Abstract
The mitochondrial RNA binding complex 1 (MRB1) is a recently discovered complex of proteins associated with the TbRGG1 and TbRGG2 proteins in Trypanosoma brucei. Based on the phenotype caused by down-regulation of these two proteins, it was proposed to play an unspecified role in RNA editing. RNAi silencing of three newly characterized protein subunits, guide RNA associated proteins (GAPs) 1 and 2 as well as a predicted DExD/H-box RNA helicase, show they are essential for cell growth in the procyclic stage. Furthermore, their down-regulation leads to inhibition of editing in only those mRNAs for which minicircle-encoded guide (g) RNAs are required. However, editing remains unaffected when the maxicircle-encoded cis-acting gRNA is employed. Interestingly, all three proteins are necessary for the expression of the minicircle-encoded gRNAs. Moreover, down-regulation of a fourth assayed putative MRB1 subunit, Nudix hydrolase, does not appear to destabilize gRNAs, and down-regulation of this protein has a general impact on the stability of maxicircle-encoded RNAs. GAP1 and 2 are also essential for the survival of the bloodstream stage, in which the gRNAs become eliminated upon depletion of either protein. Immunolocalization revealed that GAP1 and 2 are concentrated into discrete spots along the mitochondrion, usually localized in the proximity of the kinetoplast. Finally, we demonstrate that the same mtRNA polymerase known to transcribe the maxicircle mRNAs may also have a role in expression of the minicircle-encoded gRNAs.
Keywords: RNA editing, guide RNA, mitochondrion, trypanosome
INTRODUCTION
Kinetoplastid flagellates, the most studied of which are causative agents of African sleeping sickness, Chagas disease, and leishmaniases in humans, have a strikingly complex mitochondrial (mt) DNA, also termed kinetoplast (k) DNA. It is composed of two types of molecules—maxicircles and minicircles. In the single mitochondrion, maxicircles are present in about a dozen copies and represent the equivalents of classical mtDNA, bearing genes for subunits of respiratory complexes, mitoribosomal RNAs, and one mitoribosomal subunit (for a recent review, see Simpson et al. 2004; Lukeš et al. 2005; Stuart et al. 2005). A unique feature of these flagellates is that the transcripts of some of the maxicircle-encoded genes are post-transcriptionally altered by additions and/or deletions of uridylate (U) residues by a process termed RNA editing (Benne et al. 1986).
The function of kDNA minicircles had remained mysterious for decades. These molecules are concatenated in a network, number in the thousands, and their heterogeneous sequence make up the bulk of the mt genome (Liu et al. 2005). The seminal discovery that they encode genes for small RNA molecules called guide (g) RNAs revealed their biological role as a genetic reservoir complementing the maxicircles (Sturm and Simpson 1990). It is these gRNAs that provide information for multiple and specific insertions and deletions of uridines into pre-edited maxicircle mRNAs. Each gRNA is composed of three regions, differing in their function and content. The 5′ anchor region hybridizes to a complementary sequence of the pre-edited mRNA, while the downstream information region of the RNA molecule provides the template for U insertions/deletions in the cognate mRNA via both Watson-Crick and noncanonical G:U base-pairing. The 3′ end of the gRNA is comprised of the post-transcriptionally added oligo(U) tail (Blum and Simpson 1990). While the identity of the RNA components of editing was established almost 20 years ago, the identification of the myriad of proteins involved in mtRNA metabolism remains an ongoing endeavor.
The core RNA editing process has been ascribed to the 20S editosome, a multiprotein complex composed of 20 proteins (Simpson et al. 2004; Lukeš et al. 2005; Stuart et al. 2005). It has been firmly established by in vitro and in vivo experiments that editing is carried out by this complex, which contains all catalytic activities necessary for the cleavage of the pre-mRNA, addition or deletion of the U(s), and ligation of the processed RNA molecules. The 20S editosome appears to be represented by a population of at least three dynamic protein complexes slightly differing in their composition and specific role in this process (Panigrahi et al. 2006; Carnes et al. 2008).
Mitochondrial RNA binding proteins have been a focus of intense research. The MRP1/2 complex is a heterotetrameric structure composed of the MRP1 and MRP2 proteins, with positively charged residues concentrated at one face, to which the gRNAs anneal (Schumacher et al. 2006). Data obtained with the in vivo purified as well as in vitro reconstituted MRP1/2 complexes are consistent with a role in matchmaking the gRNAs with cognate pre-edited mRNAs (Müller et al. 2001; Zíková et al. 2008). RBP16 is another RNA binding protein that appears to have an in vivo role in editing (Pelletier and Read 2003), perhaps via its demonstrated ability to stimulate insertion editing in vitro (Miller et al. 2006).
The initial characterization of TbRGG1, so-called because it possesses the RGG RNA binding motif, suggested the existence of another protein complex with a role in RNA editing (Vanhamme et al. 1998). The down-regulation of TbRGG1 in the procyclic (insect) stage of Trypanosoma brucei caused an overall decline of edited mRNAs, but the never-edited transcripts were unaffected (Hashimi et al. 2008). This protein is stably associated with a putative protein complex composed of about 14 different subunits (Hashimi et al. 2008; Panigrahi et al. 2008), provisionally named the mitochondrial RNA binding complex 1 (MRB1 complex). Several of its components have features that point to RNA processing activities associated in an RNA-mediated manner (Hashimi et al. 2008; Panigrahi et al. 2008). Another component of this complex, TbRGG2 (also known as TbRGGm), has affinity for poly(U), and its depletion leads to a dramatic decrease in edited mRNAs (Fisk et al. 2008). Interestingly, a recent report on the newly discovered KPAP1 protein, poly(A) polymerase that polyadenylates maxicircle transcripts, shows that it is associated with a complex of proteins of overlapping composition with the MRB1 complex (Etheridge et al. 2008; Weng et al. 2008).
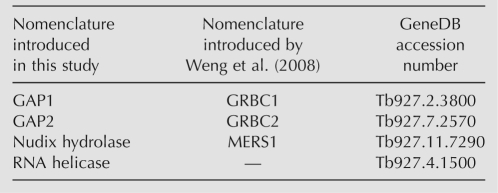
From these data, the role of the MRB1 complex is not obvious. However, its importance is underscored by the fact that orthologs of its subunits are found in all trypanosomatid genomes sequenced thus far. Here, we attempt to ascertain the function of the MRB1 complex by analysis of four subunits of this complex by RNAi silencing in the procyclic stage. The examined subunits are summarized in Table 1, including a nomenclature introduced in a similar study performed by Weng et al. (2008). Two of these subunits, Tb927.2.3800 and Tb927.7.2570, are paralogs that have no known protein motifs or domains (Hashimi et al. 2008). They will be referred to herein as guide RNA associated proteins (GAP) 1 and 2, respectively, since we provide evidence for their participation in the biogenesis of these molecules. The third subunit, Tb927.4.1500, is predicted to be an ∼240-kDa protein with DExD/H-box RNA helicase domains (Hashimi et al. 2008), and is referred to as such in this paper. The last studied subunit, Tb927.11.7290, is annotated as a Nudix hydrolase due to its conservation to other such proteins. Moreover, we show that mtRNA polymerase (mtRNAP), known to transcribe the maxicircle-encoded protein-coding genes (Grams et al. 2002), appears to have a similar role for minicircle-encoded gRNAs.
TABLE 1.
