Abstract
Drosha is a type III RNase, which plays a critical role in miRNA biogenesis. Drosha and its double-stranded RNA-binding partner protein Pasha/DGCR8 likely recognize and cleave miRNA precursor RNAs or pri-miRNA hairpins cotranscriptionally. To identify RNAs processed by Drosha, we used tiling microarrays to examine transcripts after depletion of drosha mRNA with dsRNA in Drosophila Schneider S2 cells. This strategy identified 137 Drosha-regulated RNAs, including 11 putative pri-miRNAs comprising 15 annotated miRNAs. Most of the identified pri-miRNAs seem extremely large, >10 kb as revealed by both the Drosha knock-down strategy and by RNA PolII chromatin IP followed by Drosophila tiling microarrays. Surprisingly, more than a hundred additional RNAs not annotated as miRNAs are under Drosha control and are likely to be direct targets of Drosha action. This is because many of them encode annotated genes, and unlike bona fide pri-miRNAs, they are not affected by depletion of the miRNA processing factor, dicer-1. Moreover, application of the evofold analysis software indicates that at least 25 of the Drosha-regulated RNAs contain evolutionarily conserved hairpins similar to those recognized by the Drosha–Pasha/DGCR8 complex in pri-miRNAs. One of these hairpins is located in the 5′ UTR of both pasha and mammalian DGCR8. These observations suggest that a negative feedback loop acting on pasha mRNA may regulate the miRNA-biogenesis pathway: i.e., excess Drosha cleaves pasha/DGCR8 primary transcripts and leads to a reduction in pasha/DGCR8 mRNA levels and Pasha/DGCR8 synthesis.
Keywords: Drosha processing, miRNAs, Pasha
INTRODUCTION
miRNAs are small noncoding RNAs that regulate gene expression post-transcriptionally (Du and Zamore 2007; Matranga and Zamore 2007; for review, see Bartel 2004). Although minor differences exist between species, the RNA processing pathway that generates mature miRNAs is highly conserved. These small RNAs are produced in two sequential cleavage steps by two sets of heterodimeric complexes; both contain one RNase III family member and a dsRNA binding protein (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Lee et al. 2003; Liu et al. 2003; Denli et al. 2004; Gregory et al. 2004; Han et al. 2004; Forstemann et al. 2005; Saito et al. 2005). A RNA Polymerase II (Pol II) primary transcript, referred to as the pri-miRNA, is first processed within the nucleus into a 70 base-pair (bp) stem–loop pre-miRNA by Drosha, the RNase III family member, and its partner protein Pasha/DGCR8. The pre-miRNA product is transported by Exportin 5 from the nucleus to the cytoplasm (Yi et al. 2003; Lund et al. 2004), where it is further cleaved into a short duplex by a cytoplasmic complex that contains the RNase III Dicer-1, Ago1, and loqs in Drosophila (Du and Zamore 2005). One of the two strands is then chosen as the miRNA and loaded into the RISC complex (Khvorova et al. 2003; Schwarz et al. 2003), which then serves principally to target the 3′ untranslated region (3′ UTR) of many mRNAs and drive translational repression (Filipowicz et al. 2008).
Although a few miRNAs are processed by the miRtron pathway rather than by Drosha, the synthesis of most miRNAs appears to be Drosha dependent (Okamura et al. 2007; Ruby et al. 2007a); the prominent stem–loop in pri-miRNAs is recognized by Drosha together with its partner Pasha/DGCR8. Indeed, Pasha/DGCR8 is thought to bind preferentially at the junction between the stem and the more inflexible loop, and this process can be cotranscriptional (Kim and Kim 2007). This binding then positions Drosha midway up the stem so that it is properly positioned to make a pair of staggered breaks to generate the ∼70 bp pre-miRNA.
To identify the 5′ and 3′ ends of miRNA primary transcripts and to determine more generally the direct targets of the Drosha–Pasha/DGCR8 processing complex, we used tiling arrays to perform a genome-wide analysis in Drosophila S2 cells. We compared RNA from S2 cells treated with drosha dsRNA with RNA from cells treated with a control dsRNA. As expected, the drosha dsRNA led to accumulation of many miRNA precursors or pri-miRNAs; surprisingly a large fraction of these were very long (>10 kb) as assayed by tiling arrays. RNA pol II density in these putative pri-miRNA regions mirrors the RNA expression data from the Drosha-depleted S2 cells. Even more surprising was the identification of more than a hundred additional RNAs under drosha control. The effect of drosha is likely to be direct rather than an indirect consequence of inhibition of the miRNA pathway. This is because the steady-state concentrations of these RNAs were not altered when dicer-1 was depleted by RNAi. Using evofold hairpin predictions, we found strongly conserved structural hairpins in 25 of these 137 putative Drosha–Pasha/DGCR8 targets. Five of these hairpins are located in well-annotated mRNAs, suggesting that some coding genes are targets of Drosha processing. Remarkably, one resides in the 5′ UTR of pasha, the partner of Drosha, and there is an evolutionarily conserved hairpin in the 5′ UTR of DGCR8, its human counterpart. This finding suggests that some mRNAs contain Drosha cleavage sites and that a repressive feedback loop may operate in the miRNA-biogenesis pathway.
