Summary
The mutagenic activity of transposable elements (TEs) is suppressed by epigenetic silencing and small interfering RNAs (siRNAs), especially in gametes that would transmit transposed elements to the next generation. In pollen from the model plant Arabidopsis, we show that TEs are unexpectedly reactivated and transpose, but only in the pollen vegetative nucleus, which accompanies the sperm cells but does not provide DNA to the fertilized zygote. TE expression coincides with down-regulation of the heterochromatin remodeler DECREASE IN DNA METHYLATION 1 and of most TE siRNAs. However, 21 nucleotide siRNA from Athila retrotransposons is generated in pollen and accumulates in sperm, indicating that siRNA from TEs activated in the vegetative nucleus can target silencing in gametes. We propose a conserved role for reprogramming in germline companion cells, such as nurse cells in insects and vegetative nuclei in plants, to reveal intact TEs in the genome and regulate their activity in gametes.
Introduction
Transposable elements (TEs) are the targets of small interfering RNA (siRNA)-mediated silencing in animals, fungi and plants. In the model plant Arabidopsis thaliana, siRNAs involved in post-transcriptional silencing (such as microRNAs and trans-acting siRNAs) are 21 nucleotides (nt) in length, while siRNAs involved in heterochromatin formation are 24nt(Hamilton et al., 2002) and target TEs(Kasschau et al., 2007). Each class of siRNA is bound by different Argonaute proteins, of which 10 are found in Arabidopsis. In animals, but apparently not in plants, additional 25–29nt piRNAs are produced and utilized by the PIWI clade of Argonaute proteins. piRNAs arise from clusters of repetitive DNA in a Dicer-independent mechanism(Aravin et al., 2007; Grivna et al., 2006; Lau et al., 2006). In Drosophila, piRNAs target TEs in the male and female germlines, and PIWI proteins participate in heterochromatic silencing(Brennecke et al., 2007; Brower-Toland et al., 2007; Lau et al., 2006). In rodents, piRNAs produced in the male germline silence TEs and directly or indirectly target these regions for DNA methylation(Carmell et al., 2007), a silencing signal that is potentially heritable through subsequent cell divisions after piRNAs are no longer present.
Unlike animals, plants do not set aside their germline cells early in development. Instead, plant germ cells differentiate from somatic cells in the adult. Male meiocytes in the developing anther give rise to four haploid microspores. Microspores undergo a subsequent mitotic division, producing a binucleate pollen grain, with one smaller cell completely enclosed within the larger. The smaller (generative) cell further divides into two sperm cells (SCs). In Arabidopsis, the mature pollen grain shed by the anther houses two identical SCs that provide the paternal genetic contribution to the zygote and the endosperm, respectively, in double fertilization. The delivery of the sperm is controlled by the pollen vegetative nucleus (VN), which has decondensed chromatin compared to the compact sperm, and does not contribute DNA to the fertilized zygote nor to the endosperm (McCormick, 1993).
Microarray expression profiling of Arabidopsis mature pollen (Becker et al., 2003; Honys and Twell, 2004; Pina et al., 2005; Schmid et al., 2005) has revealed that relatively few genes are expressed, the lowest number of any tissue examined(Becker and Feijo, 2007; Schmid et al., 2005), and that many of the genes involved in siRNA biogenesis and silencing are either not expressed in pollen or expressed at low levels(Pina et al., 2005). We have found that the chromatin remodeling ATPase DECREASE IN DNA METHYLATION 1 (DDM1) accumulates only in SCs and not in the VN. DDM1 is the master regulator of TE activity in Arabidopsis, as most TEs lose DNA methylation, repressive histone modifications and 24nt siRNAs in ddm1 mutants (Gendrel et al., 2002; Lippman et al., 2004; Lippman et al., 2003). Consistent with the loss of DDM1, most 24nt TE siRNAs are lost from mature pollen, co-incident with the accumulation of diverse TE transcripts as well as TE transposition. However this transposition is limited to the pollen VN so that transposed elements are not transmitted to the next generation. Comparison of siRNA sequences from pollen, purified SCs and ddm1 mutant plants reveals that the loss of DDM1 from the VN and reactivation of TEs results in the accumulation of 21nt siRNAs from the Athila family of gypsy-class retrotransposons, which are transported and preferentially accumulate in SCs. By this mechanism, activation of retrotransposons in the VN reveals their presence in the genome, and presumably targets their silencing in SCs, preventing transposition and promoting trans-generational TE silencing.
