Abstract
A 2,2,7-trimethylguanosine (TMG) cap is a signature feature of eukaryal snRNAs, telomerase RNAs, and trans-spliced nematode mRNAs. TMG and 2,7-dimethylguanosine (DMG) caps are also present on mRNAs of two species of alphaviruses (positive strand RNA viruses of the Togaviridae family). It is presently not known how viral mRNAs might acquire a hypermethylated cap. Mimivirus, a giant DNA virus that infects amoeba, encodes many putative enzymes and proteins implicated in RNA transactions, including the synthesis and capping of viral mRNAs and the promotion of cap-dependent translation. Here we report the identification, purification, and characterization of a mimivirus cap-specific guanine-N2 methyltransferase (MimiTgs), a monomeric enzyme that catalyzes a single round of methyl transfer from AdoMet to an m7G cap substrate to form a DMG cap product. MimiTgs, is apparently unable to convert a DMG cap to a TMG cap, and is thereby distinguished from the structurally homologous yeast and human Tgs1 enzymes. Nonetheless, we show genetically that MimiTgs is a true ortholog of Saccharomyces cerevisiae Tgs1. Our results hint that DMG caps can satisfy many of the functions of TMG caps in vivo. We speculate that DMG capping of mimivirus mRNAs might favor viral protein synthesis in the infected host.
Keywords: 2,2,7-trimethylguanosine; 2,7-dimethylguanosine; AdoMet; RNA 5′ processing
INTRODUCTION
The m7G cap structure of eukaryal mRNA promotes translation initiation and protects mRNA from untimely decay. All eukaryal species and many genera of eukaryal viruses share a three-step capping pathway in which: (1) an RNA triphosphatase removes the γ-phosphate of the primary transcript; (2) an RNA guanylyltransferase transfers GMP from GTP to the 5′-diphosphate RNA to form a GpppRNA cap; and (3) a cap-specific RNA (guanine-N7) methyltransferase adds a methyl group from AdoMet to the cap guanine to form the m7GpppRNA structure (Shuman 2002).
The largest eukaryal DNA viruses—baculoviruses, African swine fever virus, poxviruses, Chlorella viruses, Coccolithovirus, certain iridoviruses, and mimivirus—encode some or all of the enzymes responsible for synthesis and capping of viral mRNAs (Shuman 2002; Benarroch et al. 2008). Mimivirus has the largest genome (1.2 Mb) and proteome (911 predicted proteins) of any known virus (Raoult et al. 2004, 2007; Claverie et al. 2006). Like poxviruses, mimivirus specifies its own mRNA synthetic machinery, which includes a multisubunit RNA polymerase and a trifunctional mRNA capping enzyme. Mimivirus and poxvirus capping enzymes are modular polypeptides composed of N-terminal RNA triphosphatase, central guanylyltransferase, and C-terminal (guanine N-7) methyltransferase domains (Myette and Niles 1996; Gong and Shuman 2003; De la Peña et al. 2007; Benarroch et al. 2008; Zheng and Shuman 2008).
A subset of capped cellular RNAs contain additional methyl groups attached to the exocyclic N2 of the cap guanosine. A 2,2,7-trimethylguanosine (TMG) cap is found on noncoding eukaryal RNAs such as small nuclear (sn) and small nucleolar (sno) RNAs and telomerase RNA (Busch et al. 1982; Seto et al. 1999) and on nematode mRNAs that undergo trans-splicing of a 5′-capped leader sequence (Liou and Blumenthal 1990). A 2,7-dimethylguanosine (DMG) cap is present in the mRNAs of two RNA viruses: Sindbis virus and Semliki Forest virus (HsuChen and Dubin 1976; Van Duijn et al. 1986). Whereas TMG synthesis has attracted interest because of the involvement of snRNAs in pre-mRNA splicing, there has been little attention paid to potential roles for DMG and TMG caps in virus biology since such structures were described in Sindbis virus mRNAs more than 30 years ago.
The discovery and characterization of Tgs1 as the enzyme responsible for TMG capping in budding and fission yeasts (Mouaikel et al. 2002; Hausmann et al. 2007) provided key insights to the pathway of cap hypermethylation and opened up a genetic approach to analysis of TMG function in cellular RNA metabolism (Lemm et al. 2006; Franke et al. 2008; Gallardo et al. 2008; Hausmann et al. 2008). Schizosaccharomyces pombe Tgs1, a 239-amino acid monomeric enzyme, catalyzes two successive methyltransfer reactions from AdoMet to the N2 atom of guanosine via a distributive mechanism (Hausmann and Shuman 2005a). Tgs1 activity is strictly dependent on prior guanine-N7 methylation, thereby confining its activity to RNAs that already have an m7G cap. Tgs1 is nonessential for growth of budding and fission yeasts (Mouaikel et al. 2002; Hausmann et al. 2007), a surprising finding given that TMG caps decorate many important cellular RNAs. However, Tgs1 is essential for growth in mutant Saccharomyces cerevisiae cells that lack certain proteins involved in snRNP biogenesis or pre-mRNA splicing (Hausmann et al. 2008).
Tgs1 homologs are distributed widely among primitive and higher eukarya. Cap-specific guanine-N2 methyltransferase activity has been documented for the Giardia lamblia Tgs2 and human Tgs1 proteins (Hausmann and Shuman 2005b; Hausmann et al. 2008) and recently for the Tgs homologs from Trichomonas vaginalis and Trypanosoma brucei (Simoes-Barbosa et al. 2008). Here, we identify a Tgs1 homolog encoded by mimivirus (hereafter named MimiTgs) as the first example of a viral cap-specific guanine-N2 methyltransferase. We present a biochemical characterization of MimiTgs, which catalyzes a single round of methyl transfer from AdoMet to m7G-containing nucleotides to form a DMG product. We show genetically that MimiTgs is a true ortholog of S. cerevisiae Tgs1. Alanine scanning of MimiTgs identified amino acids essential for methyltransferase activity in vitro and in vivo.