Nomenclature of examined MRB1 complex proteins
RESULTS
Subunits of the MRB1 complex are essential for the procyclic stage
Comparison of the growth of noninduced and RNAi-induced GAP1, GAP2, RNA helicase, and Nudix hydrolase cell lines revealed that these proteins are essential for the growth of T. brucei procyclics. In these cell lines, growth inhibition is apparent 3 to 4 d after addition of the RNAi-induction agent tetracycline (Fig. 1). In the case of the GAP knockdowns, the cells became increasingly resistant to the dsRNA and recovered around day 12 (Fig. 1A,B), as previously reported in the T. brucei system (Pelletier and Read 2003). However, the RNA helicase and Nudix hydrolase knockdowns did not recover their wild-type growth over the 14-d time course. Based on these data, the time points after 3 to 4 d of RNAi induction were selected for all subsequent experiments with the knockdowns.
FIGURE 1.
Subunits of the MRB1 complex are essential for growth of procyclic stage trypanosomes. Cell densities (cells/mL) that were measured every 24 h are plotted on a logarithmic scale on the y-axis over 14 d. Cells were diluted to 2 × 106 cells/mL every 2 d. Cells grown in the presence or absence of the RNAi-induction agent 1 μg/mL tetracycline are indicated by gray (tet+) or black (tet−) lines, respectively. Growth curves of knockdowns of (A) GAP1, (B) GAP2, (C) RNA helicase, and (D) Nudix hydrolase are shown.
Stability of GAP1 and 2 is mutually dependent
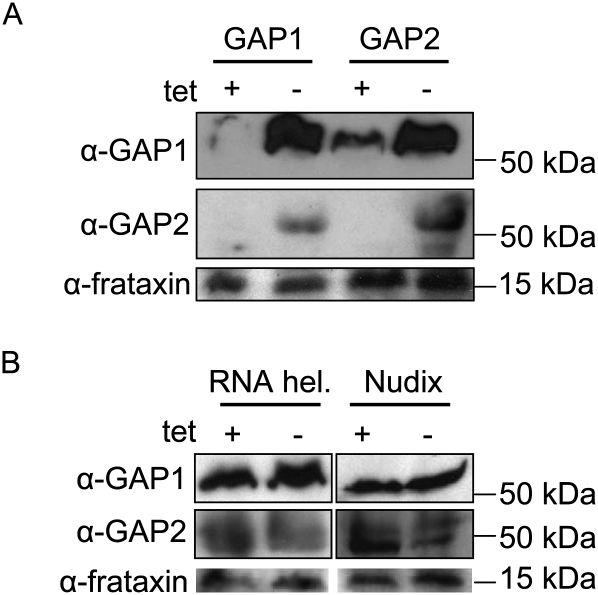
To confirm the mutual association of the GAP proteins as predicted by the mass spectrometry analysis (Hashimi et al. 2008), the mt lysates of cells with a single down-regulated GAP were probed with polyclonal antibodies against both proteins. The specificity of each α-GAP antibody for the appropriate target protein was verified by probing recombinant GAPs overexpressed in Escherichia coli (data not shown). Indeed, elimination of the target protein by RNAi was followed by the disappearance of the other GAP protein (Fig. 2A), testifying to their mutual dependence. Furthermore, we checked whether the GAPs are also destabilized upon silencing of either the RNA helicase or Nudix hydrolase. In either of these backgrounds, both proteins persisted (Fig. 2B).
FIGURE 2.
The stability of the GAP proteins is dependent on their mutual association. (A) Crude mt lysates from 108 cells were loaded per lane on a SDS-PAGE gel and transferred onto a PVDF membrane for immunodecoration with either α-GAP1 or α-GAP2 polyclonal antibody, as indicated on the left. Antibody against the mitochondrial protein frataxin was used as a loading control. Lysates from cells grown for 3 d in the presence or absence of 1 μg/mL tetracycline are indicated at the top (“+” or “−”) for either the GAP1 or GAP2 RNAi knockdown. Fifty-kDa and 15-kDa protein markers are indicated on the right. (B) Samples from the RNA helicase and Nudix hydrolase RNAi knockdown cells grown for 4 d in the presence and absence of 1 μg/mL tetracycline were prepared and labeled as described above.
RNAi silencing of MRB1 complex subunits affects maxicircle transcripts
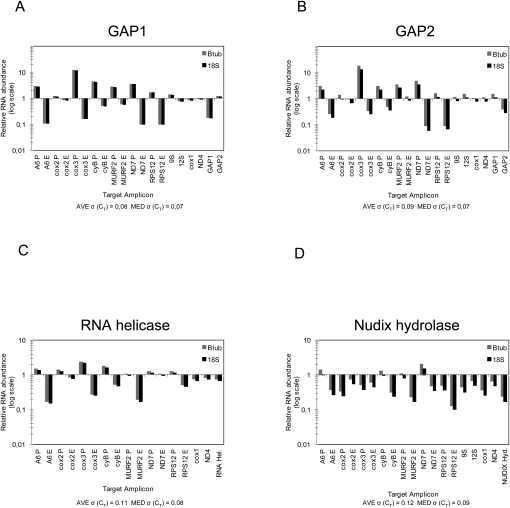
Since TbRGG1 and TbRGG2 from the MRB1 complex were shown to be essential for RNA editing (Fisk et al. 2008; Hashimi et al. 2008), we decided to investigate in detail the effect of down-regulation of the studied subunits in procyclic cells on the stability and editing of mt-encoded mRNAs. The levels of several pre-edited, edited, and never-edited RNAs were measured by a quantitative real-time (q) PCR-based assay, using cDNA from RNAi-induced and noninduced cells. Primers amplifying the pre-edited and edited mt mRNAs of ATPase subunit 6 (A6), cytochrome oxidase subunits 2 and 3 (cox2 and cox3), cytochrome reductase subunit B (cyB), maxicircle unknown reading frame 2 (MURF2), NADH dehydrogenase subunit 7 (ND7), and mitoribosomal protein S12 (RPS12) were used. The levels of two never-edited mRNAs (ND4 and cox1) and, in some cases, the 9S and 12S mitoribosomal RNAs were also measured, as were the mRNAs targeted by RNAi. The obtained values were normalized to the measured levels of the cytoplasmic β-tubulin and 18S rRNA transcripts, since they are unaffected by RNAi. All qPCR reactions with a given primer pair were done in triplicate, and the average and median standard deviations of the measured cycle threshold values are given in Figure 3.
FIGURE 3.
Effects of RNAi silencing of the MRB1 complex subunits on maxicircle transcripts. Quantitative real-time PCR analysis of pre-edited, edited, and never-edited mRNAs and rRNAs was performed on knockdowns of (A) GAP1, (B) GAP2, (C) RNA helicase, and (D) Nudix hydrolase. The reaction for each amplicon was done in triplicate on cDNA generated from cells grown in the presence or absence of the RNAi-inducer tetracycline. The relative abundance of each of the examined RNAs upon the synthesis of interfering dsRNA was plotted on a logarithmic scale: 1 represents the wild-type levels, while a value above or below this number signifies an increase or decrease of a given RNA, respectively. These values were normalized to the cytosolic transcripts β-tubulin or 18S rRNA, whose levels are not affected by RNAi. The average [AVE σ(CT)] and median standard deviations [MED σ(CT)] of the measured cycle threshold (CT) values for all the reactions performed for each knockdown are indicated under each bar graph. The following pre-edited (P) and edited (E) RNAs were assayed: ATPase subunit 6 (A6), cytochrome oxidase subunits 2 (cox2) and 3 (cox3), cytochrome reductase subunit b (cyB), maxicircle unknown reading frame 2 (MURF2), NADH dehydrogenase subunit 7 (ND7), and ribosomal protein S12 (RPS12). The following never-edited RNAs were assayed: 9S RNA, 12S RNA, cox1, and ND4. The appropriate cytoplasmic mRNA targeted by RNAi were assayed for each knockdown and are indicated on the right.