Results
Transcriptional reactivation of TEs in mature pollen
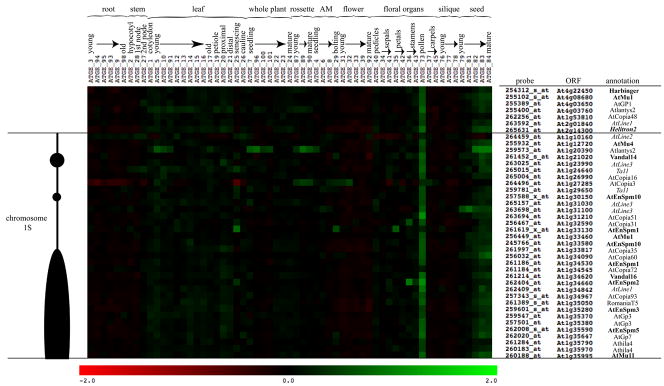
To determine the developmental expression profile of diverse types of TEs, we assayed TE expression using Affymetrix Ath1 microarray data during plant development, assembled in AtGenExpress (Schmid et al., 2005). By comparing the Ath1 microarray gene list to the Repbase repeat database, we identified probe sets that detect ORFs from seven known TEs, including at least one putatively autonomous full-length element for each class: DNA transposon, LTR-retrotransposon, non-LTR retrotransposon and helitron (Supplemental Table 2; Experimental Procedures). We found that the developmental expression patterns of diverse TEs were highly similar to each other and that for the majority of tissues and stages TEs were either not expressed or expressed at a very low level, indicating they are generally silenced in Arabidopsis (Figure 1). However, the seven full-length TEs examined were coordinately expressed in mature pollen (top portion, Figure 1). These expression patterns were verified by RT-PCR (Supplemental Figure 1). Weak expression of several TEs in developing seeds was also detected by microarray (Figure 1), though not by RT-PCR (Supplemental Figure 1).
Figure 1. TEs are expressed in mature pollen.
Microarray profiles of TEs reveal changes in expression during development. Changes in expression are indicated as up regulation (green), or down regulation (red) compared to the row-normalized expression for each ORF across array experiments (Schmid et al., 2005). The scale (bottom) represents the log2 change over the mean of all values for a given ORF. The scale has been set from −2 to +2 to emphasize small changes in gene expression. The microarray experiment identifiers are listed across the top, as well as their corresponding developmental tissue and stage (AM is apical meristem). Probes and their corresponding ORFs and Repbase annotation are shown on the right. Repbase annotations are indicated as LTR retrotransposons, non-LTR retrotransposons (italic), DNA transposons (bold) and helitron elements (bold and italic). Probe sets are generally unique to an individual ORF. However, probe names that include ‘s’ detect two or more paralogs, while probes with an ‘x’ represent theoretical genes from paralogous groups. Shown on the left is a representation of the short arm of chromosome 1 defining the location and chromatin context for each TE. The centromere is at the bottom, near ORF At1g35995. Gene-rich regions are depicted as thin black lines, while TE-rich heterochromatic regions are black ovals.
The observed co-regulation of TEs could potentially depend on their chromosomal location and context. The expression patterns and locations of 34 diverse TEs from the short arm of chromosome 1 (see Experimental Procedures) are depicted in Figure 1. The coordinated up regulation of TEs in mature pollen was observed for 30 of 34 elements (Figure 1), independent of their location on the chromosome: unique probes located in gene-rich regions were regulated in exactly the same way as unique probes located in heterochromatic regions. Therefore, the unexpected coordinated expression of diverse TEs in mature pollen does not depend on (cis-acting) position, but rather on the loss of trans-acting silencing factors.
