Abstract
The RNA component of telomerase (telomerase RNA; TER) varies substantially both in sequence composition and size (from ∼150 nucleotides [nt] to >1500 nt) across species. This dramatic divergence has hampered the identification of TER genes and a large-scale comparative analysis of TER sequences and structures among distantly related species. To identify by phylogenetic analysis conserved sequences and structural features of TER that are of general importance, it is essential to obtain TER sequences from evolutionarily distant groups of species, providing enough conservation within each group and enough variation among the groups. To this end, we identified TER genes in several yeast species with relatively large (>20 base pairs) and nonvariant telomeric repeats, mostly from the genus Candida. Interestingly, several of the TERs reported here are longer than all other yeast TERs known to date. Within these TERs, we predicted a pseudoknot containing U-A·U base triples (conserved in vertebrates, budding yeasts, and ciliates) and a three-way junction element (conserved in vertebrates and budding yeasts). In addition, we identified a novel conserved sequence (CS2a) predicted to reside within an internal-loop structure, in all the budding yeast TERs examined. CS2a is located near the Est1p-binding bulge-stem previously identified in Saccharomyces cerevisiae. Mutational analyses in both budding yeasts S. cerevisiae and Kluyveromyces lactis demonstrate that CS2a is essential for in vivo telomerase function. The comparative and mutational analyses of conserved TER elements reported here provide novel insights into the structure and function of the telomerase ribonucleoprotein complex.
Keywords: telomere, telomerase RNA, pseudoknot, phylogenetic analysis, secondary structure, yeast, Candida
INTRODUCTION
Telomeres are nucleoprotein structures protecting eukaryotic chromosome ends (for review, see Bertuch and Lundblad 2006; Hug and Lingner 2006). Nuclear telomeres are predominantly maintained by telomerase, a ribonucleoprotein (RNP) complex that can add short DNA repeats onto chromosomal ends and thus compensate for losses caused by incomplete replication or degradation (for reviews, see Smogorzewska and de Lange 2004; Autexier and Lue 2006; Hug and Lingner 2006). The essential core components of this specialized enzyme are telomerase RNA (TER; TLC1 in Saccharomyces cerevisiae) and telomerase reverse transcriptase (TERT; Est2p in budding yeasts), which can repeatedly copy a small portion of TER (termed the template) onto the telomere 3′ end. Other proteins that associate with the telomerase complex in budding yeasts include Est1p and Est3p, both of which are essential for telomerase action in vivo, the Sm proteins, and the yKu complex (Bertuch and Lundblad 2006; Hug and Lingner 2006).
TERs are highly divergent in sequence and length, even among closely related species. Phylogenetic covariation was used to predict common secondary structure models for TERs within ciliates (Romero and Blackburn 1991; ten Dam et al. 1991; Lingner et al. 1994), vertebrates (Chen et al. 2000), and Saccharomyces sensu stricto and Kluyveromyces budding yeast species (Chappell and Lundblad 2004; Dandjinou et al. 2004; Lin et al. 2004; Zappulla and Cech 2004; Brown et al. 2007). However, only a limited similarity in the general architecture of these models was observed (Chen et al. 2000; Lin et al. 2004). The known yeast TERs are relatively large–from 930 to 1540 nucleotides (nt) in K. nonfermentans and C. albicans, respectively (Tzfati et al. 2003; Hsu et al. 2007a)—and highly divergent in sequence. While significant portions of these TERs are dispensable for telomerase function, other nontemplate regions appear to be important (Roy et al. 1998; Livengood et al. 2002; Zappulla et al. 2005). Several conserved elements were found to contribute crucial functions such as specifying the template boundary (Tzfati et al. 2000; Seto et al. 2003) or recruiting Est1, the yKu complex, and the Sm proteins (Seto et al. 1999, 2002; Peterson et al. 2001). A unique pseudoknot element containing a major-grove triple-helix is crucial for K. lactis, S. cerevisiae, and human telomerase function (Theimer et al. 2005; Shefer et al. 2007; Qiao and Cech 2008). It was recently suggested that the triple helix functions in positioning the 3′ end of the telomere at the catalytic site (Qiao and Cech 2008). Another critical domain in vertebrate TERs, CR4-CR5, was suggested to interact with TERT (Mitchell and Collins 2000; Bachand et al. 2001). Within this domain, a small stem–loop, P6.1, is essential for the catalytic activity of the enzyme. Stem IV of ciliate TERs was suggested to be the functional homolog of the vertebrate P6.1 (Mason et al. 2003), and in Saccharomyces sensu stricto and Kluyveromyces TERs, a three-way junction element shows remarkable conservation with P6.1 (Brown et al. 2007).
Phylogenetic analysis of TER sequences derived from species that are close in terms of evolutionary distance has proven very helpful for identifying conserved functional elements and predicting their structure. However, to reveal universal features of telomerase, it is necessary to compare these conserved elements across more distant groups of species. Therefore, we identified putative TER sequences from several Candida, Debaryomyces, Lodderomyces, and Pichia species, analyzed them, and compared their conserved elements with those of other yeast and vertebrate TERs. The recently identified TER gene of Schizosaccharomyces pombe (Leonardi et al. 2008; Webb and Zakian 2008) could not be included in our analysis because fission yeast is relatively distant to the budding yeasts studied here and no other TER homologs from evolutionarily close fission yeast species are available in the databases. Our comparative analysis revealed sequence and structure elements that not only are conserved but also appear to be essential for function in budding yeast TERs. Moreover, certain elements are also conserved in vertebrates and ciliates.
RESULTS AND DISCUSSION
Identification of putative TER loci in yeast with large telomeric repeats
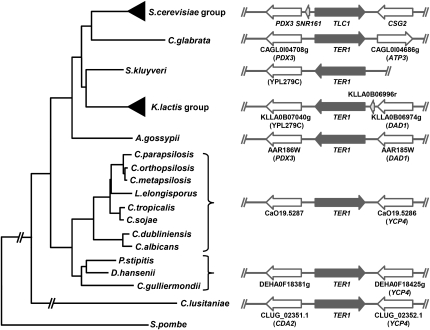
A hint for the identification of TER genes is provided by the sequence of the telomeric repeats copied from the template within TER. This information is not sufficient in cases where the telomeric repeats are too short and/or variable in nature, such as those of S. cerevisiae and S. pombe. Yet, most yeast species have relatively large (>20 base pairs [bp]) and nonvariant repeat units (McEachern and Hicks 1993; McEachern and Blackburn 1994; see Table 1). Importanly, complete or partial genome sequences of several yeast species are publicly available (Fig. 1; Table 1). We identified putative TER genes in the available sequence data for budding yeast species as described below. In addition, on the basis of gene synteny of TERs and neighboring ORFs, we cloned and analyzed TER genes from three other closely related species: C. metapsilosis, C. orthopsilosis, and C. sojae.
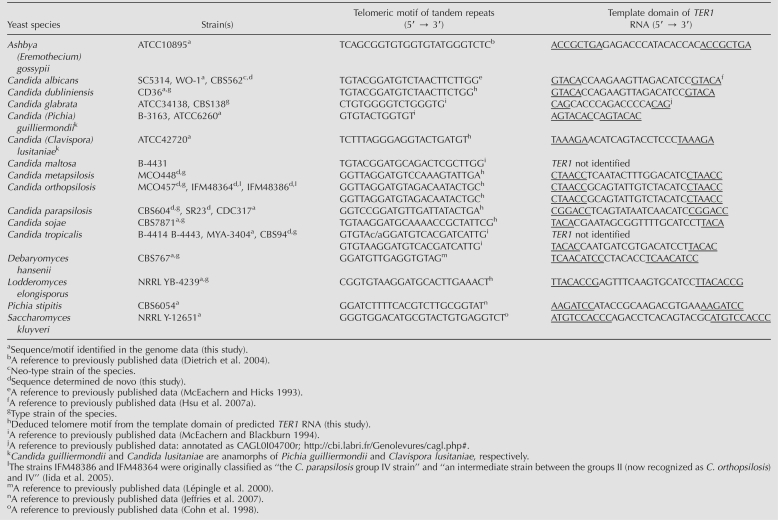
TABLE 1.
List of yeast species, the sequences of their telomeric repeats and the predicted template domains in TER
FIGURE 1.