RESULTS
DROSHA knockdown leads to pri-miRNA accumulation
We performed a genome-wide analysis to identify potential targets of the RNA-cleaving enzyme Drosha. We treated Drosophila S2 cells with dsRNA against either drosha or a control mRNA, isolated total RNA from these cells, and generated a probe for hybridization to drosophila oligonucleotide tiling microarrays (Affymetrix). Unlike standard expression arrays, tiling arrays contain high density probes for intergenic and intronic regions as well as exonic regions. In principle, therefore, these arrays should be able to identify many targets of the Drosha–Pasha/DGCR8 complex, e.g., pri-miRNAs that accumulate upon knockdown of the nuclear processing complex (Fig. 1A).
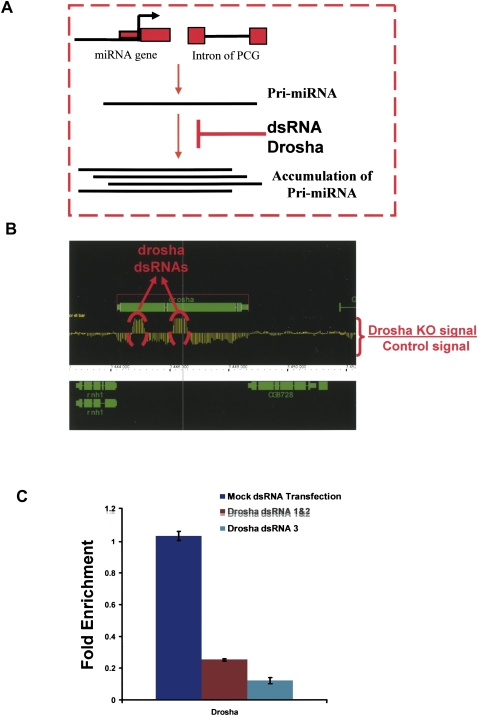
FIGURE 1.
Knockdown of drosha in S2 cells. (A) Schematics of the utilized strategy. Pri-miRNAs are transcribed from intergenic regions (miRNA genes) or contained in introns of protein coding genes (PCG). These pri-miRNAs are cleaved by a complex containing drosha and pasha. Inhibition of this pathway by the use of drosha dsRNA should result in pri-miRNA accumulation. (B) Incubation with drosha dsRNA reduces drosha mRNA levels. The graph represents the difference of expression between S2 cells treated with control (luciferase) or drosha dsRNA in the genomic region that contains drosha. Two samples for each condition were performed. The dotted red circles indicate the region of drosha to which the dsRNAs were directed. (C) Quantitative RT-PCR using primers for drosha shows that the level of drosha RNA decreases significantly in S2 cells upon treatment with two different combinations of dsRNAs against drosha.
As expected, incubation with two different dsRNA for drosha (either dsRNA1 and 2 together or dsRNA 3 alone) dramatically reduced drosha mRNA levels as assayed by microarray (Fig. 1B) or qRT-PCR (Fig. 1C). Since random primers were used to amplify total RNA and to label the microarray probe, all RNA molecules should be detectable. These include nonadenylated RNAs such as ribosomal RNA and transfer RNAs as well as the two dsRNAs used to knock down drosha mRNA levels in these experiments. Indeed, the addition of the dsRNAs caused a dramatic increase in the signal from these two subregions, compared to the prominent decrease in most of the mRNA (Fig. 1B).
Previous studies indicate that depletion of drosha mRNA leads to pri-miRNA accumulation (Lee et al. 2003; Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). Indeed, we easily detected precursors for 15 annotated miRNAs: 14 located in intergenic regions and one pri-mRNA within the intron of CG7033 (note that these 15 regions are defined based on the analysis described below) (Fig. 2A; Supplemental Fig. 1A). The data are consistent with previous reports, as all 15 pri-miRNAs have been previously described as Drosha dependent (Okamura et al. 2007; Ruby et al. 2007a). They also all harbor miRNAs known to be highly expressed in S2 cells (Ruby et al. 2007b). These 15 miRNAs were found to be up-regulated using a stringent algorithm (P-value = 1 × 10−5; Supplemental Table 1).
