Abstract
Despite numerous structural and biochemical investigations, the catalytic mechanism of hairpin ribozyme self-cleavage remains elusive. To gain insight into the coupling of active site dynamics with activity of this small catalytic RNA, we analyzed a total of ∼300 ns of molecular dynamics (MD) simulations. Our simulations predict improved global stability for an in vitro selected “gain of function” mutation, which is validated by native gel electrophoretic mobility shift assay. We observe that active site nucleobases and water molecules stabilize a geometry favorable to catalysis through a dynamic hydrogen bonding network. Simulations in which A38 is unprotonated show its N1 move into close proximity of the active site 2′-OH, indicating that A38 may act as a general base during cleavage, a role that has generally been discounted due to the longer distances observed in crystal structures involving inactivating substrate analogs. By contrast, simulations in which N1 of A38 is protonated place N1 in close proximity to the 5′-oxygen leaving group, which supports the proposal that A38 serves as a general acid. In analogy to protein enzymes, we discuss a plausible mechanism in which A38 acts bifunctionally and shuttles a proton directly from the 2′-OH to the 5′-oxygen. Furthermore, our simulations suggest an important role for protonation of N1 of A38 in promoting a favorable geometry similar to that observed in transition-state analog crystal structures, and support previously proposed roles of A38, G8, and long residency water molecules in transition-state stabilization.
Keywords: RNA conformational change, hydrogen bonding network
INTRODUCTION
Over two decades of investigations into RNA catalysis have revealed numerous structural and dynamic properties of RNA that give rise to site-specific phosphoryl transfer. For example, the first ribozymes to be discovered, RNase P (Guerrier-Takada et al. 1983) and the self-splicing group I introns (Kruger et al. 1982), utilize specifically bound metal ions as cofactors in their catalytic mechanisms (Stahley and Strobel 2005), a strategy also used by protein-based RNases (Vogel et al. 2005; Mandel et al. 2006). Positioning of organic cofactors by the glmS ribozyme (Winkler et al. 2004), and the group I intron, is an additional means of specifying the reactive phosphate. In analogy, early hypotheses proposed that the small self-cleaving ribozymes use specifically bound divalent metal ions as the necessary base and acid catalysts. Subsequently it was recognized that the catalytic competence of several small self-cleaving RNAs (the hairpin, hammerhead, and Varkud satellite ribozymes) in a wide range of ionic conditions, including in the presence of monovalent ions alone, supports the notion that nucleobase functional groups directly support reaction chemistry (Murray et al. 1998). These naturally occurring ribozymes are now generally considered to rely, at least partially, on nucleobase facilitated proton transfer to carry out their cleavage and ligation function.
High-resolution crystal structures exist for the HDV (Ke et al. 2004, 2007), hammerhead (Martick and Scott 2006; Lee et al. 2008), glmS (Klein and Ferré-D'Amaré 2006; Cochrane et al. 2007), and hairpin ribozymes (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002; Alam et al. 2005; Salter et al. 2006; Torelli et al. 2007; MacElrevey et al. 2008; Torelli et al. 2008). Based on these structures only the hairpin ribozyme's active site is unambiguously devoid of stably bound metal ions that could potentially participate in catalysis with the possible exception of electrostatic stabilization, which could influence chemistry from outside the active site. Furthermore, the hairpin ribozyme is highly active in cobalt(III) hexammine, which can only form chemically inert outer-sphere complexes, while in contrast the HDV ribozyme is competitively inhibited by it (Hampel and Cowan 1997; Ke et al. 2004). This limited potential for a catalytic role of metal ions makes the hairpin ribozyme especially useful in probing the direct role of nucleobases in RNA catalysis. In addition, its small size has allowed a wide range of modifications and base substitutions to be incorporated, facilitating investigations into structure, function, and dynamics (Pinard et al. 2001; Lebruska et al. 2002; Ryder and Strobel 2002; Bokinsky et al. 2003; Kuzmin et al. 2004; Rueda et al. 2004; Kuzmin et al. 2005; Lambert et al. 2006; Cottrell et al. 2007). Despite intense study, much remains unclear about the chemical mechanism and the structural dynamics of the catalytically competent global conformation. Here we present the results of ∼300 ns of explicit solvent molecular dynamics (MD) simulations on the hairpin ribozyme in which we examine the active site geometry together with the role of both nucleobases and structural water molecules in facilitating RNA catalysis.
MD simulation is a computational method describing the dynamics of solvated nucleic acids using classical empirical potential force fields. The outcome of simulations is determined by the quality of the force field, by the affordable simulation time scale and by the starting structures. Therefore, MD simulations are a tool primarily suitable to provide complementary analyses of known RNA structures, including testing the effects of base substitutions, modifications, protonation, and deprotonation on structural dynamics and hydration. Although such a classical simulation does not allow bond breaking and creating, it can evaluate possible catalytic mechanisms by assessment of the conformational dynamics observed in the simulation (Cheatham 2004; Li et al. 2006; Auffinger and Hashem 2007; Deng and Cieplak 2007; McDowell et al. 2007; Musselman et al. 2007; Razga et al. 2007). This was our goal here.
Biochemical and structural data have implicated G8 and A38 as participants in the catalysis of cleavage and ligation. The conserved guanine G8 in loop A is positioned near the active site and mutation or deletion at this site impairs activity 100- to 1000-fold without significantly disrupting the ribozyme's global structure (Pinard et al. 2001; Kuzmin et al. 2004). The position of G8 near the reactive 2′-OH in crystal structures suggests the possibility that an unprotonated N1 acts as a general base (Pinard et al. 2001; Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002; Bevilacqua 2003; Salter et al. 2006). By contrast, exogenous nucleobase rescue experiments suggest that the roles of G8 in catalysis lie in charge stabilization of the transition state and/or alignment of the reactive groups (Lebruska et al. 2002; Kuzmin et al. 2004; Walter 2007; Cochrane and Strobel 2008). Our MD simulations are consistent with the latter model as they provide evidence that G8 facilitates catalysis through stabilizing both the developing charge on the scissile phosphate and the strained backbone conformations adopted along the reaction pathway.
Recent crystallographic studies of a transition state analog of the hairpin ribozyme have suggested that localized water molecules also play a role in transition state charge stabilization (Torelli et al. 2007), which is also supported by our MD simulations. Transition-state analog crystal structures place N1 of the invariant A38 close to the 5′-oxygen leaving group of the cleavage site, implicating A38 as the general acid (Rupert et al. 2002; Torelli et al. 2007; MacElrevey et al. 2008; Torelli et al. 2008). Abasic substitution of A38 impairs catalysis >10,000-fold and exogenous nucleobase rescue experiments indicate that the protonation state of A38(N1) plays a direct role in chemistry (Kuzmin et al. 2005). It has also been suggested that the catalytic roles of A38 are to align reactive groups and to stabilize negative charge in the transition state (Kuzmin et al. 2005; Cottrell et al. 2007). Our MD simulations offer support for multiple roles of A38 as the general acid, in aligning reactive groups, and in stabilizing negative charge. Contrary to the prevailing opinion, however, our results also implicate A38 as the general base in the cleavage reaction, leading us to consider a previously undescribed type of proton transfer mechanism for RNA involving a single nucleobase as bifunctional general base and acid catalyst, in analogy to protein enzymes, such as serine-carboxyl peptidases, and similar to roles proposed for glucose-amine-6-phosphate in the glmS ribozyme mechanism.
RESULTS
Overview of our simulations
We performed 15 charge neutralized MD simulations of the hairpin ribozyme for a combined total of ∼300 ns using explicit solvent, either with or without five crystallographically resolved interdomain water molecules and either with or without a single Mg2+ ion in a well-resolved metal binding site, all based on the 2.05–2.65 Å resolution crystal structures of junctionless forms of the hairpin ribozyme in complex with a noncleavable substrate analog (Table 1). We simulated three different sequences based on the available crystal structures for each variant; the native sequence containing a U39 (PDB ID 2D2K), a G8A mutant (in the following termed A8) that inhibits cleavage (1ZFV), and a U39C mutant (in the following termed C39) that increases activity (Fig. 1A, 2OUE; Berzal-Herranz et al. 1993; Joseph and Burke 1993). We also performed three simulations in which an active site adenosine was protonated at the N1 position (in the following termed A38H+), based on recent evidence for catalytic relevance of the ionized form (Kuzmin et al. 2005; Cottrell et al. 2007). Where present, a catalytically inactivating 2′-O-methyl modification of A-1 at the cleavage site was replaced with the natural 2′-OH.
TABLE 1.
MD simulations performed for the current study
FIGURE 1.
