoxycholate; 0.1% [v/v] SDS).
TABLE 1.
Ratios of plasmid transfected into COS7 cells
Viral transfection
The hCCCs were cultured in monolayer in T25 flasks and infected for 18 h in serum-free Dulbecco's modified Eagle medium (DMEM) with AdRibo56 at MOIs of 0, 50, 75, 85, 95, 100, 110, 150, and 200. PSACH patient chondrocytes were infected using MOIs of 0, 50, 75, and 100. The medium was replaced with DMEM + 20% FBS and 1% Ab/Am and cells were incubated at 37°C for a total of 96 h. COS7 cells were cultured in 6-well tissue culture dishes and grown to confluency. Cells then were infected for 18 h using 1.5 mL of serum-free DMEM containing either AdWT- or AdMT-COMP at an MOI of 1 and AdR56 with increasing MOIs to give a ratio of 0× to 32×. The medium was replaced with DMEM + 10% FBS and 1% Ab/Am.
RNA isolation and purification from cultured cells
RNA was isolated from both plasmid and viral transfected COS7 cells and hCCCs after 72 or 96 h using 3 mL (for 6-well) or 4 mL (T25 flask) of Trizol. The Trizol-cell mixture then was processed using the PureLink Micro- to Midi-RNA isolation kit (Invitrogen) following the manufacturer's protocol. The RNA was quantitated using a NanoDrop spectrophotomer (Thermo Fisher Scientific). Purified samples were stored at −80°C.
Northern analysis
Northern analysis was performed as described in Sambrook and Russell (2001). A radioactive probe was made using the Stratagene Prime-It RmT Random primer labeling kit following the manufacturer's protocol. The DNA templates were made from COMP cDNA excised from a pcDNA3.1+ vector and gel purified using the Qiaquick gel extraction kit (Qiagen) following the manufacturer's protocol. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) template obtained from Origene (GenBank: NM_002046) was excised and gel purified as described above. The radioactive probe was hybridized at 55°C overnight. The washed membrane was sealed and exposed to X-ray film. Films were scanned and processed by the software GeneSnap (Syngene) and analyzed for densitometry using the GeneTools software (Syngene).
Protein isolation and Western analysis
Protein was isolated from transfected COS7 cells and chondrocytes by using 500 μl (for 6-well) or 1 mL (T25 flask) of RIPA buffer. Western analysis was performed as described in Sambrook and Russell (2001). Total protein (30 μg) was electrophoresed through a NuPAGE 10% (w/v) Bis-Tris gel (Invitrogen). The gel was transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore). The membrane was incubated with primary rabbit (rab) polyclonal antibody for COMP (1:10,000) (Kamiya Biomedical) or the FLAG epitope (1:5,000) (Sigma Chemical Co) and incubated with horseradish peroxidase (HRP) goat anti-rabbit IgG (1:7,500) (GE Healthcare). To ensure uniform loading, β-actin levels were determined with a β-actin rabbit primary conjugated with HRP (1:75,000) (Abcam). Visualization of the complex was accomplished using the ECL Plus kit (GE Healthcare) following the manufacturer's protocol. Films were scanned and processed using GeneSnap (Syngene) and analyzed for densitometry using GeneTools software (Syngene).
Statistical analysis
The results of each replicate experiment were compared for each concentration of Ribo56 by dividing the pixel score of COMP by the pixel score of GAPDH (RNA) or β-actin (protein) for the corresponding lane (data not shown). A two-tailed t-test (P < 0.05) was used to assess the differences between the COMP mRNA levels and the protein levels for the different experiments for each concentration ratio by comparing them individually to the control COMP levels. The control COMP mRNA levels and protein levels were normalized to a density value of 1, and the mean value was established in comparison for each Ribo56 concentration and displayed in a bar graph. All error bars represent the standard error of sample means (SEM). Asterisk (*) denotes mean values that are significantly (P-value < 0.05) different from the control.