RESULTS
A mimivirus cap guanine-N2 methyltransferase
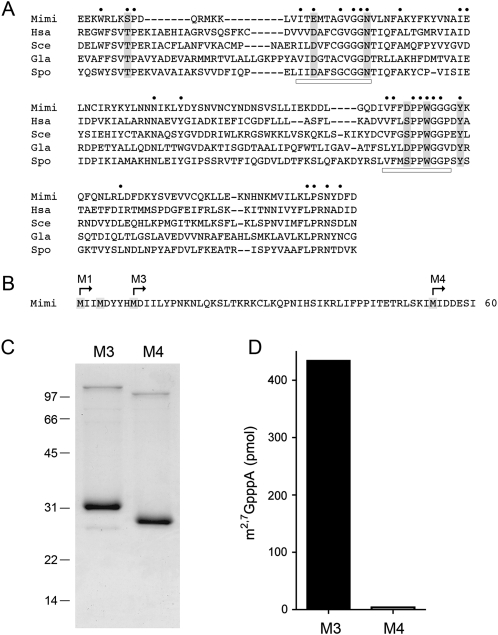
The L320 open reading frame of the mimivirus genome encodes a predicted 256-aa polypeptide that resembles the Tgs enzymes of fungi (S. cerevisiae, S. pombe), humans, and Giardia. The mimivirus Tgs-like protein (MimiTgs) shares 27 positions of side-chain identity/similarity with the four cellular Tgs enzymes to which it is aligned in Figure 1A. The conserved residues concentrate within two peptide motifs that comprise the binding sites for the AdoMet methyl donor and the m7G methyl acceptor (underscored in Fig. 1A by horizontal bars). To assess what biochemical activities, if any, are associated with the mimivirus Tgs homolog, we produced, in bacteria, several versions of recombinant MimiTgs that differed with respect to the position of the N-terminal methionine (Fig. 1B). The predicted L320 polypeptide contains three closely spaced methionines within the N-terminal decapeptide (which we will refer to as M1, M2, and M3), any one of which could plausibly be the translation start site. (The viral RNA transcript encoding MimiTgs is uncharacterized, so we do not know the location of its 5′ end.) The next methionine (M4) is located 45 amino acids downstream from M3 (Fig. 1B). We constructed pET-based plasmids designed to express MimiTgs polypeptides starting from M1, M3, or M4 as N-terminal His10Smt3 fusions. Only the M3 and M4 derivatives yielded soluble proteins in quantities sufficient for purification and biochemical characterization. The M3 and M4 proteins were isolated from the soluble bacterial extract by Ni-affinity chromatography. The His10Smt3 tag was removed with the Smt3-specific protease Ulp1 (Mossessova and Lima 2000) and the tagless M3 and M4 proteins were separated from the tag by a second round of Ni-agarose chromatography. SDS-PAGE analysis highlighted the purity of the M3 and M4 preparations and the expected difference in mobility of the M4 polypeptide (calculated mass of 29 kDa) versus M3 (24 kDa) (Fig. 1C).
FIGURE 1.
Mimivirus Tgs. (A) The amino acid sequence of MimiTgs (Mimi) from amino acids 91–229 is aligned to the sequences of the homologous cap guanine-N2 methyltransferases of Homo sapiens Tgs1 (Hsa), Saccharomyces cerevisiae Tgs1 (Sce), Giardia lambia Tgs2 (Gla), and Schizosaccharomyces pombe Tgs1 (Spo). Gaps are indicated by dashes. (●) Positions of amino acid side-chain identity/similarity in all five proteins. The AdoMet-binding motif and a putative m7G-binding motif are denoted by horizontal bars. Positions in MimiTgs that were targeted for mutational analysis in the present study are highlighted in shaded boxes. (B) The N-terminal 60 amino acids of the predicted MimiTgs polypeptide are shown, with methionines highlighted in shaded boxes. The translation start sites for the recombinant MimiTgs proteins M1, M3, and M4 are indicated by arrows. (C) MimiTgs purification. Aliquots (5 μg) of the purified tag-free M3 and M4 proteins were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (in kDa) of marker polypeptides are indicated on the left. (D) Methyltransferase reaction mixtures (10 μL) containing 1 mM m7GpppA, 50 μM [3H-CH3]AdoMet, and 0.5 μg of purified M3 or M4 protein were incubated for 15 min at 37°C. The products were analyzed by ascending TLC in 0.05 M (NH4)2SO4. The extents of 3H-methyl transfer to the cap dinucleotide are shown.
A methyltransferase activity of the M3 version of MimiTgs was demonstrated by incubating 0.5 μg of the protein with 50 μM [3H-CH3]-AdoMet and 1 mM m7GpppA for 15 min at 37°C, which resulted in 87% label transfer from AdoMet to the m7GpppA acceptor (Fig. 1D) to form a 3H-labeled product that was separated from the labeled AdoMet by PEI-cellulose TLC. The labeled m2,7GpppA product migrated immediately ahead of the input m7GpppA substrate, which was visualized by UV illumination of the chromatogram. In contrast, the recombinant M4 version of MimiTgs transferred <1% of the label from AdoMet to the cap dinucleotide (Fig. 1D). Henceforth, we will refer to the active M3 protein as “wild-type” MimiTgs.
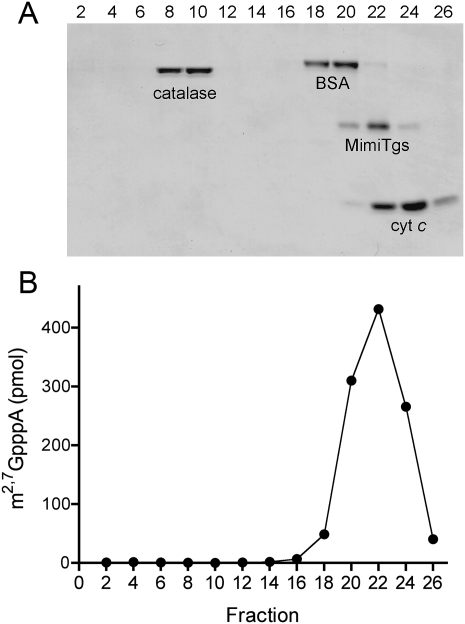
The quaternary structure of MimiTgs was examined by zonal velocity sedimentation in a 15%–30% glycerol gradient (Fig. 2). Marker proteins catalase (native size 248 kDa), bovine serum albumin (66 kDa), and cytochrome c (12 kDa) were included as internal standards. The MimiTgs polypeptide sedimented as a discrete peak (fraction 22) between bovine serum albumin and cytochrome c (Fig. 2A). The methyltransferase activity profile paralleled the abundance of the MimiTgs polypeptide (Fig. 2B). We surmise from these results that the methyltransferase activity is intrinsic to MimiTgs and that the enzyme is a monomer in solution.