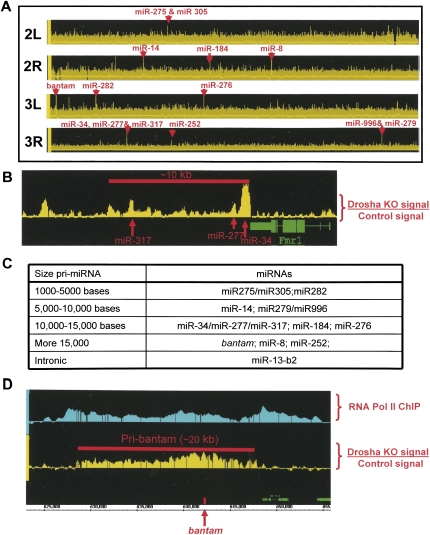
FIGURE 2.
Drosha knockdown leads to pri-miRNA accumulation. (A) The plot represents the difference of expression between S2 cells treated with control (luciferase) or drosha dsRNA among the second and third chromosomes of Drosophila. Red arrows indicate the coincidence of a significant peak (P-value < 1 × 10−5) with the location of a known miRNA. (B) The plot represents the difference of expression between S2 cells treated with control (luciferase) or drosha dsRNA in the gene region containing miR34, miR277, and miR317. Red arrows indicate the position of the mature miRNAs. (C) Table summarizing the length of the identified pri-miRNAs. (D) Image of the presumptive pri-bantam. The top graph represents the signal from the RNA Pol II Chromatin Immunoprecipitation (ChIP); the bottom graph represents the difference in expression between S2 cells treated with control (luciferase) or drosha dsRNA in the genomic region that contains bantam. The red arrow indicates the genomic location of the mature miRNA.
We noticed that depletion of Drosha causes the pri-miRNA signal to present a local intensity maximum (“bump”) around the location of the known miRNA (Fig. 2B). We also detected “bumps” not coincident with any known miRNAs (Fig. 2B), suggesting that these regions may also contain miRNAs, which are not yet described.
The tiling arrays analysis visualizes pri-miRNAs at high resolution. This allows an estimate of the transcription units, the 3′ as well as the 5′ limits of these pri-miRNA molecules. This obviously defines their sizes as well as their putative promoter regions. The 14 known nonintronic miRNAs are located in 10 transcription units. Surprisingly and despite the short size of the Drosha-excised product (around 70 bases), the pri-miRNA have a remarkable size (Fig. 2C): Over half of them are longer than 10 kb. Among the longest pri-miRNAs is the one harboring bantam (Fig. 2D), which is ∼20 kb long.
To confirm the size of these putative pri-miRNAs, we assayed RNA polymerase II density by chromatin immunoprecipitation (ChIP) followed by Drosophila tiling arrays. In the regions harboring the putative pri-miRNAs, the RNA polymerase II pattern mirrors the pri-miRNA signals revealed by the drosha dsRNA depletion (Fig. 2D; Supplemental Fig. 1B, 2A,B). This indicates that the pri-miRNA transcripts indeed reflect transcriptional units, which are longer by several orders of magnitude than the mature miRNAs they contain.
Genomic-wide identification of additional Drosha-processed RNAs
In addition to pri-miRNA accumulation, the drosha dsRNA causes many dramatic changes in RNA abundance. We identified at least 323 genomic regions upregulated upon drosha knockdown (P-value = 1 × 10−5) (Fig. 3A). We suspected that most of these were not a direct consequence of defective Drosha processing, because inhibition of the miRNA pathway has profound indirect effects on gene expression in S2 cells (Rehwinkel et al. 2006). We therefore compare the drosha knockdown results with those obtained with dsRNA against dicer-1 mRNA. Comparisons of the two tiling array profiles, i.e., depletion of Drosha versus Dicer-1, should distinguish between a direct effect, due to inhibition of Drosha processing, and an indirect effect, due to inhibition of miRNA production and therefore miRNA function. This is because incubation with dicer-1 dsRNA similarly disrupts miRNA biogenesis but does not lead to pri-miRNA accumulation (Du and Zamore 2005).
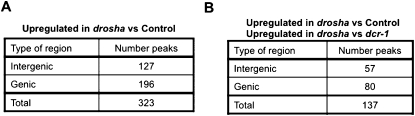
FIGURE 3.
Annotation of up-regulated peaks in drosha knockdown. (A) The table describes the location of the 323 peaks that are up-regulated in RNA samples from S2 cells treated with drosha dsRNA relative to S2 cells treated with a control dsRNA (luciferase). P-value threshold = 1e-05. (B) The peaks identified as up-regulated in the drosha knockdown were then compared against the ones obtained in the drosha–dicer analysis. This comparison resulted in 137 RNAs that were up-regulated in the drosha knockdown and not affected in the dicer 1 knockdown. Their location is shown in the table.