The qPCR assay revealed a virtually identical phenotype for both GAP1 and 2 knockdowns (Fig. 3A,B). With a single but important exception (see below), all examined pre-edited mRNAs (A6, cox3, cyB, MURF2, ND7, and RPS12) were up-regulated between 1.5-fold and up to 20-fold, as compared to RNA from the noninduced cells. Importantly, this up-regulation was correlated to the decrease of the respective edited transcripts. No effect was observed on the never-edited mRNAs (cox1 and ND4) and rRNAs (9S and 12S) (Fig. 3A,B). Significantly, pre-edited and edited cox2 mRNAs were also not affected by RNAi-mediated elimination of either GAP. This transcript is unique in that its single gRNA is contained in its 3′-untranslated region, acting as a guide for insertion editing in cis (Golden and Hajduk 2005). The assay also shows that dsRNAs specifically target only the intended GAP RNA (Fig. 3A,B).
This assay revealed a similar effect on maxicircle transcripts in cells with silenced RNA helicase: the combined increase and decrease of pre-edited and edited transcripts, respectively, with the exception of cox2 transcripts (Fig. 3C). However, it should be noted that while never-edited transcripts were unaffected, so were the levels of pre-edited and edited ND7. In contrast, silencing of Nudix hydrolase showed a general effect on almost all maxicircle-encoded transcripts (Fig. 3D). Most never-edited, pre-edited, and edited mRNAs and rRNAs are down-regulated upon depletion of this protein. This phenotype represents a highly significant departure from the otherwise relatively uniform phenotype of the other examined knockdowns.
The MRB1 complex has a role in gRNA expression
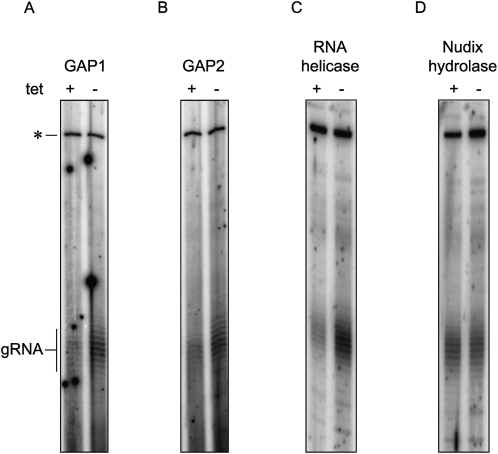
The disruption of RNA editing, observed in cells in which either GAP1, GAP2, or RNA helicase is down-regulated, appears to be independent of the core editing activities of the 20S editosome since cox2 mRNA editing proceeds normally. Another component of this process is the small 30–60-nt gRNAs. To see whether these primary transcripts are affected, they were capped with guanylyltransferase and [α-32P]GTP and visualized on a high-resolution denaturing acrylamide gel. An upper band corresponding to a cytoplasmic RNA is used as a loading control. Indeed, repression of GAP1 and 2 as well as RNA helicase in the procyclic cells leads to a decrease of the steady-state level of gRNAs (Fig. 4A–C). In contrast, neither the abundance nor the 3′-oligo(U) tail of gRNAs is affected upon depletion of Nudix hydrolase (Fig. 4D).
FIGURE 4.
Effects of RNAi silencing of the MRB1 complex subunits on minicircle transcripts. The total population of minicircle-encoded gRNAs were labeled in knockdowns of (A) GAP1, (B) GAP2, (C) RNA helicase, and (D) Nudix hydrolase. Labeling reactions were performed with guanylyltransferase, [α-32P]GTP and either (A,B) 2.5 μg or (C,D) 5 μg of RNA. The gRNAs, appearing as a ladder mainly because of their heterogeneous 3′-oligo(U) tails, are indicated on the left. The top band marked with the “*” is a cytosolic RNA that is also labeled in this reaction and is used as a loading control.
GAP1 assembly into macromolecular complexes is affected in the knockdowns for subunits of the MRB1 complex
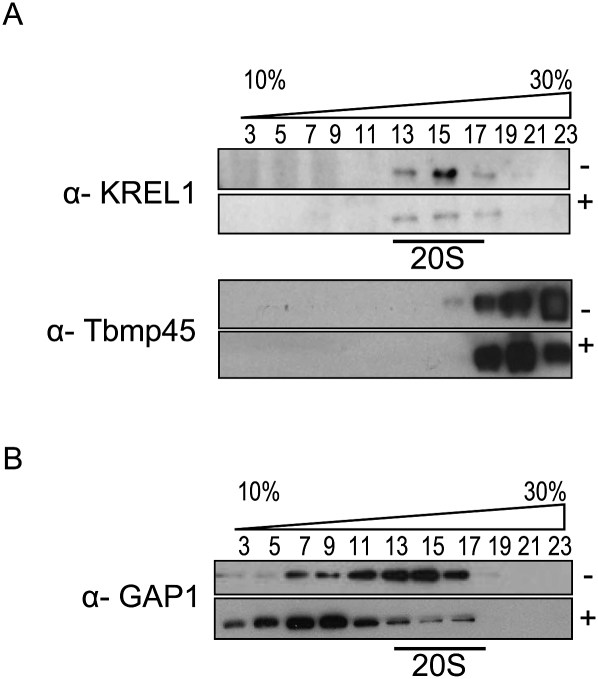
To further study the sedimentation properties of GAP1 in cells with down-regulated MRB1 subunits, lysates from hypotonically isolated mitochondria from noninduced and RNAi-induced cultures were loaded onto 10% to 30% glycerol gradients. Gradient fractions were first probed with monoclonal antibodies against three of the core editosome proteins, KREPA2, REL1, and KREPA3 (Panigrahi et al. 2001), used as markers for 20S. As an additional control, the fractions were immunodecorated with an antibody binding Tbmp45, a protein previously labeled REAP1, as a marker for 40S sedimentation (Madison-Antenucci et al. 1998; Fisk et al. 2008). A representative immunoblot performed on GAP1-induced and noninduced samples is in agreement with previous studies, as all proteins recognized by these antibodies localized to fractions 13 thru 17, the peak of 20S editosome sedimentation (Fig. 5A, top panel). Furthermore, it has been demonstrated that this complex is not disrupted when substrate RNAs are absent since it is held together by protein–protein interactions (Domingo et al. 2003; Fisk et al. 2008; Hashimi et al. 2008). The Tbmp45 protein was found to be localized in the lowest fractions, as expected (Fig. 5A, bottom panel). The other gradients depicted in Figure 5B were also validated in this fashion (data not shown).
FIGURE 5.
GAP1 assembly into macromolecular complexes is affected in the knockdowns for subunits of the MRB1 complex. Cleared lysates from hypotonically isolated mitochondria are separated by sedimentation in glycerol gradients. Odd fractions from the gradients were run on SDS-PAGE gels and transferred onto PVDF membranes. (A) All gradients were verified by immunodecoration with monoclonal antibodies against editosome subunits (top panel) KREPA2, KREL1, and KREPA3, as well as (bottom panel) Tbmp45. A representative blot for GAP1 samples is shown here in which the appropriate antibody signals are indicated on the left. The presence (+) or absence (−) of tetracycline is specified on the right. (B) α-GAP1 antibody was used to probe the glycerol gradient fractions from knockdown cell lines grown in the absence (−) or presence (+) of tetracycline as indicated on the right. The GAP1, GAP2, RNA helicase, and Nudix hydrolase cell lines are indicated on the left.
In the separated mt lysates from the noninduced cell lines, GAP1 is distributed throughout the gradient, although with some variation in the pattern as revealed by Western analysis (Figs. 5B; see, also, Fig. 7B, below). A broad distribution of GAP2 has been previously reported (Panigrahi et al. 2008). As expected, GAP1 is reduced upon interference of expression of either GAP subunit (Fig. 5B). A dramatic shift of the S-value of GAP1 is observed in the RNA helicase-silenced cells, in which it is concentrated in the lighter fractions (Fig. 5B). Such a striking difference in GAP1 sedimentation is not observed in the induced and noninduced Nudix hydrolase knockdowns (Fig. 5B). In the hydrolase-silenced sample, a reduction of GAP1 immunopositive signal in the dense fractions is apparent (Fig. 5B).