To determine the scope of TE reactivation in pollen, we assayed TE activity in maize pollen. We have shown that a silenced TE is transiently reactivated in maize pollen (Supplemental Figure 2), and additionally TE expression has been noted in the pollen of rice (Nobuta et al., 2007). Therefore, the transient reactivation of silenced TEs in pollen is conserved among flowering plants.
Location and timing of TE expression in pollen
To characterize the expression pattern of TEs in pollen, we identified 12 enhancer trap (ET) and 12 gene trap (GT) insertions into a variety of TEs in the Landsberg erecta (Ler) Arabidopsis accession (Supplemental Table 3). Upon staining for GUS reporter gene activity, most ET and GT TE lines lacked staining in the plant body, but all lines had strongly stained mature pollen (Figure 2, Supplemental Figure 3A, and Supplemental Table 3). Under the same staining conditions, mature pollen of Ler Arabidopsis does not stain (Figure 2A). Thus, we have shown the transcriptional reactivation of diverse TEs in mature pollen using two different techniques in two different Arabidopsis strains (Columbia and Ler).
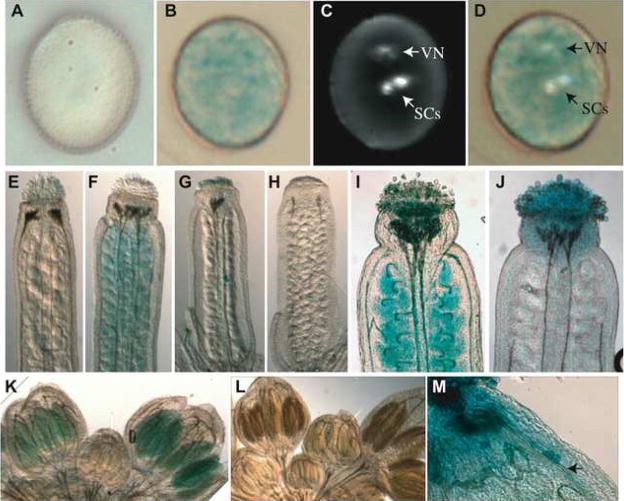
Figure 2. TE expression is confined to the pollen cytoplasm.
(A) GUS staining of non-transgenic Ler mature pollen has no staining. (B) GUS staining of the representative GT7390 inserted into an AtCopia LTR retrotransposon has diffuse cytoplasmic GUS staining. (C) DAPI staining of the same pollen grain in (B) shows the location of the vegetative nucleus (VN) and two sperm cells (SCs). (D) Merged images show that the GUS staining does not correspond to the SCs. (E) GT7390 shows no GUS staining in a mature unfertilized carpel. (F) As a positive control, a GT insertion into the PROLIFERA gene (GT148) stained in the ovules. (G–H) Clearing of immature carpels fails to detect GUS staining in the developing megagametophytes for GT7390. Stray pollen grains from GT7390 can be seen in the center of the carpel (G), and serve as a positive control. (I) The control GT148 shows ovule staining post-fertilization, while the representative GT7390 as well as others from Supplemental Table 3 do not show staining of mature ovules post-fertilization (J). (K) GUS staining of flower buds from the representative GT7390 show GUS staining beginning at floral developmental stages 10–11, while Ler flower buds do not show GUS staining at this stage (L). (M) GUS staining pollen tubes (arrow) from the representative GT7390 are detected within the carpel of a self-fertilized GT7390 flower.
Using the representative gene trap GT7390 inserted into an AtCopia LTR retrotransposon, we examined TE expression within the pollen grain. GUS staining in GT7390 was detected throughout the pollen cytoplasm (Figure 2B), which is maintained by the pollen VN. This pollen grain was counter-stained with DAPI to locate the VN and SCs (Figure 2C). SCs do not accumulate GUS stain (Figure 2D), and since proteins such as GFP produced in the SCs do not freely pass into the pollen grain cytoplasm (Engel et al., 2005), this suggests the SCs are not the site of TE reactivation. To determine if TE transcripts were produced from the SCs (in addition to the VN), we looked for but could not detect TE transcripts in the Ath1 microarray expression profile of purified sperm (Supplemental Figure 4)(Borges et al., 2008). Thus TE transcripts are expressed specifically from the VN.