ACKNOWLEDGMENTS
This study was supported by grants from NIH HL68116 (J.L.A.), NIH P20RR016458 (M.C.F.-C.), Shriners Hospital for Children (J.T.H.), and the Leah Lewis Family Foundation (J.T.H.). We thank Drs. Shea Posey and S. Shahrukh Hashmi for helpful suggestions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1335909.
REFERENCES
- Adams J., Tucker R.P., Lawler J. The thrombospondin gene family. Springer; New York: 1995. [Google Scholar]
- Andronescu M., Zhang Z.C., Condon A. Secondary structure prediction of interacting RNA molecules. J. Mol. Biol. 2005;345:987–1001. doi: 10.1016/j.jmb.2004.10.082. [DOI] [PubMed] [Google Scholar]
- Blount K.F., Uhlenbeck O.C. The hammerhead ribozyme. Biochem. Soc. Trans. 2002;30:1119–1122. doi: 10.1042/bst0301119. [DOI] [PubMed] [Google Scholar]
- Briggs M.D., Chapman K.L. Pseudoachondroplasia and multiple epiphyseal dysplasia: Mutation review, molecular interactions, and genotype to phenotype correlations. Hum. Mutat. 2002;19:465–478. doi: 10.1002/humu.10066. [DOI] [PubMed] [Google Scholar]
- Briggs M.D., Hoffman S.M., King L.M., Olsen A.S., Mohrenweiser H., Leroy J.G., Mortier G.R., Rimoin D.L., Lachman R.S., Gaines E.S., et al. Pseudoachondroplasia and multiple epiphyseal dysplasia due to mutations in the cartilage oligomeric matrix protein gene. Nat. Genet. 1995;10:330–336. doi: 10.1038/ng0795-330. [DOI] [PubMed] [Google Scholar]
- Cameron F.H., Jennings P.A. Specific gene suppression by engineered ribozymes in monkey cells. Proc. Natl. Acad. Sci. 1989;86:9139–9143. doi: 10.1073/pnas.86.23.9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C.B., Lawler J., Mosher D.F. Structures of thrombospondins. Cell. Mol. Life Sci. 2008;65:672–686. doi: 10.1007/s00018-007-7484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.L., Stevens J.W., Cole W.G., Hecht J.T., Vertel B.M. Cell-type specific trafficking of expressed mutant COMP in a cell culture model for PSACH. Matrix Biol. 2004;23:433–444. doi: 10.1016/j.matbio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Chen F.H., Thomas A.O., Hecht J.T., Goldring M.B., Lawler J. Cartilage oligomeric matrix protein/thrombospondin 5 supports chondrocyte attachment through interaction with integrins. J. Biol. Chem. 2005;280:32655–32661. doi: 10.1074/jbc.M504778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R.R., Ponseti I.V., Maynard J.A. Pseudoachondroplasia dwarfism. A rough-surfaced endoplasmic reticulum disorder. J. Bone Joint Surg. Am. 1973;55A:475–484. [PubMed] [Google Scholar]
- DiCesare P., Hauser N., Lehman D., Pasumarti S., Paulsson M. Cartilage oligomeric matrix protein (COMP) is an abundant component of tendon. FEBS Lett. 1994;354:237–240. doi: 10.1016/0014-5793(94)01134-6. [DOI] [PubMed] [Google Scholar]
- Duke J., Montufar-Solis D., Underwood S., Lalani Z., Hecht J.T. Apoptosis staining in cultured pseudoachondroplasia chondrocytes. Apoptosis. 2003;8:191–197. doi: 10.1023/a:1022926811397. [DOI] [PubMed] [Google Scholar]
- Fedor M.J., Uhlenbeck O.C. Substrate sequence effects on “hammerhead” RNA catalytic efficiency. Proc. Natl. Acad. Sci. 1990;87:1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fife R.S., Brandt K.D. Identification of a high-molecular-weight (>400,000) protein in hyaline cartilage. Biochim. Biophys. Acta. 1984;802:506–514. doi: 10.1016/0304-4165(84)90370-2. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W.L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W.L. Simple RNA enzymes with new and highly specific endoribonuclease activities. 1988. Biotechnology. 1992;24:264–269. [PubMed] [Google Scholar]