FIGURE 7.
GAP1 assembly into large macromolecular complexes is reduced in cells with RNAi-silenced mtRNAP. Western blot analysis of glycerol gradients was performed and labeled as described in Figure 5. (A) Western blots of fractions were verified using the (top panel) α-KREL1 or (bottom panel) α-Tbmp45 monoclonal antibodies or (B) probed using the α-GAP1 polyclonal antibody.
mtRNA polymerase appears to transcribe minicircles
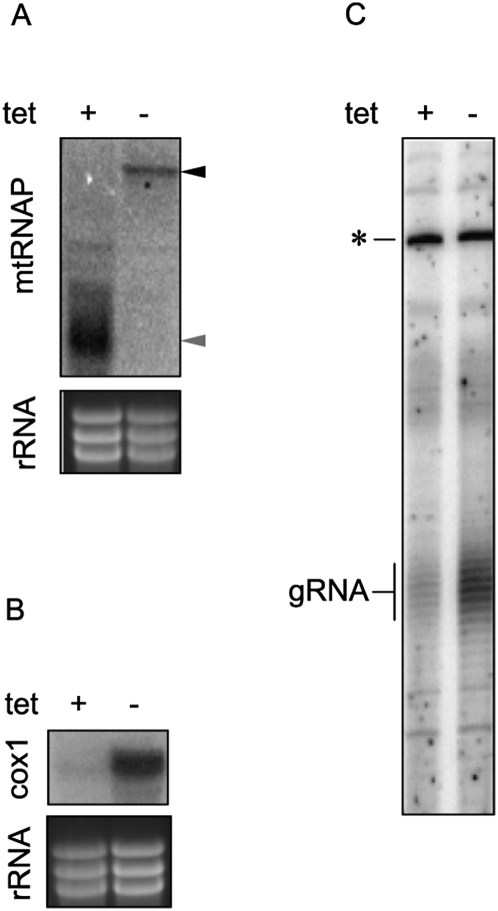
The participation of a multiprotein complex in the transcription of minicircle gRNA genes, independent of mtRNAP, has been hypothesized (Grams et al. 2002). In the inducible mtRNAP knockdowns (kindly provided by P.T. Englund, Johns Hopkins University), we have first verified the RNAi effect by Northern analysis, as antibodies against the T. brucei mtRNAP are not available. As shown in Figure 6A, the mtRNAP mRNA is undetectable 3 d after tetracycline induction, the time point at which growth inhibition is first observed (Grams et al. 2002). Next, we have hybridized blots containing mtRNA with a probe against cox1 mRNA. In agreement with the previous study (Grams et al. 2002), this never-edited transcript is undetectable 3 d after RNAi is triggered (Fig. 6B). To test whether this polymerase is responsible for the transcription of minicircles, the initial step in expression of gRNA molecules, we have performed the guanylyltransferase labeling assay on total RNA isolated from noninduced and induced knockdowns for mtRNAP. Clearly, the gRNA population is decreased in cells lacking the enzyme (Fig. 6C).
FIGURE 6.
Evidence that mtRNAP transcribes kDNA minicircles. (A) Effect of RNAi induction on the mtRNAP mRNA was followed by Northern blot analysis. Its levels were analyzed by blotting 10 μg of total RNA extracted from noninduced cells (−) and cells 3 d after RNAi induction (+) with a probe hybridizing to the gene fragment used for generation of dsRNA. The position of (black arrowhead) the targeted mRNA and (gray arrowhead) the inducibly synthesized dsRNA are indicated. To visualize characteristic rRNA bands, used here as a loading control, the gel was stained with ethidium bromide. (B) The same RNA was hybridized with a labeled probe for never-edited mRNA encoding cytochrome oxidase subunit 1 (cox1). (C) Guanylyltransferase labeling of gRNAs using the same RNA samples as used in A and B are performed and marked as in Figure 4. Five micrograms of total RNA were used per lane.
MRB1 complex is disrupted upon down-regulation of mtRNA polymerase
The similarity in the effect on gRNA expression of the knockdowns for mtRNAP and MRB1 subunits GAP1, GAP2, and RNA helicase inspired us to investigate the integrity of this complex in the absence of the polymerase. For this purpose, total lysates prepared from noninduced cells as well as from cells in which mtRNAP was targeted by RNAi for 3 d were loaded on glycerol gradients, and the obtained fractions were screened with the α-GAP1 antibody. The quality of the gradient was tested by Western blot analysis, using antibodies against the editosome KREL1 and Tbmp45 (Fig. 7A). GAP1 is broadly distributed in the noninduced cells in fractions 7–17 (Fig. 7B). Indeed, an apparent shift toward the lighter density fractions (with a marked peak in fractions 7 and 9) of the gradient in the GAP1 distribution occurs when mtRNAP is down-regulated. This result indicates that in the absence of mtRNAP, the MRB1 complex is disrupted in the same manner as in the absence of RNA helicase (Fig. 5B).
GAP1 and 2 and gRNAs are essential for the bloodstream stage
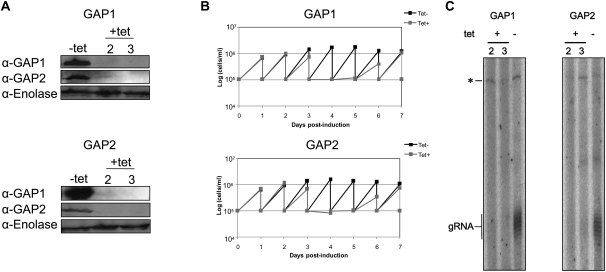
Since RNA editing is essential also in the bloodstream stage (Schnaufer et al. 2001), we decided to test whether the absence of GAP1 and 2, and consequently gRNA expression, are also lethal in this stage. The constructs containing the same GAP1 or 2 gene fragments as those used for RNAi in the procyclics were transfected into the bloodstream stage. Western blot analysis revealed that in the GAP1 and 2 knockdowns, the targeted protein was undetectable after 2 d of induction (Fig. 8A). Moreover, as in the procyclic stage, the ablation of GAP1 leads to the reduction of GAP2, and vice versa, testifying to their mutual dependence.
FIGURE 8.
GAP1 and GAP2 are essential for gRNA biogenesis in the bloodstream stage. (A) Whole cell lysates from 1.5 × 107 cells were immunoblotted with either the α-GAP1 or α-GAP2 antibody for each of the (top) GAP1 and (middle) GAP2 cell lines. Cells grown in the absence (−) or presence (+) of 1 μg/mL tetracycline are indicated, as well as the days post-induction in the latter samples. (Bottom) An antibody decorating the cytosolic enolase was used as a loading control. All antibodies are indicated on the left. (B) RNAi silencing of the GAP proteins in the bloodstream stage results in growth inhibition. Growth curves over 7 d are shown for the (top) GAP1 and (bottom) GAP2 knockdown cell lines and are labeled as in Figure 1. Cells were diluted every 24 h to a density of 105 cells/mL. (C) Guide RNA levels are diminished in the GAP-silenced bloodstream stage. The guanylyltransferase assay was performed on 5 μg of total RNA as described in Figure 4. The gels shown for (left) GAP1 and (right) GAP2 are also labeled as in Figure 4, with the days post-induction by tetracycline indicated below the “+.”
Down-regulation of either GAP1 or 2 results in growth inhibition, indicating that these proteins are also essential in this life cycle stage. When compared with the noninduced cells, the growth of the induced clones decreases at day 3 and stops at day 4 (Fig. 8B). Both induced cell lines recovered from the effect of RNAi at day 7 post-induction. Labeling of total RNA collected at days 2 and 3 post-induction with guanylyltransferase revealed that steady-state levels of gRNAs are decreased when either GAP1 or 2 is silenced in the bloodstream stage (Fig. 8C). This result indicates that the function of these proteins is conserved between the two major life stages of T. brucei, and that gRNAs are also required for survival of the infective blood forms.