To examine TE expression in the female gametophyte, we stained carpels of ET and GT lines inserted into TEs, and found no GUS expression in the mature carpel (Figure 2E and Supplemental Figure 3). Staining of immature ovules (summarized in Figure 2G–H) and mature pollinated carpels (summarized in Figure 2J) failed to detect any GUS expression in carpels or ovules for GT7390, or other ETs or GTs inserted into TEs (Supplemental Table 3). Thus expression of at least these TEs is elevated in the male gametophyte to a significantly greater extent than in the female. To determine the developmental timing of TE pollen expression, developing flowers from GT7390 were stained for GUS activity (Figure 2K). Pre-meiotic anthers at floral stage 8–9 did not stain, while stage 10–11 anthers stained weakly. At floral stages 10–11, meiosis has occurred and the pollen grain is beginning to differentiate(Goldberg et al., 1993). Post-pollination, GUS positive pollen tubes were observed in GT7390 carpels (Figure 2M), and we detect elevated TE transcripts in microarray data from growing pollen tubes, as well as desiccated and hydrated mature pollen grains(Wang et al., 2008). Both the timing and location of TE activation in pollen demonstrates that the VN is the origin of TE expression.
TEs are mobile in pollen
Transcriptional activation of TEs in mature pollen could lead to TE mobility, so we performed AFLP transposable element display to detect new insertions of TEs in pollen DNA. This technique detects restriction site polymorphisms generated upon insertion. Using nested PCR primer sets from the AtMu1 DNA transposon, Tag2 DNA transposon, and AtLine1 non-LTR retrotransposon families, we detected new restriction site polymorphisms in the pollen sample (Supplemental Figure 5), suggesting these TEs are mobile in wild-type plants at this stage of development.
Only a subset of AtMu1 family elements are capable of transposition when activated(Singer et al., 2001), which belong to the AtMu1a and AtMu1b subfamilies. Under more stringent PCR conditions that would only detect the highly similar AtMu1a and AtMu1b sub-families, three new polymorphisms found in pollen (Figure 3A) were excised from the gel, cloned and sequenced. These new polymorphisms correspond to new AtMu1a insertions, two of which are located near the transcriptional start of protein-coding genes, a known preference for insertion of Mutator family TEs(Lisch, 2002). Thus TEs that are epigenetically silent throughout most of development are transiently reactivated and transpose in mature pollen.
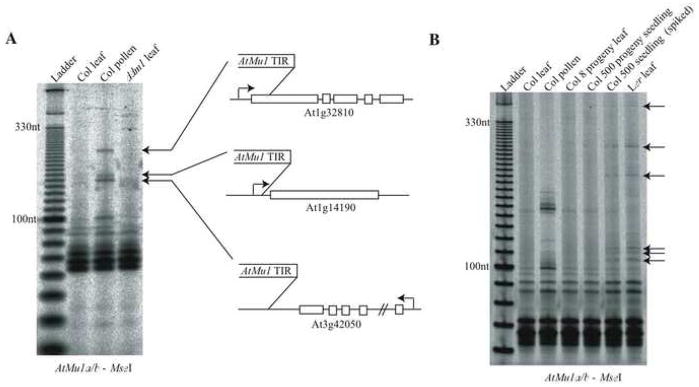
Figure 3. The mobilization and inheritance of a TE in pollen.
(A) Transposable element display was performed under stringent conditions to amplify only AtMu1 family members AtMu1a and AtMu1b. DNA was extracted from either the pooled leaves or pooled pollen from the same 96 Columbia reference strain wild-type plants. These DNA samples are the same as in Supplemental Figure 5. New transpositions were detected in pollen compared to the leaf sample. These new polymorphisms were sequenced and the insertion sites of three new AtMu1a insertions are shown.
