Abstract
In the last decade short interfering RNA (siRNA) became an important means for functional genomics and the development of gene-specific drugs. However, major technical hurdles in the application of siRNA include its cellular delivery followed by its intracellular trafficking and its release in order to enter the RNA interference (RNAi) machinery. The novel phosphorothioate-stimulated cellular uptake of siRNA contrasts other known delivery systems because it involves a caveosomal pathway in which large amounts of siRNA are delivered to the perinuclear environment, leading to measurable though moderate target suppression. Limited efficacy seems to be related to intracellular trapping of siRNA. To study the role of intracellular trafficking of siRNA for biological effectiveness we studied whether a signal peptide for trans-membrane transport of bacterial protein toxins, which is covalently attached to siRNA, can promote its release from the perinuclear space into the cytoplasm and thereby enhance its biological effectiveness. We show that attachment of the peptide TQIENLKEKG to lamin A/C-directed siRNA improves target inhibition after its PS-stimulated delivery. This is related to increased efflux of the siRNA–peptide conjugate from the ER-specific perinuclear sites. In summary, this study strongly suggests that intracellular release of siRNA leads to increased biological effectiveness. Thus covalent peptide–siRNA conjugates are proposed as new tools to study the relationship between intracellular transport and efficacy of siRNA.
Keywords: conjugation, delivery, phosphorothioate, subcellular, trafficking
INTRODUCTION
RNA interference (RNAi) is a gene silencing process that is triggered by double-stranded RNA (dsRNA), which is converted to small interfering RNA (siRNA) (Rana 2007). Such processed siRNA is loaded into the RNA-induced silencing complex (RISC); it guides this complex to the target RNA, and it directs sequence-specific degradation of complementary target RNA or inhibition of translation of partly complementary target RNA (Hannon 2002; Zamore and Haley 2005). The use of siRNA as tools and drugs to specifically suppress gene expression has moved from basic biological research into the focus of applied molecular biology and molecular medicine (Dorsett and Tuschl 2004; Bumcrot et al. 2006; Haney 2007). The increasing importance of siRNA as a potential therapeutic, first shown in an animal disease model in 2003 (Song et al. 2003), is reflected by the fact that three clinical phase I studies have already been completed (Song et al. 2003; de Fougerolles et al. 2007). For siRNA as well as for other oligomeric nucleic acid-based drugs, major technical problems for in vivo applications still include the efficient delivery to target cells and sufficient intracellular bioavailability.
Silencing RNA can be delivered via a variety of transfection methods, including cell penetrating peptides (Turner et al. 2007; Deshayes et al. 2008; Lebleu et al. 2008), cholesterol conjugation (Soutschek et al. 2004; Cheng et al. 2006), proteamine–antibody fusion proteins (Song et al. 2005; Vornlocher 2006), atelocollagen (Minakuchi et al. 2004; Takeshita et al. 2005), stable nucleic acid–lipid particles (SNALPs; Morrissey et al. 2005; Santel et al. 2006), or polyethyleneimine-mediated uptake (Ge et al. 2004; Tan et al. 2005; Grzelinski et al. 2006). Even though these findings provide promising delivery concepts, the optimization of cellular delivery and bioavailability is still the key technical problem for therapeutic application of siRNA. Since RNAi is thought to be a cytoplasmic process in most cases (Zeng and Cullen 2002), siRNA molecules need to be released from compartments where they are captured in order to enter the RNAi pathway. Almost all of the known delivery modes of nucleic acid-based tools and drugs are based on a combination of adsorptive and fluid-phase endocytosis (Opalinska and Gewirtz 2002). One fundamentally alternative mode of delivery is the phosphorothioate (PS)-mediated uptake of naked siRNA by mammalian cells (Overhoff and Sczakiel 2005), which makes use of the caveosomal uptake pathway rather than the endosomal pathway. Despite the PS-mediated delivery of surprisingly large amounts of siRNA to perinuclear sites, however, the extent of target suppression is limited (Mescalchin et al. 2007).
As a proof-of-concept study to promote intracellular release, we covalently attached a signal peptide derived from bacterial protein toxins to siRNA in order to overcome the apparently low bioavailability of this mode of delivery. The peptide TQIENLKEKG was identified by Ratts et al. (2005) as a conserved 10 amino acid motif in several bacterial toxins, e.g., diphtheria toxin, anthrax edema factor, anthrax lethal factor, and botulinum toxins. This motif is part of a transmembrane helix within the toxin that facilitates translocation of the respective catalytic domains from transport vesicles into the cytoplasm where the toxin exerts its harmful effects (Falnes and Sandvig 2000). Bacterial toxin-derived peptides might be a promising tool when conjugated to siRNA since they mediate direct vesicular release into the cytoplasm (Falnes and Sandvig 2000), bypassing the natural route of newly synthesized proteins from the endoplasmatic reticulum (ER) via the Golgi to secretory or lysosomal vesicles (Bonifacino and Glick 2004). Circumvention of the secretory pathway might be of advantage for the delivery of siRNA, since siRNA might be directly released into the cytoplasm from uptake vesicles. Thus, the TQIENLKEKG peptide was chosen as a promising candidate for conjugation to siRNA.
The aim of this study was to investigate whether the covalent conjugation of TQIENLKEKG to siRNA had any effect on its intracellular localization and release and thereby on the extent of siRNA-mediated target inhibition. Here, we provide strong experimental evidence for increased RNAi due to increased intracellular release and bioavailability of this siRNA–peptide conjugate. This work shows that intracellular transport is crucial for effectiveness of PS-delivered siRNA and suggests the consideration of siRNA–peptide conjugates as tools to study the role of intracellular trafficking and localization for the biological effectiveness of siRNA.
RESULTS
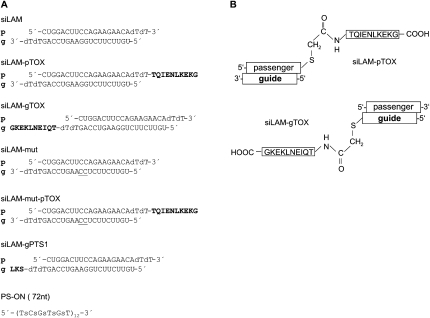
The PS-stimulated uptake of siRNA is a new and mechanistically alternative pathway to deliver siRNA to cells. The main hurdle in this uptake mechanism is the low bioavailability of the delivered siRNA. We speculate that siRNA is primarily delivered to perinuclear structures, which function as traps, thereby preventing the availability of siRNA for the RNAi machinery (Mescalchin et al. 2007). To overcome this problem we attached a signal peptide sequence to a very potent lamin A/C-directed siRNA termed siLAM (Elbashir et al. 2002) and measured the bioactivity of this siRNA–peptide conjugate in comparison to parental siRNA and controls (Fig. 1A,B).