GAP1 and GAP2 have a novel localization within the mitochondrion
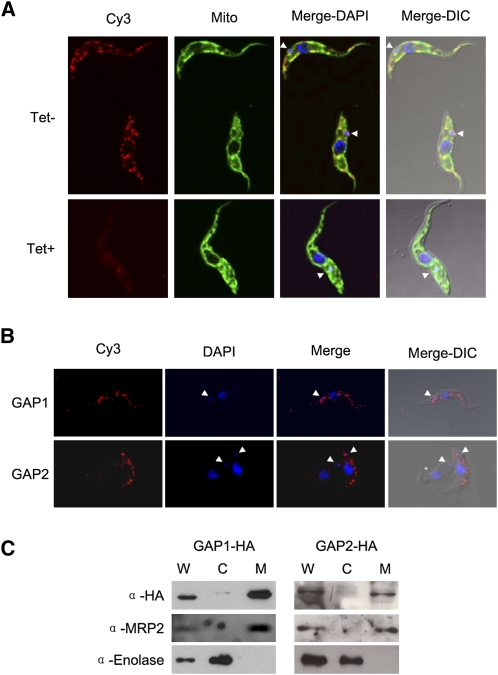
Previous data provided evidence for localization of the MRB1 complex in the mitochondrion (Hashimi et al. 2008; Panigrahi et al. 2008). We therefore have resorted to indirect immunofluorescence experiments to determine where these proteins are located in the organelle. Using the α-GAP1 antibody, we show that in the procyclic cells, the protein is confined to several discrete foci, distributed throughout the reticulated mitochondrion (Fig. 9A, top row), often in the proximity of the kDNA disk. Specificity of the used antibody was confirmed by its failure to detect target protein in RNAi-silenced GAP1 cells (Fig. 9A, bottom row). The polyclonal α-GAP2 antibodies proved to be less useful in the immunolocalization studies.
FIGURE 9.
The GAP proteins are localized in punctuate loci along the reticulated mitochondrion of procyclic T. brucei. (A) Immunolocalization using rabbit α-GAP1 antibody in the (top row, labeled Tet−) GAP1 noninduced and (bottom row, Tet+) RNAi-induced cells. Signal from cy3-conjugated α-rabbit antibody (Cy3), MitoTracker Green FM (Mito), the two images merged with DAPI staining of nucleus, and kDNA (Merge-DAPI) and a composite image with a differential interference contrast image (Merge-DIC) are indicated along the top. (White arrowheads) Position of kDNA. (B) Immunolocalization of the HA3-tagged versions (top row) GAP1 and (bottom row) GAP2, which is immunodecorated with rabbit polyclonal antibody against the epitope. Signal from cy3-conjugated α-rabbit antibody (Cy3), DAPI staining of nucleus and kDNA (DAPI), a merged image of the two (Merge), and a composite image with a differential interference contrast image (Merge-DIC) are indicated along the top. (White arrowheads) Position of kDNA. (C) Immunoblot analysis of the localization of overexpressed HA3-tagged (left) GAP1 and (right) GAP2 proteins. (W) Whole cell lysates and digitonin extracted (C) cytosolic and (M) mitochondrial fractions of these cell lines grown for 2 d in the presence of 1 μg/mL tetracycline. Antibody against the HA3-tag was used as well as those against the MRP2 and enolase proteins, serving as mitochondrial and cytosolic markers, respectively. Ten micrograms of protein of each digitonin fraction were loaded per lane.
To corroborate this unexpected observation, cells were transfected with a construct for tetracycline-inducible expression of either GAP1 or 2 proteins bearing an HA3-epitope tag. Immunocytochemistry and DAPI staining performed on cells grown in the presence of the antibiotic for 2 d confirmed that both epitope-tagged proteins are confined to multiple discrete punctuate loci unevenly distributed throughout the organelle (Fig. 9B). As observed with the α-GAP1 antibody, an immunopositive signal is also often observed near the kinetoplast.
Although attachment of the HA3 tag to the C terminus of the protein should not interfere with the mt import signal located at the N terminus, possible extra-mitochondrial localization had to be ruled out. Therefore, the 29-13 procyclics containing the HA3-tagged GAP1 or 2 were fractionated by extraction with increasing concentrations of digitonin. Indeed, probing the obtained cytosolic and mt fractions, as well as the total cell lysate, with antibodies against the HA3 tag showed that both tagged GAP proteins are properly imported into the organelle (Fig. 9C).
DISCUSSION
Since the discovery of RNA editing in 1986 (Benne et al. 1986), considerable efforts have been directed at understanding the molecular mechanisms underlying what was initially an enigmatic biological process. A major breakthrough was the discovery of gRNAs, which revealed that the information for U insertion/deletion events resides in these molecules, and a function of the thousands of minicircles is to encode them (Blum and Simpson 1990). The establishment of an in vitro editing assay led to the validation that proteins provided the enzymatic machinery for RNA editing (Seiwert et al. 1996). The isolation and characterization of the 20S editosome have dissected how a multi-protein complex can orchestrate the catalytic steps required for the maturation of most mtRNAs in trypanosomes (Simpson et al. 2004; Lukeš et al. 2005; Stuart et al. 2005).
Studies focusing on identifying proteins involved in other aspects of mtRNA metabolism have uncovered the MRB1 complex in three different instances (Etheridge et al. 2008; Hashimi et al. 2008; Panigrahi et al. 2008). Although the subunit composition in these studies is not identical, the degree of overlap is quite significant. The MRB1 complex contains up to 14 proteins, some of which possess known motifs and domains involved in RNA processing or protein interactions (Hashimi et al. 2008; Panigrahi et al. 2008). Two components of the MRB1 complex have been already characterized to some extent, namely, TbRGG1 and TbRGG2, the latter of which was suggested to be a transiently associated editing factor required for the processivity of RNA editing (Fisk et al. 2008). RNAi silencing of both proteins leads to a down-regulation of edited transcripts, suggesting some kind of a role in RNA editing (Fisk et al. 2008; Hashimi et al. 2008). However, these data do not adequately address a possible role of the MRB1 complex.
Here we addressed the question of function by examining the phenotype resulting from RNAi silencing of four subunits of this complex: GAP1, GAP2, RNA helicase, and Nudix hydrolase. We have shown that a subset of the MRB1 complex, GAP1, GAP2 and RNA helicase, is involved in gRNA biogenesis. This notion is supported by the absence of gRNAs when any of these proteins are depleted as a consequence of RNAi. Furthermore, editing of the maxicircle-encoded RNAs is subsequently reduced, with the notable exception of cox2. This transcript contains its own cis-gRNA in the 3′-untranslated region, thus circumventing the requirement for minicircle-encoded gRNAs acting in trans (Golden and Hajduk 2005). The independence of cox2 mRNA editing, in the trans-acting gRNA-deficient background, on these MRB1 proteins clearly indicates that they are not involved in core RNA editing activities.
The question is specifically what role do these proteins play in the expression of gRNAs? It was hypothesized that a multiprotein complex may be involved in the transcription of minicircle-encoded genes, independent of the canonical mtRNAP involved in maxicircle transcription (Grams et al. 2002). Here we provide evidence that mtRNAP is the most likely candidate for the transcription of gRNA genes as well as maxicircle genes. This observation indicates that the MRB1 complex may have a distinct role in gRNA synthesis, since mtRNAP was not demonstrated to be associated with this complex in any of the published reports (Etheridge et al. 2008; Hashimi et al. 2008; Panigrahi et al. 2008). Furthermore, the possibility that the mtRNAP transcribes both minicircle-encoded gRNAs and maxicircle-encoded mRNAs is surprising given the requirement of the multiple DNA ligases and DNA polymerases for kDNA maintenance and replication (Klingbeil et al. 2002; Sinha et al. 2004; Liu et al. 2005).