FIGURE 2.
Zonal velocity sedimentation. An aliquot (50 μg) of the tag-free wild-type MimiTgs protein was mixed with catalase (50 μg), BSA (50 μg), and cytochrome c (50 μg). The mixture was applied to a 5 mL 15%–30% glycerol gradient containing 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, 2 mM DTT, 1 mM EDTA, 0.05% Triton X-100. The gradient was centrifuged at 50,000 rpm for 17 h at 4°C. Fractions (∼0.18 mL) were collected from the bottom of the tube. (A) Aliquots (20 μL) of the even-numbered gradient fractions were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown, with the bottom (heaviest) gradient fraction at left and the top (lightest) fraction at right. The identities of the polypeptides are indicated. (B) Aliquots (1 μL) of the even-numbered fractions were assayed for cap guanine-N2 methyltransferase activity. The extents of methyl transfer from 50 μM [3H-CH3]AdoMet to 1 mM m7GpppA are plotted.
MimiTgs is a cap-specific guanine-N2 methyltransferase
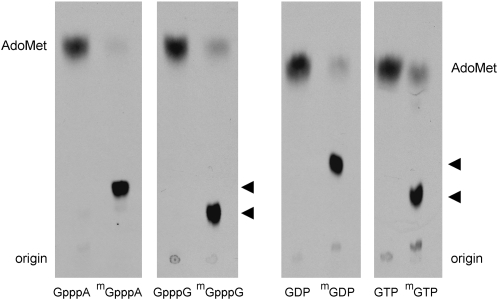
Various nucleotides were tested as methyl acceptors at 1 mM concentration. MimiTgs catalyzed label transfer from AdoMet to the cap dinucleotides m7GpppG and m7GpppA to form unique products (m2,7GpppG and m2,7GpppA) that were resolved from AdoMet by PEI-cellulose TLC (Fig. 3). The labeled nucleotide products migrated immediately ahead of the respective input unlabeled cap dinucleotides m7GpppG and m7GpppA (data not shown). MimiTgs formed no labeled product when reacted with unmethylated cap dinucleotides GpppG and GpppA (Fig. 2). We also tested MimiTgs activity with guanine nucleoside diphosphates and triphosphates. The enzyme catalyzed methyl transfer from AdoMet to m7GDP and m7GTP to form labeled products corresponding to m2,7GDP and m2,7GTP (Fig. 3). No labeled products were formed in the presence of GDP or GTP (Fig. 3). Thus, mimivirus guanine-N2 methyltransferase activity is stringently dependent on prior guanine-N7 methylation. In this respect, MimiTgs resembles its human, fungal, and Giardia homologs.
FIGURE 3.
Methyl acceptor specificity of MimiTgs. Reaction mixtures (10 μL) containing 50 mM Tris-acetate (pH 6.0), 5 mM DTT, 50 μM [3H-CH3]AdoMet, 1 mM methyl acceptor as specified, and 1 μg MimiTgs were incubated for 60 min at 37°C. Aliquots (4 μL) were spotted on PEI-cellulose TLC plates, which were developed either with 0.05 M (NH4)2SO4 (for reactions with GpppA, m7GpppA, GpppG, and m7GpppG as methyl acceptors) or 0.1 M (NH4)2SO4 (for reactions with GDP and m7GDP as methyl acceptors) or 0.2 M (NH4)2SO4 (for reactions with GTP, and m7GTP as methyl acceptors). The chromatograms were treated with Enhance and 3H-labeled material was visualized by autoradiography. The methyltransferase reaction products m2,7GpppA, m2,7GpppG, m2,7GDP, and m2,7GTP are denoted by arrowheads at the right of the chromatograms.
MimiTgs adds one methyl group at N2 of m7GDP
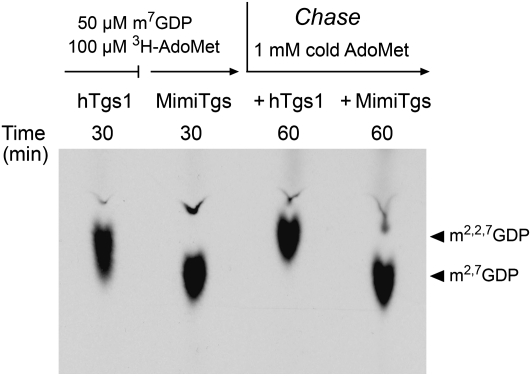
The reaction of MimiTgs with 100 μM [3H-CH3]-AdoMet and 50 μM m7GDP for 30 min yielded a single product that comigrated during TLC with m2,7GDP (Fig. 4). In contrast, human Tgs1 synthesized a mixture of m2,7GDP and m2,2,7GDP products, with TMG comprising the major species. TMG migrates slightly ahead of DMG in the TLC analysis (Fig. 4). The absence of a trimethylated m2,2,7GDP product of the MimiTgs reaction under conditions of AdoMet excess implies that either: (1) MimiTgs is exclusively a DMG synthase (like the Giardia homolog); or (2) MimiTgs does catalyze a second methylation reaction, but we are precluded from detecting it because the AdoMet donor is limiting functionally.
FIGURE 4.
MimiTgs catalyzes a single methyl addition at cap guanine-N2. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 5 mM DTT, 100 μM [3H-CH3]AdoMet, 50 μM m7GDP, and either 1.8 μM purified recombinant human Tgs1(576–853) (hTgs1) or 0.8 μM MimiTgs were incubated for 30 min at 37°C. The MimiTgs reaction mixture was then split in half and supplemented with 1 mM “cold” AdoMet and either 7 μM hTgs1 or 7 μM MimiTgs, and incubation was continued for another 30 min. Aliquots were removed after the 30-min pulse-labeling phase and again after the cold AdoMet chase phase. The products were analyzed by PEI cellulose TLC in 0.1 M ammonium sulfate. The chromatogram was treated with Enhance and the 3H-labeled material was visualized by autoradiography.
To address this issue, the pulse-labeled MimiTgs reaction mixture was “chased” for 30 min by adding 1 mM cold AdoMet plus either human Tgs1 or fresh MimiTgs. TLC analysis showed that whereas all of the 3H-labeled m2,7GDP formed during the MimiTgs pulse phase was subsequently converted by human Tgs1 to m2,2,7GDP, MimiTgs was unable to effect this conversion (Fig. 4). We surmise that MimiTgs is a dimethylguanosine synthase that performs just one methyl addition reaction at the N2 atom of m7GDP.