FIGURE 1.
Structures of siRNA and conjugates used in this study. (A) Schematic presentation of names and sequences and modifications of siRNAs used here. (B) Structure of covalent linkage between siLAM and the bacterial peptide TQIENLKEKG. (p) Passenger strand; (g) guide strand; (dT) deoxyribothymidine; (s in PS-ON depicted in panel A) internucleotide phosphorothioate; one-letter code for amino acids indicated in bold; the two consecutive underlined C residues at positions 10 and 11 of siLAM-mut and siLAM-mut-pTOX indicate mutations of the parental guide strand; (TOX) TQIENLKEKG; (PTS1) peroxisomal targeting signal 1, SKL.
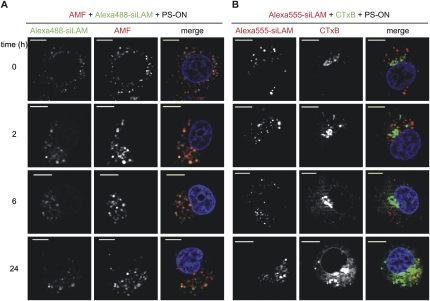
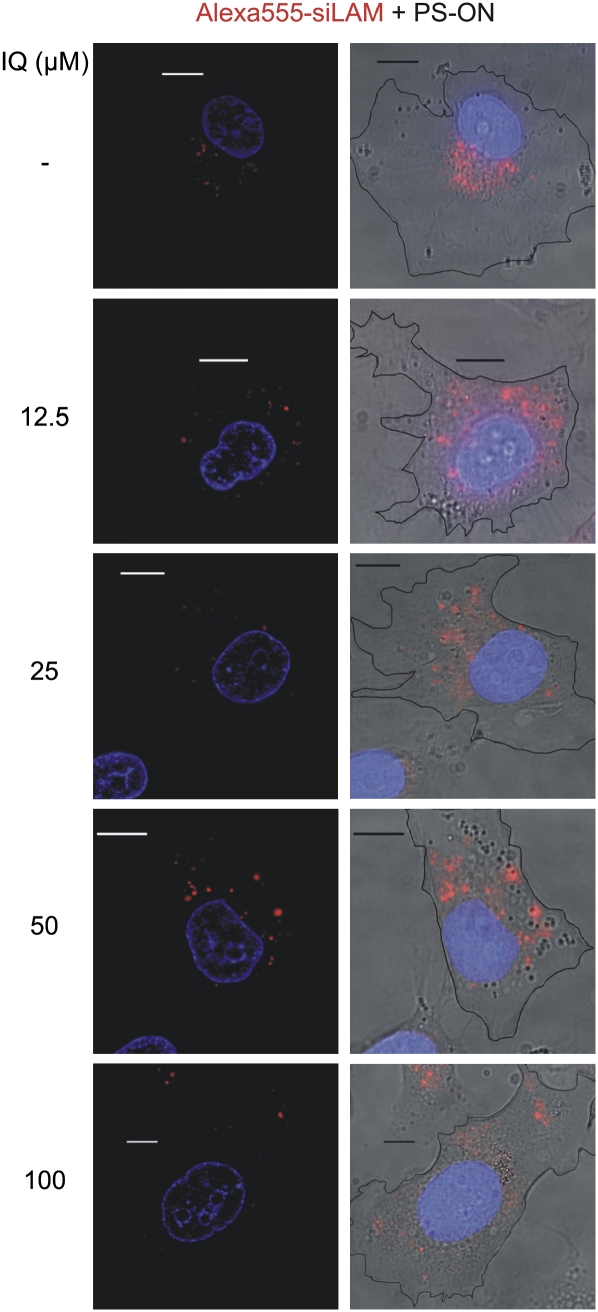
Since fluorescence microscopy showed a strong signal of fluorescently labeled siRNA in the perinuclear environment after PS-stimulated cellular siRNA uptake, we hypothesized that intracellular trapping of siRNA occurred in perinuclear organelles, including the ER and the Golgi apparatus (Mescalchin et al. 2007). Therefore we performed colocalization studies using the Golgi-specific marker CTxB and AMF, which is located in the smooth ER (Le and Nabi 2003). Both are model cargos of caveolin-dependent endocytotic internalization. These experiments suggest that trapping of PS-delivered siRNA occurs perinuclearly in a dot-like fashion at sites that seem to better coincide with the ER than the Golgi (Fig. 2A,B). The model of captured siRNA is strongly supported by the apparent release of siRNA and by increased siRNA-mediated target suppression when cells are treated with ilimaquinone (Fig. 3; data not shown). This substance is known to transiently destroy Golgi membranes and, at higher concentrations, can also disrupt membranes of the ER (Takizawa et al. 1993; Wang et al. 1997). Consequently, it became attractive to consider the concept of attaching a trans-membrane transport signal to siRNA in order to release siRNA to cellular sites where it can act against its target mRNA.
FIGURE 2.
siLam delivered by PS-ON colocalizes with the smooth ER marker AMF but not with the Golgi marker CTxB. ECV-304 cells were simultaneously treated with 1 μM PS-ON 72mer, 1 μM siLam-Alexa488 (green) or -Alexa555 (red), and 10 μg/mL AMF-Cy3 (red; A) or 0.5 μg/mL CTxB-FITC (green; B), respectively, for 60 min. Afterward, cells were extensively washed with Opti-MEM and nuclei (blue) were stained with Hoechst 33342. Optical sections were acquired 0, 2, 6, or 24 h after the treatment. Bars, 10 μm.
FIGURE 3.
Ilimaquinone leads to a dispersion of PS-ON delivered siLam-Alexa555 from the perinuclear space into the cytoplasm. ECV-304 cells were incubated with 1 μM siLam-Alexa555 and 1 μM PS-ON for 60 min at 37°C. After five washes with Opti-MEM, cells were incubated with 12.5–100 μM ilimaquinone for 30 min. Nuclei (blue) were stained with Hoechst 33342. (Left) optical sections; (right) overlay of conventional fluorescence images and the respective brightfield image showing the cell borders as black outlines. Bars, 10 μm.