The GAPs and RNA helicase do appear to be involved in some aspect of gRNA processing and/or stability. The most established processing event in the biogenesis of a gRNA is its post-transcriptional addition of the 3′-oligo(U) tail (Blum and Simpson 1990) by the KRET1 enzyme (Aphasizhev et al. 2003). It was also proposed that functional gRNAs are spliced from the 5′ ends of polycistronic minicircle transcripts (Grams et al. 2000). However, a corresponding accumulation of these large precursor RNAs was not detected in the GAP knockdowns (data not shown), suggesting that if this processing step does exist, it is not mediated by these proteins.
Another proposed feature of gRNA is the presence of two intramolecular hairpin structures with single-stranded ends (Schmid et al. 1995; Hermann et al. 1997). The presence of a predicted DExD/H-box RNA helicase in the gRNA-expression component of the MRB1 complex raises the possibility that these proteins may play a role in facilitating the formation of this secondary structure in these small transcripts. Perhaps these thermodynamically unfavorable loops (Schmid et al. 1995) are not formed in the absence of the GAPs or RNA helicase, leading to the apparent destabilization of the gRNAs. However, this speculative model is tempered by the involvement of the DExD/H box RNA helicases in diverse roles in RNA metabolism not restricted just to processing, such as transcription regulation and RNA turnover (for review, see Fuller-Pace 2006).
We show that the GAP proteins are essential for the bloodstream stage, where they appear to have the same role in gRNA processing/stability as they do in the procyclic stage. Although an active RNA metabolism has been established as a requirement for survival of this infective stage (Schnaufer et al. 2001; Fisk et al. 2008), to our knowledge, this result is the first to indicate that the gRNA biogenesis is required in this part of the life cycle as well. This result is significant in light of the existence of naturally occurring dyskinetoplastic and akinetoplastic trypanosomes (Lai et al. 2008), which must circumvent the lack of the requisite repertoire of gRNAs for the expression of normally essential mt-encoded genes, such as the ATPase subunit 6 (Schnaufer et al. 2005). Apparently, in both stages, all kDNA is transcribed, and any regulation occurs at the post-transcriptional level, a situation strikingly similar to what is known for the expression of nuclear genes in trypanosomes and other kinetoplastid flagellates (Haile and Papadopoulou 2007).
We also establish here that the group of proteins that we designate to the MRB1 complex do not share an overall function. While GAP1, GAP2, and RNA helicase are involved in gRNA expression, TbRGG1 and 2 clearly are not (Fisk et al. 2008; Hashimi et al. 2008). Furthermore, we show here that the Nudix hydrolase is also not involved in gRNA expression, since these molecules persist in a background deficient of this protein. The family of Nudix hydrolases has a diverse array of functions in the cell, such as decapping mRNAs (Cohen et al. 2005) and clearance of oxidized nucleosides (for review, see McClennan 2006). Designation of a specific role of this hydrolase is still not feasible based on the observed destabilization of most of the maxicircle transcripts upon its depletion. However, these results counter those of Weng et al. (2008) indicating a specific decrease in edited RNAs.
The array of phenotypes affecting different aspects of mtRNA metabolism upon down-regulation of various putative subunits of the MRB1 complex, along with the assortment of associating proteins, is quite striking (Etheridge et al. 2008; Fisk et al. 2008; Hashimi et al. 2008; Panigrahi et al. 2008; Weng et al. 2008). These observations raise the question whether it represents a single bona fide complex, held together by protein–protein interactions, or a collection of monomers and/or smaller complexes connected by substrate RNAs. Indeed, the latter situation has been proposed (Weng et al. 2008), and incorporation of both TbRGG proteins into large complex(es) appears to be RNase sensitive, while the 20S editosome complex remains intact (Fisk et al. 2008; Hashimi et al. 2008). Interestingly, RNAi silencing of either RNA helicase or mtRNAP clearly disrupts the GAP1 assembly into macromolecular complexes. This result may reflect a genuine interaction of these proteins with GAP1 or be a result of the consequent decrease of RNA molecules via which these proteins seem to associate.
We conclude by discussing the unique localization of the GAP proteins to discrete points along the reticulated mitochondrion, often proximal to the kDNA network. To our knowledge, such a pattern has not been reported for other proteins involved in mt nucleic acid metabolism (Vanhamme et al. 1998; Klingbeil et al. 2002; Liu et al. 2005; Etheridge et al. 2008). Panigrahi et al. (2008) have previously reported a more uniform distribution of different subunits of the MRB1 complex throughout the mitochondrion. However, given the aforementioned diversity of RNAi-silencing phenotypes for the MRB1 complex proteins, it is possible that they may also exhibit diverse yet overlapping sites in the organelle as well. Nevertheless, as the GAP proteins have a demonstrated role in gRNA biogenesis, we imagine these loci may represent centers where gRNAs are processed and distributed for their participation in RNA editing as catalyzed by the 20S editosome.
MATERIALS AND METHODS
Generation of RNAi-knockdown and transgenic cell lines
Primers for generation of 400–600-base pair (bp) gene fragments for cloning into the p2T7-177 vector were designed using the RNAit online tool available on the TrypanoFAN website (http://trypanofan.path.cam.ac.uk/software/RNAit.html). The following primer pairs (with restriction site underlined and indicated in parentheses) were used: GAP1 gene, GAP1-F (5′-CTCCTCGAGCCTTTCAGC-3′) (XhoI) and GAP1-R (5′-GGCAAGCTTCTGCGAATGTAG-3′) (HindIII), amplified a 533-bp-long fragment; GAP2 gene, GAP2-F (5′-CGAGGATCCACAACGGCATT-3′) (BamHI) and GAP2-R (5′-CATCATCAGCAAGCTTTATGATG-3′) (HindIII), amplified a 580-bp-long fragment; Hel1-F (5′-GGATCCGTAGGAACTGGCAGAGACGC-3′) (BamHI) and He1-R (5′-CTCGAGGCTACTGATTGCACGCAAAA-3′) (XhoI), amplified a 453-bp-long fragment; and Hyd1-F (5′-GGATCCCGTAAGGTGTCAGGACCGAT-3′) (BamHI) and Hyd1-R (5′-CTCGAGTCTACGGTAATGCCCGTTTC-3′) (XhoI) amplified a 467-bp-long fragment. All amplified fragments were cloned into the p2T7-177 vector creating constructs, which were upon linearization electroporated into the procyclic T. brucei strain 29-13, and the clones were obtained by limiting dilution at 27°C as described elsewhere (Vondrušková et al. 2005). Synthesis of dsRNA was induced by the addition of 1 μg/mL tetracycline to six clonal cell lines for each knockdown. For further experiments, clones were selected based on the tightness of tetracycline expression of dsRNA and the robust elimination of target mRNA, as determined by Northern blot analysis using the appropriate gene fragment as a probe. The procyclic knockdown generated by cloning a fragment of the mtRNAP gene in the pZJM vector (Grams et al. 2002) was kindly provided by P.T. Englund (Johns Hopkins University, Baltimore).
Linearized p2T7-177 vectors containing the GAP1 and 2 gene fragments were electroporated using the Amaxa Nucleofector II electroporator into the 427 bloodstream T. brucei strain. About 3 × 107 cells of exponentially growing culture were used for transfection, following the protocol of Vassella et al. (2001) with minor modifications. Transfectants were selected with 1.25 μg/mL phleomycin and cloned by limiting dilution. RNAi was triggered and assessed in six clonal cell lines by the addition of 1 μg/mL tetracycline using the same strategy as that described for the procyclics.