Phylogenetic tree of selected yeast species with identified TER genes. The tree, derived from the sequences of the D1/D2 domain of 26S rRNA genes, was calculated by the MEGA 4 software package (Tamura et al. 2007). The Schizosaccharomyces pombe sequence was used as an outgroup. The right panel shows the chromosomal context of the TER1 loci. S. cerevisiae homologs are shown in parentheses. We mapped the TER1 locus on the chromosome I (∼3 Mbp) in C. parapsilosis strains CBS604 and SR23 (data not shown).
In order to identify telomerase RNA genes in yeast with canonical telomeric repeats, we first performed a BlastN search of the C. albicans genome database (http://www.candidagenome.org/) using the telomeric repeat sequence 5′-ACGGATGTCTAACTTCTTGGTGT-3′ as a query. Adding a random 12-bp flanking sequence on both sides of the telomeric repeat yielded a single hit with a perfect match to the repeat. The identified sequence lies at nucleotides 6792–6813 of Contig19-2285 and is identical to the recently published TER1 sequence (Hsu et al. 2007a). In silico analysis of 2500 bp of flanking sequences of the putative template region (TR) revealed two open reading frames (ORFs): orf19.5286 (3′ side), a putative flavodoxin homologous to the YCP4 gene of S. cerevisiae, and orf19.5287 (5′ side), with no homology with any known protein (Fig. 1). Next we performed a similar analysis with the telomeric repeat unit of C. tropicalis 5′-AAGGATGTCACGATCATTGGTGT-3′ (again with random 12-bp flanking sequences) as a query and searched the C. tropicalis genome database (http://www.broad.mit.edu/annotation/genome/candida_tropicalis/Home.html). As in the case of C. albicans, we obtained a single hit with a perfect match to the telomeric repeat. The identified sequence lies at nucleotides 1260787–1260763 of supercontig 3.1. Importantly, sequence analysis of the region revealed the presence of two ORFs highly homologous to the orf19.5286 and orf19.5287 of C. albicans. The orientation of both ORFs was the same as in C. albicans, demonstrating synteny in this region (Fig. 1). A combination of similar search criteria using the corresponding known TR sequences with the observation of the (at least partial) synteny within the region led to the identification of the putative TER loci in additional species, namely C. dubliniensis, C. guilliermondii, and D. hansenii. In addition, inspection of the corresponding genome regions containing the orf19.5286 and orf19.5287 homologs in C. parapsilosis, C. lusitaniae, L. elongisporus, and P. stipitis and subsequent alignment with the TER loci of other species resulted in the identification of putative TRs and thus the putative TER genes and the predicted telomeric repeats in these yeast species. For other species lacking available genomic sequences (C. metapsilosis, C. orthopsilosis, C. sojae), putative TER loci were amplified by PCR with primers derived from conserved regions of the flanking ORFs, cloned into a plasmid vector and sequenced.
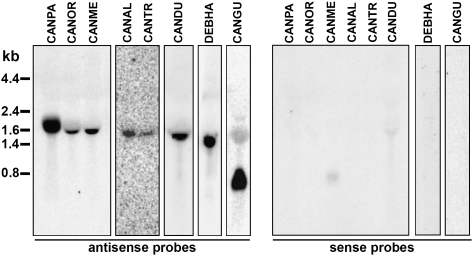
To demonstrate that the putative TER loci are transcribed from the predicted strand, we performed Northern blot analysis of total RNA isolated from several yeast species using oligonucleotides derived from the region surrounding the TR as probes (Supplemental Table S1). This analysis revealed that the TER loci are transcribed from one strand, producing RNA molecules with sizes between 800 nt (C. guilliermondii) to ∼1800 nt (C. parapsilosis) (Fig. 2). Hybridization with the oligonucleotides derived from the sense strand of the TER did not yield any significant signal (Fig. 2). The identification of putative TER loci for the related yeast species described above served as a starting point for comparative analysis of their sequences and structures.
FIGURE 2.
The putative TER loci are transcribed. Northern blot analysis of TER1 transcripts from C. parapsilosis CBS604T (CANPA), C. orthopsilosis MCO457T (CANOR), C. metapsilosis MCO448T (CANME), C. albicans CBS562NT (CANAL), C. tropicalis CBS94T (CANTR), C. dubliniensis CBS7987T (CANDU), D. hansenii CBS767T (DEBHA), and C. guilliermondii CBS2030T (CANGU), using the antisense (left panel) or sense (right panel) oligonucleotides as probes.
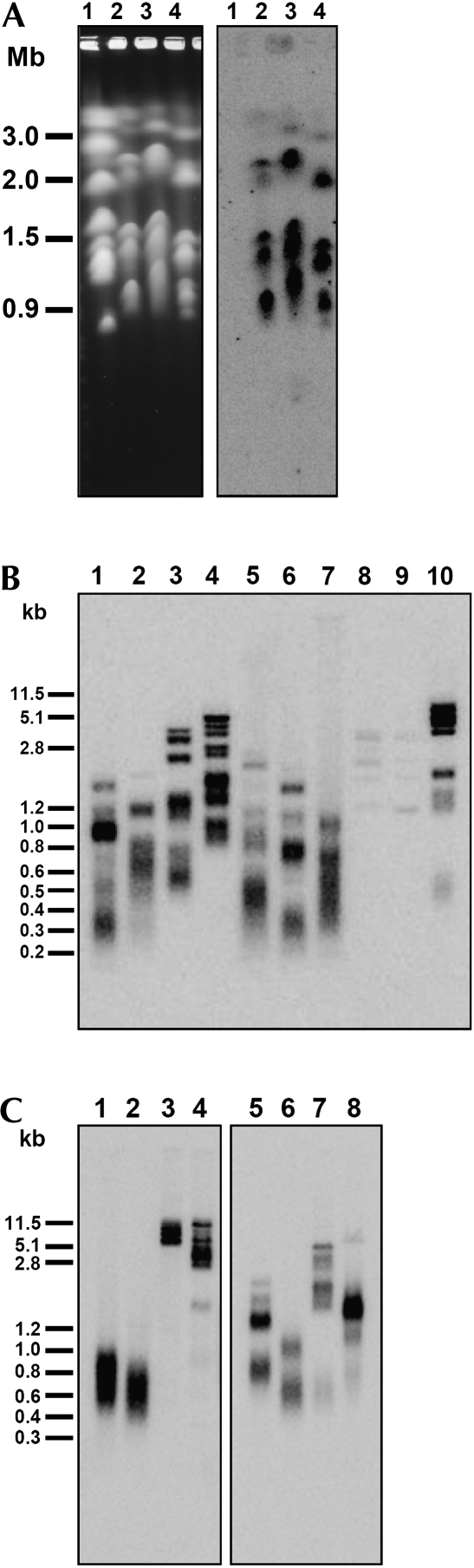
Next we asked if the predicted repetitive sequences are present at the ends of the chromosomes. To address this question, we performed two experiments. First, we separated the chromosomes of three strains of C. parapsilosis and one of the closely related species L. elongisporus by pulsed-field gel electrophoresis (PFGE) and performed a Southern blot hybridization using a trimer of the predicted C. parapsilosis repeat as a probe (Supplemental Table S1). The results demonstrate that the target sequence is present at all C. parapsilosis chromosomes (Fig. 3A, lanes 2–4). In contrast, none of the chromosomes of L. elongisporus hybridized to the telomeric oligonucleotide probe, indicating that the telomeric repeat is species specific (Fig. 3A, lane 1). In the second experiment, the genomic DNA (gDNA) of C. parapsilosis was digested with a battery of restriction endonucleases and subjected to a Southern blot analysis using the same C. parapsilosis telomeric oligonucleotide probe as in Figure 3A. Although the digestion of gDNA with different enzymes resulted in a different hybridization pattern, in each case we observed smeared bands typical for heterogeneous populations of telomeric fragments (Fig. 3B, lanes 1–7,10), whose sizes indicate that the length of the telomeric tracts ranges between 300 and 600 bp. Importantly, the digestion of the C. parapsilosis gDNA with AvaII and MspI, which have a recognition site within the predicted telomeric repeat, resulted in an almost complete loss of the hybridization signal (Fig. 3B, lanes 8,9). Analogous results were obtained with the C. metapsilosis and C. orthopsilosis gDNAs hybridized to their predicted telomeric repeat sequences as probes (Fig. 3C).
FIGURE 3.