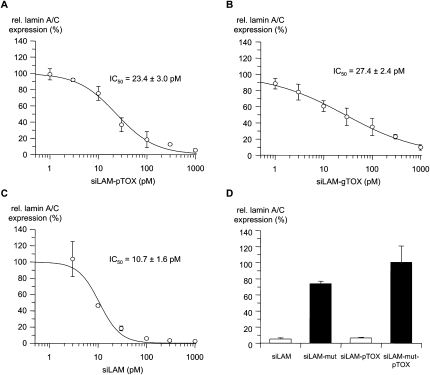
In principle, the chosen peptide TQIENLKEKG can be covalently attached by a stable thioether linkage to the guide strand or the passenger strand of siRNA, respectively (Fig. 1B). First, we synthesized and studied both possible 3′-modified variants of siRNA–peptide conjugates. Since the cellular uptake of siRNA and subsequent steps giving rise to incorporation of the guide strand into RISC followed by target suppression are, in summary, complex processes including intracellular translocation steps, we compared key characteristics of both conjugates. This study shows no difference between the guide strand conjugate and the passenger strand conjugate, including their cellular uptake and localization, their stability, and their biological effectiveness, i.e., the extent of inhibition of lamin A/C expression after lipofectamine-mediated delivery (Fig. 4A,B; data not shown). This finding allows us to further modify either strand of siRNA, e.g., by covalent peptide conjugation and, further, suggests that the siRNA stays in a double-stranded form during all of the relevant intracellular translocation steps before strand separation and RISC loading occur. For further experiments we chose the siLAM-gTOX conjugate (Fig. 1B).
FIGURE 4.
Potency of siLAM and siLAM–peptide conjugates after lipofectamine-mediated delivery. Dose–response curves and IC50 values of inhibition of lamin A/C expression by (A) siLAM-pTOX, (B) siLAM-gTOX, or (C) siLAM after transfection. ECV-304 cells were treated with the indicated concentrations of siLAM, siLAM-gTOX, or siLAM-pTOX using lipofectamine as transfectant. After 24 h in culture, cells were collected, their total RNA was extracted and reverse transcribed, and lamin A/C mRNA levels were determined by quantitative PCR. (D) Sequence-specificity of lamin A/C mRNA suppression by siLAM and siLAM-pTOX. ECV-304 cells were transfected with 1 nM of siLAM or siLAM-pTOX using lipofectamine. Lamin-A/C mRNA levels were determined as described. (g) Guide strand; (p) passenger strand; (mut) G → C transversion at positions 10 and 11 of the guide strand; (TOX) TQIENLKEKG.
Nuclease resistance of siLAM-TQIENLKEKG conjugates
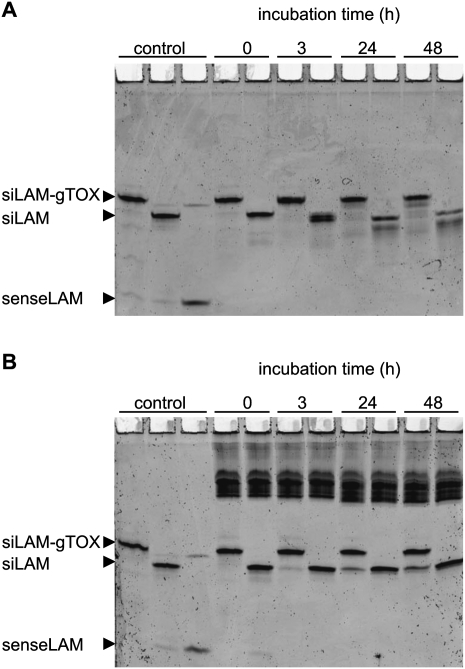
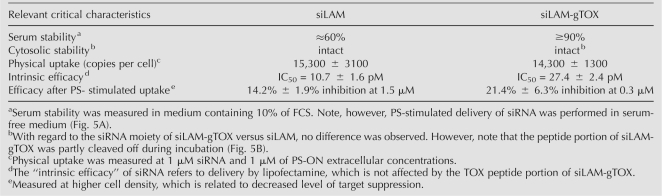
The resistance against degradation of parental siLAM and siLAM-gTOX was measured in medium containing 10% serum as well as in cytosolic extracts, i.e., the S100 fraction. In the case of serum stability, the peptide moiety seems to protect the siLAM-gTOX conjugate to a minor extent compared to unconjugated siLAM (Fig. 5A; Table 1). In case of cytosolic stability no measurable difference was found after up to 48 h of incubation, which is the relevant time range in standard experiments carried out here. It is noteworthy that the peptide moiety of siLAM-gTOX seems to be cleaved off in moderate amounts after 24 h in cytosolic extracts while the siRNA portion remains double-stranded and almost undegraded over the time period observed (Fig. 5B; Table 1). It has to be considered that the coincubation time of siRNA and derivatives with ECV-304 cells occurs for up to 24 h in serum-free medium before this is replaced by complete medium. In summary, these experiments indicate that there is no relevant altered stability of siLAM-gTOX versus siLAM influencing this study.
FIGURE 5.
Stability of siLAM and siLAM-gTOX. Both siRNAs (f.c. 0.5 μM) were incubated in (A) medium 199 containing 10% (v/v) FCS or (B) cytosolic extracts (S100 fraction) at 37°C. Aliquots withdrawn at indicated time points were rapidly frozen using liquid nitrogen and stored at −80°C. Samples were analyzed on a 20% (w/v) acrylamide nondenaturing PAGE, and RNA was detected using a PhosphorImager after staining with Sybr Gold dye. (g) guide strand; (TOX) TQIENLKEKG.
TABLE 1.
Summary of characteristics of siLAM and siLAM-gTOX relevant for the relationship between cellular delivery and effectiveness
Cellular uptake of siRNA and cell toxicity
In order to study whether the TOX moiety influences the cellular uptake step in addition to the presumed intracellular trans-membrane translocation step, we compared the amount of PS-delivered parental siRNA, siLAM-gTOX, and an unrelated siRNA–peptide conjugate, siLAM-gPTS1. A sensitive nuclease protection assay (Overhoff et al. 2004) with total RNA isolated from an intracellular fraction of treated cells shows no distinguishable difference of the amount of ∼15,000 copies per cell in all of these three cases (Table 1; data not shown). These experiments indicate that the covalent conjugation of TQIENLKEKG does not influence characteristics of the conjugates that are relevant for events prior to their intracellular transport.