- Hashimoto Y., Tomiyama T., Yamano Y., Mori H. Mutation (D472Y) in the type 3 repeat domain of cartilage oligomeric matrix protein affects its early vesicle trafficking in endoplasmic reticulum and induces apoptosis. Am. J. Pathol. 2003;163:101–110. doi: 10.1016/S0002-9440(10)63634-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht J.T., Nelson L.D., Crowder E., Wang Y., Elder F.F., Harrison W.R., Francomano C.A., Prange C.K., Lennon G.G., Deere M., et al. Mutations in exon 17B of cartilage oligomeric matrix protein (COMP) cause pseudoachondroplasia. Nat. Genet. 1995;10:325–329. doi: 10.1038/ng0795-325. [DOI] [PubMed] [Google Scholar]
- Hecht J.T., Deere M., Putnam E., Cole W., Vertel B., Chen H., Lawler J. Characterization of cartilage oligomeric matrix protein (COMP) in human normal and pseudoachondroplasia musculoskeletal tissues. Matrix Biol. 1998a;17:269–278. doi: 10.1016/s0945-053x(98)90080-4. [DOI] [PubMed] [Google Scholar]
- Hecht J.T., Montufar-Solis D., Decker G., Lawler J., Daniels K., Duke P.J. Retention of cartilage oligomeric matrix protein (COMP) and cell death in redifferentiated pseudoachondroplasia chondrocytes. Matrix Biol. 1998b;17:625–633. doi: 10.1016/s0945-053x(98)90113-5. [DOI] [PubMed] [Google Scholar]
- Hecht J.T., Makitie O., Hayes E., Haynes R., Susic M., Montufar-Solis D., Duke P.J., Cole W.G. Chondrocyte cell death and intracellular distribution of COMP and type IX collagen in the pseudoachondroplasia growth plate. J. Orthop. Res. 2004;22:759–767. doi: 10.1016/j.orthres.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Hecht J.T., Hayes E., Haynes R., Cole W.G. COMP mutations, chondrocyte function and cartilage matrix. Matrix Biol. 2005;23:525–533. doi: 10.1016/j.matbio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Hendry P., McCall M.J., Lockett T.J. Design of hybridizing arms in hammerhead ribozymes. In: Turner P.C., editor. Ribozyme protocols. Humana Press; Totowa, NJ: 1997. pp. 254–264. [DOI] [PubMed] [Google Scholar]
- Holden P., Meadows R.S., Chapman K.L., Grant M.E., Kadler K.E., Briggs M.D. Cartilage oligomeric matrix protein interacts with type IX collagen, and disruptions to these interactions identify a pathogenetic mechanism in a bone dysplasia family. J. Biol. Chem. 2001;276:6046–6055. doi: 10.1074/jbc.M009507200. [DOI] [PubMed] [Google Scholar]
- James H.A., Turner P.C. Ribozyme Protocols. Humana Press; Totowa, NJ: 1997. Ribozymes; pp. 1–9. [Google Scholar]
- Kempson G.E., Freeman M.A., Swanson S.A. Tensile properties of articular cartilage. Nature. 1968;220:1127–1128. doi: 10.1038/2201127b0. [DOI] [PubMed] [Google Scholar]
- Kennedy J., Jackson G.C., Barker F.S., Nundlall S., Bella J., Wright M.J., Mortier G.R., Neas K., Thompson E., Elles R., et al. Novel and recurrent mutations in the C-terminal domain of COMP cluster in two distinct regions and result in a spectrum of phenotypes within the pseudoachondroplasia–multiple epiphyseal dysplasia disease group. Hum. Mutat. 2005;25:593–594. doi: 10.1002/humu.9342. [DOI] [PubMed] [Google Scholar]