The full-length open reading frames of the GAP1 and 2 genes were cloned into the pJH54 plasmid containing the C-terminal HA3-tag (a gift from Christine Clayton, University of Heidelberg). The PCR-amplified insert was digested with either HindIII and XbaI (GAP1) or just XbaI (GAP2). The orientation of the latter insert was verified by diagnostic restriction digests and sequencing. The utilized PCR primers were (with restriction site underlined): GAP1-HA-F (5′-AAGCTTATGCTGCGCGCGCGCCTG-3′) and GAP1-HA-R (5′-TCTAGAGTATGCCGAAACGGCAGT-3′); GAP2-HA-F (5′-TCTAGAATGCTTCGCTTATTGCGG-3′) and GAP2-HA-R (5′-TCTAGACAACTTCGCCTCACAGCC-3′).
Generation of antibodies
Affinity-purified polyclonal rabbit antibodies against GAPs 1 and 2 were raised against synthetic oligopetides by GenScript. The GAP1 synthetic oligopeptide (EYGDWGAEPGFEDRC) corresponds to the 137–150-amino-acid region of the T. brucei protein, while the GAP2 oligopeptide (TIKRSKDQLHVDLDC) is derived from the 297–310 amino acid region.
Western and Northern blot analyses
For Western blot analysis, cell lysates corresponding to 5 × 106 procyclics/lane or 1 × 107 bloodstreams/lane were separated on a 12% SDS-PAGE gel. The polyclonal rabbit antibodies against the MRP2 protein (Vondrušková et al. 2005), enolase (provided by P.A.M. Michels), and frataxin (Long et al. 2008) were used at 1:2000, 1:150,000, and 1:1000 dilutions, respectively. The antibodies against GAP1 and GAP2 were used at the respective dilutions 1:2000 and 1:500. Monoclonal antibodies against KREPA2, KREL1, KREPA3, and Tbmp45 were used as described elsewhere (Madison-Antenucci et al. 1998; Panigrahi et al. 2001). Commercial antibodies against the HA3 tag were used according to the manufacturer's protocol (Sigma).
For Northern blot analysis, ∼10 μg per lane of total RNA from procyclic or bloodstream cells were loaded on a 1% formaldehyde agarose gel, blotted, and cross-linked as published elsewhere (Vondrušková et al. 2005). After pre-hybridization in NaPi solution (0.25 M Na2HPO4 and 0.25 M NaH2PO4 at pH 7.2, 1 mM EDTA, 7% SDS) for 2 h at 55°C, hybridization was performed overnight in the same solution at 55°C. A wash in 2× SSC + 0.1% SDS for 20 min at room temperature was followed by two washes in 0.2× SSC + 0.1% SDS for 20 min each at 55°C.
Growth curves
Following Northern blot analysis of several clones for each knockdown, one clone was selected for further experiments. Growth curves were obtained over a period of 14 d in the presence or absence of the RNAi-induction agent tetracycline for transfected procyclics or 8 d for transfected bloodstreams. Cell density was measured every 24 h using the Beckman Z2 Cell Counter.
Cell fractionation, glycerol gradients, digitonin fractionation
For separation on glycerol gradients, mitochondria from 109 procyclic cells were purified following the protocol detailed in Hashimi et al. (2008). Digitonin fractionation and glycerol gradient sedimentation were performed as described elsewhere (Smíd et al. 2006).
Quantitative real-time PCR and guanylyltransferase labeling
RNA for quantitative real-time (q) PCR and guanylyltransferase labeling were collected and DNased as described in Hashimi et al. (2008). After checking the integrity by running an aliquot on a formaldehyde gel, 4.5 μg of RNA was used as a template for cDNA synthesis for subsequent qPCR analysis (for details, see Hashimi et al. 2008). A parallel cDNA synthesis reaction was set up with the same amount of RNA but without the reverse transcriptase to serve as a control for effective DNase treatment. Primers for maxicircle mRNAs, as well as the housekeeping β-tubulin and 18S rRNA transcripts serving as reference genes, are listed in Carnes et al. (2005) and Hashimi et al. (2008). New primer pairs amplifying cDNAs corresponding to transcripts of the subunits of the MRB1 complex are GAP1-qPCR-Fw (5′-AACGTATTGCGGATGCTTAC-3′) and Rv (5′-GAACCACACGCTCACAACAG-3′); GAP2-qPCR-Fw (5′-CGGCTCATATTCCTGCCAATG-3′) and Rv (5′-CGAAGTCCTCAGCAACCAACC-3′); Hel-qPCR-Fw (5′-ATCGCGTTAGGTGAAGCAGT-3′) and Rv (5′-AAATGGGGATCCCTAAGGTG-3′); and Hyd-qPCR-Fw (5′-ATTTTTCACCCTTGCACGTC-3′) and Rv (5′-TCGATGGATTTGTTGTCACC-3′). Relative abundances of maxicircle transcripts were calculated using the Pfaffl method (Pfaffl 2001).
Guanylyltransferase reactions were performed on 2.5–5 μg of total RNA to cap gRNAs with [α-32P]GTP as previously described (Hashimi et al. 2008). The recombinant enzyme was either purchased from Ambion or provided by Ruslan Aphasizhev (University of California, Irvine). The reactions were separated on a denaturing 12% acrylamide–8 M urea gel.
Immunolocalization
In order to visualize mitochondria, 5 × 106 to 1 × 107 cells were incubated in SDM-79 media supplemented with 1 mM of MitoTracker Green FM (Molecular Probes) for 20 min at 27°C. Cells were subsequently washed 3× in phosphate-buffered saline (PBS) and then spread on slides, fixed, and permeabilized as described in Klingbeil et al. (2002). All subsequent steps were carried out in a humid chamber. The slides were first blocked for 1 h at room temperature with 10% goat serum diluted in PBS. Afterward, the primary rabbit antibody was applied to the slides at the appropriate dilution in 10% goat serum + PBS for 90 min. After washing 3× for 10 min each in PBS, the slides were incubated with secondary goat α-rabbit antibody conjugated to the cy3 fluorophore (Jackson Immunoresearch) at a 1:800 dilution for 1 h. The slides were then washed 3× for 10 min each in PBS and mounted in Vectashield containing DAPI. The α-GAP1 antibody was used at a dilution of 1:100, and the α-HA3 antibody was used according to the manufacturer's protocol (Sigma). Samples probed with the latter antibody were not pre-treated with MitoTracker. Cells were examined on the Olympus Fluoview FV1000 confocal microscope using the accompanying Fluoview v1.7 software. A composite of the Z-stack of the fluorescent images was rendered using the ImageJ software and processed using Adobe Photoshop v6.0.
ACKNOWLEDGMENTS
We thank Lucie Hanzálková and Julius Lukeš IV for help with some experiments, and Paul T. Englund (Johns Hopkins University), Paul A.M. Michels (Université catholique de Louvain), Ken Stuart (Seattle Biomedical Research Institute), Stephen Hajduk (University of Georgia), Christine Clayton (Heidelberg University), and Ruslan Aphasizhev (University of California at Irvine) for sharing the mtRNA polymerase knockdowns, the α-enolase antibody, antibodies against 20S editosome subunits, α-Tbmp45 antibody, pJH54 plasmid, and recombinant guanylyltransferase, respectively. We also thank Aswini Panigrahi and Alena Zíková (Seattle Biomedical Research Institute) for critical reading of the manuscript. This work was supported by the Grant Agency of the Czech Republic 204/09/1667, Grant Agency of the Czech Academy of Sciences A500960705, and the Ministry of Education of the Czech Republic (LC07032 and 2B06129) to J.L. and the Ph.D. student grant 524/03/H133 to H.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1411809.