As potential cell toxicity during delivery studies is an important parameter, we determined cell viability for all siRNA-based constructs studied here as well as for PS-ON at 24 and 40 h post-treatment of ECV-304 cells. The results do not indicate any toxicity at the concentrations used here (data not shown).
Measurement of the “quasi-intrinsic” potency of siRNA–peptide conjugates
In the case of the PS-stimulated delivery of siLAM, high amounts of intracellular siRNA in the order of up to 15,000 copies per cell can be determined at low efficacy of maximally 40% target inhibition (Overhoff and Sczakiel 2005). Conversely, delivery of siLAM by lipofectamine indicates 50% target suppression at ∼300 copies of siRNA per cell (Mescalchin et al. 2007), indicating that in this case intracellular release of siRNA is very efficient or does not even affect biological activity significantly. Thus, it is reasonable to assume that lipofectamine-mediated delivery of siLAM and siLAM-TOX conjugates monitors the activity of these constructs without major influences on endogenous transport steps by the peptide portion of siLAM-TOX, which we term here “quasi-intrinsic” potency.
It is controversially discussed whether the position of conjugation affects siRNA activity. Therefore we compared siRNA conjugates in which the TOX peptide moiety has been covalently attached to the 3′-terminus of either the guide strand or the passenger strand of siLAM with the parental siLAM by determing the IC50 values after transfection of ECV-304 cells using lipofectamine. We observed a threefold decrease of effectiveness, from an IC50 value of 10.7 ± 1.6 pM for parental siLAM to 27.4 ± 2.4 pM for siLAM-gTOX (where the guide strand is conjugated) and 23.4 ± 3.0 pM for siLAM-pTOX (where the passenger strand is conjugated), respectively (Fig. 4A–C; Table 1). So the reducing effect of the peptide covalently attached to either the guide strand or the passenger strand, respectively, is quite minor although presumably significant if one considers the standard deviation of the mean values. However, even though the potency of siLAM-TOX conjugates is reduced compared to the parental siLAM, they remain a very potent suppressor of lamin A/C expression.
In order to study whether the siLAM-TOX conjugates exert their suppression of lamin A/C expression via a RNAi pathway, we included a mismatch control for siLAM and siLAM-pTOX, respectively (Figs. 1A, 4D) and a scrambled siRNA control (siscr3; Kretschmer-Kazemi Far and Sczakiel 2003) as well as the peptide alone in the lipofectamine-mediated delivery (data not shown). In neither case did we observe any target suppression. In summary, these findings support the view that the biological effectiveness of siLAM-TOX conjugates is not due to effects other than RNAi (Elbashir et al. 2001).
On the technical level it should be noted that the role of “cell density” is important. In general, cell culture conditions have to be optimized for each mode of delivery. In these experiments, we used 100,000 ECV-304 cells per well of a 12-well plate, which corresponds to 70%–80% cell confluence. This is optimal for lipofectamine-mediated siRNA transfection and should not be further reduced due to cytotoxic side effects. Conversely, for PS-mediated siRNA delivery, as described in the next paragraph, we used 80,000 ECV-304 cells per well of a 12-well plate, which is related to 50%–60% cell confluence at the beginning of the experiment, i.e., when PS-ON and siRNA are added to cells. It is important to note that in this case discrimination of siRNA-mediated suppression of lamin A/C between constructs of different effectiveness is highest, although the extent of inhibition is reduced when compared to lower cell density. In other words, these experimental conditions are best for comparative studies but not for achieving maximal inhibitory effects.
Suppression of lamin A/C expression by siLAM-gTOX after PS-stimulated delivery
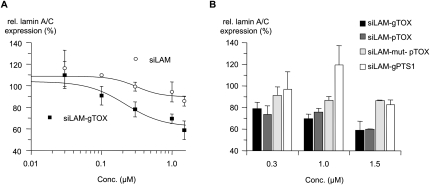
To investigate whether the peptide moiety influences the bioavailability and thereby efficacy of siLAM-gTOX-mediated target suppression after PS-stimulated uptake, we measured the levels of lamin A/C mRNA in ECV-304 cells. As shown in Figure 6A, cells treated at 0.3 μM and higher concentrations of siLAM-gTOX showed significant suppression of lamin A/C mRNA levels compared to untreated cells. In the case of PS-stimulated uptake of parental siLAM, only at an extracellular concentration of 1.5 μM was a minor reduction of lamin A/C expression observed (Fig. 6A; Table 1). In these experiments, the amount of siRNA inside treated cells was ∼15,000 siRNA copies per cell (Table 1). If one considers that in the case of transfection with lipofectamine, ∼300 copies per cell are detectable at half-maximal inhibition (Mescalchin et al. 2007), then one might hypothesize that the limited effectiveness in the case of PS-stimulated delivery is due to very low intracellular bioavailability. It is noteworthy that both siRNA–peptide conjugates, siLAM-pTOX and siLAM-gTOX, display a comparable extent of target suppression after PS-mediated delivery whereas the control siRNA–peptide conjugate siLAM-PTS1, which carries the peroxisomal targeting signal 1 SKL, as well as a mutated form of siLAM-pTOX show no biological activity. These controls indicate the importance of the TOX peptide moiety (Fig. 6B). In summary, all controls are consistent with the view that the siLAM-TOX conjugate exerts its biological effects via a TOX peptide-promoted step and according to RNAi mechanisms.
FIGURE 6.
Potency of siLAM and siLAM-peptide conjugates after PS-ON-stimulated delivery. (A) Increased dose dependent suppression of lamin A/C in ECV-304 cells after PS-stimulated delivery of siLAM-gTOX versus unconjugated siLAM. ECV-304 cells were incubated with the indicated concentrations of siLAM or siLAM-gTOX in the presence of 1 μM PS-ON 72mer for 24 h. After another 16 h in culture, cells were collected, their total RNA was extracted and reverse transcribed, and lamin A/C mRNA levels were determined by quantitative PCR. (B) Potency of siLAM-peptide conjugates to suppress lamin A/C mRNA is restricted to the conjugation of the TOX–peptide but independent of the siRNA strand carrying it. (g) Guide strand; (p) passenger strand; (mut) G → C transversion at positions 10 and 11 of the guide strand; (TOX) TQIENLKEKG; (PTS1) peroxisomal targeting signal 1, SKL.