- Khan A.U. Ribozyme: A clinical tool. Clin. Chim. Acta. 2006;367:20–27. doi: 10.1016/j.cca.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Kipnes J., Carlberg A.L., Loredo G.A., Lawler J., Tuan R.S., Hall D.J. Effect of cartilage oligomeric matrix protein on mesenchymal chondrogenesis in vitro. Osteoarthritis Cartilage. 2003;11:442–454. doi: 10.1016/s1063-4584(03)00055-4. [DOI] [PubMed] [Google Scholar]
- Liu R., Li W., Karin N.J., Bergh J.J., Adler-Storthz K., Farach-Carson M.C. Ribozyme ablation demonstrates that the cardiac subtype of the voltage-sensitive calcium channel is the molecular transducer of 1, 25-dihydroxyvitamin D(3)-stimulated calcium influx in osteoblastic cells. J. Biol. Chem. 2000;275:8711–8718. doi: 10.1074/jbc.275.12.8711. [DOI] [PubMed] [Google Scholar]
- Liu R., Rohe B., Carson D.D., Farach-Carson M.C. A rapid and simple nonradioactive method for in vitro testing of ribozyme activity. Antisense Nucleic Acid Drug Dev. 2002;12:283–288. doi: 10.1089/108729002320351601. [DOI] [PubMed] [Google Scholar]
- Mann H.H., Ozbek S., Engel J., Paulsson M., Wagener R. Interactions between the cartilage oligomeric matrix protein and matrilins. Implications for matrix assembly and the pathogenesis of chondrodysplasias. J. Biol. Chem. 2004;279:25294–25298. doi: 10.1074/jbc.M403778200. [DOI] [PubMed] [Google Scholar]
- McCall M.J., Hendry P., Jennings P.A. Minimal sequence requirements for ribozyme activity. Proc. Natl. Acad. Sci. 1992;89:5710–5714. doi: 10.1073/pnas.89.13.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt T.M., Alcorn J.L., Haynes R., Hecht J.T. Expression of mutant cartilage oligomeric matrix protein in human chondrocytes induces the pseudoachondroplasia phenotype. J. Orthop. Res. 2006;24:700–707. doi: 10.1002/jor.20100. [DOI] [PubMed] [Google Scholar]
- Merritt T.M., Bick R., Poindexter B.J., Alcorn J.L., Hecht J.T. Unique matrix structure in the rough endoplasmic reticulum cisternae of pseudoachondroplasia chondrocytes. Am. J. Pathol. 2007;170:293–300. doi: 10.2353/ajpath.2007.060530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirog-Garcia K.A., Meadows R.S., Knowles L., Heinegard D., Thornton D.J., Kadler K.E., Boot-Handford R.P., Briggs M.D. Reduced cell proliferation and increased apoptosis are significant pathological mechanisms in a murine model of mild pseudoachondroplasia resulting from a mutation in the C-terminal domain of COMP. Hum. Mol. Genet. 2007;16:2072–2088. doi: 10.1093/hmg/ddm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey K.L., Hayes E., Haynes R., Hecht J.T. Role of TSP-5/COMP in Pseudoachondroplasia. Int. J. Biochem. Cell Biol. 2004;36:1005–1012. doi: 10.1016/j.biocel.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Posey K.L., Yang Y., Veerisetty A.C., Sharan S.K., Hecht J.T. Model systems for studying skeletal dysplasias caused by TSP-5/COMP mutations. Cell. Mol. Life Sci. 2008;65:687–699. doi: 10.1007/s00018-007-7485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg K., Olsson H., Morgelin M., Heinegard D. Cartilage oligomeric matrix protein shows high affinity zinc-dependent interaction with triple helical collagen. J. Biol. Chem. 1998;273:20397–20403. doi: 10.1074/jbc.273.32.20397. [DOI] [PubMed] [Google Scholar]
- Ruffner D.E., Stormo G.D., Uhlenbeck O.C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Schmidt M.B., Mow V.C., Chun L.E., Eyre D.R. Effects of proteoglycan extraction on the tensile behavior of articular cartilage. J. Orthop. Res. 1990;8:353–363. doi: 10.1002/jor.1100080307. [DOI] [PubMed] [Google Scholar]