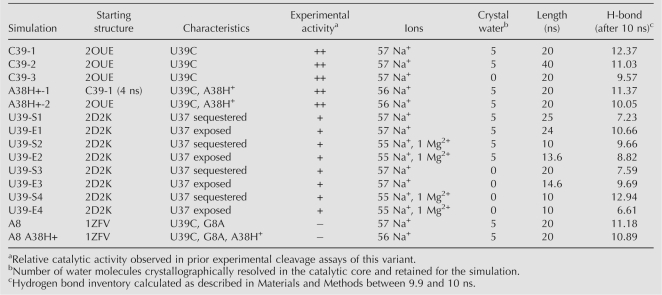
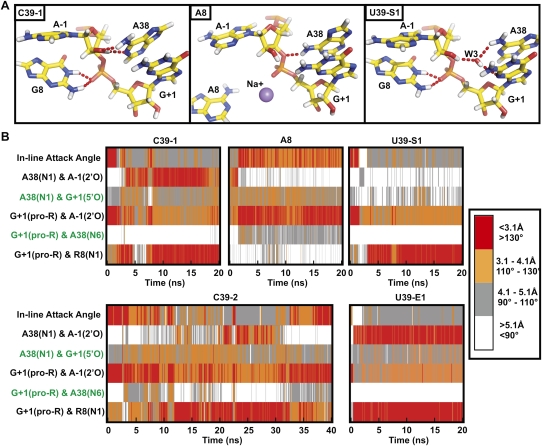
Backbone conformational dynamics and global stability observed in MD simulations agree well with experimental data. (A) Secondary structure schematic of the minimal hairpin ribozyme used in our simulations. The cleavage site between A-1 and G+1 is indicated by a red arrow. Important interdomain hydrogen bond interactions are indicated in blue (ribose zipper), purple (U42 binding pocket), and orange (G+1 binding pocket). The S-turn and E-loop motifs in loop B are indicated with red and yellow boxes, respectively. (B) Analysis of a highly contorted region of the backbone using the software Suitename (Richardson et al. 2008), whereby regions of the backbone that fall within one of the inferred low energy conformations are identified by their corresponding two character codes and outliers are identified by “!!.” Crystal structures and corresponding simulations are aligned to compare backbone conformations. Two regions of the backbone that are likely more accurately represented in the simulations than the corresponding crystal structures are highlighted in yellow. Backbone conformations of simulations were determined using coordinates averaged over the last 10 ns of each simulation. The degree of conformity of the entire backbone to conformations experimentally observed for RNA, in general, is indicated by the average suiteness given in parentheses. Suiteness can range from 0 to 1, where 1 signifies that all suites are exactly equal to the mean dihedral values associated with their respective conformation. (C) Histograms of the number of C39 (black) and U39 (gray) simulations with a given number of interdomain hydrogen bonds observed between 9.9 and 10 ns into the simulation. (D) Snapshots of the U42 binding pocket and part of the ribose zipper from the end of three simulations as indicated (averaged from 19.9 to 20 ns) with key hydrogen bonds indicated by dashed red tubes. (E) Density plots of the interatomic distances involved in key interdomain hydrogen bonds (color coded as in panel A) as tracked over the first 20 ns of each simulation. Distances are color coded as indicated in the legend, i.e., red indicates atoms within hydrogen bonding distance. (F) Experimental EMSA analysis of a two-way junction form of the hairpin ribozyme indicates greater abundance of predominantly docked (D) relative to predominantly undocked (U) conformers for the C39 gain-of-function mutant compared to the U39 wild type.
MD simulations properly predict backbone conformations as defined by their suiteness and provide indirect evidence for protonation of A38 in crystal structures
Given the large number of torsion angles in the RNA backbone, a particular challenge of MD simulations is to correctly predict backbone conformations over extended simulation times (McDowell et al. 2007). To evaluate the validity of our simulations, screen for potential force-field artifacts, and rapidly identify regions of interest we therefore applied the recently developed program Suitename (Davis et al. 2007; Richardson et al. 2008) to the backbone torsion angles in all our simulations. Overall, we observe that the conformity of the experimentally derived crystal structures with empirically inferred low-energy conformations is largely maintained in their corresponding simulations (Fig. 1B; Supplemental Fig. 2). Generally, backbone conformations that fall outside the empirically identified low energy conformations during the simulation also do so in the starting crystal structure, and therefore do not appear to arise from limitations of the parm99 (see Materials and Methods) force-field. In fact, we observe that some regions of the backbone are more accurately represented in the simulations. For example, the suite following U37 consistently shifts from 1g in the U39 crystal structure, usually observed as part of GNRA tetra loops, to the more conserved 2o suite observed in the higher resolution C39 crystal structures (highlighted in Fig. 1B). This result is consistent with regularization of the RNA backbone in the course of previous simulations of the HDV ribozyme (Krasovska et al. 2006). In general, backbone conformations in the hairpin ribozyme depart from empirically identified low energy structures more significantly in the U39 simulations than in the C39 containing simulations (Fig. 1B), potentially indicating increased dynamics of the U39 containing ribozyme.
Simulations in which N1 of A38 [A38(N1)] is not protonated diverge from the crystallographic backbone conformations more significantly than simulations in which N1 is protonated (Fig. 1B; Supplemental Fig. 2), providing indirect evidence that A38(N1) (unperturbed solution pK a = 3.5 [Bloomfield et al. 2000]) may be protonated in the crystal (pH = 6.0). The observed similarities between simulations of the G8A mutant in which A38 is protonated and the corresponding crystal structure, such as in overall backbone conformation and 2′O-P′-5′O in-line attack angle (IAA) of the scissile phosphate (simulation average = 107°; crystal = 112°), are consistent with protonation of A38(N1) in the G8A crystal also. Taken together, our results support previous biochemical (Kuzmin et al. 2005), structural (Rupert et al. 2002; Torelli et al. 2007), and computational evidence (Tang et al. 2007) for a functionally relevant shift in the pK a of A38.
MD simulations correctly predict enhanced docking stability of the C39 gain-of-function mutant
We have previously observed that MD simulations are effective at predicting the stability of the active docked state of the hairpin ribozyme. To assess stability, we used hydrogen bond inventories that add up all fractional hydrogen bonds as derived from the fraction of time that the heavy-atom distances of these hydrogen bonds are ≤3.1 Å and that the X–H–Y angle is within 60° of linearity. When taken at 10 ns after equilibration such hydrogen bond inventories were found to correlate linearly with the experimentally observed decreased stability of mutants, yielding a reasonable ΔΔG of 2.4 kcal/mol per hydrogen bond lost during the simulation (Rhodes et al. 2006). Hydrogen bond inventories of our current simulations predict that the inactive A8/C39 double mutant (Table 1, A8) should form a docked state equal in stability to the C39 gain-of-function mutant, consistent with previous electrophoretic mobility shift assays (EMSA) (Pinard et al. 2001). Additionally, our hydrogen bond inventories suggest that the U39 sequence is less stably docked than the C39 mutants due to a loss of, on average, ∼2 hydrogen bonds (Fig. 1C). To pinpoint this observation more specifically, we monitored the integrity of the U42 binding pocket, ribose zipper, and the G+1 binding pocket, three interactions known to be essential to docking stability (Fig. 1D,E; Bokinsky et al. 2003). The U39 wild-type sequence exhibits a relative loss of hydrogen bonding interactions particularly in the U42 binding pocket and the ribose zipper, which involves the 2′-OH functional groups of A10, G11, A24, and C25 (Fig. 1D,E; Supplemental Fig. 3; Table 1). These observations suggest that the C39 gain-of-function mutant increases activity indirectly by stabilizing the catalytically active docked state through tertiary contacts distal from C39, attesting to a significant long-range effect of the mutation.
To further test the validity of our MD simulations, we sought to experimentally assess the relative stabilities of the U39 wild type and C39 gain-of-function mutant by subjecting a common two-way junction form of the ribozyme to EMSA. We find that the C39 mutant is characterized by two bands of distinct mobility, whereas the U39 ribozyme lacks the more rapidly migrating band (Fig. 1E). It has been previously shown that mutations that disturb global stability result in the loss of the more rapidly migrating band, which contains a predominantly docked and thus compact species (Hampel and Burke 2001; Pinard et al. 2001; Gaur et al. 2008). Our EMSA results therefore suggest that indeed the U39 wild type is less stably docked than the C39 gain-of-function mutant, as also predicted by our simulations.
MD simulations corroborate an experimentally observed conformational heterogeneity in the U39 wild-type ribozyme
Crystal structures of the U39 wild-type ribozyme reveal two distinct conformations (termed “U37-sequestered” and “U37-exposed”) at position 37 within the S-turn of loop B (Fig. 1A; Alam et al. 2005). We performed MD on both conformations of the U39 ribozyme (Table 1) and based on hydrogen bond inventories do not observe any clear differences in stability between the two conformations (Table 1). Remarkably, after 19 ns of one of our four U37-sequestered simulations the ribozyme undergoes a conformational change in which U37 moves away from the G+1 binding pocket and adopts a conformation similar to the U37-exposed conformation; extending this simulation for an additional 5 ns following this conformational change did not result in a return to the U37-sequestered conformation (Supplemental Fig. 4). By contrast, in over 100 ns of U39 simulations, the U37-exposed conformation is never observed to adopt the U37-sequestered conformation, nor is the U37-exposed conformation observed in any of our C39 simulations. These results provide evidence, within the framework of a necessarily limited set of simulations, that the U39 wild-type sequence can transition from the U37-sequestered to the U37-exposed conformation within a single docking event, whereas the C39 mutation suppresses this conformational change, consistent with the observed crystallographic heterogeneity of the U39 wild type and its absence in the C39 gain-of-function mutant (Alam et al. 2005). It has been speculated that the enhanced conformational stability of the adjacent S-turn leads to, at least in part, the higher metal ion binding affinity and catalytic activity of the C39 mutant (Alam et al. 2005). Our data support this notion and further implicate increased docking stability as a source of catalytic enhancement of the C39 mutant.