Inhibition by AdoHcy and sinefungin
Methylation of 1 mM m7GpppA in the presence of 50 μM [3H-CH3]-AdoMet was inhibited in a concentration-dependent fashion by AdoHcy; the apparent IC50 for AdoHcy was 28 μM (data not shown). This result indicates that MimiTgs has slightly higher affinity for its product AdoHcy than for its substrate AdoMet. (Product inhibition complicated our efforts to measure steady-state kinetic parameters for MimiTgs.) Sinefungin is a natural product analog of AdoMet that has a C–NH3 amine in lieu of the S–CH3 sulfonium center. Sinefungin also inhibited methyl transfer from 50 μM [3H-CH3]-AdoMet to m7GpppA in a concentration-dependent manner (data not shown). The apparent IC50 was 5 μM sinefungin, implying that MimiTgs bound sinefungin about 10-fold and fivefold more avidly than it bound AdoMet and AdoHcy, respectively. In contrast, Giardia Tgs2 was less sensitive to the same inhibitors, displaying apparent IC50 values of 75 μM AdoHcy and 45 μM sinefungin in the presence of 50 μM [3H-CH3]-AdoMet substrate (Hausmann and Shuman 2005b).
Alanine scanning identifies amino acids essential for methyltransferase activity
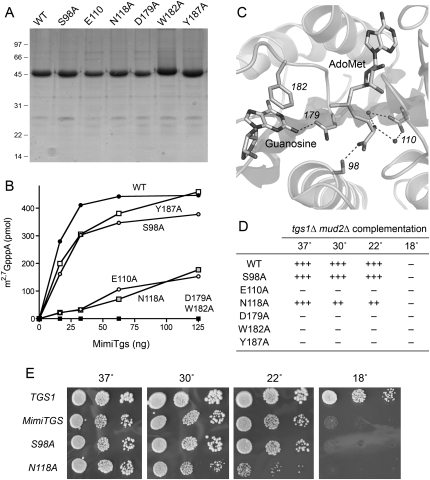
Amino acid sequence comparisons of Tgs-like proteins (Fig. 1A) and reference to the atomic structure of an rRNA guanine-N2 methyltransferase enzyme RsmC (Demirci et al. 2008) lead to prediction of a canonical AdoMet binding site in MimiTgs composed of the 108ITEMTAGVGGN118 peptide and a putative methyl acceptor site formed by the conserved proline/glycine-containing motif, 176VFFDPPWGGG185 (Fig. 1A). Prior mutational analyses of S. pombe, S. cerevisiae, Giardia, and human Tgs enzymes identified individual side chains within these motifs as essential for guanine-N2 methyltransferase activity (Hausmann and Shuman 2005b; Hausmann et al. 2007, 2008). Here we conducted a limited mutational study of MimiTgs by targeting five conserved positions within or near these two motifs for alanine scanning: Glu110, Asn118, Asp179, Trp182, and Tyr187. We also targeted the conserved Ser98 residue located upstream of the AdoMet-binding motif. The MimiTgs-Ala proteins were produced in bacteria as His10Smt3 fusions, purified by Ni-agarose chromatography in parallel with the wild-type His10Smt3-MimiTgs (Fig. 5A), and then assayed by protein titration for methyltransferase activity with m7GpppA as substrate (Fig. 5B). Wild-type MimiTgs formed ∼590 pmol of methylated product per pmol of input enzyme during the 15-min reaction. Two of the mutants—D179A and W182A—had <1% of wild-type specific activity. These findings are concordant with previously reported mutational effects at the equivalent amino acids in Giardia Tgs2 (Hausmann et al. 2007). The likely functions of Asp179 and Trp182 in binding the m7G substrate are illuminated by reference to the active site of the ternary complex of RsmC with AdoMet and guanosine (Fig. 5C). The RsmC equivalents of Asp179 and Trp182 are asparagine and phenylalanine, respectively. The essential aromatic side chain forms a π stack on the guanine base, which would be a π-cation interaction with m7G in the case of MimiTgs. The guanine-N2 atom is pointed toward the AdoMet methyl group, consistent with an in-line mechanism of nucleophilic attack on the methyl carbon. The equivalent of MimiTgs Asp179 accepts a hydrogen bond to Oδ from guanine-N2, suggesting a role for Asp179 in properly orienting of the N2 nucleophile and, perhaps, as a general base catalyst.
FIGURE 5.
Effects of alanine mutations on MimiTgs activity. (A) Aliquots (5 μg) of the nickel-agarose preparations of wild-type His10Smt3-MimiTgs and the indicated alanine mutants were analyzed by SDS-PAGE. Polypeptides were visualized by staining with Coomassie Blue dye. The positions and sizes (in kDa) of marker proteins are indicated on the left. (B) Reaction mixtures (10 μL) containing 50 mM Tris-acetate (pH 6.0), 5 mM DTT, 50 μM [3H-CH3]AdoMet, 1 mM m7GpppA, and proteins as specified were incubated for 15 min at 37°C. The extents of methyl transfer are plotted as a function of input enzyme. (C) Active site of Thermus thermophilus RNA guanine-N2 methyltransferase RsmC in complex with AdoMet and guanosine (from PDB 3DMH). Highlighted are the RsmC side chains that contact the methyl donor and acceptor and that have putative counterparts among the MimiTgs residues that were subjected to alanine scanning. Hydrogen bonding interactions are denoted by dashed lines. The residue numbers refer to the homologous positions in MimiTgs. (D,E) The ability of 2μ LEU2 plasmids bearing wild-type MimiTGS and the indicated alanine mutants to complement growth of a yeast tgs1Δ mud2Δ strain was tested by plasmid shuffle as described by Hausmann et al. (2008). Lethal mutations of MimiTGS were those that failed to support growth on medium containing 0.75 mg/mL 5-fluoroorotic acid (FOA) at any of the temperatures tested (scored as –). The tgs1Δ mud2Δ MimiTGS strains that grew on FOA at 30°C were tested for growth on rich medium by spotting serial 10-fold dilutions of liquid cultures (grown in SD-Leu medium) on YPD agar plates, which were then incubated at 18, 22, 30, or 37°C. Yeast tgs1Δ mud2Δ p(CEN LEU2 TGS1) cells were processed in parallel as a positive control. Photographs of the plates are shown in E. Growth of the viable MimiTGS-Ala strains was scored in D as follows: (+++) colony size indistinguishable from a strain bearing wild-type MimiTGS; (++) reduced colony size; (–) no growth.