- Schmitz M., Niehoff A., Miosge N., Smyth N., Paulsson M., Zaucke F. Transgenic mice expressing D469Delta mutated cartilage oligomeric matrix protein (COMP) show growth plate abnormalities and sternal malformations. Matrix Biol. 2007;27:67–85. doi: 10.1016/j.matbio.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Shimayama T., Nishikawa S., Taira K. Generality of the NUX rule: Kinetic analysis of the results of systematic mutations in the trinucleotide at the cleavage site of hammerhead ribozymes. Biochemistry. 1995;34:3649–3654. doi: 10.1021/bi00011a020. [DOI] [PubMed] [Google Scholar]
- Shiota M., Sano M., Miyagishi M., Taira K. Ribozymes: Applications to functional analysis and gene discovery. J. Biochem. 2004;136:133–147. doi: 10.1093/jb/mvh119. [DOI] [PubMed] [Google Scholar]
- Smith R.K., Zunino L., Webbon P.M., Heinegard D. The distribution of cartilage oligomeric matrix protein (COMP) in tendon and its variation with tendon site, age and load. Matrix Biol. 1997;16:255–271. doi: 10.1016/s0945-053x(97)90014-7. [DOI] [PubMed] [Google Scholar]
- Svensson L., Aszodi A., Heinegard D., Hunziker E.B., Reinholt F.P., Fassler R., Oldberg A. Cartilage oligomeric matrix protein-deficient mice have normal skeletal development. Mol. Cell. Biol. 2002;22:4366–4371. doi: 10.1128/MCB.22.12.4366-4371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler M., Sczakiel G. Design and production of asymmetric hammerhead ribozymes. Methods Mol. Biol. 1997;74:141–149. doi: 10.1385/0-89603-389-9:141. [DOI] [PubMed] [Google Scholar]
- Thur J., Rosenberg K., Nitsche D.P., Pihlajamaa T., Ala-Kokko L., Heinegard D., Paulsson M., Maurer P. Mutations in cartilage oligomeric matrix protein causing pseudoachondroplasia and multiple epiphyseal dysplasia affect binding of calcium and collagen I, II, and IX. J. Biol. Chem. 2001;276:6083–6092. doi: 10.1074/jbc.M009512200. [DOI] [PubMed] [Google Scholar]
- Timpe J.M., Verrill K.C., Trempe J.P. Effects of adeno-associated virus on adenovirus replication and gene expression during coinfection. J. Virol. 2006;80:7807–7815. doi: 10.1128/JVI.00198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger S., Hecht J.T. Pseudoachondroplasia and multiple epiphyseal dysplasia: New etiologic developments. Am. J. Med. Genet. 2001;106:244–250. [PubMed] [Google Scholar]
- Urban J.P., Maroudas A., Bayliss M.T., Dillon J. Swelling pressures of proteoglycans at the concentrations found in cartilaginous tissues. Biorheology. 1979;16:447–464. doi: 10.3233/bir-1979-16609. [DOI] [PubMed] [Google Scholar]
- Welting T.J., Mattijssen S., Peters F.M., van Doorn N.L., Dekkers L., van Venrooij W.J., Heus H.A., Bonafe L., Pruijn G.J. Cartilage-hair hypoplasia-associated mutations in the RNase MRP P3 domain affect RNA folding and ribonucleoprotein assembly. Biochim. Biophys. Acta. 2008;1783:455–466. doi: 10.1016/j.bbamcr.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Wickham T.J., Roelvink P.W., Brough D.E., Kovesdi I. Adenovirus targeted to heparin-containing receptors increases its gene delivery efficiency to multiple cells. Nat. Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- Xu K., Zhang Y., Ilalov K., Carlson C.S., Feng J.Q., Di Cesare P.E., Liu C.J. Cartilage oligomeric matrix protein associates with granulin-epithelin precursor (GEP) and potentiates GEP-stimulated chondrocyte proliferation. J. Biol. Chem. 2007;282:11347–11355. doi: 10.1074/jbc.M608744200. [DOI] [PubMed] [Google Scholar]