MD simulations properly predict crystallographically resolved water molecules that are part of an extended interdomain hydrogen bonding network
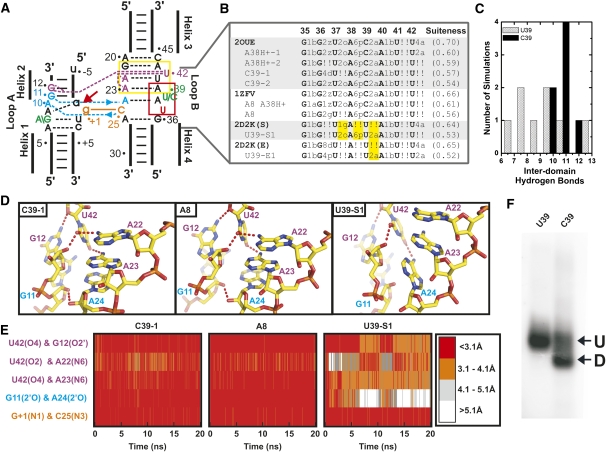
Consistent with previous x-ray crystallography (Alam et al. 2005; Salter et al. 2006) and MD studies (Rhodes et al. 2006), we observe long residency water molecules in an interdomain cavity near the catalytic core (Fig. 2). The two most stable structural water molecules are observed at the active site with residency times longer than can be characterized by our typically 20 ns simulations, while an additional set of three to five water molecules is present and remains relatively mobile within the interdomain cavity, but only slowly exchanges with bulk solvent (Fig. 2). All of these water molecules are an integral part of an extensive interdomain hydrogen bonding network (Fig. 2A), and notably are most stable in simulations of the C39 sequence that experimentally is most active and yields the crystal structures of highest resolution in which water is clearly identified (Salter et al. 2006). The behavior of explicit water in our simulations is therefore generally consistent with the crystal structures and previous MD simulations, providing collective evidence for an essential structural and potentially catalytic role for long residency water molecules in the hairpin ribozyme catalytic core.
FIGURE 2.
Long residency interdomain waters are an integral part of the interdomain hydrogen bonding network. (A) The positions of interdomain waters typical of our simulations are shown. (B) Specific interdomain waters were identified 10 ns into the simulation and interatomic distances of their hydrogen bonding partners were tracked over the entire length of three representative simulations. Distances are color coded as in Figure 1E. (C) The crystal structure of the noncleavable ribozyme–substrate analog complex (Pre, gray) (2OUE), as well as snapshots from two simulations (C39-1 and A38H+-1, color) overlaid with a vanadate TSA crystal structure (green) (2P7E). In the TSA crystal structure, a crystallographically resolved active-site water molecule in the precursor structure (W52) is displaced by the nonbridging pro-S oxygen, while flanking waters (W3 and W5) remain in nearly identical positions. In contrast, during simulations in which A38 is unprotonated (such as C39-1) the 2′-OH of A-1 takes the place of W52, while flanking waters remain in nearly identical positions. Similarly to the TSA structure, during simulations in which A38 is protonated (such as A38H+-1) the nonbridging pro-S oxygen takes the place of W52, while flanking waters remain in nearly identical positions.
More specifically, water molecules W3 and W5 in our simulations occupy nearly identical positions to water molecules resolved in crystal structures of vanadate transition state analog and noncleavable ribozyme-substrate analog complex (2P7E and 2OUE) (Fig. 2C). The stable binding of W5 bridging A9 and G+1 throughout our simulations regardless of protonation state of A38 and the resulting stable hydrogen bond donated to the pro-R nonbridging oxygen upon protonation of A38 support the notion of a role of structural water molecules in stabilizing the developing negative charge of the scissile phosphate in the transition state (Salter et al. 2006). In contrast, W52 is present in the noncleavable ribozyme-substrate analog complex, but absent in the vanadate transition state analog and, accordingly, is lost during our simulations (Fig. 2C). In our simulations the site occupied by W52 is instead occupied by A-1(2′OH) when A38 in unprotonated, and by the pro-R nonbridging oxygen in the protonated A38H+ simulations. The fact that the same water molecule (W52) is also experimentally observed to be replaced by the same pro-R nonbridging oxygen in crystal structures of both vanadate and 2′,5′-phosphodiester transition state analogs (TSAs) (2P7E and 2P7F) links A38 protonation to the ability to access the transition state geometry.
To test the impact of initial water placement on our MD simulations we performed five simulations in which the crystallographic water molecules were removed and all water was placed using the LEAP module of AMBER-8; these simulations began without any intercavity water (Table 1). During the course of each simulation water moved into the interdomain cavity. Differences between simulations were most pronounced at early time points with behavior converging as the simulations progressed, thus reflecting a longer equilibration period as observed previously (Rhodes et al. 2006). Accordingly, we noted a tendency for a sugar pucker flip at A-1 (discussed in detail below) to occur more slowly when experimentally resolved waters were excluded from the starting structure.
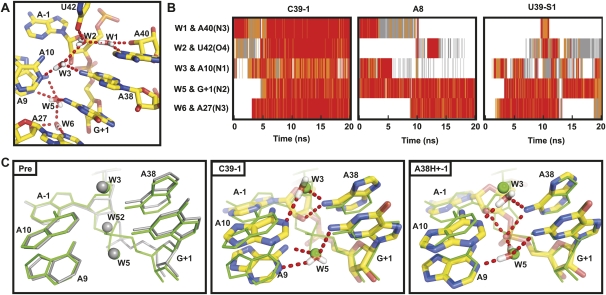
MD simulations reproduce experimentally observed metal ion binding sites
Generally, we observe good agreement between the available experimental data and the behavior of metal ions in our MD simulations, as previously noted (Rhodes et al. 2006). For example, we find the highest Na+ ion density at two sites within the E-loop (E1 and E2) of loop B, along the major groove of domain A (M), and in some simulations at the S-turn (S) in domain B (Fig. 3A). The Na+ density in the E-loop corresponds to two crystallographically observed calcium binding sites in the four-way junction crystal structures (Fig. 3B; Pinard et al. 2001) and, consequently, is strongly competed during our MD simulations upon placement of a Mg2+ ion into the E-loop (Fig. 3C). Notably, site E2 is only observed when the IAA is favorable and is lost upon formation of the hydrogen bond from A-1(2′OH) to A38(N1) discussed below. When present, monovalent ion density observed at the S-turn (Fig. 3A) corresponds to a high-affinity metal binding site identified through foot printing (Walter et al. 2000). In our A8 simulations (Table 1) we observe additional high Na+ density in close proximity to the nonbridging oxygens of the scissile phosphate.
FIGURE 3.
Monovalent ion behavior shows agreement with experimentally determined metal ion binding sites. (A) Regions of high Na+ density observed in our simulations are shown in green. High density is observed in the major grove MA and in the E-loop at site E1 in domain B in all of our simulations. High density is observed at site E2 only when the IAA is favorable. The crystallographically observed cobalt(III) hexammine binding site (blue) (2OUE) and an ion binding site observed through solution foot printing (red) (Walter et al. 2000) are indicated for comparison. High density at site S is only occasionally observed in our simulations. (B) Sites E1 and E2 in the E-loop are overlaid with the corresponding calcium binding sites (yellow sticks) and the crystallogaphically resolved calcium ions (transparent yellow spheres)) from the four-way junction hairpin ribozyme precursor crystal structure (1M5K) (Rupert et al. 2002). (C) The region of high Mg2+ density is shown in magenta, high sodium density is not observed at this site when Mg2+ is present. The Mg2+ binding site is overlaid with the corresponding cobalt(III) hexammine binding site (red) and the crystallographically resolved cobalt(III) hexamine (orange) from the crystal structure of the junctionless noncleavable ribozyme-substrate analog complex (2OUE).
Most of our simulations were carried out in charge-neutralizing monovalent cations alone due to the limitations in the parameters used for divalent ions (McDowell et al. 2007) and the catalytic proficiency of the hairpin ribozyme in monovalents alone (Rhodes et al. 2006). In four of our simulations we placed a single Mg2+ ion at a well-characterized multivalent outer-sphere metal ion binding site within the E-loop motif of Loop B (Fig. 3C; Alam et al. 2005), and again charge neutralized with Na+ ions (Table 1). We do not observe significant differences in either global stability or active site architecture upon inclusion of this Mg2+, consistent with observations from previous simulations (Rhodes et al. 2006). We only note that the outer-sphere bound Mg2+ ion quickly loses a water molecule of its inner coordination sphere and then directly chelates the RNA at U41 (Fig. 3C), consistent with the known bias toward inner-shell binding of Mg2+ ions in RNA simulations due to force field approximations (Reblova et al. 2006).