MimiTgs mutants S98A and Y187A were 75% as active as the wild-type enzyme (Fig. 5B), signifying that neither side chain is essential for catalysis. A possible equivalent of Ser98 in RsmC donates a hydrogen bond from Oδ to the AdoMet carboxylate (Fig. 5C). Different Tgs homologs rely, to different extents, on this conserved hydroxyamino acid. For example, alanine substitution of the corresponding Thr40 residue in Giardia Tgs2 reduced guanine-N2 methyltransferase activity to <1% of wild type, whereas the analogous T674A mutant of human Tgs1 retained 13% of wild-type activity.
MimiTgs mutants E110A and N118A displayed feeble methyltransferase activity: 14% and 8% of wild type, respectively (Fig. 5B). The RsmC acidic side chain equivalent to MimiTgs Glu110 makes two water-mediated contacts to the AdoMet amine (Fig. 5C); this interaction is a shared feature of many AdoMet-dependent methyltransferases, including eukaryal cap guanine-N7 methyltransferase (Fabrega et al. 2004). The RsmC counterpart of MimiTgs Asn118 is a leucine, which is an aliphatic isostere of the conserved Asn/Asp residue present in the Tgs clade and cellular cap guanine-N7 methyltransferases. In the latter enzyme, the Oδ atom of this residue makes another water-mediated contact to the AdoMet amine (Fabrega et al. 2004).
MimiTgs is a functional ortholog of S. cerevisiae Tgs1
Budding yeast can serve as a surrogate genetic system to demonstrate the bioactivity of viral enzymes that synthesize a m7G mRNA cap (Ho et al. 2000; Saha et al. 2003; Schwer et al. 2006; Zheng and Shuman 2008). Adapting this strategy to heterologous Tgs-like proteins is less straightforward, insofar as the endogenous Tgs1 is not essential for growth of budding yeast (Mouaikel et al. 2002). However, Tgs1 is required for viability when the yeast gene encoding the pre-mRNA splicing factor Mud2 is deleted (Hausmann et al. 2008). We have exploited the synthetic lethality of the tgs1Δ mud2Δ strain to develop a genetic complementation assay for the function of heterologous Tgs1 enzymes. We thereby demonstrated the in vivo activity of human Tgs1 in yeast (Hausmann et al. 2008). Here we queried whether expression of MimiTgs could complement the tgs1Δ mud2Δ double mutation.
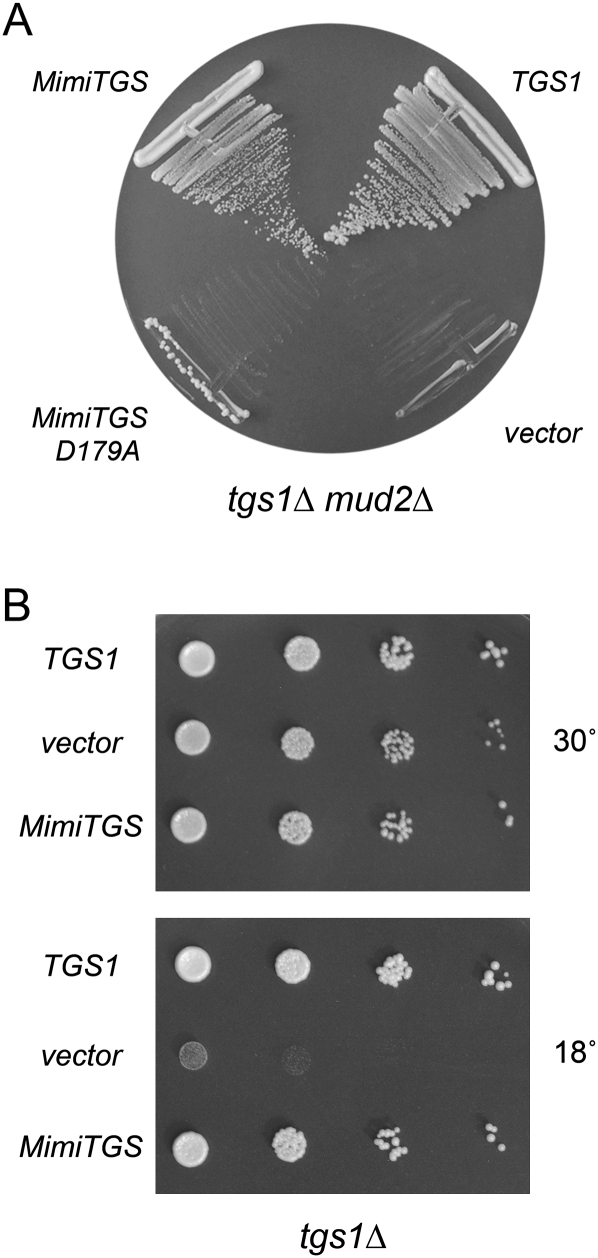
A yeast 2μ LEU2 plasmid expressing wild-type MimiTGS under the control of a constitutive yeast TPI1 promoter was tested for complementation of tgs1Δ mud2Δ by the plasmid shuffle procedure (Hausmann et al. 2008). A CEN LEU2 plasmid bearing wild-type S. cerevisiae TGS1 served as a positive control; the empty vector provided a negative control. We found that yeast TGS1 and MimiTGS were both capable of supporting growth on FOA-containing medium at 30°C, whereas the vector was not (Fig. 6A). Thus, MimiTgs, a DMG synthase, could fulfill the essential functions of yeast Tgs1 in the mud2Δ background. We compared growth of the tgs1Δ mud2Δ TGS1 and tgs1Δ mud2Δ MimiTGS strains on YPD agar at 18, 22, 30, and 37°C (Fig. 5E). The notable finding was that MimiTGS cells failed to grow at 18°C.
FIGURE 6.