Analysis of deduced telomeric repeats by Southern blot. (A) Chromosomal DNA samples from L. elongisporus CBS5301 (lane 1) and C. parapsilosis CBS2194 (lane 2), SR23 (lane 3), and CBS604T (lane 4) were separated by pulsed-field gel electrophoresis and subjected to Southern blot analysis using a 5′-end-labeled oligonucleotide consisting of three copies of the putative C. parapsilosis telomeric repeat as a probe (Supplemental Table S1). Ethidium bromide staining of this gel is shown on the left and the autoradiogram on the right. (B) Southern blot analysis of C. parapsilosis SR23 telomeric restriction fragments obtained by digestion of gDNA with AluI (lane 1), Csp6I (lane 2), HaeIII (lane 3), HinPI (lane 4), MseI (lane 5), Sau3A (lane 6), TaqI (lane 7), MspI (lane 8), AvaII (lane 9), and EcoRI (lane 10). Note that AvaII and MspI have a recognition site within the C. parapsilosis telomeric repeat. (C) Southern blot analysis of telomeric DNA fragments obtained by digestion of gDNA from C. metapsilosis MCO448T (lanes 1–4) and C. orthopsilosis MCO457T (lanes 5–8) with AluI (lanes 1,5), Csp6I (lanes 2,6), MspI (lanes 3,7), and Sau3A (lanes 4,8). 5′-end-labeled oligonucleotides consisting of three copies of the putative telomeric repeats from C. metapsilosis (lanes 1–4) or C. orthopsilosis (lanes 5–8) were used as probes (Supplemental Table S1).
Sequence conservation of Candida spp. TERs
Unlike the highly conserved and structured ribosomal RNA, telomerase RNA appears to include only small elements conserved in sequence and/or structure, embedded within much larger nonconserved regions. Given the large size and sequence divergence of the Candida spp. TERs, we assumed that these TERs may include even larger nonconserved and dispensable regions than those in other species. Indeed, an initial attempt to align the seven Candida spp. TERs using the Clustal X program (Thompson et al. 1997) failed to yield a reliable sequence alignment. Therefore, we focused our efforts on the identification of small functional elements that are conserved among budding yeast TERs, as described below. The identified conserved sequences were named CS2–CS7 according to the Kluyveromyces terminology (Tzfati et al. 2003). With these elements identified and since they were positioned in the same order within each TER sequence, it was possible to use them as anchors and improve the alignment as follows: The TER sequences were divided into fragments defined by the conserved elements (upstream of the template, from the template to CS2, CS2 to CS3, CS3 to CS5, CS5 to CS6, and downstream from CS6) and each fragment was aligned separately by Clustal X. Subsequently, these alignments were merged into a full-length alignment (Supplemental Fig. S1). Based on the already mapped 5′ end of the C. albicans TER (Hsu et al. 2007a) and the relatively reliable alignment of the sequence close to the 5′ end, we predicted the 5′ ends of the rest of the aligned TERs. In addition, we expected the 3′ end to be located close (downstream) to the consensus Sm binding site (CS7), as previously reported for Kluyveromyces and Saccharomyces TERs (Tzfati et al. 2003; Dandjinou et al. 2004; Lin et al. 2004).
Mapping of the 5′ and 3′ ends of the TER transcripts
In order to map the ends of the TER transcripts we used the following approach (see Materials and Methods for details). Total RNA was poly(A) tailed and then reverse transcribed using an oligo dT primer. The cDNA was circularized by intramolecular ligation using CircLigase (Epicentre Biotechnologies). A fragment containing the ligated 5′ and 3′ ends was amplified by two subsequent PCR reactions with nested primers and sequenced from both ends (Polidoros et al. 2006). Alternatively, the 3′ ends were mapped by a protocol described by Lingner and Keller (1993) and Tzfati et al. (2003). Based on our results the 3′ ends of C. parapsilosis, C. dubliniensis, C. orthopsilosis, C. metapsilosis, L. elongisporus, D. hansenii, C. guilliermondii, and C. tropicalis TERs were mapped to be located within 8–40 nt downstream from the Sm site and upstream (in most species immediately upstream) of CS8 (Supplemental Fig. S2A; see below). We assume that the ends of the rest of the species are at a similar position. The 5′ ends of the C. parapsilosis, C. orthopsilosis, and C. dubliniensis TERs were mapped exactly as predicted by the alignment to the C. albicans TER, and that of C. tropicalis, 3 nt downstream (Supplemental Fig. S2A). As reported previously, there was some variation, mostly in the 3′ ends (Hsu et al. 2007a). Interestingly, the longest possible transcripts of the experimentally mapped TERs were 1817 nt (C. parapsilosis), 1779 nt (L. elongisporus), 1659 nt (C. orthopsilosis), 1581 nt (C. tropicalis), and 1519 nt long (C. dubliniensis). The C. parapsilosis TER is the longest mapped yeast telomerase RNA reported to date. Furthermore, based on our experimental data, the C. guilliermondii TER (779 nt) is the shortest among yeast TERs known to date. Altogether, these results are consistent with the ends predicted by the sequence alignment (Supplemental Fig. S1) and the results of the Northern blot (Fig. 2).
Sm site and downstream splicing consensus sequences
A conserved binding site for the Sm proteins was identified in all the new TERs (Supplemental Figs. S1, S2A). This sequence (GAU3–4G) is different from the conserved Sm sites found in the Kluyveromyces (ANU5G) or Saccharomyces (AAU5G) TERs. However, it is consistent with the budding yeasts Sm consensus sequence (AU3–6G; Riedel et al. 1987). Interestingly, two additional conserved sequences, termed CS8 and CS9, were identified downstream from the Sm site (Supplemental Figs. S1, S2A). Based on the 3′ ends mapped in this work and the sequence alignment, CS8 and CS9 are not included in the mature TER. Rather, the 3′ ends of most of the TERs were mapped immediately upstream of CS8. Interestingly, it was recently found that the 3′ end of the S. pombe TER is processed by a partial splicing reaction (Box et al. 2008). CS8 fits perfectly well with the yeast consensus sequence for the 5′ splice site, GTATGT (with only one exception of GTACGT in C. guilliermondii). CS9 is a perfect match of the TACTAAC box located at intron branch sites. Both of these elements are known to be essential for splicing in budding yeasts (Langford et al. 1984). The identification of these perfectly conserved splicing elements in the Candida TERs strongly suggests that Candida species, just like S. pombe, utilize a partial splicing reaction for the processing of the 3′ ends of TERs. However, such splicing consensus sequences are not apparent at the 3′ ends of the Kluyveromyces and Saccharomyces TER genes, and there is evidence for a different mechanism used for this purpose (S. Larose, S. Abou-Elela, and R.J. Wellinger, unpubl.).