A 2′-OH substitution at the cleavage site restores an active site architecture perturbed in 2′-O-methylated crystal structures
The starting coordinates for our MD simulations were derived from high-resolution crystal structures of the minimal hairpin ribozyme in complex with a noncleavable substrate analog, solved with a cleavage blocking 2′-O-methyl substitution in place of the nucleophilic 2′-OH of residue A-1 (Alam et al. 2005; Salter et al. 2006). For our simulations we reverted back to the native 2′-OH, allowing us to test for any impact that the 2′-O-methyl substitution may have on active site architecture. In all but one of our simulations, we observe a flip in the A-1 sugar pucker from the 2′-endo/3′-exo of the ribozyme-substrate analog complex to a 2′-exo/3′-endo conformation (Fig. 4A), usually within the first few nanoseconds of the simulation and consistent with our previous observations (Rhodes et al. 2006). An analysis of a total of 16 MD simulations (including four previously reported simulations [Rhodes et al. 2006]) yields a single-exponentially distributed rate constant for this irreversible conformational change of k flip = 0.3 × 10−9 s−1 (Fig. 4B). The change in sugar pucker is accommodated by a slight reorientation of the whole nucleotide and frequently results in a hydrogen bond from the A-1(2′-OH) to A38(N1) (Fig. 4A). The formation of this additional, partially occupied (Fig. 5B; Supplemental Fig. 5), interdomain hydrogen bond is consistent with the increased stability of the cleavable ribozyme–substrate complex [containing A-1(2′-OH)] relative to the noncleavable ribozyme–substrate analog complex [containing A-1(2′-O-methyl)] as observed by single-molecule FRET (1.8 kcal/mol) (Liu et al. 2007). This hydrogen bond is precluded from forming in the crystal structure due to the presence of the 2′-O-methyl modification, which cannot donate a hydrogen bond and is not easily accommodated sterically by the active site when the A-1 sugar adopts the MD simulated 2′-exo/3′-endo conformation.
FIGURE 4.
Replacement of the inactivating A-1(2′-O-methyl) modification with the native A-1(2′OH) results in a local, functionally relevant conformational change. (A) Active site geometry of a C39 gain-of-function mutant simulation (color) overlaid with the noncleavable ribozyme-substrate analog complex crystal structure (PDB ID 2OUE, gray) illustrates a change in sugar pucker of A-1 that results in a hydrogen bond from A-1(2′OH) to A38(N1) and hydrogen bonds from G8 to the nonbridging pro-R oxygen (red dashed tubes). (B) The number of simulations that change sugar pucker after a given time (open circles) is well fit with a single-exponential (line).
FIGURE 5.
Active site dynamics provide insight into plausible roles of the functionally critical active site nucleobases G8 and A38. (A) Snapshots of the active site from the end of the three indicated simulations (averaged from 19.9 to 20 ns), in which N1 of A38 is unprotonated. Key hydrogen bonds are indicated by red dash tubes. (B) In-line attack angle and key interatomic distances over the course of each simulation, with distances and angles color coded as indicated in the legend. Close proximity of A-1(2′OH) and A38(N1) is frequently observed for ribozymes known to be catalytically active (C39 and U39) and is correlated with an unfavorable in-line attack angle. Hydrogen bonds observed in TSA crystal structures (2P7E, 2P7F, and 1M50, indicated in green) generally are not observed in these simulations.
Notably, four high-resolution precursor crystal structures containing the native 2′-OH together with inactivating mutations exist (Salter et al. 2006), none of which exhibit the 2′-endo/3′-exo sugar pucker observed in the ribozyme–substrate analog crystal structures. Two of these four structures adopt the 2′-exo/3′-endo conformation observed in our simulations, while the other two are assigned an unusual planar sugar ring (possibly representing an average of 2′-endo and 3′-endo sugar puckers), thus further supporting our proposal that a 2′-O-methyl modification of A-1 distorts the backbone geometry in the catalytic core by suppressing the favorable 2′-exo/3′-endo sugar pucker of A-1.
The simulated active site conformation rationalizes the experimentally observed higher importance of A38 compared to G8 in catalysis
The prevailing conformational change in our simulations toward a 2′-exo/3′-endo A-1 sugar pucker moves the 2′-OH away from G8, thus eliminating the intra-domain G8(N1) to A-1(2′O) hydrogen bond observed in the crystal structures (Figs. 4A, 5A, 6A). This finding contradicts proposals that G8 either acts as a general base or plays a direct role in the positioning and pK a suppression of the 2′-OH of A-1 for catalysis (Pinard et al. 2001; Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002; Salter et al. 2006). Instead, the observed new hydrogen bond from A-1 (2′-OH) to A38(N1) supports the notion that A38 plays the role of general base and hydrogen bonds from A38(N6) and A10(N6) to the A-1(2′-OH) provides an alternative means of pK a suppression of the 2′-OH. This emphasis of A38 over G8 is consistent with the significantly larger catalytic rate decrease (>10,000-fold) measured upon mutation or deletion of A38 compared to that observed upon mutation or deletion of G8 (only 100- to 1,000-fold) (Pinard et al. 2001; Kuzmin et al. 2004; Kuzmin et al. 2005; Cottrell et al. 2007), as well as the particularly critical role observed for the Watson–Crick face of A38 in shaping cleavage site architecture (MacElrevey et al. 2008; Torelli et al. 2008).
FIGURE 6.
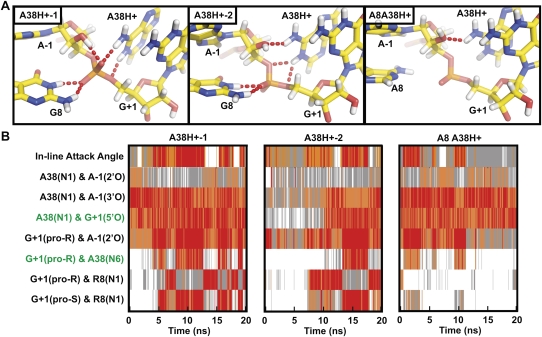
Protonation of A38(N1) results in a conformational rearrangement around the active site that establishes structural features predicted to be present within the transition state. (A) Snapshots of the active site from three simulations (averaged either from 9.9 to 10 ns for A38H+-1 or from 19.9 to 20 ns for A38H+-2 and A8A38H+) in which N1 of A38 is protonated. Key hydrogen bonds are indicated by red dash tubes. (B) In-line attack angle and important interatomic distances are tracked over the course of the simulation, with distances and angles color coded as in Figure 4. Proximity of the cleavage site G+1(5′O) and A38(N1) is frequently observed in all three simulations. A favorable IAA coincides with rotation of the nonbridging oxygens such that of G8(N1) loses its hydrogen bonding with the pro-R nonbridging oxygen in favor of a hydrogen bond with the pro-S nonbridging oxygen, as illustrated in the A38H+-1 snapshot of (A). Hydrogen bonding partners observed in TSA crystal structures (2P7E, 2P7F, and 1M50, indicated in green) are more frequently observed in these simulations.
In simulations with a charge neutral (unprotonated) A38(N1) we find that the 2′-OH of A-1 is the most common hydrogen bond donor to A38(N1), implicating A38 as a general base in the cleavage reaction. We note, however, that this hydrogen bond is observed only when the IAA of 2′O-P′-5′O is quite unfavorable (<130° or even lower; Fig. 5; Supplemental Fig. 5), far removed from the 180°, thought to be optimal for nucleophilic substitution (Soukup and Breaker 1999; Min et al. 2007), or the 162° predicted in the transition state for model reactions of RNA backbone cleavage (Lopez et al. 2006). A conformational change would thus be needed upon deprotonation of A-1(2′-OH) for cleavage to proceed, a notion that we address below. In addition to the prevalent hydrogen bond to A38(N1), we find that A-1(2′-OH) alternatively donates a hydrogen to the nonbridging pro-R oxygen, or to a long residency interdomain water bridging the A-1(2′-OH) and A38(N1) (Fig. 5; Supplemental Fig. 5). These two additional hydrogen bond acceptors have also been proposed to play a role in catalysis based on earlier simulations (Rhodes et al. 2006; Nam et al. 2008a,b).
Coincident with the change in A-1 sugar pucker, G8(N1) moves within hydrogen bonding distance of the nonbridging phosphate oxygens, specifically the pro-R oxygen, thus supporting the previous proposal that G8 stabilizes the increased negative charge on the scissile phosphate in the transition state (Lebruska et al. 2002; Kuzmin et al. 2004; Walter 2007). The 5′-oxygen leaving group is hydrogen bond acceptor to several transiently bound water molecules from bulk solvent that could potentially serve as specific acid catalysts in the cleavage reaction.
Protonation of A38 drives the active site toward the expected transition-state geometry
The significant biochemical and structural evidence for a shift in the pK a of A38 in the context of the docked ribozyme (Rupert et al. 2002; Kuzmin et al. 2005; Cottrell et al. 2007; Torelli et al. 2007) prompted us to carry out simulations in which N1 of A38 is protonated (A38H+), complementing our simulations with the charge neutral (unprotonated) A38 (Table 1). In striking contrast to the charge neutral A38 simulations, A38(N1H+) now primarily donates a hydrogen bond to the 5′-oxygen of G+1, the cleavage reaction's leaving group (and, to an extent, to the 3′-oxygen; Fig. 6), placing A38 in a position to serve as the general acid during catalysis. Furthermore, we observe that proximity of A38(N1) and G+1(5′O) frequently coincides with a favorable (>130°) IAA (Fig. 5). G8 moves again within hydrogen bonding distance of the nonbridging scissile phosphate oxygens, but now we observe alternating proximity to both the pro-S and the pro-R oxygens, with proximity to the pro-S oxygen strongly correlated with the ribozyme adopting a favorable IAA. These observations further support a primary role of G8 in stabilizing the negative charge of the scissile phosphate, rather than as a general base catalyst. Additionally, the nonbridging pro-R oxygen is the most common hydrogen bond acceptor of the A-1(2′-OH), consistent with the idea that it may be a suitable general base catalyst (Nam et al. 2008a,b).