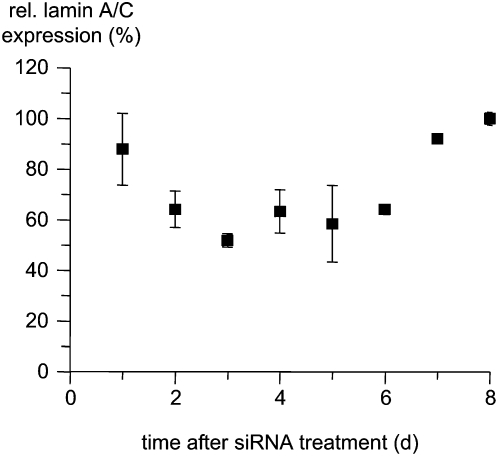
To further characterize this core finding we measured the time course of suppression of lamin A/C after PS-stimulated delivery of siLAM-pTOX, which reaches its maximal extent at day 3 after delivery and lasts until day 6 after delivery (Fig. 7). Further, we tested primary human endothelial HUVEC cells by using the PS-stimulated approach and observed ∼50% suppression of lamin A/C expression at 1.5 μM siLAM-pTOX (data not shown), which indicates that this mode of delivery is not restricted to ECV-304 cells but might be useful for a variety of cell types.
FIGURE 7.
Duration of siLAM-pTOX-mediated lamin A/C suppression after PS-ON-stimulated delivery. ECV-304 cells were incubated with 1.5 μM siLAM-pTOX in the presence of 1 μM PS-ON 72mer for 24 h at 37°C. Cells were kept in culture, and at the indicated time points cells were collected, their total RNA was extracted and reverse transcribed, and lamin A/C mRNA levels were determined by quantitative PCR. The expression of lamin A/C was normalized to the parental siLAM, which was set to 100%. (p) Passenger strand; (TOX) TQIENLKEKG.
Subcellular localization of siLAM-gTOX after PS-stimulated delivery
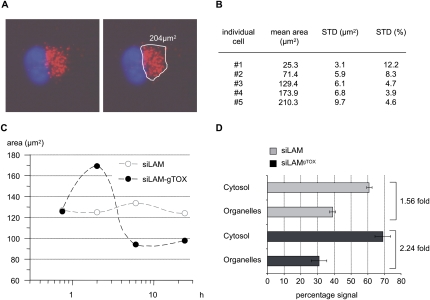
Next, we investigated whether the increased target suppression of siLAM-gTOX versus siLAM after PS-stimulated delivery could be due to intracellular release, possibly promoted by the toxin portion of siLAM-gTOX. Therefore we studied the endogenous localization of siLAM-gTOX by fluorescence microscopy with siLAM-gTOX to which a fluorescent group (Alexa555) was attached at the 3′ end of the passenger strand. Cells were incubated with PS-ON and siLAM-gTOX-Alexa555, and the fluorescent areas of living cells at distinct time points were quantified (Fig. 8A). The signal variability among individual cells was averaged over 40 cells. The error made by defining the fluorescent area for each cell was estimated to range between 3.9% and 12.2% of mean values in a size-dependent manner (Fig. 8B). Initially, the vesicular pattern in the perinuclear space is the same for Alexa555-siLAM and Alexa555-siLAM-gTOX (data not shown). This indicates that immediately after incubation of cells with PS-ON and siRNA the primary intracellular site of accumulation of both constructs is indistinguishable, implying that the peptide portion of Alexa555-siLAM-gTOX does not affect initial uptake steps, which is consistent with the fact that the conjugated peptide does not influence the amount of internalized siRNA copies per cells (Table 1). Two hours later, a spread of the fluorescence signal is observed for Alexa555-siLAM-gTOX but not for Alexa555-siLAM (Fig. 8C). At this time point, analyzed cells (n = 40) showed a 35% greater area for siLAM-gTOX versus the unconjugated siLAM. For Alexa555-siLAM the area of fluorescence signals above threshold levels stayed almost constant (Fig. 8C), which we interpret as a stable captured state. By contrast, the fluorescent area for Alexa555-siLAM-gTOX peaks at 2 h post-incubation and decreases at later time points. We assume this is due to a decrease of the fluorescence signal below detection limits caused by release of the conjugate and quasi-homogenous intracellular distribution.
FIGURE 8.
Fluorescence microscopy and cytosol/organelle separation indicate time-dependent release of siLAM-gTOX from perinuclear compartments. (A) Example for defining signal boundaries (white line) and for quantification of areas containing fluorescently labeled siRNA (red). Nuclei are stained with Hoechst 33342 and are shown in blue. (B) Area measurement of five individual cells differing in total fluorescent area. Determination of signal boundaries was repeated 10 times for each cell. Standard deviations of this area measurement are indicated as total area and as percentage of the mean. (C) The mean area of siRNA signals measured in ECV-304 cells (n = 40) at the indicated time points shows an initial increase in areas and a subsequent decrease for siLAM-gTOX but constant areas for siLAM. (D) The distribution of radiolabeled siLAM or siLAM-gTOX in organelles and cytosolic cell fraction shows increased cytosolic availability of the siLAM–peptide conjugate versus siLAM. Radioactively labeled siRNA was delivered by PS stimulation in ECV-304 cells. Radioactive signals in cytosolic and organelle fractions were quantified by liquid scintillation counting. The total radioactivity was set to 100%. For siLAM-gTOX the cytosolic amount was 2.24-fold greater than the organelle-associated amount versus a factor of 1.56 observed for siLAM. (g) Guide strand; (TOX) TQIENLKEKG.
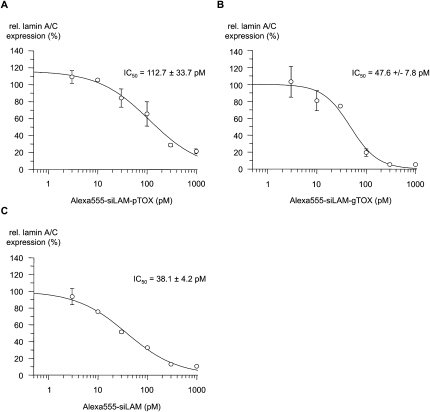
In order to investigate the influence of the fluorescence label Alexa555 on the biological activity of labeled siRNAs additional dose–response measurements were performed with Alexa555-labeled parental siLAM, siLAM-gTOX, and siLAM-pTOX (Fig. 9). The IC50 values indicate a stronger loss of activity for siLAM-pTOX (112 pM) and a moderate loss for siLAM-gTOX (38 pM). When compared to the effectiveness of the unlabeled homologous siRNAs (Fig. 4), it turns out that the covalent attachment of the Alexa555 moiety at the guide strand of Alexa555-siLAM-pTOX has a stronger negative effect on efficacy than its attachment to the 3′-end of the passenger strand in case of Alexa555-siLAM-gTOX.