Pseudoknot
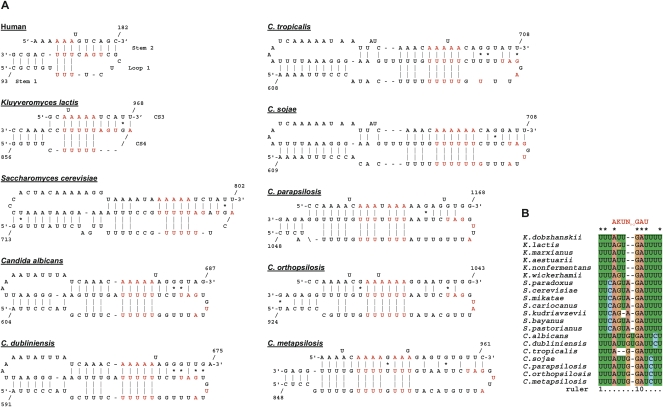
A pseudoknot element is crucial for telomerase function in ciliates, vertebrates, and Kluyveromyces species (ten Dam et al. 1991; Chen et al. 2000; Tzfati et al. 2003). Recently, the human, K. lactis, S. cerevisiae, and ciliate pseudoknots were proposed to contain U-A·U base triples (Theimer et al. 2005; Ulyanov et al. 2005, 2007; Shefer et al. 2007; Qiao and Cech 2008). Given the low sequence conservation among the Candida spp. TERs and the limited ability of existing computer programs to predict pseudoknots, we took the following approach. We first designed a specific computer algorithm to search for sequence parameters that are compatible with the conserved features of yeast and vertebrate TER pseudoknots: stem 1 of at least 4 bp, stem 2 of at least 6 bp, with no more than two bulged nucleotides or two mismatched base pairs, no more than four unpaired nucleotides between stem 1 and stem 2, and the potential of stem 2 and loop 1 to form at least three consecutive U-A·U base triples. Candidate sequences identified by this algorithm were then examined for their conservation in sequence and position within TER by Clustal X alignment. When this approach was tested on the Kluyvermyces TERs, pseudoknot elements composed of CS3 and CS4, identical to the published models, were predicted in all of the six sequences (Tzfati et al. 2003). When the Saccharomyces sensu stricto TERs were analyzed, a pseudoknot consistent with the published model was predicted (Lin et al. 2004; Qiao and Cech 2008), which includes eight potential U-A·U base triples (Fig. 4A). This algorithm was applied for each of the Candida spp. TER sequences, identifying a number of candidate pseudoknot elements. Then, two multiple sequence alignments (one of C. albicans, C. dubliniensis, C. tropicalis, and C. sojae TERs and another of C. orthopsilopsis, C. metapsilopsis, and C. parapsilopsis TERs) were searched with the candidate sequences to identify an element that is conserved in both sequence and position (with a freedom for misalignment of up to 4 nt). All the Candida spp. TERs contained sequences compatible with pseudoknot elements that can potentially form 8–11 U-A·U base triples (Fig. 4A). Additional C-G·U and C-G·C base triples may also form in some of the species. C-G·U and C-G·C base triples were predicted to maintain the triple helix formation (Theimer et al. 2005; Shefer et al. 2007; Qiao and Cech 2008). Since we did not test the predicted Candida spp. pseudoknot models experimentally, we do not know which of the potential base triples actually form. However, the remarkable conservation of sequences that are compatible with such a structure in the highly divergent Candida spp. TERs supports the model and suggests that at least some of these putative base triples in each sequence do actually form.
FIGURE 4.
Telomerase RNA pseudoknots. (A) Schematic representations of pseudoknot elements from human (based on Theimer et al. 2005), K. lactis (based on Shefer et al. 2007), S. cerevisiae (based on Qiao and Cech 2008), and putative schemes for the Candida pseudoknots predicted in this study. Indicated in red are the A and U residues potentially forming base triples and CS3 nucleotides conserved in all budding yeast TERs examined. (B) An alignment of the conserved CS3 residues from 20 Kluyveromyces, Saccharomyces, and Candida species. The consensus sequence (AKUN0–2GAU, where K indicates U or G) is indicated on top.
Interestingly, the alignment of the CS3 sequences revealed that not only the stretches of U residues that participate in the triple helix formation are conserved, but also the sequence within loop 1 and/or in the double-stranded part of stem 2. All the Kluyveromyces, Saccharomyces sensu stricto, and Candida spp. TERs contain the consensus sequence A(G/U)UN0–2GAU in this part of the pseudoknot (Fig. 4B). The functional significance of this consensus sequence is yet to be explored.
Est1-binding site
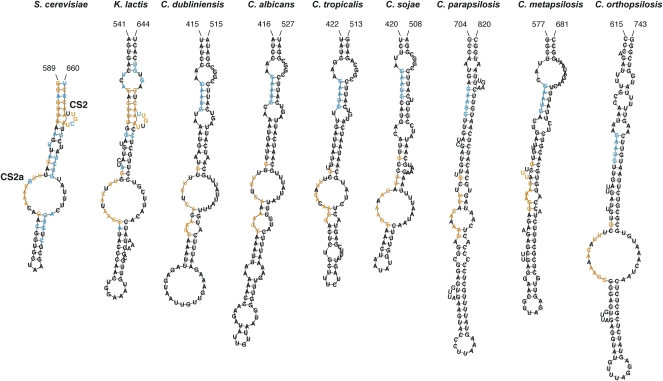
A conserved bulged stem in Saccharomyces and Kluyveromyces TERs was identified as a binding site for Est1p (Seto et al. 2002). Since no similar conserved sequence was apparent in the initial Clustal X alignment, we employed the computer program MEME (Bailey et al. 2006) to search for additional sequence motifs conserved in the Candida spp. TERs, independently of sequence alignment. This search identified a conserved sequence that resembles the sequence of the Est1 binding site reported previously (termed CS2 in Kluyveromyces TERs) (Supplemental Fig. S1; Seto et al. 2002; Tzfati et al. 2003). However, its predicted conformation is not conserved among the Candida spp. TERs. Interestingly, another conserved sequence, termed CS2a, was found ∼30–50 nt upstream of CS2. Secondary structure prediction of TER fragments containing CS2 and CS2a using the computer program RNAfold (Hofacker 2003) revealed a stem–loop structure with several internal loops (Fig. 5). The newly identified CS2a was predicted to be mostly in a single-stranded conformation, usually within a large internal loop in all the 20 Candida, Kluyveromyces, and Saccharomyces species examined.
FIGURE 5.
Conservation of the Est1p binding domain. Shown are secondary structure predictions made by the computer program RNAfold (Hofacker 2003) for K. lactis, S. cerevisiae, and seven Candida TERs. Indicated in blue are residues conserved in all species within each group of six Kluyveromyces, seven Saccharomyces, or seven Candida TERs. Indicated in orange are residues conserved in at least 18 of 20 yeast TERs (>90%).
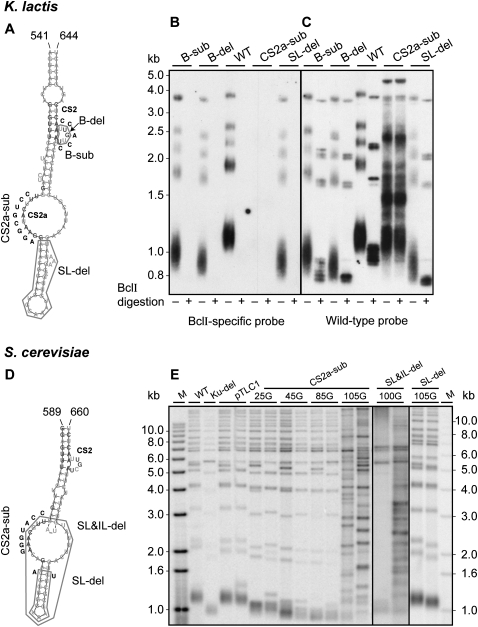
To test the importance of the CS2a and CS2 for telomerase function, we introduced several mutations into the K. lactis TER1 gene, as shown in Figure 6A. We replaced the wild-type (WT) TER1 gene in K. lactis cells with the mutant alleles using a plasmid shuffling system previously described (Roy et al. 1998) and analyzed telomere length and telomerase activity in vivo. An additional single-nucleotide mutation was introduced into the TER template, resulting in the incorporation of BclI restriction sites into the telomeric repeats. This mutation is otherwise apparently silent and thus can be used to mark the nascent repeats incorporated by telomerase and analyze its in vivo activity (Roy et al. 1998). The BclI repeats can be identified by Southern analysis of telomeric fragments hybridized to a BclI-specific telomeric probe (see Fig. 6B) or by BclI restriction endonuclease digestion of the telomeric fragments and hybridization to a wild-type probe (see Fig. 6C). Interestingly, the substitution or deletion of the CS2 bulge had impaired but not abolished the activity of telomerase in vivo, as revealed by the short but stable telomeres of the resulting mutant strains (Fig. 6B,C, B-sub, B-del). In contrast, the transition substitution of 8 nt in CS2a completely abolished telomerase activity, as revealed by the lack of BclI repeat incorporation (Fig. 6B, IL-sub) and the deregulated telomere length typical to the recombination-based alternative lengthening of telomeres (ALT) pathway (Fig. 6C, IL-sub). In addition, the deletion of the apical stem–loop severely reduced the length of the telomeres (Fig. 6B,C, SL-del), revealing that this stem–loop is also important for telomerase function. Analogous experiments were also carried out in S. cerevisiae (Fig. 6D,E). When both the internal loop and the apical stem–loop were deleted from the TLC1 gene (Fig. 6E, SL&IL-del), effectively deleting CS2a but leaving the CS2 bulge intact, the cells grew for about 60 generations before most of the cells on plates died (data not shown). Occasional colonies growing up on that plate were further propagated and DNA isolated after 100 generations. The DNA of those growing cultures displays typical patterns of terminal restriction fragments for survivors (Fig. 6E, SL&IL-del; type I and II for clone 1 and 2, respectively; Larrivée and Wellinger 2006). Furthermore, the transition substitution of 7 nt in CS2a also resulted in gradual telomere shortening, senescence, and the generation of survivors (Fig. 6E, CS2a-sub; type II). In contrast, no such growth problems and only slight telomere shortening were noted when only the apical stem was deleted (Fig. 6E, SL-del). Altogether, these results indicate that CS2a is essential for telomerase function, in both K. lactis and S. cerevisiae, consistent with its conservation in all budding yeast TERs examined. Whether CS2a contributes to the binding site for Est1p or has another role is yet to be tested experimentally. The importance of several elements relatively distant within the secondary structure model of this domain suggests that either they form tertiary interactions and a more complex structure that is required for the function of this domain or that different sequence elements provide different functions. The high-resolution structure and precise function of this domain are yet to be investigated in more detail.