The presence of a hydrogen bond from A38(N1H+) to the G+1(5′O) leaving group of the cleavage site and the adoption of favorable in-line attack angles both coincide with a rotation of the scissile phosphate, thus maximizing the number of intramolecular hydrogen bonds to the nonbridging oxygens, as expected for transition state stabilization. In particular the scissile phosphate rotates away from the precursor structure resulting in the pro-R oxygen accepting alternating hydrogen bonds from the exocyclic amines of A9 and A38, A-1(2′OH), and the highly stable specifically bound water molecule W5, while the pro-S oxygen accepts a bifurcated hydrogen bond from N1 and N3 of G8 (Figs. 2C, 6). Taken together, this dynamic conformation results in a hydrogen bonding network very similar to that observed in crystal structures of transition state analogs (TSAs) (Figs. 2C, 6; Rupert et al. 2002; Torelli et al. 2007). While this TSA-like conformation is observed ∼20% of the time in A38H+ simulations, it is not present in any of our simulations with a neutral A38, further supporting the catalytic relevance of the molecular dynamics observed in our A38H+ simulations.
Active site nucleobases stabilize strained backbone conformations
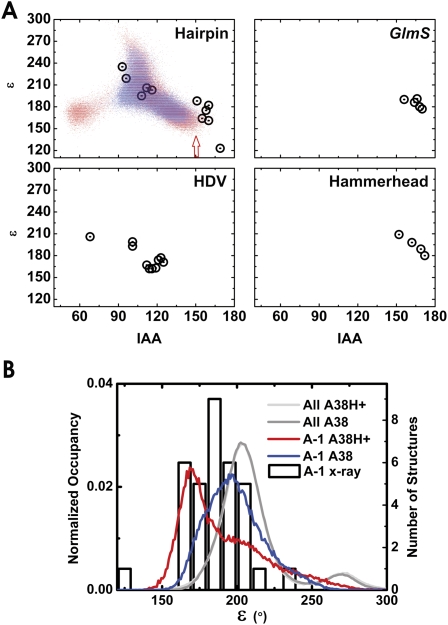
Not surprisingly, examination of the backbone conformations in our simulations and the starting crystal structures reveals that the cleavage site does not conform to any of the established RNA backbone conformational families (Supplemental Fig. 2; Richardson et al. 2008). Specifically the epsilon (C4′-C3′-O-P) dihedral is significantly smaller than that typically observed experimentally (Richardson et al. 2008). In our MD simulations a decreased epsilon angle correlates with an increased IAA (Fig. 7A), and inspection of multiple hairpin ribozyme crystal structures reveals the same correlation in experimental data (Fig. 7A). In order to determine how general this trend is in backbone cleavage, we evaluated all naturally occurring self-cleaving ribozymes for which high resolution crystal structures are available (hairpin, hammerhead, HDV, and glmS ribozymes) and find the same trend in all four ribozymes (Fig. 7A). The general trend of lower epsilon angles accompanying the high IAA angles required for catalysis suggests that distortion of this dihedral plays an important role in defining the cleavage site in self-cleaving RNA. Importantly, protonation of A38 results in a broader range of epsilon values at the cleavage site during our simulations (Fig. 7B), resulting in sampling of both lower epsilon and higher IAA angles than when A38 is not protonated (Fig. 7A). Regardless of A38 protonation state the distributions of epsilon angles at the cleavage site favor smaller values than all other regions of the backbone, consistent with the distribution range of epsilon angles that is experimentally observed at the cleavage site of self-cleaving RNAs (Fig. 7B).
FIGURE 7.
The cleavage site dynamics of the RNA backbone of our simulations share conformational features common among the small self-cleaving RNAs. (A) Cleavage site dihedral angle epsilon as a function of the IAA for all crystal structures with <3.1 Å resolution of the hairpin ribozyme, the glmS ribozyme bound to glucose amine 6-phosphate, the HDV ribozyme, and the full-length hammerhead ribozyme (open circles). Epsilon as a function of IAA is also shown for the A38H+ simulations (red dots), as well as the C39-1 and C39-2 simulations (blue dots). The A38H+ simulations sample a wider range of values in these plots; notably, the region of both lowest epsilon and highest IAA (red arrow) is more frequently sampled in the A38H+ simulations. (B) Probability densities of the cleavage site (A-1) epsilon angles as well as all backbone epsilon angles in our simulations (left axis and lines) and cleavage site (A-1) epsilon distributions among the available ribozyme crystal structures (right axis and open boxes). The epsilon angles at the cleavage site during our simulations and in the crystal structures both favor low values relative to the distribution observed in our simulations for all epsilon angles in the entire backbone. Moreover, simulations in which A38 is protonated (red line) favor even lower epsilon values than simulations where A38 is unprotonated (blue line).
DISCUSSION
A number of catalytic mechanisms have been proposed for the hairpin ribozyme. Cleavage is generally thought to proceed through a mechanism analogous to that carried out by RNase A where two histidine residues of near-neutral pK a act as separate base and acid catalysts and a protonated lysine residue neutralizes the increased negative charge of the scissile transition state phosphate (Doudna and Lorsch 2005; Fedor and Williamson 2005; Walter 2007; Cochrane and Strobel 2008). While the solution pK a's of RNA functional groups are far from neutrality, there is substantial evidence for pK a up-shifting in the context of the strong, long-range, negative electrostatic field of structured RNA (Knitt et al. 1994; Kuzmin et al. 2005; Gong et al. 2007; Tang et al. 2007; Torelli et al. 2007). Biochemical and computational approaches have suggested that in the catalytically active hairpin ribozyme the pK a's of one or all of the adenosines A10 (Ryder et al. 2001; Tang et al. 2007), A22 (Tang et al. 2007), and A38 (Rupert et al. 2002; Kuzmin et al. 2005; Tang et al. 2007; Torelli et al. 2007) are significantly perturbed toward neutrality. Our MD simulations indicate that A38 is positioned to concomitantly fulfill multiple catalytic functions potentially including general base catalysis (when unprotonated), transition state stabilization, and general acid catalysis (once protonated), consistent with the extraordinarily detrimental impact its replacement with an abasic site has on catalytic activity (Kuzmin et al. 2005).
MD simulations are generally known to have significant limitations in the attainable sampling (i.e., short simulated time spans) and underlying force fields (McDowell et al. 2007), however, they uniquely reveal detailed dynamics at an atomistic level that allow one to test and derive structure-based mechanistic hypotheses. Overall, we find surprisingly good agreement between the available experimental data and our multiple, independent, explicit-solvent MD simulations of the hairpin ribozyme (Table 1), suggesting that the AMBER-8 (parm99) (see Materials and Methods) force-field performs well in such an application. In particular, the backbone conformations generally remain consistent with those observed in the underlying crystal structures or make them more canonical; the C39 gain-of function mutant is correctly predicted to dock more stably and to show less conformational heterogeneity than the U39 wild type (Fig. 1); and our simulations reproduce the binding sites of crystallographically resolved water molecules and cations (Figs. 2, 3). We therefore focus our discussion on aspects of our MD simulations that provide unexpected insights into structural dynamics and catalysis of the hairpin ribozyme.
Crystallographically observed atomic distances have been used to propose that A38 and G8 function as the general acid and general base catalysts, respectively, in hairpin ribozyme catalysis (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002; Bevilacqua 2003; Alam et al. 2005; Salter et al. 2006; Torelli et al. 2007; MacElrevey et al. 2008). However, our simulations reveal relevant local distortions introduced into the crystal structures by the presence of a catalysis blocking 2′-O-methyl modification, and suggest alternative roles for these nucleobases in backbone cleavage. Most strikingly, the stable A-1(O2′H) to A38(N1) hydrogen bond, masked in crystal structures with A-1(2′-O-methyl) modification, but stably observed in our MD simulations (Fig. 4), opens the possibility that A38 acts as the general base. If instead G8 were the general base, as proposed previously (Pinard et al. 2001; Rupert and Ferré-D'Amaré 2001; Bevilacqua 2003; Thomas and Perrin 2006; Wilson et al. 2006), the catalytically relevant form of G8 would need to be deprotonated at position N1 (or be in a rare enol tautomeric form). However, exogenous nucleobase rescue experiments suggest that it is the N1 protonated (charge-neutral) state that activates catalysis (Lebruska et al. 2002; Kuzmin et al. 2004). Furthermore, based on comparison of precursor crystal structures of the U39C and G8A mutants it has been suggested that the role of G8 is to facilitate a favorable IAA by donating a hydrogen bond from G8(N1) and/or G8(N2) to A-1(O2′), and to lower the pK a of the 2′-OH (Salter et al. 2006). However, this hydrogen bond is lost in our simulations and we observe a hydrogen bond with the nonbridging oxygens instead. This is consistent with the observation that activity of a ribozyme with an abasic substitution at position 8 is rescued by a cationic exogenous nucleobase (Kuzmin et al. 2004). In addition, deprotonation of G8(N1) seems unlikely considering the pocket of deep negative electrostatic potential at the catalytic center of the ribozyme, whereas participation of the G8 tautomer is inconsistent with QM/MM simulations (Nam et al. 2008b). Overall, our data support a catalytic role of G8 in facilitating the reactive conformation through its interaction with the nonbridging oxygens and in stabilizing negative charge in the transition state, but not in general base catalysis. This leaves A38 as a prime suspect to remove the proton of A-1(O2′H). The exocyclic amines of A10 and A38 donate hydrogen bonds to the 2′OH in our simulations, which, similar to the role previously proposed for G8, should help to lower the pK a of the 2′-OH.