MimiTgs is a functional ortholog of S. cerevisiae Tgs1. (A) Yeast tgs1Δ mud2Δ p (CEN URA3 TGS1) cells were transformed with a CEN LEU2 plasmid bearing wild-type S. cerevisiae TGS1 (positive control), an empty 2μ LEU2 vector (negative control), and 2μ LEU2 plasmids expressing either wild-type MimiTGS or MimiTGS-D179A. Leu+ transformants were selected at 30°C and then streaked to agar medium containing FOA. The plate was photographed after 3 d at 30°C. (B) Yeast tgs1Δ cells were transformed with a CEN LEU2 plasmid bearing wild-type S. cerevisiae TGS1 (positive control), an empty 2μ LEU2 vector (negative control), and a 2μ LEU2 plasmid expressing MimiTGS. Leu+ transformants were selected at 30°C and then tested for growth at 30°C and 18°C by spotting serial 10-fold dilutions of liquid cultures (grown at 30°C in SD-Leu medium) on Leu– agar plates. The plates were photographed after incubation for 3 d at 30°C or 7 d at 18°C.
We then queried whether MimiTgs methyltransferase catalytic activity was required to sustain growth of tgs1Δ mud2Δ cells by performing the plasmid-shuffle complementation test with MimiTGS-Ala mutants that we had characterized biochemically. We found that the catalytically defective D179A mutant failed to complement the mud2Δ tgs1Δ strain at 30°C (Fig. 6A) or any other temperature tested (Fig. 5D). The catalytically defective W182A mutant was also lethal in yeast (Fig. 5D). E110A displayed reduced methyltransferase activity in vitro and was lethal in yeast. The N118A mutant displayed a similarly reduced level of activity in vitro, yet was capable of sustaining normal growth of yeast tgs1Δ mud2Δ cells at 37°C (as gauged by colony size) (Fig. 5E). However, the N118A strain grew slowly at 30 and 22°C (Fig. 5D,E). In contrast, the catalytically active S98A mutant supported yeast growth as well as wild-type MimiTgs (Fig. 5D). Thus, for these five alanine mutants, there was a good correlation between the in vivo growth phenotype and methyltransferase function in vitro.
The exceptional case was Y187A, a mutant that retained substantial methyltransferase activity in vitro, yet was unconditionally lethal in vivo in yeast (Fig. 5). Any of several scenarios could account for this disparity: (1) The steady-state level of the Y187A mutant protein in yeast is inadequate to sustain viability; (2) the protein is produced but mislocalized; or (3) Tyr187 might interact with the RNA chain of the m7G-capped RNA substrates and assist in docking the cap in place to serve as a methyl acceptor, in which case the assays of cap methylation in vitro using a m7GpppA substrate would not be sensitive to alterations of a putative RNA docking site. Further studies and additional reagents (e.g., anti-MimiTgs antibodies) will be needed to resolve these issues. It is worth noting that Tyr187 is conserved among the members of the Tgs clade (Fig. 1A), but has no apparent counterpart in RsmC.
Distinctions in vivo between a DMG synthase and a TMG synthase
The fact that tgs1Δ mud2Δ MimiTGS cells failed to grow at 18°C raises an interesting question about possible differences in vivo between Tgs enzymes that catalyze one versus two cap guanine-N2 methylation steps. Before invoking such distinctions, we sought to test whether MimiTgs activity in yeast was intrinsically cold sensitive. To do this, we took advantage of the cold-sensitive growth phenotype of the singly deleted yeast tgs1Δ mutant (Mouaikel et al. 2002; Hausmann et al. 2008). Growth of tgs1Δ at restrictive temperature (18°C) was restored by a plasmid-bearing wild-type yeast TGS1, but not by the empty vector (Fig. 6B). Analyses of tgs1 active-site mutants confirmed that complementation of the tgs1Δ cold-sensitive phenotype depended on yeast Tgs1 methyltransferase activity (Mouaikel et al. 2002, 2003; data not shown). The instructive finding was that expression of MimiTgs complemented the tgs1Δ strain at 18°C (Fig. 6B), signifying that MimiTgs was active at the cold temperatures in vivo. This result rules out a trivial explanation for the cold sensitivity of the tgs1Δ mud2Δ MimiTGS strain. It suggests instead that yeast cells are uniquely reliant on a TMG synthase for growth at 18°C when the Mud2 splicing factor is absent. Otherwise, a viral DMG synthase appears to suffice.
DISCUSSION
The present study provides the first demonstration of a virus-encoded cap-specific guanine-N2 methyltransferase. Our characterization of MimiTgs discloses its similarity to Giardia Tgs2, which, like MimiTgs, is competent for synthesis of a DMG cap, but apparently not for a second round of methyl addition to form TMG. MimiTgs is thereby distinguished from the S. pombe and human Tgs1 enzymes, which are capable of two methyl additions in vitro to form a TMG product.
Available knowledge of mimivirus biology and gene expression strategies is sparse. We showed previously that mimivirus encodes a trifunctional mRNA capping enzyme with autonomous triphosphatase-guanylyltransferase and guanine-N7 methyltransferase domains (Benarroch et al. 2008). This enzyme could suffice to form monomethyl cap structures on mimivirus mRNAs during a natural infection. We now find that mimivirus has, in principle, the capacity to convert those m7GpppN caps to m2,7GpppN cap structures. Because it is not yet feasible to genetically engineer mimivirus, the role of MimiTgs in the virus replication cycle cannot be evaluated. However, we can speculate on the potential role of DMG caps in viral mRNAs given that: (1) they are known to be present in certain RNA virus transcripts (HsuChen and Dubin 1976; Van Duijn et al. 1986); (2) DMG caps confer an advantage in mRNA translation vis a vis an otherwise identical reporter mRNA containing a standard m7G cap (Darzynkiewicz et al. 1988; Cai et al. 1999); and (3) this advantage is a specific feature of a DMG cap, i.e., a TMG cap renders mRNA severalfold less translatable than a standard m7G-capped transcript (Darzynkiewicz et al. 1988; Cai et al. 1999). An appealing scenario is that mimivirus chooses to synthesize DMG-modified capped mRNAs in order to achieve preferential translation of viral polypeptides versus those of the host cell, Acanthamoeba polyphaga. Available evidence indicates that the gain in translatability of DMG-capped synthetic mRNAs correlates with their increased affinity for the cap-binding translation initiation factor eIF4E (Grudzien et al. 2004). Thus, it is surely no coincidence that mimivirus encodes its own eIF4E homolog (Raoult et al. 2004). Indeed, mimivirus is unique among viruses in having its own cap DMG synthase and cap-binding translation factor. We suspect that mimivirus eIF4E might have a distinctive specificity for DMG caps versus the eIF4E of the host, thereby enforcing viral versus host protein synthesis.