FIGURE 6.
CS2a is essential for budding yeast telomerase function in vivo. (A) Deletions of the bulge (B-del) or the apical stem–loop (SL-del) or transition substitutions of the bulge (B-sub) or CS2a in the internal loop (IL-sub) were introduced into the Est1 binding arm of K. lactis TER1. Indicated in dark gray are residues conserved in at least 18 of 20 yeast TERs (>90%). Inserted nucleotides are in black and deletions are boxed. The rest of the sequence is in light gray. The wild-type and mutant TER1 genes, both containing the BclI template mutation (see main text), were introduced into K. lactis cells on CEN-ARS plasmids, replacing the WT TER1 gene, as described previously (Roy et al. 1998). gDNA was prepared from the sixth passage (∼90–120 generations). Two micrograms of gDNA samples were digested with EcoRI or EcoRI+BclI, electrophoresed on a 1% agarose gel, vacuum blotted onto a membrane, and hybridized first with a BclI-specific telomere probe (B) and then with a WT telomeric probe (C), as described previously (Shefer et al. 2007). (D) Deletions of the apical stem–loop and the internal loop (SL&IL-del) or only the apical stem–loop (SL-del) or transition substitution of 7 nt in CS2a (CS2a-sub) were introduced into an otherwise WT S. cerevisiae TLC1 gene on a plasmid (pTLC1TRP; Bah et al. 2004). The resulting plasmids were used to replace the WT TLC1 gene by plasmid shuffling. (E) Southern blot of gDNA derived from the resulting S. cerevisiae strains. DNA was isolated after the indicated numbers of generations of outgrowth, digested with XhoI, subjected to Southern blotting, and probed with an S. cerevisiae specific telomeric repeat DNA probe. DNAs used were as follows: M: end-labeled MW marker DNA; WT: strain RWY10 harboring pAZ1 (wtTLC1) before the plasmid shuffle; Ku-del: a S. cerevisiae strain harboring a deletion of the YKU70 gene as a control for short terminal restriction fragments; pTLC1: two independent clones of RWY10 harboring pTLC1 (WT) after the plasmid shuffle and grown for 100 generations; CS2a-sub: RWY10 harboring pTLC1-CS2a-sub and grown for 25, 45, 85, and 105 generations; SL&IL-del: RWY10 harboring pTLC1-SL&IL-del and grown for 100 generations; and SL-del: RWY10 harboring pTLC1-SL-del and grown for 105 generations. Where two independent clones are shown, clones 1 and 2 are on the left and right lanes, respectively.
Three-way junction
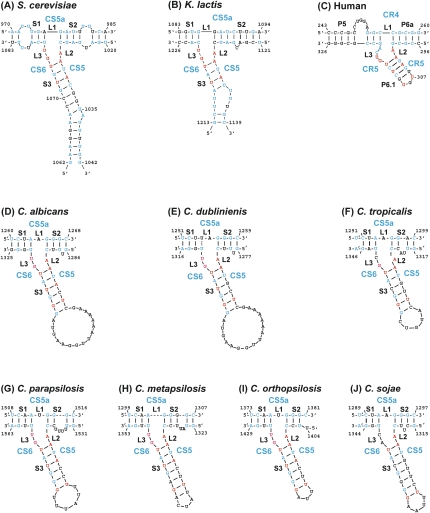
A conserved three-way junction (TWJ) similar to the P6.1 element in vertebrate TERs was identified recently in Kluyveromyces and Saccharomyces TERs (Brown et al. 2007). Mutational analysis in K. lactis confirmed that this element (and in particular the residues conserved across yeast and vertebrates) is critical for telomerase activity. We used phylogenetic covariation to identify the TWJ in the Candida spp. TERs. Since the initial multiple sequence alignment of the seven Candida spp. TERs did not seem to be reliable for the fragment between CS4 and the Sm site, we first aligned these TER fragments separately in two groups of species that are closer within each group. One includes C. parapsilosis, C. orthopsilosis, and C. metapsilosis, and the other includes C. albicans, C. dubliniensis, C. tropicalis, and C. sojae. We then subjected each of the two multiple sequence alignments to secondary structure prediction by the computer program RNAalifold, which combines free energy calculation and phylogenetic covariation (Hofacker et al. 2002). The predicted consensus structures revealed TWJ elements close to the 3′ ends of each structure. The conformation parameters of the predicted consensus structure were used to constrain Mfold to predict the structure of this domain for each of the Candida spp. TERs. The same structures were also predicted by RNAfold (Hofacker 2003) even without constraints, when given the correct sequence fragments (Fig. 7). Strikingly, these structures revealed a high degree of conservation of the TWJ among the yeast and vertebrate TERs. In particular, the linker L2 (a single A residue) and the first base pair in stem 3 (A-U) are absolutely conserved in all 55 vertebrate and budding yeast TERs examined. The linker L3 is GU in most yeast species, while in vertebrates, it is GUCA. Finally, either the closing base pair in stem 3 or the one before it is a U-G pair in all Candida spp. TERs, reminiscent of the conserved and essential U and G in P6.1. Altogether, the conservation of this element across vertebrates and yeast TERs suggests that it has a conserved role in telomerase function. Such a role, providing a binding site for TERT/Est2p, was suggested for the human P6.1 (Mitchell and Collins 2000) and supported by the analysis of the K. lactis TWJ (Brown et al. 2007).
FIGURE 7.
Conservation of the three-way junction element. Secondary structure models for the TWJ of human, K. lactis, S. cerevisiae, and seven Candida spp. TERs. Indicated in blue are nucleotides conserved in at least six of seven Saccharomyces spp., five of six Kluyveromyces spp., six of seven Candida spp., or 33 of 35 vertebrate TERs. Indicated in red are 9 nt conserved across yeast and vertebrates in at least 51 of the 55 TER sequences examined (>93%).
Comparative analysis of telomerase proteins
Using in silico analysis we have identified putative ORFs encoding Est2p, the catalytic subunit of telomerase, in all examined species (Supplemental Fig. S4A). Although the deduced protein sequences exhibit relatively weak similarity at the level of whole amino acid sequences, the regions involved in RNA binding and reverse transcription are well conserved, as demonstrated previously (Xia et al. 2000; Kelleher et al. 2002; Autexier and Lue 2006).
The situation with Est1p is more complicated. Whereas each of the genomes of C. albicans (Hsu et al. 2007b), C. tropicalis, C. dubliniensis, C. guilliermondii, C. lusitaniae, P. stipitis, and D. hansenii contain a single putative gene encoding a protein from the Est1p/Ebs1p family, we were unable to identify candidate EST1 genes in genomic sequences of C. parapsilosis and L. elongisporus by Blast search using Est1 sequences from different species as queries. Detailed inspection of the DNA sequences of C. parapsilosis and L. elongisporus corresponding by synteny to the genomic loci of the putative EST1 genes in the other Candida and Debaryomyces spp. indicated that the whole locus had been lost in these two species (Supplemental Fig. S4B). The Est1 proteins of C. albicans and C. tropicalis exhibit a relatively high degree of homology with each other (55% identity and 75% similarity). In addition to the Est One Homology (EOH) domain (Snow et al. 2003), the identified Est1 protein sequences exhibited higher sequence similarity within the region corresponding to amino acids 485–531 in S. cerevisiae Est1p (Supplemental Fig. S4C). However, even if these conserved domains were used as queries, we were unable to identify Est1p homologs in C. parapsilosis and L. elongisporus. Although we cannot rule out that EST1 in C. parapsilosis and L. elongisporus is located in a different genomic region and/or the sequence of Est1p is too distant from its counterparts to be detected by Blast, it is possible that the species belonging to a narrow branch of the phylogenetic tree (Fig. 1) have lost this protein component of the telomerase complex and its role is provided by another telomerase component.