Indeed, most of the regions up-regulated due to drosha knockdown were also up-regulated in response to dicer-1 knockdown, suggesting that they are due to miRNA depletion. However, we identified 137 genomic regions up-regulated upon drosha knockdown and not affected or affected significantly less by depletion of dicer-1 mRNA (Fig. 3B; Supplemental Table 2). These 137 regions passed a significance test in the drosha versus dicer knockdown comparison as well as in the drosha versus control dsRNA comparison (both have a P value = 1 × 10 = 1e−5) and are putative direct targets of the Drosha–Pasha/DGCR8 RNase complex. These changes are not consequence of different drosha and dicer-1 knockdown efficiencies. This is because quantification of the tiling array data revealed that the changes in drosha and dicer-1 mRNA levels upon the incubation with the specific dsRNAs are equivalent (data not shown).
Identification of hairpins in Drosha-processed candidate transcripts
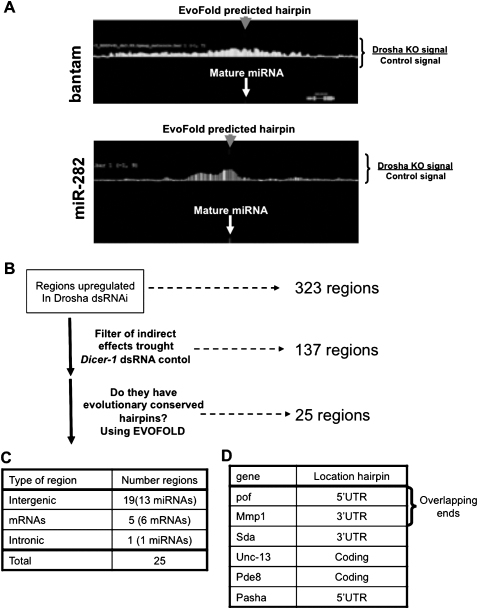
To test whether these 137 RNAs contain common features of drosha-cleaved RNAs, we searched for hairpins with secondary structure and conservation patterns similar to those of previously annotated pre-miRNAs. We utilized the published evofold hairpin predictions (Pedersen et al. 2006; Stark et al. 2007) to search for hairpins in these 137 putative drosha targets. For each candidate transcript, we also determined whether it contains a predicted hairpin that is evolutionarily conserved across the 12 sequenced Drosophila species (Stark et al. 2007). The search routine was validated by successfully locating the described hairpin structure coincident with the annotated microRNA of all 15 miRNAs identified in this study (Fig. 4A; data not shown). Of the 137 RNAs up-regulated upon drosha knockdown, 25 contain evofold-predicted conserved hairpins (Fig. 4B,C). This includes several mRNAs, including one that encodes the partner of Drosha, the dsRNA binding protein Pasha/DGCR8 (Fig. 4D).
FIGURE 4.
Prediction of hairpins with secondary structure in drosha-regulated RNAs. (A) The graph represents the difference in expression between S2 cells treated with control (luciferase) or drosha dsRNA in the genomic region that contains bantam (top) or miR-282 (bottom). The white arrows indicate the genomic location of the mature miRNAs. The gray arrows indicate the genomic location of the hairpins predicted by evofold for these genomic regions. (B) Schematics of the analysis performed (see Results and Materials and Methods). (C) Genomic location of the 25 putative direct drosha targets obtained after the analysis described in B. (D) Six mRNAs listed are putative direct drosha targets and contain an evofold predicted hairpin in their mature transcript.
Pasha/DGCR8 processing by Drosha: A mechanism to regulate miRNA processing?
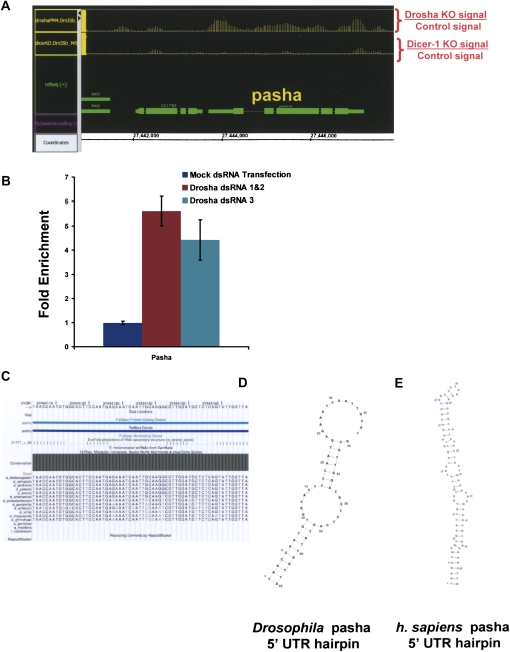
Pasha mRNA accumulates upon incubation of S2 cells with drosha dsRNA but not with dcr-1 dsRNA (Fig. 5A). We verified this result with a different dsRNA against drosha. Incubation with a new dsRNA amplicon (Fig. 5B, dsRNA 3) as well as the drosha dsRNAs utilized for the tiling array (Fig. 5B, dsRNA 1&2) caused a strong decrease in drosha mRNA levels as assayed by RT-PCR (Fig. 1C). Moreover, both strategies significantly increased the levels of pasha mRNA (Fig. 5B) in agreement with the tiling array results.