Alternatively, recent QM/MM simulations implicate the nonbridging scissile phosphate oxygens as reasonable general base candidates in hairpin ribozyme self-cleavage (Wilson et al. 2006; Nam et al. 2008a,b). The frequent hydrogen bonds between the A-1(2′OH) and the nonbridging oxygen observed in our MD simulations are consistent with such a role (Figs. 5, 6). Active site nucleobases such as G8, A9, A10, or A38 are in a position to potentially participate indirectly in catalysis by promoting proton transfer to these nonbridging oxygens (Figs. 5, 6). The proximity of A38(N1) and G+1(O5′) in several crystal structures of reaction precursor, TSA, and cleaved forms lend strong support to the proposal that A38 acts as general acid in the cleavage reaction (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002; Alam et al. 2005; Salter et al. 2006; Torelli et al. 2007; MacElrevey et al. 2008), and our simulations further emphasize the stability of this juxtaposition within hydrogen bonding distance, despite rearrangement of the A-1 sugar pucker. Biochemical evidence also suggests that A38 plays a vital role in stabilizing the catalytically productive active site architecture in a way that is dependent upon the protonation state of N1 (Kuzmin et al. 2005; Cottrell et al. 2007). Strikingly, our simulations begin with coordinates from an inactivated precursor but, over the course of simulations in which A38(N1) is protonated, undergo significant local structural changes in the active site to adopt characteristics exhibited by the experimentally determined TSAs (Figs. 2C, 6; Rupert et al. 2002; Torelli et al. 2007). In contrast, in simulations with an unprotonated A38(N1) a similar transition of the nonbridging oxygen to a TSA-like conformation is not observed (Fig. 5; Supplemental Fig. 5). Moreover, in simulations with the inactivating G8A mutation the transition is far less pronounced regardless of the A38 protonation state (Figs. 5, 6).
Taken together, our observations support a vital structural role of A38 protonation in stabilizing the catalytic geometry and provide further support for A38 as the general acid catalyst. The structural importance of A38 may explain why previous QM/MM calculations that imposed constraints on the hairpin ribozyme's active site geometry that were external to the standard force field unexpectedly predicted a similar free energy barrier to catalysis regardless of the protonation state of A38 (Nam et al. 2008b), despite strong experimental evidence to the contrary (Kuzmin et al. 2005; Cottrell et al. 2007). Our results indicate that maintenance of a favorable IAA requires protonation of A38 so that the protonation state of A38 is expected to significantly impact the overall energy barrier to catalysis in a way that can only be fully assessed if the dynamic ground state structure is kept unrestrained. The greater consistency between the conformations in our A38H+ simulations and the crystal structures than the unprotonated simulations further suggests that the protonated A38H+ is the predominant ionization state at the crystallographic pH of 6.0, consistent with the notion that its pK a is perturbed from its solution value of 3.5. It should be noted, however, that the ribozyme is most active at even higher pH [with a pK a,app for cleavage of 6.1 that varies among mutants with the identity of the functional group in the position that A38(N1) occupies in the wild type (Kuzmin et al. 2005)]; our simulations therefore may not reflect on the predominant protonation state at optimal cleavage conditions.
Our MD simulations depict the unprotonated A38(N1) as receiving a hydrogen bond from A-1(O2′H) (Figs. 4A, 5; Supplemental Fig. 5), whereas the protonated A38(N1H+) donates a hydrogen bond to the 5′-oxygen (Fig. 6). A recent TSA crystal structure containing a 2′,5′-linkage at the cleavage site exhibits an apparently bifurcated hydrogen bond between N1 of A38 and both the 2′- and 5′-oxygens of the cleavage site (Torelli et al. 2007), supporting the mechanistic relevance of our observations of both hydrogen bonds in MD simulations. Several mechanisms are consistent with these observations (Fig. 8). First, as previously proposed A38 may serve as the general acid, with A9, A10, water, or the nonbridging oxygens acting as general base (Fig. 8A). In the context of this mechanism, the A-1(2′OH) to A38(N1) hydrogen bond and associated poor IAA may be attributed to an unproductive ground state, wherein a perturbed pK a allows A38 to accept a solvent proton and subsequently aid adoption of the catalytically active conformation. Second, we propose a mechanism in which A38 acts bifunctionally as both general base and acid by shuttling a proton directly from A-1(O2′H) to G+1(O5′) (Fig. 8B). Such a mechanism is analogous to the role of, for example, an active site Glu in the first step of the reaction carried out by serine-carboxyl peptidases (Fig. 8C; Wlodawer et al. 2004; Xu et al. 2007). Here, the A-1(2′OH) to A38(N1) hydrogen bond would be essential to the mechanism; following transfer of the proton from A-1(2′OH) to A38(N1) the protonated A38H+ again promotes formation of the active geometry. This mechanism requires a certain degree of conformational flexibility in that proton exchange between 2′-OH and A38(N1) must be followed by a rapid transition of the deprotonated 2′-oxyanion to a position more favorable to in-line attack, as well as by a ∼0.4 Å motion of the now protonated A38(N1H+) toward the 5′-oxygen (Fig. 8B). We can infer that such dynamics are indeed possible as we observe that the protonated A38(N1H+) occasionally approaches the 2′-OH, while the unprotonated A38(N1) makes rapid excursions toward the 5′-oxygen on a picosecond time scale (Figs. 5B, 6B), however we do not formally attempt to simulate the actual 2′-oxyanion, but rather maintain the 2′OH at all times. Consistent with a bifunctional A38 base-acid mechanism, the asymmetric pH activity profile observed particularly in studies involving the more structurally stable, native four-way junction form of the hairpin ribozyme suggests the presence of a single titratable group (Kuzmin et al. 2005). In contrast, proton inventories are consistent with two protons in flight during the transition state as indicated in the first mechanism, however, the inventory data are sufficiently ambiguous that a one-proton mechanism cannot be discounted (Pinard et al. 2001). In addition, the interpretation of proton inventories is complicated by the fact that changes in hydrogen bonding alone can give rise to the underlying kinetic solvent isotope effects (Tinsley et al. 2003).
FIGURE 8.
Plausible mechanisms for the reversible cleavage/ligation by the hairpin ribozyme consistent with the active site dynamics observed during MD. (A) Mechanism in which protonation of A38 is required to drive the active site toward the catalytic geometry and the protonated A38H+ acts as the general acid, while A9, A10, a water, or a nonbridging oxygen could serve as the base in the cleavage reaction. (B) Mechanism in which A38 acts as both general base and acid by shuttling a proton (gray sphere) directly from the 2′-OH of A-1 to the 5′-oxygen of G+1 for the cleavage reaction. In both mechanistic proposals G8, A10, A38, and water 5 stabilize the negative charge accumulated in the transition state. (C) Mechanism proposed for the protein-based serine-carboxyl peptidase kumamolisin-As (Wlodawer et al. 2004; Xu et al. 2007), analogous to the mechanism in panel B. A glutamate side chain with acidic solution pK a (4.3) acts as both general base and acid by shuttling a proton directly from the serine nucleophile with solution pK a of (∼13) to the leaving group during formation of the acylated enzyme.
In conclusion, our results further support the importance of nucleobase functional groups in ribozyme catalysis, and the utility of MD simulations in exploring their impact in atomistic detail. Most significantly, our simulations expose the significant structural consequences of the protonation state of the active site A38 nucleobase, providing a detailed rationalization for previous mechanistic proposals based on experimental data of the hairpin ribozyme. Finally, our simulations reveal that in addition to transition state stabilization and general acid catalysis A38 should be considered a plausible candidate for general base catalysis.
MATERIALS AND METHODS
Initial MD structures, water, and ion placement
Table 1 summarizes all 15 (10–40 ns long) simulations performed for this study. Simulations of the C39 ribozyme were started from the 2.05 Å resolution crystal structure of the junctionless ligated hairpin ribozyme (PDB ID 2OUE). For Simulations C39-1 and C39-2 five interdomain water molecules were placed based on their crystallographic coordinates, a third simulation (C39-3) was initiated without these core water molecules. Eight simulations of the U39 sequence were started from the 2.65 Å resolution crystal structure of the same ribozyme (2D2K), using either the U37 sequestered (U39-S) or U37 exposed (U39-E) conformation. Half of the U39 simulations were run with a single Mg2+ placed based on the coordinates of a crystallographically resolved cobalt(III) hexammine binding site (Table 1; Alam et al. 2005). Another half of the U39 simulations (encompassing half of the Mg2+ containing simulations) were initiated with five interdomain water molecules, placed by superimposing the C39 structure (for which interdomain waters could be clearly resolved) on the U39 structure. A G8A mutant ribozyme was simulated based on the corresponding 2.4 Å resolution crystal structure (1ZFV), with interdomain water molecules placed by superimposing the C39 structure onto the G8A structure. An additional three simulations were performed in which A38 was protonated at atom N1 starting from either the junctionless C39 crystal structure (2OUE) or the G8A structure (1ZFV), and all three used the crystallographically placed interdomain water molecules. The LEAP module from the AMBER-8 software package was used to place additional bulk water molecules and a charge neutralizing number (55–57) of Na+ ions at points of favorable electrostatic surface potential.