CONCLUSIONS
We have identified TER genes from several budding yeast species, all of which harbor a long and precise telomeric repeat. Among them, seven Candida species belonging to two subgroups were evolutionarily close enough to allow phylogenetic analysis: One group includes C. albicans, C. dubliniensis, C. tropicalis, and C. sojae and the other includes C. orthopsilopsis, C. metapsilopsis, and C. parapsilopsis. First, multiple sequence alignments and identification of conserved elements were achieved within each group. Then, the alignments were merged and the conserved elements were compared with those identified in Saccharomyces, Kluyveromyces, and vertebrate TERs.
Interestingly, we predicted two elements that are conserved in their sequence and structural features across budding yeasts and vertebrates, suggesting that they provide conserved telomerase functions. The first, a pseudoknot element with a major groove U-A·U triple helix, was proposed previously in human, K. lactis, S. cerevisiae, and ciliate TERs (Theimer et al. 2005; Ulyanov et al. 2005, 2007; Shefer et al. 2007; Qiao and Cech 2008). We have now shown that sequences potentially forming such a structure are conserved in all other budding yeasts examined. The Candida spp. and Saccharomyces spp. pseudoknots are similarly large with 7–11 potential U-A·U base triples. In other species, the size of the pseudoknot is smaller: five potential triples in Kluyveromyces, three in vertebrates, and one to three in ciliates. It was recently suggested that the triple helix functions in positioning the 3′ end of the telomere at the catalytic site (Qiao and Cech 2008). This function is presumably conserved in all telomerases. The second element is a three-way junction highly conserved between vertebrate and yeast TERs. In ciliate TERs, stem IV was suggested to be the functional homolog of this domain (Mason et al. 2003). In human telomerase, P6.1 (the homolog of stem 3 of the yeast TWJ) was suggested to serve as a binding site for TERT (Mitchell and Collins 2000; Bachand et al. 2001) and to interact with the template domain (Ueda and Roberts 2004). Whether the yeast stem 3 provides similar protein–RNA and RNA–RNA interactions is yet to be tested.
A bulged-stem binding site for the telomerase protein Est1p was identified previously in S. cerevisiae TER (Seto et al. 2002), but we failed to identify this bulged stem in the Candida spp. TERs. Est1p was found to be essential for C. albicans telomerase function (Hsu et al. 2007b) and we identified additional Est1p homologs in several species (Supplemental Fig. S4B,C). However, we were unable to identify a putative Est1p encoding sequence in the genomes of C. parapsilosis and L. elongisporus. Interestingly, comparing the TER domain corresponding to the Est1p binding site in Saccharomyces, Kluyveromyces, and Candida spp. revealed a novel conserved sequence, CS2a, located within an internal loop structure in all species. Mutational analysis in K. lactis and S. cerevisiae revealed that this element is essential for telomerase activity in vivo, validating the general utility of our phylogenetic approach. However, it is currently unknown whether it is required for Est1p-binding or has a different role.
Using a reiterative folding procedure, preliminary secondary structure predictions for some of the identified TERs could be derived (Supplemental Fig. S3). Remarkably, a structure with a central core area with the respective template and from which several long stems emanate could be identified. Visually, these structures do resemble the ones predicted for the Saccharomyces and Kluyveromyces spp. (Dandjinou et al. 2004; Brown et al. 2007), and also further support the hypothesis that in these fungi, the global TER structure may be quite flexible (Zappulla and Cech 2004). While only exhaustive covariation analyses combined with biochemical mappings on many more variant sequences of Candida spp. TERs will be able to confirm these predictions, the data presented here suggest that indeed, all fungal TER RNAs may adopt similar functionally related structures. Importantly, within the global structures, small functional elements are much more conserved, reflecting their still largely unknown roles in telomerase function.
MATERIALS AND METHODS
Yeast strains
The following yeast strains were used in this work: Candida albicans CBS562NT, Candida dubliniensis CBS7987T, Candida guilliermondii CBS2030T, Candida metapsilosis MCO448T, Candida orthopsilosis MCO457T, Candida orthopsilosis IFM48364, Candida orthopsilosis IFM48386, Candida parapsilosis CBS604T CBS2194, and SR23, Candida sojae CBS7871T, Candida tropicalis CBS94T, Lodderomyces elongisporus CBS2605T and CBS5301, and Debaryomyces hansenii CBS767T. T and NT indicate Type and Neo-Type (respectively) strains of the species. The yeast cultures were grown in liquid YPD media (1% yeast extract, 1% peptone, 2% glucose) with constant shaking at 28°C until late exponential phase.
For the mutational analysis by plasmid shuffling in K. lactis, strain yJR27 (ter1Δ ura3-1 his2-2 trp1), based on the parental strain 7B520, was used, as described (Roy et al. 1998). For the plasmid shuffling experiments in S. cerevisiae, strain RWY10 (MATα, ura3-52, lys2-801, ade2-101, trp1-Δ63, his3-Δ200, leu2-Δ1, VR-ADE2-T, tlc1Δ∷LEU2), containing pAZ1 was used (Gravel and Wellinger 2002). pAZ1 is a URA3-based plasmid containing the complete wild-type genomic locus of TLC1. Plasmid pTLC1TRP (Bah et al. 2004) was used as a base vector for introducing the mutations in the Est1p binding domain. The shuffle was performed with RWY10 strains containing both pAZ1 and a pTLC1TRP (carrying WT or mutated TLC1 gene) vector. Cells were plated on FOA for growth without pAZ1 and subsequently restreaked on fresh plates for evaluation of telomere maintenance.
Identification of TER1 loci
We identified TER1 loci in the complete genomes of the following species: Ashbya (Eremothecium) gossypii ATCC 10895 (Dietrich et al. 2004), C. albicans SC5314 (Jones et al. 2004), D. hansenii CBS767T (Lépingle et al. 2000), and P. stipitis CBS6054 (Jeffries et al. 2007). We also analyzed unpublished genome sequences of C. albicans WO-1, C. guilliermondii ATCC6260T, C. lusitaniae ATCC42720T, C. tropicalis MYA-3404, and L. elongisporus NRRL YB-4239T downloaded from the sequencing projects of the Broad Institute (http://www.broad.mit.edu) and the sequence data of C. parapsilosis CDC317 and C. dubliniensis CD36T from the Sanger Institute (http://www.sanger.ac.uk). The locus of S. kluyveri was identified in the GenBank entry AACE02000358 from the whole genome shot-gun project (Washington University School of Medicine). In addition, we determined and analyzed TER1 loci in C. albicans CBS562NT, C. metapsilosis MCO448T, C. orthopsilosis MCO457T, Candida orthopsilosis IFM48364, Candida orthopsilosis IFM48386, C. parapsilosis CBS604T and SR23, C. sojae CBS7871T, and C. tropicalis CBS94T.
PCR amplification, cloning, and sequencing of TER1 loci
Based on the synteny within the TER1 loci identified in the complete genome sequences of species from the genus Candida, we amplified the corresponding region from the gDNA of selected yeast species by PCR and primers (Supplemental Table S1) derived from conserved motifs in the ORFs flanking TER1, using the GeneAmp High Fidelity PCR system (Applied Biosystems). Single PCR products were extracted from agarose gels and sequenced. When sequencing revealed the presence of two different alleles, the products were cloned using a PCR cloning kit into pDrive vector (Qiagen). At least two clones were then sequenced to identify both alleles. DNA sequences were determined using the BigDye Terminator v3.1 Cycle Sequencing kit and ABI3100-Avant Genetic Analyzer (Applied Biosystems). The sequence assembly and analysis were done using the Vector NTI Advance v. 10.1.1 software package (Invitrogen).
DNA sequence accession numbers
The sequences of TER1 loci determined in this work were deposited in the GenBank data library under the following accession numbers: EU477488, EU477489 (C. albicans CBS562NT), EU477484, EU477485 (C. metapsilosis MCO448T), EU477482, EU477483 (C. orthopsilosis MCO457T), FJ377369 (C. orthopsilosis IFM48364), FJ377368 (C. orthopsilosis IFM48386), EU477481 (C. parapsilosis CBS604T), EU477480 (C. parapsilosis SR23), EU477486, EU477487 (C. sojae CBS7871T), and EU477490 (C. tropicalis CBS94T).