FIGURE 5.
Pasha/DGCR8 is a target of drosha. (A) Pasha mRNA is up-regulated upon knockdown of drosha, but not upon knockdown of dcr-1. The plot represents the difference of RNA abundance in the genomic region surrounding pasha produced by treatment of S2 cells with drosha dsRNA (top) or dicer dsRNA (bottom). (B) dsRNA treatment against drosha with two different combinations of dsRNAs leads to the accumulation of pasha mRNA. The graph represent the quantification pasha mRNA in S2 cells treated without dsRNA, with the combinations of drosha dsRNAs used in the previous experiments (dsRNA 1&2) or with another dsRNA against drosha (dsRNA3). (C) The plot illustrates the conservation of the evofold predicted hairpin located in the 5′ UTR of pasha. The structure is highly conserved among 12 fly genomes. (D) Secondary structure of Drosophila pasha hairpin as predicted by evofold (see Materials and Methods). (E) Secondary structure of human DGCR8/pasha hairpin located in the 5′ UTR as predicted by evofold. Structures were plotted as described in Materials and Methods.
The evofold analysis also predicts a strong hairpin in the pasha 5′ UTR, which is extensively conserved throughout the 12 sequenced Drosophila species (Fig. 5C,D). To examine further this putative Drosha substrate, we plotted the secondary structure of the Evofold-predicted hairpin with VARNA (RNAViz/RNAMovies). It compares favorably with hairpins in known miRNA drosha targets and is therefore a good candidate for a direct Drosha/Pasha target (Fig. 5D).
To examine whether this type of mechanism might be conserved in more distant species, we searched for conserved hairpins in the 5′ UTR of human DGCR8, the mammalian ortholog of pasha. We found a strong and conserved hairpin in the DGCR8 5′ UTR, suggesting that the function of the Drosophila hairpin is conserved and present in the common ancestor of flies and mammals (Fig. 5E).
DISCUSSION
Drosha is a type III RNase involved in miRNA biogenesis. With the aim of determining the targets of the Drosha–Pasha/DGCR8 processing complex, we performed a genome-wide analysis by knocking down drosha mRNA and identifying RNAs that accumulate with Drosophila tiling microarrays. Using this strategy, we identified 11 pri-miRNAs harboring 15 miRNAs. Our study also led to the identification of more than a hundred additional RNAs putatively under Drosha control.
Surprisingly, we found that most of the identified miRNA precursors are much larger than the miRNAs they harbor. For example, the precursor of bantam appears ∼20 kb long despite containing only one miRNA 23 nt in length (Fig. 2D). Although we cannot rule out the possibility that pri-miRNAs are much shorter and heterogeneous, the fact that the entire regions are transcriptionally active and up-regulated upon drosha knockdown is consistent with single, large transcription units. This interpretation also agrees with the observed profile of RNA pol II density in these regions as assayed by ChIP.
Only a single intronic miRNA, within CG7033, was identified as a Drosha target signal in the drosha versus dicer-1 comparison. Many intronic miRNAs reported to be expressed in S2 cells (Ruby et al. 2007b) were detectable by eye but not statistically significant, perhaps because of our stringent criteria. Other intronic miRNAs gave no signal. In some cases this is because the intronic pri-miRNAs were also up-regulated by the dicer-1 knockdown (data not shown). We therefore suspect that many transcripts within which these miRNAs are expressed have 3′ UTRs that are down-regulated via miRNA function and suggest that the miRNA within CG7033 differs quantitatively rather than qualitatively from other intron-containing miRNAs.
In a similar vein, the 10 nonintronic pri-miRNAs differ quantitatively from other intergenic pri-miRNAs. These 14 miRNAs are among the miRNAs with highest expression in S2 cells (Ruby et al. 2007b). We also could detect by eye inspection an increase in the levels of another 25 pri-miRNAs upon drosha knockdown (not statistically significant; Supplemental Table 1). These signals are too low, however, to estimate pri-miRNA sizes. It is therefore unclear whether the key feature of the 10 identified intergenic pri-miRNAs, a single known miRNA in ∼5–20 kb, is characteristic of other intergenic pri-miRNAs.