(B) Transposable element display performed on the progeny of the plants used in part A. If new insertions in pollen were inherited by the next generation, then new insertions would segregate at a low frequency in the self-fertilized progeny of the 96 plants used for pollen collection in part A. The “Col leaf” and “Col pollen” samples are the same samples as in part A. The “Col 8 progeny leaf” and “Col 500 progeny seedling” samples are pools of 8 and 500 individuals that are the self-fertilized progeny of the plants used for leaf and pollen collection in part A. The “Col 500 seedling (spiked)” sample represents 500 Columbia seedlings pooled together with 5 Ler seedlings prior to DNA extraction. This sample serves as a positive control for the sensitive detection of rare polymorphisms in a DNA sample. The polymorphisms found in the spiked control are from Ler, as shown in the “Ler leaf” sample (arrows).
To determine if new transpositions in pollen were inherited by the next generation, 500 progeny of the 96 plants used for pollen collection in Figure 3A were analyzed in a pool. No new insertions were detected in this pool of progeny seedlings (Figure 3B). This suggests that transpositions do not occur in the somatic lineage leading to meiocytes, because insertions would be present in the SCs and passed to the next generation. Additionally, TE mobility could not have occurred before the mitotic divisions that produce the haploid VN and SCs. Instead, the TE insertions in pollen must have taken place in the VN of the pollen grain, which does not contribute DNA to the fertilized zygote. Therefore, both TE expression and mobility are generated by the VN of wild-type pollen.
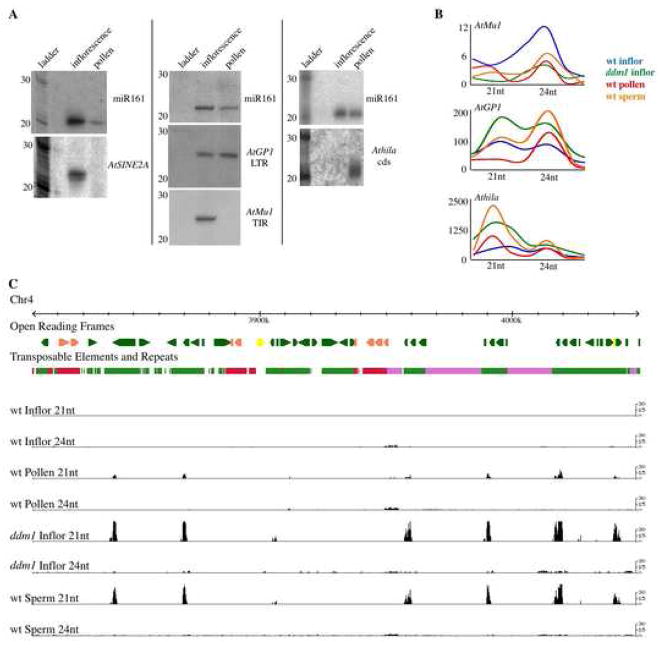
TEs have reduced DNA methylation in mature pollen
Epigenetic reactivation of TEs is often associated with loss of DNA methylation, for example in DNA methyltransferase mutants (Lippman et al., 2003). To monitor DNA methylation, we converted all unmethylated cytosine to thymine using bisulfite conversion. We followed the bisulfite conversion by sequencing a non-methylated gene as a control, along with the 5′ terminal inverted repeat (TIR) of the DNA transposon AtMu1a (At4g08680) and the 5′ long terminal repeat (LTR) of the retrotransposon AtGP1 (At4g03650), from mature pollen as well as wild-type leaf DNA (Supplemental Figure 6 and Figure 4). After bisulphite treatment unmethylated cytosines are converted to thymine, while methylated cytosines are unchanged. Both AtMu1a and AtGP1 retained some DNA methylation in mature pollen in all three sequence contexts (CG, CHG and CHH). For AtGP1, the LTR as a whole had similar levels of methylation compared to the leaf, although methylation was lost from specific cytosines close to the promoter (Supplemental Figure 6). In contrast, for AtMu1, asymmetric DNA methylation and some CHG symmetric DNA methylation were reduced on both the sense and antisense strands of the TIR (Figure 4). Since asymmetric DNA methylation is targeted by siRNAs, the reduction of asymmetric DNA methylation from the AtMu1 TIR suggests that this region is not targeted by siRNAs in mature pollen.