RNA isolation and Northern blot analysis
Total cellular RNA from yeast cultures (C. albicans CBS562NT, C. dubliniensis CBS7987T, C. guilliermondii CBS2030T, C. metapsilosis MCO448T, C. orthopsilosis MCO457T, C. parapsilosis CBS604T, C. tropicalis CBS94T, and D. hansenii CBS767T) grown to early stationary phase in YPD medium was extracted by the standard phenol-chloroform method using glass beads (Cross and Tinkelenberg 1991). The RNA samples were then electrophoretically separated in 1.2% agarose/1.11% formaldehyde gels and capillary transferred overnight onto a Hybond N+ membrane (Amersham) with a Turboblotter Transfer System (Schleicher and Schuell) in DEPC treated 10× SSC (1.5 M NaCl, 0.15 M Na-citrate). RNA was fixed to the nylon membrane by a combination of baking for 1 h at 80°C and UV (312 nm) for 1 min (each side). Strips of membranes were prehybridized for 1–1.5 h at 47°C (C. albicans, C. dubliniensis, C. guilliermondii, C. metapsilosis, C. orthopsilosis, C. parapsilosis, and C. tropicalis) or 50°C (D. hansenii) in hybridization buffer (5× SSC [0.75 M NaCl, 0.075 M Na citrate], 5× Denhardt's solution [0.1% Ficoll 400, 0.1% polyvinylpyrrolidone, 0.1% BSA fraction V], 0.5% SDS) and subsequently hybridized under the same conditions with antisense oligonucleotide probes derived from the TER1 loci (Supplemental Table S1) labeled on their 5′ ends with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). In a control experiment, the blots were probed with the corresponding complementary (sense) oligonucleotides. The membranes were rinsed once with washing buffer (2× SSC [0.3 M NaCl, 0.03 M Na citrate], 0.1% SDS) for 5 min at room temperature and then twice in the same buffer for 15 min at 37°C (C. albicans, C. dubliniensis, C. guilliermondii, C. metapsilosis, C. orthopsilosis, C. parapsilosis, and C. tropicalis) or 40°C (D. hansenii). The analysis of the blots was done by Personal Molecular Imager (Bio-Rad).
Mapping of the ends of the TER transcripts
The 5′ ends of C. parapsilosis and C. tropicalis and the 3′ end of C. tropicalis were mapped using a modified protocol described by Polidoros et al. (2006). Approximately 1 × 107 to 5 × 107 yeast cells obtained from exponential growth phase cultures were pelleted, resuspended in 2 mL of lysis buffer (1 M sorbitol, 0.1 M EDTA, 0.1% 2-mercaptoethanol, 50 U zymolyase-20T) and incubated for 30 min at 30°C with occasional shaking. The total cellular RNA was extracted using the Ultraspec RNA isolation system (Biotecx Laboratories, Inc.). Isolated RNA (5–8 μg) was polyadenylated for 20 min at 37°C in a 10-μL reaction containing 1 mM ATP, 50 mM Tris-HCl (pH 7.9), 250 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, and 5 U of Escherichia coli poly(A) polymerase (New England Biolabs). An aliquot (2 μL; 1–1.6 μg RNA) of the polyadenylation reaction was mixed with 120 pmol of phosphorylated BamHI(T18) DNA primer, incubated for 5 min at 80°C, and then cooled down slowly to 42°C. The samples were then supplemented with ImProm-II Reaction Buffer (Promega), 3 mM MgCl2, 0.5 mM of all four dNTP (Fermentas), 20 U of RiboLock RNase Inhibitor (Fermentas), and 1 μL of ImProm-II Reverse Transcriptase (Promega) in a total volume of 20 μL and incubated for 5 min at 42°C and then 55 min at 55°C. The enzyme was inactivated by additional incubation for 15 min at 70°C. The samples were then treated for 20 min at 37°C with 5 U of RNase H (Fermentas), and the resulting cDNA was concentrated by ethanol precipitation. Purified cDNA was circularized by intramolecular ligation using CircLigase ssDNA ligase (Epicentre Biotechnologies) in a total volume of 10 μL. One third of each ligation reaction was used to amplify a fragment containing the ligated 5′ and 3′ ends in two subsequent PCR reactions with nested primers (Supplemental Table S1). PCR products were purified by gel extraction using the Wizard SV Gel and PCR Clean-Up System (Promega) and sequenced from both ends using oligonucleotides from the second nested PCR as primers.
The 5′ ends of C. orthopsilosis, C. metapsilosis, C. guillermondii, D. hansenii, L. elongisporus, and C. dubliniensis were mapped using the Ligation-Anchored PCR strategy described by Ansari-Lari et al. (1996). The 3′ ends of all TERs (with the exception of C. tropicalis) were mapped according to Lingner and Keller (1993) with modifications as described by Tzfati et al. (2003).
Pulsed-field gel electrophoresis
Chromosomal DNA samples from L. elongisporus CBS5301 and C. parapsilosis CBS2194, SR23, and CBS604T, were separated by PFGE in contour-clamped orthogonal field (CHEF) configuration (Pulsaphor, Pharmacia-LKB) in 0.8% agarose gel prepared in 0.5× TBE. The separation program included three steps of pulse switching (linear interpolation): (1) 10–200 sec for 48 h, (2) 200–400 sec for 16 h, and (3) 400–600 sec for 48 h at 100 V and 9°C, throughout.
Southern blot analysis of telomeric fragments
Gels were blotted onto a charged nylon membrane (Hybond N+, Amersham) and probed with the corresponding telomeric oligonucleotide (Supplemental Table S1) labeled with T4 polynucleotide kinase and [γ-32P]ATP. The hybridization in Figure 3A was carried out overnight at 60°C in 5× SSC, 5× Denhardt's solution, 0.5% SDS, washed once with 2× SSC, 0.1% SDS for 5 min at room temperature and two times for 15 min at 50°C. The hybridizations in Figure 3B,C were performed under the same conditions except that hybridization temperatures were 50°C for C. parapsilosis and 55°C for C. orthopsilosis and C. metapsilosis. The Southern blot analyses in Figure 6 were carried out as described previously (Gravel and Wellinger 2002; Shefer et al. 2007). The blots were visualized using a Personal Molecular Imager (Bio-Rad) or exposed to X-ray film.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank the members of our laboratories for stimulating discussions, L. Kovac (Department of Biochemistry, Comenius University, Bratislava) for helpful comments and continuous support, and Hiroshi Fukuhara (Institut Curie, Paris-Orsay, France), A. Vaughan-Martini (University of Perugia, Perugia, Italy), and Yuzuru Mikami (Research Center for Pathogenic Fungi, Chiba University, Chiba, Japan) for providing some of the yeast strains. Our work is supported by grants from the Fogarty International Research Collaboration Award (2-R03-TW005654-04A1 to L.T.), Howard Hughes Medical Institute (55005622 to J.N.), the Slovak grant agencies APVT (20-001604 and VVCE-0064-07 to L.T. and 0024-07 to J.N.) and VEGA (1/3247/06; 1/0132/09 to L.T. and 1/0219/08 to J.N.), Comenius University (UK/236/06 to S.G.), a grant from the Canadian Institutes for Health Research (FRN74438 to R.J.W.), the Laboratoire de Génomique Fonctionnelle de l'Université de Sherbrooke to R.J.W., and the United States–Israel Binational Science Foundation Grant 2005088, and the German-Israeli Foundation grant I-849-253.13/2004 to Y.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1194009.