At least for the ∼20 kb bantam pri-miRNA, four results suggest that it may not contain additional functional miRNAs: (1) Some bantam deletion phenotypes can be rescued by expressing a much smaller pri-miRNA (Brennecke et al. 2003), (2) a similar conclusion was reported for phenotypes produced by overexpression of bantam (Bilen et al. 2006), (3) there are no additional known Drosophila miRNAs that map within this 20 kb (data not shown), and (4) evofold detected only a single hairpin within the bantam pri-miRNA region (Fig. 4A; data not shown). It is of course possible that these large pri-miRNAs still harbor additional as yet unidentified hairpins and miRNAs, of lower conservation, very low abundance, and/or only expressed in particular tissues.
It is perhaps noteworthy that bantam is highly abundant in S2 cells. This may be related to high transcription levels. Indeed, the present study should lead to a test of this hypothesis, by comparing the strength of pri-miRNA promoter regions of bantam with those of less abundant miRNA genes. Because of the large size of these transcripts, it would be impossible to find the promoter sequence of these intergenic pri-miRNAs from the genomic position of the mature miRNA.
It is also possible that high levels of bantam are related to the transcript size. A long primary transcript would provide more opportunity for the Drosha/Pasha complex to recognize a target hairpin cotranscriptionally, with or without pol II tethering of the processing complex (Kim and Kim 2007). As this processing step is nuclear restricted, rapid cytoplasmic transport after adenylation of shorter pri-miRNA transcripts may compete with Drosha/Pasha recognition and cleavage in the nucleus. As introns are probably retained in the nucleus by their snRNP association, cytoplasmic transport of intron-containing pri-miRNAs may not compete with nuclear processing of the hairpins by Drosha–Pasha.
In addition to annotated miRNAs, our study has identified new putative Drosha–Pasha targets. Surprisingly most of these mRNAs were not up-regulated in a previous study examining gene expression of S2 cells treated with dsRNA against Drosha (Rehwinkel et al. 2006). The differences can be attributed to (1) the fact that the previous study used expression arrays and oligo dT priming of cDNA synthesis rather than tiling arrays and random priming and (2) nonidentical S2 cell lines.
Of the 137 RNAs/peaks regulated by Drosha and not by Dicer-1, we selected a group of 25 based on the presence of conserved hairpin predictions by the program evofold (Pedersen et al. 2006). These hairpins are presumably processed directly by Drosha at some frequency, which lowers the percentage of intact primary transcripts that eventually access the cytoplasm. A decrease in Drosha activity, with drosha dsRNA for example, should therefore increase mRNA abundance. Twenty-five is a minimal number, since a substantial fraction of the other 112 RNAs may also be Drosha targets. It is also possible that several of these other putative Drosha targets are not well conserved in related species, as previously observed for several miRNA hairpins (Ruby et al. 2007b).
The selected 25 RNA regions include 19 intergenic regions, 11 of which do not include annotated genes. These may encode new pri-miRNAs, which may be present at very low levels in S2 cells. This would explain their absence even from deep sequencing assays (Ruby et al. 2007b). In addition, there may be cell-type-specific regulation in the cytoplasm, either miRNA turnover or processing by the cytoplasmic dicer-1 complex. In the case of these hairpins, this would result in highly cell-type-specific miRNAs despite more promiscuous transcription.
Five of these 25 regions encode mRNAs that may be Drosha targets. Two of them (Mmp1 and pof) have overlapping ends. Recent studies have shown that genes with overlapping ends can be substrates for the dsRNA processing machinery (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008). However, careful examination of the up-regulated genomic regions upon knockdown of drosha suggests that the effect is asymmetric and principally in Mmp1 (data not shown). The other four candidate mRNAs all have conserved hairpins in their 5′ UTRs, 3′ UTRs, or coding sequences.
One of these hairpins is located in the 5′ UTR of pasha, and drosha mRNA depletion probably leads to pasha mRNA accumulation by inhibiting cleavage of the pasha hairpin and as a result its 5′ UTR. The hairpin is extremely conserved among Drosophila species, and a strong hairpin is also present in the 5′ UTR of human homolog of pasha, DGCR8. The data suggest an interesting possibility: The miRNA-biogenesis pathway could be autoregulated by a negative feedback loop. Excess Drosha and perhaps Pasha/DGCR8 should then decrease pasha/DGCR8 mRNA levels. Similarly, a decrease in Drosha and Pasha/DGCR8 levels should cause an increase in pasha/DGCR8 mRNA levels. Although the main effect of Drosha is probably degradation of pasha/DGCR8 mRNA, it is possible that the cleaved hairpin is further processed by Dicer-1 to produce a miRNA.
In summary, this preliminary genomic level characterization of the cleavage targets of the Drosha–Pasha complex suggests that it not only processes pri-miRNAs but also specific mRNAs. This may constitute a new mRNA regulatory pathway, which functions independently of Dicer-1 within the nucleus.