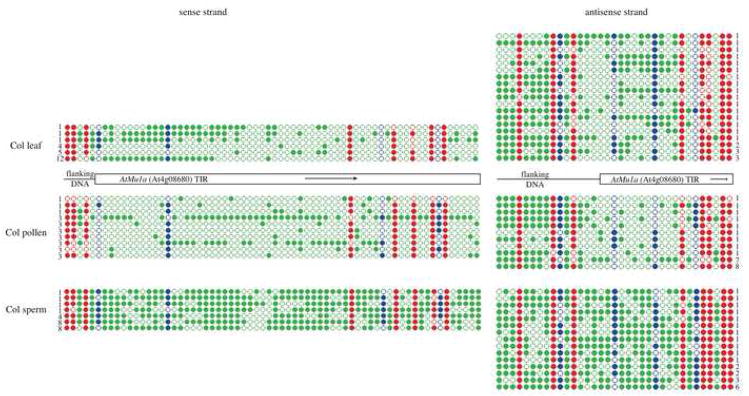
Figure 4. DNA methylation of the AtMu1a TE in pollen and sperm.
The AtMu1a element (At4g08680) was amplified from bisulfite converted DNA and sequenced from leaf tissue, mature pollen and purified SCs. PCR primers in TE-flanking DNA were used to anchor the analysis to one particular TE copy. The location of the DNA transposon AtMu1 terminal inverted repeat (TIR) is shown as a rectangle, while the flanking DNA (a helitron element into which AtMu1 has inserted) is shown as a line. Twenty-four clones were sequenced for each strand (sense or antisense) of each sample (leaf, pollen or sperm), and only unique “non-sister” individual bisulfite clones are shown. The number of times each unique clone was sequenced is shown next to the clone. The methylation state of each cytosine is shown as an opened circle (not methylated) or closed circle (methylated). The sequence context of the cytosine is denoted by the color of the circle (red = CG, blue = CHG, green = CHH). All cytosines in the DNA sequence are shown. Bisulfite sequencing of a control gene and the AtGP1 retrotransposon are shown in Supplemental Figure 6.
The analysis of DNA methylation in mature pollen is confounded by the presence of two very different nuclei (the VN and SCs). We used DNA isolated from purified SCs (Borges et al., 2008) to assay the DNA methylation levels of the AtMu1 TIR in sperm. Cytosines in all sequence contexts were highly methylated on both sense and antisense strands in SCs, and particularly CHG and CHH methylation were increased compared to whole pollen and leaf (Figure 4). This data demonstrates that the location of reduced DNA methylation in the pollen grain must be in the pollen VN. Furthermore, in the total pollen bisulfite sequencing data, individual clones specific to either the VN (reduced methylation) or SCs (increased methylation) could be distinguished (Figure 4). This suggests that the reduced DNA methylation in pollen is not the result of all DNA strands losing some DNA methylation, but rather the sperm-derived DNA retains high levels of DNA methylation in all sequence contexts, while the VN-derived DNA lacks most asymmetrical and some CHG methylation, corresponding to the reactivation and mobilization of TEs.
Genes required for heterochromatic silencing are down-regulated in pollen
We sought an explanation for the activation and de-methylation of TEs in pollen by examining the expression of genes known to regulate TEs. Many genes involved in siRNA-mediated silencing and heterochromatin regulation have reduced expression in mature pollen (Pina et al., 2005)(Supplemental Table 4). RT-PCR analysis of these genes demonstrates that the 24nt siRNA-mediated heterochromatin-formation pathway is down regulated in mature pollen (Supplemental Figure 7) while expression is normal for the trans-acting siRNA pathway genes, as well as the microRNA specific Dicer DCL1, suggesting that the microRNA and tasiRNA pathways are functional in the pollen grain. Interestingly, the TE master-regulator gene DDM1 is expressed in pollen and is highly enriched in SCs (Borges et al., 2008).