FIGURE 9.
Potency of Alexa555-labeled siLAM and siLAM-peptide conjugates after lipofectamine-mediated delivery. Dose–response curves and IC50 values of inhibition of lamin A/C expression by (A) Alexa555-siLAM-pTOX, (B) Alexa555-siLAM-gTOX, or (C) Alexa555-siLAM after transfection. ECV-304 cells were treated with the indicated concentrations of Alexa555-siLAM-pTOX, Alexa555-siLAM-gTOX, or Alexa555-siLAM using lipofectamine as transfectant. After 24 h in culture, cells were collected, their total RNA was extracted and reverse transcribed, and lamin A/C mRNA levels were determined by quantitative PCR. (g) Guide strand; (p) passenger strand; (TOX) TQIENLKEKG.
As an independent assay to study whether siRNA is contained within organelles or distributed within the cytoplasm, we separated both fractions by ultracentrifugation of lysates of ECV-304 cells 2 h after treatment with PS-ON and 32P-labeled siLAM or 32P-labeled siLAM-gTOX, respectively (Fig. 8D). At this time point fluorescence microscopy showed most pronounced effects (Fig. 8C). This experiment showed increased amounts of radioactivity in cytosolic fractions when siLAM-gTOX was used compared to the use of siLAM. There is no detectable degradation of siRNA and siRNA conjugates during the time period of this experiment (Fig. 5B; data not shown). Thus, we conclude that greater amounts of siLAM-gTOX have been released from capturing organelles than siLAM.
In summary, the experiments described here indicate strongly that the peptide moiety of siLAM-gTOX promotes the intracellular release from perinuclear organelles to the cytoplasm, where RNAi is thought to occur.
A semi-quantitative view on the importance of intracellular release of siLAM
The apparent extent of inhibition of lamin A/C expression is a function of a sum of characteristics of the mode of delivery and of intrinsic properties of siRNA (Table 1). Here, we focused on the importance of the endogenous release of siRNA from perinuclear compartments, which are the primary location of siRNA after PS-stimulated delivery, and we considered all steps that are thought to influence the overall extent of target suppression listed in Table 1. For reasons of completeness of data, we also measured the stability of siRNAs in medium containing 10% FCS. However, in this analysis we did not consider this parameter, since all experiments were performed in serum-free medium.
The quasi-intrinsic potency of siLAM-gTOX was compared to parental siLAM by lipofectamine-mediated delivery, since this mode is not affected by the peptide portion of siLAM-gTOX (see above and Mescalchin et al. 2007). When considering the approximately threefold loss of activity of the siLAM–peptide conjugate (IC50 = 27 pM) (Table 1) versus siLAM (IC50 = 11 pM) (Table 1) and, further, considering the indistinguishable stability of siLAM and siLAM-gTOX in cytosolic extracts and their comparable physical intracellular amounts, then a promoting effect by increased intracellular release of siLAM-gTOX of greater than one order of magnitude can thus be derived (Table 1). This view is consistent with the apparently increased intracellular release of siLAM-gTOX versus siLAM from the perinuclear space (Fig. 8).
DISCUSSION
In summary, we showed clearly a higher bioavailability and therefore bioactivity for siLAM-gTOX compared to siLAM when delivered by PS-ON. The higher biological activity comes along with a bigger area of located siRNA within the cells, which probably is a sign for the efflux of siRNAs out of the perinuclear vesicles after 2 h of incubation. This assumption is strengthened by the fact that the area is decreased at later time points, which means that the amount of the remaining siRNA is equally well decreased. In addition to these findings, ultracentrifugation studies show that the signal ratio cytosol/organelles was higher for siLAM-gTOX compared to siLAM, which is also a sign of a higher amount of free siRNA within the cytosol and therefore for the higher bioavailability of siLAM-gTOX. The efflux of siRNA out of the perinuclear region and the higher amount of siLAM-gTOX in the cytosol explains the increased bioactivity of siLAM-gTOX after PS-stimulated uptake. Together with the observation that siLAM-gTOX has a decreased potency compared to siLAM after lipofectamine transfection, we assume that the attachment of the bacterial toxin sequence TQIENLKEKG to siLAM has a positive influence on the bioavailability of siLAM. In addition to reporting on the influence of this “leader peptide” on the bioavailability of siRNA after PS-stimulated uptake, this work introduces siRNA–peptide conjugates as tools to study the role of intracellular trafficking and localization for the biological effectiveness of siRNA in general and of exploiting peptide-steered intracellular transport pathways for siRNA.
It is reasonable to assume that this approach is also relevant for other classes of oligonucleotide-based tools and drugs, including microRNA, antisense oligonucleotides, or aptamer-based molecules. Further, this study suggests the value of investigating siRNA conjugates with other signal peptides that are known to signal the translocation to other specific cellular compartments, such as, for example, KDEL, which serves as a retrieval signal for proteins residing in the endoplasmic reticulum (Munro and Pelham 1987), the SV40 nuclear localization signal (PKKKRKV) that is known as a trigger for the transport through the nuclear pore with the help of the GTPase Ran (Adam and Gerace 1991), or the trans-Golgi network retention signal SXYQRL (Bos et al. 1993; Humphrey et al. 1993). Such work is in progress.
MATERIALS AND METHODS
Oligonucleotides, peptides, and plasmids
3′-thiopropyl functionalized oligonucleotides for peptide coupling were synthesized and purified by RNA-TEC. All other oligonucleotides were purchased from IBA. The N-terminally bromoacetylated peptide TQIENLKEKG was synthesized by Advanced Biomedical, and the chemical coupling of this peptide with RNA strands was carried out by J.J.T., G.D.I., and M.J.G. (for details see the section Synthesis of siRNA-peptide conjugates). Sequences and chemical structures of used oligonucleotides are depicted in Figure 1.