MATERIALS AND METHODS
Double-stranded RNA synthesis and RNAi treatment
To knock down drosha and dicer in S2 cells, we followed the S2 cell RNAi protocol previously described (Nawathean et al. 2005) with the following exceptions. Three different dsRNAs were synthesized for drosha; dsRNA 1 and 2 were used in combination and dsRNA 3 (Park et al. 2004) was used alone. Two different dsRNAs were used in combination for dicer knockdowns and one dsRNA was synthesized against luciferase and was used as a control. In all cases, 20 μg of dsRNA were added to the S2 cells and after 3 d an additional 20 μg were added. After 5 d, RNA was extracted from cells using Trizol reagent (Invitrogen) according to the manufacturer's protocol.
Tiling arrays using total RNA
cDNA synthesis and labeling were carried out using the WT double-stranded cDNA synthesis kit (Affymetrix) and the WT double-stranded DNA terminal labeling kit (Affymetrix) as described by the manufacturer. Affymetrix Droshophila Tiling 1.0R arrays were probed, hybridized, stained, and washed according to the manufacturer's protocol.
Chromatin immunoprecipitation
ChIP protocol was adapted from Andrulis et al. (2000): Cells were grown in three wells of a six-well plate for 3 d prior to harvesting and cross-linking. Cells were harvested in 1× PBS and incubated in 0.5% formaldehyde for 15 min. Cells were incubated in 50 mM Tris/Glycine (pH 7.5) for 5 min. Cells were washed twice in 1 mL 1× PBS/0.1 M Glycine/0.1 M Tris (pH 7.5) and then resuspended in 1 mL ice-cold 1× PBS. Triton X-100 was added to final concentration of 0.5% and samples were incubated on ice for 10 min with periodic mixing. Lysed cells were centrifuged at 1500g for 10 min and the pellet nuclei were resuspended in 500 μL cold ChIP Lysis Buffer (1× PBS, 50 mM HEPES/KOH at pH 7.6, 2 mM EDTA, 1% Triton X-100, 0.1% Na-Deoxycholate, 0.2 mg/mL RNase A, Complete Protease Inhibitors). Sarkosyl was added to a final concentration of 2% and the sample was placed on ice for 30 min. The samples were sonicated with Fisher Scientific Sonic Dismembrator 550 at setting 2 twice for 20 sec “ON” and 40 sec “OFF” on ice. Sonicated samples were centrifuged at 15,000g for 15 min to remove insoluble debris. The resulting supernatent—“chromatin”—was transferred to a new eppendorf tube and diluted twofold in Lysis Buffer prior to being stored at −80°C or used immediately for immunoprecipitation.
For RNA PolII ChIPs, 2 μL of a Polyclonal RNA Polymerase II antibody made against the purified RNA polymerase II core enzyme (a generous gift from A. Greenleaf, Duke University) were added to 150 μL of chromatin and incubated overnight at 4°C. We saved 150 μL of chromatin as the “Input” sample. Protein G beads were blocked with 0.1 mg/mL yeast tRNA and 1 mg/mL BSA before being incubated with the chromatin–antibody mixture for 4 h at 4°C. The IPs were washed once with 1.5 mL ChIP Wash Buffer (1× PBS, 50 mM HEPES/KOH at pH 7.6, 1 mM EDTA, 1% Triton X-100, 0.1% Na-Deoxycholate, 0.1% Sarkosyl, 0.1% BSA, 0.5 M KCl, Complete Protease Inhibitors). Then, the beads were incubated in ChIP wash buffer for 30 min at 4°C before being washed 1× with ChIP Wash Buffer, 1× with Li Wash Buffer (10 mM Tris/HCl at pH 8.0, 0.25 M LiCl, 0.5% NP-40, 0.5% Na-Deoxycholate, 1 mM EDTA) and 1× in ice-cold TE. Beads were then resuspended in 150 μL of Elution buffer (50 mM Tris/HCl at pH 8.0, 10 mM EDTA, 1% SDS, 1 mM DTT, 0.1 mg/mL proteinase K) and incubated for 2 h at 37°C. Supernatant was moved to a fresh eppendorf tube and incubated at 65°C overnight to reverse cross-links. DNA was purified using the PCR Purification Kit (Qiagen).
Chromatin IP tiling arrays
Aliquots of 10 μL from both Input and RNA Pol II IP samples were amplified, fragmented, and labeled using the Affymetrix Chromatin Immunoprecipitation Assay Protocol, as described by the manufacturer. Affymetrix Drosophila Tiling 2.0R arrays were probed, hybridized, stained, and washed according to the manufacturer's protocol.
Data analysis
All tiling array data files were analyzed with the Model-based Analysis of Tiling-array software package (Johnson et al. 2006). RNA arrays were analyzed using a bandwidth size of 150, maximum probe gap of 300, minimum probe number of 5, and a P-value threshold of 1e-05. ChIP array were analyzed using a bandwidth size of 300, maximum probe gap of 300, minimum probe number of 10, and a P-value threshold of 1e-05.