MD equilibration and simulation
All MD simulations were carried out by using the AMBER-8 software package with the parm99 Cornell et al. force field (Cornell et al. 1995; Cheatham et al. 1999; Wang et al. 2000). The initial structures were solvated in a rectangular periodic box of TIP3P water molecules extending (at least) 10 Å from the RNA surface. The Sander module of AMBER-8 was used for the equilibration and production simulations based on our standard protocols (Krasovska et al. 2005; Krasovska et al. 2006; Rhodes et al. 2006; Sefcikova et al. 2007a,b). The particle mesh Ewald method (Essmann et al. 1995) was applied with a heuristic pair list update, using a 2.0 Å nonbonded pair list buffer and a 9.0 Å cutoff. A charge grid spacing of close to 1 Å and a cubic interpolation scheme were used. The production runs were carried out at 300 K with constant-pressure boundary conditions using the Berendsen temperature coupling algorithm (Berendsen et al. 1984) with a time constant of 1.0 ps. SHAKE (Ryckaert et al. 1977) was applied with a tolerance of 10−8 to constrain bonds involving hydrogens. Parameters for the protonated form of A38 were obtained by applying the RESP (Bayly et al. 1993) fitting method to the quantum mechanically (BLY/6-31G*) derived electrostatic potential. The charges on the backbone atoms were held constant with those present in the parm99 force field, and atom types were adjusted as necessary to maintain consistency with the parm99 force field (Supplemental Fig. 1).
MD analysis
Production trajectories were analyzed using the ptraj modules of the AMBER-8 package to obtain interatomic distances, angles, pseudorotation angles, and ion density maps. To obtain a detailed inventory of all direct interdomain RNA–RNA hydrogen bonds, the ptraj module of AMBER-8 was used to track the number of hydrogen bonds formed after 9.9–10 ns of simulation time. Fractional hydrogen bonds were calculated from the fraction of time that the heavy-atom distances of these hydrogen bonds were <3.1 Å and that the X–H…Y angle was within 60° of linearity. List density plots of distances were generated from the appropriate ptraj output using Matlab (The MathWorks, Inc.). Backbone torsion angle analysis was carried out using Molprobity (Davis et al. 2007). The average backbone suiteness was determined from all complete suites, including triaged suites. Snapshots averaged over 100 ps of MD simulation time were rendered using Pymol (DeLano Scientific LLC).
Electrophoretic mobility shift assay (EMSA)
The hairpin ribozyme sequences used were Strand RzA (5′-AAAUAGAGAAACGAACCAGAGAAACACACGCCAAA-3′) with the underlined residue at position 8 mutated from a G to an A to prevent cleavage during electrophoresis, and Strand RzB (5′-AUAUAUUUGGCGUGGUAYAUUACCUGGUACCCCCUCGCAGUCCUAUUU-3′) with the underlined pyrimidine representing either a U in the wild-type sequence or a C in the gain-of-function mutant. RNA was generated in vitro from single-stranded DNA templates containing a double-stranded T7 promoter as described (Milligan et al. 1987). Strand RzA was 5′-32P labeled using γ-32P-ATP and T4 polynucleotide kinase. The two composite strands were annealed in buffer (50 mM Tris–acetate at pH 7.5, 50 mM Na–acetate) by heating to 70°C for 2 min followed by cooling to 25°C, 10% (v/v) glycerol were added, and the sample loaded onto a nondenaturing 10% (w/v) polyacrylamide (19:1 acrylamide:bisacrylamide ratio) gel containing 50 mM Tris–acetate at pH 7.5, 12 mM Mg–acetate. Gels were run at 20 V/cm and 4°C for 15 h, and radioactive bands visualized using autoradiography with phosphorscreens and quantified using a PhosphorImager Storm 840 with ImageQuant software (Molecular Dynamics).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Michal Otyepka for helpful discussions and Kamila Réblová for providing the electrostatic potential used to derive partial charges of A38H+. This work was supported by NIH grant no. GM62357 (to N.G.W.) and an NIH Molecular Biophysics Training Grant fellowship (to M.A.D.) J.Š. was supported by the Academy of Sciences of the Czech Republic, grant nos. AV0Z50040507 and AV0Z50040702, and by grant no. IAA400040802 by the Grant Agency of the Academy of Sciences of the Czech Republic.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1416709.
REFERENCES
- Alam S., Grum-Tokars V., Krucinska J., Kundracik M.L., Wedekind J.E. Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn. Biochemistry. 2005;44:14396–14408. doi: 10.1021/bi051550i. [DOI] [PubMed] [Google Scholar]
- Auffinger P., Hashem Y. Nucleic acid solvation: From outside to insight. Curr. Opin. Struct. Biol. 2007;17:325–333. doi: 10.1016/j.sbi.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Bayly C.I., Cieplak P., Cornell W., Kollman P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., Van Gunsteren W.F., Dinola A., Haak J.R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- Berzal-Herranz A., Joseph S., Chowrira B.M., Butcher S.E., Burke J.M. Essential nucleotide sequences and secondary structure elements of the hairpin ribozyme. EMBO J. 1993;12:2567–2573. doi: 10.1002/j.1460-2075.1993.tb05912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua P.C. Mechanistic considerations for general acid-base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- Bloomfield V.A., Crothers D.M., Tinoco I. Nucleic Acids—Structures, Properties, and Functions. University Science Books; Sausalito, CA: 2000. [Google Scholar]
- Bokinsky G., Rueda D., Misra V.K., Rhodes M.M., Gordus A., Babcock H.P., Walter N.G., Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc. Natl. Acad. Sci. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatham T.E., III Simulation and modeling of nucleic acid structure, dynamics, and interactions. Curr. Opin. Struct. Biol. 2004;14:360–367. doi: 10.1016/j.sbi.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Cheatham T.E., III, Cieplak P., Kollman P.A. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- Cochrane J.C., Strobel S.A. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- Cochrane J.C., Lipchock S.V., Strobel S.A. Structural investigation of the glmS ribozyme bound to its catalytic cofactor. Chem. Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- Cottrell J.W., Kuzmin Y.I., Fedor M.J. Functional analysis of hairpin ribozyme active site architecture. J. Biol. Chem. 2007;282:13498–13507. doi: 10.1074/jbc.M700451200. [DOI] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., 3rd, Snoeyink J., Richardson J.S., et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng N.J., Cieplak P. Molecular dynamics and free energy study of the conformational equilibria in the UUUU RNA hairpin. J. Chem. Theory Comput. 2007;3:1435–1450. doi: 10.1021/ct6003388. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., Lorsch J.R. Ribozyme catalysis: Not different, just worse. Nat. Struct. Mol. Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- Fedor M.J., Williamson J.R. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- Gaur S., Heckman J.E., Burke J.M. Mutational inhibition of ligation in the hairpin ribozyme: Substitutions of conserved nucleobases A9 and A10 destabilize tertiary structure and selectively promote cleavage. RNA. 2008;14:55–65. doi: 10.1261/rna.716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B., Chen J.H., Chase E., Chadalavada D.M., Yajima R., Golden B.L., Bevilacqua P.C., Carey P.R. Direct measurement of a pKa near neutrality for the catalytic cytosine in the genomic HDV ribozyme using Raman crystallography. J. Am. Chem. Soc. 2007;129:13335–13342. doi: 10.1021/ja0743893. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hampel K.J., Burke J.M. A conformational change in the “loop E-like” motif of the hairpin ribozyme is coincidental with domain docking and is essential for catalysis. Biochemistry. 2001;40:3723–3729. doi: 10.1021/bi0028385. [DOI] [PubMed] [Google Scholar]
- Hampel A., Cowan J.A. A unique mechanism for RNA catalysis: The role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol. 1997;4:513–517. doi: 10.1016/s1074-5521(97)90323-9. [DOI] [PubMed] [Google Scholar]
- Joseph S., Burke J.M. Optimization of an anti-HIV hairpin ribozyme by in vitro selection. J. Biol. Chem. 1993;268:24515–24518. [PubMed] [Google Scholar]
- Ke A., Zhou K., Ding F., Cate J.H., Doudna J.A. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- Ke A., Ding F., Batchelor J.D., Doudna J.A. Structural roles of monovalent cations in the HDV ribozyme. Structure. 2007;15:281–287. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Klein D.J., Ferré-D'Amaré A.R. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Knitt D.S., Narlikar G.J., Herschlag D. Dissection of the role of the conserved G•U pair in group I RNA self-splicing. Biochemistry. 1994;33:13864–13879. doi: 10.1021/bi00250a041. [DOI] [PubMed] [Google Scholar]