Synthesis of siRNA–peptide conjugates
Chemical coupling of the N-bromoacetyl peptide TQIENLKEKG to the 3′-thiopropyl terminus of the guide or passenger strand of siLAM (denoted in the following as siLAM-gTOX or siLAM-pTOX, respectively) was carried out as follows. 100 μL of an aqueous solution of the single-stranded RNA (170 nmol), 100 μL of an aqueous solution of bromoacetyl peptide (1000 nmol, 5.9 eq.), and 50 μL 1M Na-phosphate buffer (pH 7.0) were mixed and the reaction allowed to continue for 16 h. Conjugates were purified by a single injection on a Resource Q analytical anion exchange column eluting at 1 ml min−1 with salt gradient and other parameters as previously described, and characterized by MALDI-TOF mass spectrometry (Turner et al. 2006). siLAM-gTOX: m/e expected 7977.4, found 7975.5; siLAM-pTOX: m/e expected 8025.6, found 8009.3.
PS-stimulated uptake of siRNA
The PS-stimulated delivery of siRNA to ECV-304 cells was performed as described (Overhoff and Sczakiel 2005).
Labeling and annealing of siRNA, RNase protection, and measurement of lamin A/C-specific mRNA
All of these methods are described elsewhere (Overhoff et al. 2004; Overhoff and Sczakiel 2005).
Fluorescence microscopy
Initially 10,000 ECV-304 cells per chamber were seeded to eight-chambered Lab-Tek coverglasses (NUNC). Prior to further treatment, cells were washed three times with 500 μL of serum-free Opti-MEM medium followed by incubation with 1 μM of fluorescently labeled siRNA and PS-ON in 200 μL of Opti-MEM as described above. Cells were washed five times with 500 μL of Opti-MEM and incubated with 200 μL of Opti-MEM supplemented with 1 μg/mL of Hoechst 33342 (Invitrogen). Microscopic analyses were carried out by semi-confocal Apotome microscopy using AxioVision software (Zeiss Axiovert 200M).
Localization studies on the PS-ON-stimulated uptake of siRNA
We seeded 8,000 ECV-304 cells/well in Medium 199 supplemented with 10% FCS into eight-well LabTek chambered coverglasses (Nalgene Nunc International). After 16 h, cells were incubated with 1 μM fluorescently labeled siRNA, 1 μM PS-ON, and 10 μg/mL AMF-Cy3 or 0.5 μg/mL CTxB-FITC, respectively, in a final volume of 200 μL OptiMem/well for 60 min at 37°C, 5% CO2. After washing five times with Opti-Mem, the nuclei were stained with Hoechst 33342.
Inhibitor studies on the PS-ON-mediated siRNA uptake using ilimaquinone
A total of 8,000 ECV-304 cells were incubated with 1 μM siLam-Alexa555 and 1 μM PS-ON 72mer for 60 min as described above. After washing five times with OptiMem, the cells were incubated with 12.5 to 100 μM ilimaquinone in Opti-Mem for 30 min at 37°C, 5% CO2. After removing ilimaquinone-containing supernatant and washing five times with Opti-Mem, the nuclei were stained with Hoechst 33342.
Preparation of cytosol and organelles
A total of 200,000 cells was seeded in a 12-well plate and incubated overnight in medium 199 supplemented with 10% fetal calf serum. Subsequently, cells were washed with serum-free Opti-MEM medium and incubated with 1 μM of siRNA, 3.5 nM of radiolabeled siRNA, and 1 μM of PS-ON in a total volume of 200 μL of Opti-MEM medium for 2 h at 37°C and 5% CO2. Cells were washed four times with 1 mL of PBS, harvested by trypsin treatment, and washed again twice with PBS. Lysis of cells was carried out by adding 250 μL of TE buffer (10 mM Tris/HCl at pH 7.4; 1 mM EDTA) and by a 10-fold passage of the solution through a 100-μL pipette tip. To avoid degradation of siRNA, 40 U of RNase inhibitor (RiboLock, Fermentas) were added. Next, the cytosol was prepared by ultracentrifugation at 100,000g for 45 min at 4°C. Radioactivity in fractions containing supernatant (cytosol) or pellet (organelles) was measured in a liquid scintillation counter and the percentage of signals was calculated.
Regarding the preparation of organelle fractions one should note that in this study we aimed to study whether the conjugation of a bacterial protein toxin to siRNA is capable to initiate the trans-membrane translocation of this conjugate to the cytoplasm where RNAi is thought to take place (Zeng and Cullen 2002). This is why we simply aimed to separate membrane fractions from cytosolic fractions. We believe that a centrifugation step at 100,000g is a sort of standard protocol for preparing cytosolic extracts via segregation of all membranous particles, and this view is derived from the literature (e.g., Dignam et al. 1983; Martinez et al. 2002; Meister et al. 2004). However, we do not wish to state that the 100,000g fraction represents organelles; this protocol rather separates organelles from cytosol.
ACKNOWLEDGMENTS
This work was supported by a grant of the medical faculty of the Universität zu Lübeck to M.O. (FKZ: A28:2007). J.J.T was supported by a grant from EC Framework 5 (QLK3-CT-2002-01989).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1305209.