Selective loss of siRNA in mature pollen and ddm1 mutants
To determine whether siRNA loss is responsible for TE activation in pollen, we assayed siRNA accumulation from several TEs (Figure 5A). siRNA from the AtMu1 DNA transposon and the AtSINE2A non-autonomous non-LTR retrotransposon were both absent in mature pollen. Interestingly, loss of siRNA from the AtMu1 TIR corresponds to loss of asymmetric DNA methylation (Figure 4) from the likely promoter of AtMu1 based on the closely related Mutator family in maize (Raizada et al., 2001). In contrast, siRNAs from the epigenetically reactivated AtGP1 and Athila LTR retrotransposons (Supplemental Figure 1) increased in relative abundance in pollen (Figure 5A). Retention of AtGP1 24nt siRNA corresponds to retention of DNA methylation in the LTR (Supplemental Figure 6), and this TE is known to reactivate without loss of small RNAs (Lippman et al., 2003).
Figure 5. Pollen, sperm and ddm1 siRNAs.
A. Northern blots of mature pollen RNA were sequentially hybridized with probes that detect microRNA161 and TE siRNAs. AtGP1 and Athila are LTR retrotransposons, AtSINE2A is a non-autonomous non-LTR retrotransposon, and AtMu1 is a DNA transposon.
B. Sequencing of small RNA libraries was used to quantify changes in size distribution and relative abundance for several TE families (AtMu1, Athila and AtGP1) across several samples: wild-type inflorescence (blue), ddm1 inflorescence (green), as well as wild-type pollen (red), and purified sperm (orange) isolated by FACS. The X-axis shows siRNA size (20–26nt), while the Y-axis represents the library size normalized relative number of siRNAs sequenced per TE category (Athila, AtGP1, AtMu1).
C. Distributions of 21nt and 24nt siRNAs are shown for a section of pericentromeric heterochromatin from chromosome 4. Red represents DNA transposons, green is retrotransposon, and pink are tandem repeat islands. Bars represent library size-normalized, genome copy-corrected counts of 21 and 24nt siRNA in 100bp windows for wt inflorescence, wt pollen, ddm1 inflorescence, and wt sperm. In pericentromeric heterochromatin, large peaks of 21nt siRNAs in ddm1, pollen and sperm match Athila family retrotransposons.
We noted the similarity between the pattern of TE siRNA accumulation in pollen and that observed in ddm1 mutants (Lippman et al., 2003). This indicated that DDM1 might somehow be involved in this shift in siRNA accumulation. To test this hypothesis we sequenced small RNA libraries from mature wild-type pollen, as well as from wild-type and ddm1 mutant inflorescences. A full analysis of the sequenced siRNAs will be present elsewhere, and the data is displayed online at http://hispaniola.cshl.edu/annoj_slotkin2008_1/. Alignment of the sequenced siRNAs with TE sequences revealed a striking similarity between the siRNA in ddm1 mutants and mature pollen, each representing genomes with epigenetically reactivated TEs (Figure 5). Most TE families lose siRNA when epigenetically reactivated in either ddm1 mutant plants or wild-type pollen. For example, the AtMu1a element (At4g08680) loses siRNAs in ddm1(Lippman et al., 2003)and has significantly reduced siRNAs in pollen (Figure 5). The two families of LTR retrotransposons shown in Figure 5A are the exception and gain siRNAs when epigenetically reactivated in ddm1 mutants and wild-type pollen. The AtGP1 family retains 24nt siRNAs in both ddm1 and pollen, despite reactivation (Figure 5). This may reflect the presence of distinct classes of active and inactive AtGP1 elements in the genome. But the most striking effect in ddm1 and pollen is the massive increase in siRNA accumulation corresponding to the Athila family of LTR retrotransposons, which gain 21nt siRNAs in both ddm1 mutants and mature pollen, but have only low levels of 24nt siRNA in wild-type inflorescences (Figure 5). Therefore, the reactivation of diverse TEs in pollen is associated with the loss of most 24nt siRNAs and the dramatic gain of a novel class of 21nt TE siRNAs.