REFERENCES
- Aphasizhev R., Aphasizheva I., Simpson L. A tale of two TUTases. Proc. Natl. Acad. Sci. 2003;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R., van den Burg J., Brakenhoff J.P., Sloof P., Van Boom J.H., Tromp M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Blum B., Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell. 1990;62:391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- Blum B., Bakalara N., Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Carnes J., Trotter J.R., Ernst N.L., Steinberg A.G., Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl. Acad. Sci. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J., Trotter J.R., Peltan A., Fleck M., Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell. Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L.S., Mikhli C., Jiao X., Kiledjian M., Kunkel G., Davis R.E. Dcp2 decaps m2,2,7 GpppN-capped RNAs, and its activity is sequence and context dependent. Mol. Cell. Biol. 2005;25:8779–8791. doi: 10.1128/MCB.25.20.8779-8791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo G.J., Palazzo S.S., Wang B., Panicucci B., Salavati R., Stuart K.D. Dyskinetoplastic Trypanosoma brucei contains functional editing complexes. Eukaryot. Cell. 2003;2:569–577. doi: 10.1128/EC.2.3.569-577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge R.D., Aphasizheva I., Gershon P.D., Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk J.C., Ammerman M.L., Presnyak V., Read L.K. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J. Biol. Chem. 2008;283:23016–23025. doi: 10.1074/jbc.M801021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace F.V. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden D.E., Hajduk S.L. The 3′-untranslated region of cytochrome oxidase II mRNA functions in RNA editing of African trypanosomes exclusively as a cis guide RNA. RNA. 2005;11:29–37. doi: 10.1261/rna.7170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grams J., McManus M.T., Hajduk S.L. Processing of polycistronic guide RNAs is associated with RNA editing complexes in Trypanosoma brucei. EMBO J. 2000;19:5525–5532. doi: 10.1093/emboj/19.20.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grams J., Morris J.C., Drew M.E., Wang Z.F., Englund P.T., Hajduk S.L. A trypanosome mitochondrial RNA polymerase is required for transcription and replication. J. Biol. Chem. 2002;277:16952–16959. doi: 10.1074/jbc.M200662200. [DOI] [PubMed] [Google Scholar]
- Haile S., Papadopoulou B. Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr. Opin. Microbiol. 2007;10:569–577. doi: 10.1016/j.mib.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Hashimi H., Zíková A., Panigrahi A.K., Stuart K.D., Lukeš J. TbRGG1, a component of a novel multi-protein complex involved in kinetoplastid RNA editing. RNA. 2008;14:970–980. doi: 10.1261/rna.888808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann T., Schmid B., Heumann H., Göringer H.U. A three-dimensional working model for a guide RNA from Trypanosoma brucei. Nucleic Acids Res. 1997;25:2311–2318. doi: 10.1093/nar/25.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingbeil M.M., Motyka S.A., Englund P.T. Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol. Cell. 2002;10:175–186. doi: 10.1016/s1097-2765(02)00571-3. [DOI] [PubMed] [Google Scholar]
- Lai D.-H., Hashimi H., Lun Z.-R., Ayala F.J., Lukeš J. Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc. Natl. Acad. Sci. 2008;105:1999–2004. doi: 10.1073/pnas.0711799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.Y., Liu Y.N., Motyka S.A., Agbo E.E.C., Englund P.T. Fellowship of the rings: The replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Long S., Jirků M., Mach J., Ginger M.L., Sutak R., Richardson D., Tachezy J., Lukeš J. Ancestral roles of eukaryotic frataxin: mitochondrial frataxin function and heterologous expression of hydrogenosomal Trichomonas homologues in trypanosomes. Mol. Microbiol. 2008;69:94–109. doi: 10.1111/j.1365-2958.2008.06260.x. [DOI] [PubMed] [Google Scholar]
- Lukeš J., Hashimi H., Zíková A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci S., Sabatini R.S., Pollard V.W., Hajduk S.L. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): A novel editing complex protein with repetitive domains. EMBO J. 1998;21:6368–6376. doi: 10.1093/emboj/17.21.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClennan A.G. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.M., Halbig K., Cruz-Reyes J., Read L.K. RBP16 stimulates trypanosome RNA editing in vitro at an early step in the editing reaction. RNA. 2006;12:1292–1303. doi: 10.1261/rna.2331506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U.F., Lambert L., Göringer H.U. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 2001;20:1394–1404. doi: 10.1093/emboj/20.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Schnaufer A., Carmean N., Igo R.P., Jr, Gygi S.P., Ernst N.L., Palazzo S.S., Weston D.S., Aebersold R., Salavati R., et al. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Ernst N.L., Domingo G.J., Fleck M., Salavati R., Start K.D. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Zíková A., Halley R.A., Acestor N., Ogata Y., Myler P.J., Stuart K. Mitochondrial complexes in Trypanosoma brucei: A novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics. 2008;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- Pelletier M., Read L.K. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA. 2003;9:457–468. doi: 10.1261/rna.2160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B., Riley G.R., Stuart K., Göringer H.U. The secondary structure of guide RNA molecules from Trypanosoma brucei. Nucleic Acids Res. 1995;23:3093–3102. doi: 10.1093/nar/23.16.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A., Panigrahi A.K., Panicucci B., Igo R.P., Jr, Wirtz E., Salavati R., Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Schnaufer A., Clark-Walker G.D., Steinberg A.G., Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M.A., Karamooz E., Zíková A., Trantírek L., Lukeš J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA-binding complex reveals RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- Seiwert S.D., Heidmann S., Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- Simpson L., Aphasizhev R., Gao G., Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha K.M., Hines J.C., Downey N., Ray D.S. Mitochondrial DNA ligase in Crithidia fasciculata. Proc. Natl. Acad. Sci. 2004;101:4361–4366. doi: 10.1073/pnas.0305705101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smíd O., Horáková E., Vilímová V., Hrdý I., Cammack R., Horváth A., Lukeš J., Tachezy J. Knockdowns of mitochondrial iron-sulfur cluster assembly proteins IscS and IscU down-regulate the active mitochondrion of procyclic Trypanosoma brucei. J. Biol. Chem. 2006;281:28679–28686. doi: 10.1074/jbc.M513781200. [DOI] [PubMed] [Google Scholar]
- Stuart K.D., Schnaufer A., Ernst N.L., Panigrahi A.K. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Sturm N., Simpson L. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell. 1990;61:879–884. doi: 10.1016/0092-8674(90)90198-n. [DOI] [PubMed] [Google Scholar]
- Vanhamme L., Peréz-Morga D., Marechal C., Speijer D., Lambert L., Geuskens M., Alexandre S., Ismaili N., Göringer U., Benne R., et al. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem. 1998;273:21825–21833. doi: 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- Vassella E., Kramer R., Turner C.M.R., Wankell M., Modes C., Van den Bogaard M., Boshart M. Deletion of a novel protein kinase with PX and FYVE-related domains increases the rate of differentiation of Trypanosoma brucei. Mol. Microbiol. 2001;41:33–46. doi: 10.1046/j.1365-2958.2001.02471.x. [DOI] [PubMed] [Google Scholar]
- Vondrušková E., van den Burg J., Zíková A., Ernst N.L., Stuart K., Benne R., Lukeš J. RNA interference analyses suggest a transcript-specific regulatory role for MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J. Biol. Chem. 2005;280:2429–2438. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- Weng J., Aphasizheva I., Etheridge R.D., Huang L., Wang X., Falick A.M., Aphasizhev R. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol. Cell. 2008;32:1–12. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zíková A., Kopečná J., Schumacher M.A., Stuart K.D., Trantírek L., Lukeš J. Structure and function of the native and recombinant mitochondrial MRP1/MRP2 complex from Trypanosoma brucei. Int. J. Parasitol. 2008;38:901–912. doi: 10.1016/j.ijpara.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]