REFERENCES
- Adam S.A., Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Glick B.S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bos K., Wraight C., Stanley K.K. TGN38 is maintained in the trans-Golgi network by a tyrosine-containing motif in the cytoplasmic domain. EMBO J. 1993;12:2219–2228. doi: 10.1002/j.1460-2075.1993.tb05870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumcrot D., Manoharan M., Koteliansky V., Sah D.W. RNAi therapeutics: A potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K., Ye Z., Guntaka R.V., Mahato R.I. Enhanced hepatic uptake and bioactivity of type α1(I) collagen gene promoter-specific triplex-forming oligonucleotides after conjugation with cholesterol. J. Pharmacol. Exp. Ther. 2006;317:797–805. doi: 10.1124/jpet.105.100347. [DOI] [PubMed] [Google Scholar]
- de Fougerolles A., Vornlocher H.P., Maraganore J., Lieberman J. Interfering with disease: A progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshayes S., Morris M., Heitz F., Divita G. Delivery of proteins and nucleic acids using a noncovalent peptide-based strategy. Adv. Drug Deliv. Rev. 2008;60:537–547. doi: 10.1016/j.addr.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y., Tuschl T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Weber K., Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Falnes P.O., Sandvig K. Penetration of protein toxins into cells. Curr. Opin. Cell Biol. 2000;12:407–413. doi: 10.1016/s0955-0674(00)00109-5. [DOI] [PubMed] [Google Scholar]
- Ge Q., Filip L., Bai A., Nguyen T., Eisen H.N., Chen J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc. Natl. Acad. Sci. 2004;101:8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzelinski M., Urban-Klein B., Martens T., Lamszus K., Bakowsky U., Hobel S., Czubayko F., Aigner A. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum. Gene Ther. 2006;17:751–766. doi: 10.1089/hum.2006.17.751. [DOI] [PubMed] [Google Scholar]
- Haney S.A. Expanding the repertoire of RNA interference screens for developing new anticancer drug targets. Expert Opin. Ther. Targets. 2007;11:1429–1441. doi: 10.1517/14728222.11.11.1429. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Humphrey J.S., Peters P.J., Yuan L.C., Bonifacino J.S. Localization of TGN38 to the trans-Golgi network: Involvement of a cytoplasmic tyrosine-containing sequence. J. Cell Biol. 1993;120:1123–1135. doi: 10.1083/jcb.120.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer-Kazemi Far R., Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: A comparison with antisense oligonucleotides. Nucleic Acids Res. 2003;31:4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le P.U., Nabi I.R. Distinct caveolae-mediated endocytic pathways target the Golgi apparatus and the endoplasmic reticulum. J. Cell Sci. 2003;116:1059–1071. doi: 10.1242/jcs.00327. [DOI] [PubMed] [Google Scholar]
- Lebleu B., Moulton H.M., Abes R., Ivanova G.D., Abes S., Stein D.A., Iversen P.L., Arzumanov A.A., Gait M.J. Cell penetrating peptide conjugates of steric block oligonucleotides. Adv. Drug Deliv. Rev. 2008;60:517–529. doi: 10.1016/j.addr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska A., Urlaub H., Lührmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Dorsett Y., Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescalchin A., Detzer A., Wecke M., Overhoff M., Wünsche W., Sczakiel G. Cellular uptake and intracellular release are major obstacles to the therapeutic application of siRNA: Novel options by phosphorothioate-stimulated delivery. Expert Opin. Biol. Ther. 2007;7:1531–1538. doi: 10.1517/14712598.7.10.1531. [DOI] [PubMed] [Google Scholar]
- Minakuchi Y., Takeshita F., Kosaka N., Sasaki H., Yamamoto Y., Kouno M., Honma K., Nagahara S., Hanai K., Sano A., et al. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res. 2004;32:e109. doi: 10.1093/nar/gnh093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey D.V., Lockridge J.A., Shaw L., Blanchard K., Jensen K., Breen W., Hartsough K., Machemer L., Radka S., Jadhav V., et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Munro S., Pelham H.R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Opalinska J.B., Gewirtz A.M. Nucleic-acid therapeutics: Basic principles and recent applications. Nat. Rev. Drug Discov. 2002;1:503–514. doi: 10.1038/nrd837. [DOI] [PubMed] [Google Scholar]
- Overhoff M., Sczakiel G. Phosphorothioate-stimulated uptake of short interfering RNA by human cells. EMBO Rep. 2005;6:1176–1181. doi: 10.1038/sj.embor.7400535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhoff M., Wünsche W., Sczakiel G. Quantitative detection of siRNA and single-stranded oligonucleotides: relationship between uptake and biological activity of siRNA. Nucleic Acids Res. 2004;32:e170. doi: 10.1093/nar/gnh168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana T.M. Illuminating the silence: Understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- Ratts R., Trujillo C., Bharti A., vanderSpek J., Harrison R., Murphy J.R. A conserved motif in transmembrane helix 1 of diphtheria toxin mediates catalytic domain delivery to the cytosol. Proc. Natl. Acad. Sci. 2005;102:15635–15640. doi: 10.1073/pnas.0504937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A., Aleku M., Keil O., Endruschat J., Esche V., Fisch G., Dames S., Loffler K., Fechtner M., Arnold W., et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13:1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- Song E., Lee S.K., Wang J., Ince N., Ouyang N., Min J., Chen J., Shankar P., Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Song E., Zhu P., Lee S.K., Chowdhury D., Kussman S., Dykxhoorn D.M., Feng Y., Palliser D., Weiner D.B., Shankar P., et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Takeshita F., Minakuchi Y., Nagahara S., Honma K., Sasaki H., Hirai K., Teratani T., Namatame N., Yamamoto Y., Hanai K., et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc. Natl. Acad. Sci. 2005;102:12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P.A., Yucel J.K., Veit B., Faulkner D.J., Deerinck T., Soto G., Ellisman M., Malhotra V. Complete vesiculation of Golgi membranes and inhibition of protein transport by a novel sea sponge metabolite, ilimaquinone. Cell. 1993;73:1079–1090. doi: 10.1016/0092-8674(93)90638-7. [DOI] [PubMed] [Google Scholar]
- Tan P.H., Yang L.C., Shih H.C., Lan K.C., Cheng J.T. Gene knockdown with intrathecal siRNA of NMDA receptor NR2B subunit reduces formalin-induced nociception in the rat. Gene Ther. 2005;12:59–66. doi: 10.1038/sj.gt.3302376. [DOI] [PubMed] [Google Scholar]
- Turner J.J., Williams D., Owen D., Gait M.J. Wiley; New York: 2006. Disulfide conjugation of peptides to oligonucleotides and their analogs. Current protocols in nucleic acid chemistry, Chap. 4; pp. 4.28.21–24.28.21. [DOI] [PubMed] [Google Scholar]
- Turner J.J., Jones S., Fabani M.M., Ivanova G., Arzumanov A.A., Gait M.J. RNA targeting with peptide conjugates of oligonucleotides, siRNA and PNA. Blood Cells Mol. Dis. 2007;38:1–7. doi: 10.1016/j.bcmd.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Vornlocher H.P. Antibody-directed cell-type-specific delivery of siRNA. Trends Mol. Med. 2006;12:1–3. doi: 10.1016/j.molmed.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Wang H.J., Benlimame N., Nabi I. The AMF-R tubule is a smooth ilimaquinone-sensitive subdomain of the endoplasmic reticulum. J. Cell Sci. 1997;110:3043–3053. doi: 10.1242/jcs.110.24.3043. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Cullen B.R. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002;8:855–860. doi: 10.1017/s1355838202020071. [DOI] [PMC free article] [PubMed] [Google Scholar]