- Krasovska M.V., Sefcikova J., Spackova N., Šponer J., Walter N.G. Structural dynamics of precursor and product of the RNA enzyme from the hepatitis delta virus as revealed by molecular dynamics simulations. J. Mol. Biol. 2005;351:731–748. doi: 10.1016/j.jmb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Krasovska M.V., Sefcikova J., Reblova K., Schneider B., Walter N.G., Šponer J. Cations and hydration in catalytic RNA. Molecular dynamics of the hepatitis delta virus ribozyme. Biophys. J. 2006;91:626–638. doi: 10.1529/biophysj.105.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Kuzmin Y.I., Da Costa C.P., Fedor M.J. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- Kuzmin Y.I., Da Costa C.P., Cottrell J.W., Fedor M.J. Role of an active site adenine in hairpin ribozyme catalysis. J. Mol. Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Lambert D., Heckman J.E., Burke J.M. Cation-specific structural accommodation within a catalytic RNA. Biochemistry. 2006;45:829–838. doi: 10.1021/bi0513709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebruska L.L., Kuzmine I.I., Fedor M.J. Rescue of an abasic hairpin ribozyme by cationic nucleobases: Evidence for a novel mechanism of RNA catalysis. Chem. Biol. 2002;9:465–473. doi: 10.1016/s1074-5521(02)00130-8. [DOI] [PubMed] [Google Scholar]
- Lee T.S., Silva Lopez C., Giambasu G.M., Martick M., Scott W.G., York D.M. Role of Mg2+ in hammerhead ribozyme catalysis from molecular simulation. J. Am. Chem. Soc. 2008;130:3053–3064. doi: 10.1021/ja076529e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sengupta J., Rath B.K., Frank J. Functional conformations of the L11-ribosomal RNA complex revealed by correlative analysis of cryo-EM and molecular dynamics simulations. RNA. 2006;12:1240–1253. doi: 10.1261/rna.2294806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Bokinsky G., Walter N.G., Zhuang X. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic “fingerprinting.”. Proc. Natl. Acad. Sci. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez X., Dejaegere A., Leclerc F., York D.M., Karplus M. Nucleophilic attack on phosphate diesters: A density functional study of in-line reactivity in dianionic, monoanionic, and neutral systems. J. Phys. Chem. B. 2006;110:11525–11539. doi: 10.1021/jp0603942. [DOI] [PubMed] [Google Scholar]
- MacElrevey C., Salter J.D., Krucinska J., Wedekind J.E. Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme. RNA. 2008;14:1600–1616. doi: 10.1261/rna.1055308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel C.R., Kaneko S., Zhang H., Gebauer D., Vethantham V., Manley J.L., Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M., Scott W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell S.E., Spackova N., Šponer J., Walter N.G. Molecular dynamics simulations of RNA: An in silico single molecule approach. Biopolymers. 2007;85:169–184. doi: 10.1002/bip.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min D., Xue S., Li H., Yang W. “In-line attack” conformational effect plays a modest role in an enzyme-catalyzed RNA cleavage: A free energy simulation study. Nucleic Acids Res. 2007;35:4001–4006. doi: 10.1093/nar/gkm394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.B., Seyhan A.A., Walter N.G., Burke J.M., Scott W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- Musselman C., Al-Hashimi H.M., Andricioaei I. iRED analysis of TAR RNA reveals motional coupling, long-range correlations, and a dynamical hinge. Biophys. J. 2007;93:411–422. doi: 10.1529/biophysj.107.104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K., Gao J., York D.M. Electrostatic interactions in the hairpin ribozyme account for the majority of the rate acceleration without chemical participation by nucleobases. RNA. 2008a;14:1501–1507. doi: 10.1261/rna.863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K., Gao J., York D.M. Quantum mechanical/molecular mechanical simulation study of the mechanism of hairpin ribozyme catalysis. J. Am. Chem. Soc. 2008b;130:4680–4691. doi: 10.1021/ja0759141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinard R., Hampel K.J., Heckman J.E., Lambert D., Chan P.A., Major F., Burke J.M. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razga F., Koca J., Mokdad A., Šponer J. Elastic properties of ribosomal RNA building blocks: Molecular dynamics of the GTPase-associated center rRNA. Nucleic Acids Res. 2007;35:4007–4017. doi: 10.1093/nar/gkm245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reblova K., Lankas F., Razga F., Krasovska M.V., Koca J., Šponer J. Structure, dynamics, and elasticity of free 16s rRNA helix 44 studied by molecular dynamics simulations. Biopolymers. 2006;82:504–520. doi: 10.1002/bip.20503. [DOI] [PubMed] [Google Scholar]
- Rhodes M.M., Reblova K., Šponer J., Walter N.G. Trapped water molecules are essential to structural dynamics and function of a ribozyme. Proc. Natl. Acad. Sci. 2006;103:13380–13385. doi: 10.1073/pnas.0605090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J.S., Schneider B., Murray L.W., Kapral G.J., Immormino R.M., Headd J.J., Richardson D.C., Ham D., Hershkovits E., Williams L.D., et al. RNA backbone: Consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda D., Bokinsky G., Rhodes M.M., Rust M.J., Zhuang X., Walter N.G. Single-molecule enzymology of RNA: Essential functional groups impact catalysis from a distance. Proc. Natl. Acad. Sci. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert P.B., Ferré-D'Amaré A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Rupert P.B., Massey A.P., Sigurdsson S.T., Ferré-D'Amaré A.R. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- Ryckaert J.P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- Ryder S.P., Strobel S.A. Comparative analysis of hairpin ribozyme structures and interference data. Nucleic Acids Res. 2002;30:1287–1291. doi: 10.1093/nar/30.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder S.P., Oyelere A.K., Padilla J.L., Klostermeier D., Millar D.P., Strobel S.A. Investigation of adenosine base ionization in the hairpin ribozyme by nucleotide analog interference mapping. RNA. 2001;7:1454–1463. [PMC free article] [PubMed] [Google Scholar]
- Salter J., Krucinska J., Alam S., Grum-Tokars V., Wedekind J.E. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefcikova J., Krasovska M.V., Spackova N., Šponer J., Walter N.G. Impact of an extruded nucleotide on cleavage activity and dynamic catalytic core conformation of the hepatitis delta virus ribozyme. Biopolymers. 2007a;85:392–406. doi: 10.1002/bip.20693. [DOI] [PubMed] [Google Scholar]
- Sefcikova J., Krasovska M.V., Šponer J., Walter N.G. The genomic HDV ribozyme utilizes a previously unnoticed U-turn motif to accomplish fast site-specific catalysis. Nucleic Acids Res. 2007b;35:1933–1946. doi: 10.1093/nar/gkl1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup G.A., Breaker R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley M.R., Strobel S.A. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- Tang C.L., Alexov E., Pyle A.M., Honig B. Calculation of pKas in RNA: On the structural origins and functional roles of protonated nucleotides. J. Mol. Biol. 2007;366:1475–1496. doi: 10.1016/j.jmb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Thomas J.M., Perrin D.M. Active site labeling of G8 in the hairpin ribozyme: Implications for structure and mechanism. J. Am. Chem. Soc. 2006;128:16540–16545. doi: 10.1021/ja063942y. [DOI] [PubMed] [Google Scholar]
- Tinsley R.A., Harris D.A., Walter N.G. Significant kinetic solvent isotope effects in folding of the catalytic RNA from the hepatitis delta virus. J. Am. Chem. Soc. 2003;125:13972–13973. doi: 10.1021/ja037870b. [DOI] [PubMed] [Google Scholar]
- Torelli A.T., Krucinska J., Wedekind J.E. A comparison of vanadate to a 2′-5′ linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization. RNA. 2007;13:1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torelli A.T., Spitale R.C., Krucinska J., Wedekind J.E. Shared traits on the reaction coordinates of ribonuclease and an RNA enzyme. Biochem. Biophys. Res. Commun. 2008;371:154–158. doi: 10.1016/j.bbrc.2008.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A., Schilling O., Spath B., Marchfelder A. The tRNase Z family of proteins: Physiological functions, substrate specificity, and structural properties. Biol. Chem. 2005;386:1253–1264. doi: 10.1515/BC.2005.142. [DOI] [PubMed] [Google Scholar]
- Walter N.G. Ribozyme catalysis revisited: Is water involved? Mol. Cell. 2007;28:923–929. doi: 10.1016/j.molcel.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter N.G., Yang N., Burke J.M. Probing nonselective cation binding in the hairpin ribozyme with Tb(III) J. Mol. Biol. 2000;298:539–555. doi: 10.1006/jmbi.2000.3691. [DOI] [PubMed] [Google Scholar]
- Wang J., Cieplak P., Kollman P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- Wilson T.J., Ouellet J., Zhao Z.Y., Harusawa S., Araki L., Kurihara T., Lilley D.M. Nucleobase catalysis in the hairpin ribozyme. RNA. 2006;12:980–987. doi: 10.1261/rna.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W.C., Nahvi A., Roth A., Collins J.A., Breaker R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Li M., Gustchina A., Tsuruoka N., Ashida M., Minakata H., Oyama H., Oda K., Nishino T., Nakayama T. Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity. J. Biol. Chem. 2004;279:21500–21510. doi: 10.1074/jbc.M401141200. [DOI] [PubMed] [Google Scholar]
- Xu Q., Guo H.B., Wlodawer A., Nakayama T., Guo H. The QM/MM molecular dynamics and free energy simulations of the acylation reaction catalyzed by the serine-carboxyl peptidase kumamolisin-As. Biochemistry. 2007;46:3784–3792. doi: 10.1021/bi061737p. [DOI] [PMC free article] [PubMed] [Google Scholar]