Our finding that MimiTGS, which synthesizes only DMG caps in vitro, can complement the essential function of yeast Tgs1 (a TMG synthase) raises some interesting issues concerning the role of the cap guanine-N2 modification in cellular physiology. Taken at face value, our results indicate that DMG caps can satisfy the essential functions of TMG caps (e.g., in snRNA-mediated mRNA splicing) in a tgs1Δ mud2Δ MimiTGS strain at 25–37°C and in a tgs1Δ MimiTGS strain at 18°C. We regard it as unlikely that MimiTgs can perform a second round of cap guanine-N2 methylation in vivo when it is unable to do so in vitro. Indeed, any notion that Tgs enzymes that display exclusive DMG synthase activity with m7GpppN or m7GTP substrates might somehow display TMG synthase activity with m7G-capped RNA substrates has been overturned by the recent work of Simoes-Barbosa et al. (2008).
The availability of a yeast tgs1Δ mud2Δ strain that depends on DMG synthesis for survival opens up new avenues to study the biogenesis of cap-modified U RNAs. The cold-sensitive growth defect of the yeast tgs1Δ mud2Δ MimiTGS strain hints that not every essential aspect of TMG biology can be fulfilled by a DMG cap. We have identified 20 nonessential yeast genes in addition to MUD2 that, when deleted, elicit synthetic lethal or synthetic sick growth defects in combination with tgs1Δ (Hausmann et al. 2008). These genes cluster around the processes of pre-mRNA splicing, snRNP assembly, RNA decay, and gene transcription. It will be interesting to see which of the synthetic defects can be rescued by MimiTGS. Such studies could supply insights into when and how cells discriminate between DMG and TMG caps.
MATERIALS AND METHODS
Materials
[3H-CH3]AdoMet and Enhance were purchased from Perkin Elmer Life Sciences. m7GTP, GTP, AdoMet, AdoHcy, and sinefungin were purchased from Sigma. m7GpppA, m7GpppG, GpppA, and GpppG were purchased from New England Biolabs.
Recombinant mimivirus Tgs
The L320 open reading frame encoding MimiTgs (NCBI accession AAV50589) was PCR-amplified from mimivirus genomic DNA (a generous gift of Dr. Didier Raoult, University of Marseille) using sense-strand primers designed to introduce a BamHI restriction site immediately upstream of the translation start codon and an antisense primer that introduced a XhoI site downstream from the stop codon. The PCR products were digested with BamHI and XhoI and then inserted into the bacterial expression vector pET-His10-Smt3, which thereby encodes a His10Smt3-MimiTgs fusion protein. Single-alanine mutations S98A, E110A, N118A, D179A, W182A, and Y187A were introduced into the MimiTgs gene by two-stage PCR overlap extension with mutagenic primer oligonucleotides. The plasmid inserts were sequenced completely to exclude the acquisition of unwanted changes during PCR amplification and cloning.
The pET28b-His10Smt3-MimiTgs plasmids were transformed into Escherichia coli BL21(DE3)-RIL (Stratagene). Cultures (500 mL) derived from single transformants were grown at 37°C in Luria-Bertani medium containing 50 μg/mL kanamycin and 12.5 μg/mL chloramphenicol until the A 600 reached 0.6. The cultures were adjusted to 0.2 mM isopropyl-1-thio-β-D-galactopyranoside and 2% (v/v) ethanol and placed on ice for 2 h, after which incubation was continued for 20 h at 17°C with constant shaking. The cells were harvested by centrifugation, and the pellets were stored at –80°C. All of the subsequent steps were performed at 4°C. Thawed bacteria were resuspended in 25 mL of buffer A (50 mM Tris-HCl at pH 7.5, 500 mM NaCl, 10% glycerol). Lysozyme was added to a final concentration of 100 μg/mL. After 30 min, Triton X-100 was added to a final concentration of 0.1% and the lysate was sonicated to reduce viscosity. Insoluble material was removed by centrifugation for 45 min at 16,000 rpm in a Sorval SS34 rotor. The soluble extract was mixed for 30 min with 2 mL of Ni2+-nitrilotriacetic acid-agarose resin (QIAGEN) that had been equilibrated with buffer A. The resin was recovered by centrifugation, resuspended in buffer A, and then poured into a column and washed with 25 mL of buffer A containing 50 mM imidazole. Bound material was eluted with 500 mM imidazole in buffer A. The elution profiles were monitored by SDS-PAGE of the column fractions. The 500 mM imidazole eluate containing the His-tagged MimiTgs was dialyzed against buffer A containing purified Ulp1 (a Smt3-specific protease) at a Ulp1:MimiTgs ratio of 1:2000. The Ulp1-digested dialysates were applied to 1 mL columns of Ni2+-nitrilotriacetic acid-agarose equilibrated with buffer A. The tag-free MimTgs proteins were recovered in the flow-through fractions. The proteins were stored at –80°C and their concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin (BSA) as the standard.
Methyltransferase assay
Standard reaction mixtures (10 μL) containing 50 mM Tris-Acetate (pH 6.0), 5 mM DTT, 50 μM [3H-CH3]AdoMet, and m7GpppA and enzyme as specified were incubated for 15 min at 37°C. Aliquots (4 μL) were spotted on polyethyleneimine cellulose TLC plates, which were developed with 0.05 M (NH4)2SO4. The AdoMet- and m2,7GpppA-containing portions of the lanes were cut out and the radioactivity in each was quantified by liquid scintillation counting.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM42498. S.S. is an American Cancer Society Research Professor.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1462109.