Replicates of drosha and dicer knockdowns were each compared to luciferase RNAi control replicates, resulting in the identification of 323 and 350 peaks, respectively. In order to rule out dicer-dependent effects, drosha replicates were compared directly against dicer replicates, resulting in 479 peaks. The peaks identified in the drosha knockdown were then compared against the drosha–dicer analysis. This comparison resulted in 137 peaks that were present in both the drosha knockdown and drosha–dicer analyses. All the data have been deposited at NCBI GEO with accession number GSE14215.
Peak annotation
Peaks were annotated by taking the midpoint of the significant region and determining its location among genic or intergenic coordinates downloaded from the genome.ucsc.edu table browser for genome version dm3. In the drosha knockdown analysis, 196 of the 323 peaks had midpoints in regions covered by a gene. The other 127 peaks were located in intergenic regions. Of the 137 peaks that were found to be statistically significant in both the drosha and drosha–dicer analyses, 80 were located in regions covered by genes, while 57 were found in intergenic regions.
Conserved hairpin identification
Published conserved hairpins predicted by evofold (Stark et al. 2007) were visualized with the Affymetrix Integrated Genome Browser. Candidate peaks identified in the Drosha knockdown were then manually inspected for the presence of a hairpin. Twenty-five of the 137 peaks contained a conserved hairpin. Hairpin structures for Homo sapiens DGCR8 and Drosophila melanogaster pasha predictions were visualized using VARNA (http://www.lri.fr/∼ponty/VARNA/demo.html). The Homo sapiens hairpin prediction in DGCR8 was extracted from Pedersen et al. (2006).
Gene expression analysis by real-time PCR
Real-time PCR was performed as described previously (Kadener et al. 2008). The following primers were used:
rp49: 5′-ATCCGCCCAGCATACAG-3′, 5′-TCCGACCAGGTTACAAGAA-3′;
drosha: 5′-TCACCATCCACGAGCTAGACAT-3′, 5′-CCTTTCCATTATCTGGCAGGTC-3′; and
pasha: 5′-ACAACGTGGAACTTTGATTGG-3′, 5′-TGTTCTTCATTTTTGGCCACT-3′.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank A. Greenleaf for the RNA Pol II antibody, P. Zamore for comments on the manuscript, and P. Nawathean for helpful discussions. We also thank K. Palm for administrative assistance. The work was partly supported by NIH grants P01 NS44232, P30 NS45713, and R01 GM23549 to M.R. The microarray data have been deposited at NCBI GEO with accession number GSE14215. While this work was in progress, we learned that Kim and colleagues (Han et al. 2009) had independently identified mammalian Pasha as a putative Drosha processing substrate.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1319309.
REFERENCES
- Andrulis E.D., Guzman E., Doring P., Werner J., Lis J.T. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bilen J., Liu N., Burnett B.G., Pittman R.N., Bonini N.M. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol. Cell. 2006;24:157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J.A., Sachidanandam R., et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Du T., Zamore P.D. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Du T., Zamore P.D. Beginning to understand microRNA function. Cell Res. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Forstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M., Seitz H., Horwich M.D., Li C., Du T., Lee S., Xu J., Kittler E.L., Zapp M.L., Weng Z., et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha–DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Pederson J.S., Kwon S.C., Belair C.D., Kim Y.K., Yeom K.H., Yang W.Y., Haussler D., Blelloch R., Kim V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Johnson W.E., Li W., Meyer C.A., Gottardo R., Carroll J.S., Brown M., Liu X.S. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl. Acad. Sci. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S., Menet J.S., Schoer R., Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6:e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y., Saito K., Kin T., Ono Y., Asai K., Sunohara T., Okada T.N., Siomi M.C., Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Kim V.N. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Liu Q., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Matranga C., Zamore P.D. Small silencing RNAs. Curr. Biol. 2007;17:R789–R793. doi: 10.1016/j.cub.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Nawathean P., Menet J.S., Rosbash M. Assaying the Drosophila negative feedback loop with RNA interference in s2 cells. Methods Enzymol. 2005;393:610–622. doi: 10.1016/S0076-6879(05)93032-2. [DOI] [PubMed] [Google Scholar]
- Okamura K., Hagen J.W., Duan H., Tyler D.M., Lai E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Chung W.J., Ruby J.G., Guo H., Bartel D.P., Lai E.C. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.W., Parisky K., Celotto A.M., Reenan R.A., Graveley B.R. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl. Acad. Sci. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J.S., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Natalin P., Stark A., Brennecke J., Cohen S.M., Izaurralde E. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol. Cell. Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C.H., Bartel D.P. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007a;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007b;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Ishizuka A., Siomi H., Siomi M.C. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Stark A., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]