REFERENCES
- Ansari-Lari M.A., Jones S.N., Timms K.M., Gibbs R.A. Improved ligation-anchored PCR strategy for identification of 5′ ends of transcripts. Biotechniques. 1996;21:34–36. doi: 10.2144/96211bm06. [DOI] [PubMed] [Google Scholar]
- Autexier C., Lue N.F. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- Bachand F., Triki I., Autexier C. Human telomerase RNA–protein interactions. Nucleic Acids Res. 2001;29:3385–3393. doi: 10.1093/nar/29.16.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A., Bachand F., Clair É., Autexier C., Wellinger R.J. Humanized telomeres and an attempt to express a functional human telomerase in yeast. Nucleic Acids Res. 2004;32:1917–1927. doi: 10.1093/nar/gkh511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch A.A., Lundblad V. The maintenance and masking of chromosome termini. Curr. Opin. Cell Biol. 2006;18:247–253. doi: 10.1016/j.ceb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Box J.A., Bunch J.T., Tang W., Baumann P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature. 2008;456:910–914. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- Brown Y., Abraham M., Pearl S., Kabaha M.M., Elboher E., Tzfati Y. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res. 2007;35:6280–6289. doi: 10.1093/nar/gkm713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell A.S., Lundblad V. Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol. Cell. Biol. 2004;24:7720–7736. doi: 10.1128/MCB.24.17.7720-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.L., Blasco M.A., Greider C.W. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- Cohn M., McEachern M.J., Blackburn E.H. Telomeric sequence diversity within the genus Saccharomyces. Curr. Genet. 1998;33:83–91. doi: 10.1007/s002940050312. [DOI] [PubMed] [Google Scholar]
- Cross F.R., Tinkelenberg A.H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- Dandjinou A.T., Levesque N., Larose S., Lucier J.F., Abou Elela S., Wellinger R.J. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- Dietrich F.S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pöhlmann R., Luedi P., Choi S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Gravel S., Wellinger R.J. Maintenance of double-stranded telomeric repeats as the critical determinant for cell viability in yeast cells lacking Ku. Mol. Cell. Biol. 2002;7:2182–2193. doi: 10.1128/MCB.22.7.2182-2193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L., Fekete M., Stadler P.F. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- Hsu M., McEachern M.J., Dandjinou A.T., Tzfati Y., Orr E., Blackburn E.H., Lue N.F. Telomerase core components protect Candida telomeres from aberrant overhang accumulation. Proc. Natl. Acad. Sci. 2007a;104:11682–11687. doi: 10.1073/pnas.0700327104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M., Yu E.Y., Singh S.M., Lue N.F. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot. Cell. 2007b;6:1330–1338. doi: 10.1128/EC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug N., Lingner J. Telomere length homeostasis. Chromosoma. 2006;115:413–425. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- Iida S., Imai T., Oguri T., Okuzumi K., Yamanaka A., Moretti-Branchini M.L., Nishimura K., Mikami Y. Genetic diversity of the internal transcribed spacers (ITS) and 5.8S rRNA genes among the clinical isolates of Candida parapsilosis in Brazil and Japan. Jpn. J. Med. Mycol. 2005;46:133–137. doi: 10.3314/jjmm.46.133. [DOI] [PubMed] [Google Scholar]
- Jeffries T.W., Grigoriev I.V., Grimwood J., Laplaza J.M., Aerts A., Salamov A., Schmutz J., Lindquist E., Dehal P., Shapiro H., et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat. Biotechnol. 2007;25:319–326. doi: 10.1038/nbt1290. [DOI] [PubMed] [Google Scholar]
- Jones T., Federspiel N.A., Chibana H., Dungan J., Kalman S., Magee B.B., Newport G., Thorstenson Y.R., Agabian N., Magee P.T., et al. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher C., Teixeira M.T., Förstemann K., Lingner J. Telomerase: Biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 2002;27:572–579. doi: 10.1016/s0968-0004(02)02206-5. [DOI] [PubMed] [Google Scholar]
- Langford C.J., Klinz F.J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984;36:645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Larrivée M., Wellinger R.J. Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat. Cell Biol. 2006;8:741–747. doi: 10.1038/ncb1429. [DOI] [PubMed] [Google Scholar]
- Leonardi J., Box J.A., Bunch J.T., Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- Lépingle A., Casaregola S., Neuvéglise C., Bon E., Nguyen H., Artiguenave F., Wincker P., Gaillardin C. Genomic exploration of the hemiascomycetous yeasts: 14. Debaryomyces hansenii var. hansenii. FEBS Lett. 2000;487:82–86. doi: 10.1016/s0014-5793(00)02285-7. [DOI] [PubMed] [Google Scholar]
- Lin J., Ly H., Hussain A., Abraham M., Pearl S., Tzfati Y., Parslow T.G., Blackburn E.H. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc. Natl. Acad. Sci. 2004;101:14713–14718. doi: 10.1073/pnas.0405879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Keller W. 3′-end labeling of RNA with recombinant yeast poly(A) polymerase. Nucleic Acids Res. 1993;21:2917–2920. doi: 10.1093/nar/21.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Hendrick L.L., Cech T.R. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes & Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- Livengood A.J., Zaug A.J., Cech T.R. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D.X., Goneska E., Greider C.W. Stem–loop IV of Tetrahymena telomerase RNA stimulates processivity in trans. Mol. Cell. Biol. 2003;23:5606–5613. doi: 10.1128/MCB.23.16.5606-5613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M.J., Blackburn E.H. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc. Natl. Acad. Sci. 1994;91:3453–3457. doi: 10.1073/pnas.91.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M.J., Hicks J.B. Unusually large telomeric repeats in the yeast Candida albicans. Mol. Cell. Biol. 1993;13:551–560. doi: 10.1128/mcb.13.1.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.R., Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- Peterson S.E., Stellwagen A.E., Diede S.J., Singer M.S., Haimberger Z.W., Johnson C.O., Tzoneva M., Gottschling D.E. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- Polidoros A.N., Pasentsis K., Tsaftaris A.S. Rolling circle amplification-RACE: A method for simultaneous isolation of 5′ and 3′ cDNA ends from amplified cDNA templates. Biotechniques. 2006;41:35–36. doi: 10.2144/000112205. [DOI] [PubMed] [Google Scholar]
- Qiao F., Cech T.R. Triple-helix structure in telomerase RNA contributes to catalysis. Nat. Struct. Mol. Biol. 2008;15:634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel N., Wolin S., Guthrie C. A subset of yeast snRNA's contains functional binding sites for the highly conserved Sm antigen. Science. 1987;235:328–331. doi: 10.1126/science.2948278. [DOI] [PubMed] [Google Scholar]
- Romero D.P., Blackburn E.H. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- Roy J., Fulton T.B., Blackburn E.H. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes & Dev. 1998;12:3286–3300. doi: 10.1101/gad.12.20.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto A.G., Zaug A.J., Sobel S.G., Wolin S.L., Cech T.R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Seto A.G., Livengood A.J., Tzfati Y., Blackburn E.H., Cech T.R. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes & Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto A.G., Umansky K., Tzfati Y., Zaug A.J., Blackburn E.H., Cech T.R. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA. 2003;9:1323–1332. doi: 10.1261/rna.5570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer K., Brown Y., Gorkovoy V., Nussbaum T., Ulyanov N.B., Tzfati Y. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol. Cell. Biol. 2007;27:2130–2143. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- Snow B.E., Erdmann N., Cruickshank J., Goldman H., Gill R.M., Robinson M.O., Harrington L. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 2003;13:698–704. doi: 10.1016/s0960-9822(03)00210-0. [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- ten Dam E., van Belkum A., Pleij K. A conserved pseudoknot in telomerase RNA. Nucleic Acids Res. 1991;19:6951. doi: 10.1093/nar/19.24.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer C.A., Blois C.A., Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfati Y., Fulton T.B., Roy J., Blackburn E.H. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- Tzfati Y., Knight Z., Roy J., Blackburn E.H. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes & Dev. 2003;17:1779–1788. doi: 10.1101/gad.1099403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda C.T., Roberts R.W. Analysis of a long-range interaction between conserved domains of human telomerase RNA. RNA. 2004;10:139–147. doi: 10.1261/rna.5118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulyanov N.B., Shefer K., Gorkovoy V., Nussbaum T., Brown Y., Abraham M., Pearl S., Shalev D., Tzfati Y. Model of a pseudoknot structure with a triple-helical stem from the telomerase RNA of Kluyveromyces lactis. J. Biomol. Struct. Dyn. 2005;22:808. [Google Scholar]
- Ulyanov N.B., Shefer K., James T.L., Tzfati Y. Pseudoknot structures with conserved base triples in telomerase RNAs of ciliates. Nucleic Acids Res. 2007;35:6150–6160. doi: 10.1093/nar/gkm660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C.J., Zakian V.A. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat. Struct. Mol. Biol. 2008;15:34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Peng Y., Mian I.S., Lue N.F. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla D.C., Cech T.R. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla D.C., Goodrich K., Cech T.R. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat. Struct. Mol. Biol. 2005;12:1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]