REFERENCES
- Benarroch D., Smith P., Shuman S. Characterization of a trifunctional mimivirus mRNA capping enzyme and crystal structure of the RNA triphosphatase domain. Structure. 2008;16:501–512. doi: 10.1016/j.str.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Busch H., Reddy R., Rothblum L., Choi Y.C. SnRNAs, SnRNPs, and RNA processing. Annu. Rev. Biochem. 1982;51:617–654. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- Cai A., Jankowska-Anyszka M., Centers A., Chlebicka L., Stepinki J., Stolarski R., Darzynkiewicz E., Rhoads R.E. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry. 1999;38:8538–8547. doi: 10.1021/bi9830213. [DOI] [PubMed] [Google Scholar]
- Claverie J.M., Ogata H., Audic S., Abergel C., Suhre K., Fournier P.E. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006;117:133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz E., Stepinski J., Ekiel I., Haber D., Tahara S.M. β-globin mRNAs capped with m7G, m2,7G or m2,2,7G differ in intrinsic translation efficiency. Nucleic Acids Res. 1988;16:8953–8962. doi: 10.1093/nar/16.18.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Peña M., Kyrielis O., Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyltransferase. EMBO J. 2007;26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci H., Gregory S.T., Dahlberg A.E., Jogl G. Crystal structure of the Thermus thermophilus 16S rRNA methyltransferase RsmC in complex with cofactor and substrate guanosine. J. Biol. Chem. 2008;283:26548–26556. doi: 10.1074/jbc.M804005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrega C., Hausmann S., Shen V., Shuman S., Lima C.D. Structure and mechanism of cap (guanine-N7) methyltransferase. Mol. Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- Franke J., Gehlen J., Ehrenhofer-Murray A.E. Hypermethylation of yeast telomerase RNA by the snRNA and snoRNA methyltransferase Tgs1. J. Cell Sci. 2008;121:3553–3560. doi: 10.1242/jcs.033308. [DOI] [PubMed] [Google Scholar]
- Gallardo F., Olivier C., Dandjinoud A.T., Welinger R.J., Chartrand P. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 2008;27:748–757. doi: 10.1038/emboj.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Shuman S. Mapping the active site of vaccinia virus RNA triphosphatase. Virology. 2003;309:125–134. doi: 10.1016/s0042-6822(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Grudzien E., Stepinski J., Jankowska-Anyszka M., Stolarski R., Darzynkiewicz E., Rhoads R.E. Novel cap analogs for in vitro synthesis of mRNAs with high translational efficiency. RNA. 2004;10:1479–1487. doi: 10.1261/rna.7380904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S., Shuman S. Specificity and mechanism of RNA cap guanine-N2 methyltransferase (Tgs1) J. Biol. Chem. 2005a;280:4021–4024. doi: 10.1074/jbc.C400554200. [DOI] [PubMed] [Google Scholar]
- Hausmann S., Shuman S. Giardia lamblia RNA cap guanine-N2 methyltransferase (Tgs2) J. Biol. Chem. 2005b;280:32101–32106. doi: 10.1074/jbc.M506438200. [DOI] [PubMed] [Google Scholar]
- Hausmann S., Ramirez A., Schneider S., Schwer B., Shuman S. Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe . Nucleic Acids Res. 2007;35:1411–1420. doi: 10.1093/nar/gkl1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S., Zheng S., Costanzo M., Brost R.L., Garcin D., Boone C., Shuman S., Schwer B. Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: Functional overlap of TMG caps, snRNP components, pre-mRNA splicing factors, and RNA decay pathways. J. Biol. Chem. 2008;283:31706–31718. doi: 10.1074/jbc.M806127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.K., Martins A., Shuman S. A yeast-based genetic system for functional analysis of viral mRNA capping enzymes. J. Virol. 2000;74:5486–5494. doi: 10.1128/jvi.74.12.5486-5494.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HsuChen C.C.,, Dubin D.T. Di-and trimethylated congeners of 7-methylguanine in Sindbis virus mRNA. Nature. 1976;264:190–191. doi: 10.1038/264190a0. [DOI] [PubMed] [Google Scholar]
- Lemm I., Girard C., Kuhn A.N., Watkins N.J., Schneider M., Bordonné R., Lührmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou R.F., Blumenthal T. trans-Spliced Caenorhabditis elegans mRNA retain trimethylguanosine caps. Mol. Cell. Biol. 1990;10:1764–1768. doi: 10.1128/mcb.10.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E., Lima C.D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Mouaikel J., Verheggen C., Bertrand E., Tazi J., Bordonné R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- Mouaikel J., Bujnicki J.M., Tazi J., Bordonné R. Sequence-structure-function relationships of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res. 2003;31:4899–4909. doi: 10.1093/nar/gkg656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myette J., Niles E.G. Domain structure of the vaccinia virus mRNA capping enzyme: Expression in Escherichia coli of a subdomain possessing the RNA 5′-triphosphatase and guanylyltransferase activities and a kinetic comparison to the full-size enzyme. J. Biol. Chem. 1996;271:11936–11944. doi: 10.1074/jbc.271.20.11936. [DOI] [PubMed] [Google Scholar]
- Raoult D., Audic S., Robert C., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J.M. The 1.2-megabase genome sequence of mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Raoult D., La Scola B., Birtles R. The discovery and characterization of mimivirus, the largest known virus and putative pneumonia agent. Clin. Infect. Dis. 2007;45:95–102. doi: 10.1086/518608. [DOI] [PubMed] [Google Scholar]
- Saha N., Shuman S., Schwer B. Yeast-based genetic system for functional analysis of poxvirus mRNA cap methyltransferase. J. Virol. 2003;77:7300–7307. doi: 10.1128/JVI.77.13.7300-7307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Hausmann S., Schneider S., Shuman S. Poxvirus cap methyltransferase: Bypass of the requirement for the stimulatory subunit by mutations in the catalytic subunit and evidence for intersubunit allostery. J. Biol. Chem. 2006;281:18953–18960. doi: 10.1074/jbc.M602867200. [DOI] [PubMed] [Google Scholar]
- Seto A.G., Zaug A.J., Sobel S.G., Wolin S.L., Cech T.R. Saccharomyces cerevisiae telomerase is a small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- Simoes-Barbosa A., Louly C., Franco O.L., Rubio M.A., Alfonzo J.D., Johnson P.J. The divergent eukaryote Trichomonas vaginalis has an m7G cap methyltransferase capable of a single N2 methylation. Nucleic Acids Res. 2008;36:6848–6858. doi: 10.1093/nar/gkn706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duijn L.P., Kasperaitis M., Ameling C., Voorma H.O. Additional methylation at the N(2)-position of the cap of 26S Semliki Forest virus late mRNA and initiation of translation. Virus Res. 1986;5:61–66. doi: 10.1016/0168-1702(86)90065-1. [DOI] [PubMed] [Google Scholar]
- Zheng S., Shuman S. Structure-function analysis of vaccinia virus mRNA cap methyltransferase. RNA. 2008;14:696–705. doi: 10.1261/rna.928208. [DOI] [PMC free article] [PubMed] [Google Scholar]