The DDM1 chromatin remodeling protein is absent from the vegetative nucleus
TEs in pollen share several characteristics with TEs in ddm1 mutants: reduced DNA methylation, transcriptional reactivation, transposition, and selective loss and gain of siRNAs (Lippman et al., 2003; Singer et al., 2001). This suggests that lack of DDM1 activity could be responsible, at least in part, for TE reactivation in pollen. However, DDM1 transcripts accumulate in pollen (Supplemental Figure 7) and are enriched in SCs (Borges et al., 2008). To directly test if the DDM1 protein is present in the pollen VN, we generated GFP protein fusions to the DDM1 coding region under control of the native DDM1 promoter. Six independent transgene insertions resulted in sperm-specific fluorescence (Figure 6)(Supplemental Movies), demonstrating that the DDM1 protein accumulates in the SCs to a much higher level compared to the VN, in which DDM1 protein accumulation is undetectable.
Figure 6. DDM1 protein accumulation is sperm-specific.
GFP reporter lines monitoring DDM1 protein localization were examined by confocal microscopy. The location of the vegetative nucleus (VN) and sperm cells (SCs) were detected in the same pollen grain using DAPI (arrows). The control pollen grain without GFP has only background auto-florescence. GEX2p-GFP control pollen grains have sperm-specific protein accumulation (Engel et al., 2005), while histone H3.3-GFP pollen grains (a gift from Y. Fang and D. Spector) have VN-specific GFP fluorescence. Six independent transformation events revealed sperm-specific protein accumulation of DDM1-GFP. Three-dimensional movies of these pollen grains are included as Supplemental Data.
In ddm1 mutants, centromeric repeats lose DNA methylation and are expressed (May et al., 2005; Vongs et al., 1993). In each of the six independent transformants, the DDM1-GFP transgene resulted in centromeric and TE DNA methylation in the plant body, indicating that these transgene insertion events complement the ddm1-2 mutation (Supplemental Figure 8). However, in contrast to ddm1 mutants, centromeric repeats do not show elevated expression in pollen compared to wild-type inflorescences (Supplemental Figure 9), suggesting that centromeric satellite promoters are not active in pollen. Perhaps ddm1-induced expression of the centromeric repeats is dependent on centromere function and identity, as centromere-specific histone localization is lost in the pollen VN (Ingouff et al., 2007). We conclude that the TE-specific reactivation in the VN is regulated, in whole or in part, by the lack of DDM1 protein.
Accumulation of TE siRNAs in sperm
Epigenetic reactivation of TEs leads to the selective loss of TE siRNAs, but also to the accumulation of 21nt siRNAs from Athila retrotransposons. These siRNA begin with all 4 nucleotides (A, C, G, U), are derived from both strands, and are not excised precisely from precursor substrates (not shown), so we refer to them as siRNA rather than miRNA or tasiRNAs(Kasschau et al., 2007). The fact that these 21nt siRNAs are present in ddm1 mutants indicates they are derived from epigenetically activated retrotransposons, and therefore in wild-type pollen they must be products of TE transcripts produced by the VN. To test if these retrotransposon siRNAs are present in purified sperm, we sorted SCs using the GEX2-GFP SC-specific marker (Borges et al., 2008; Engel et al., 2005) and sequenced small RNA libraries from purified sperm. Intriguingly, 21nt Athila siRNAs were enriched in purified SCs (Figure 5), even though Athila TEs transcripts are undetectable in sperm (Supplemental Figure 4). This indicates that the 21nt Athila siRNAs produced from transcripts from the VN accumulate in purified sperm. In order to test whether small RNA could move between these compartments, we expressed an artificial microRNA (amiRNA) targeting GFP using the LAT52 promoter, which is specific to the VN (Eady et al., 1995; Engel et al., 2005). When we transformed this amiRNA-GFP construct into the GEX2-GFP sperm-specific expression line, we noted that half as many pollen grains had GFP positive sperm in primary (heterozygous) transformants, and very few if any pollen grains had GFP positive sperm in amiRNA homozygous progeny (Supplemental Figure 10). These data demonstrate that small RNA can move and/or communicate between the pollen grain cytoplasm and the SCs. Therefore, siRNAs that accumulate in SCs are generated from reactivated Athila TE transcripts from the VN.
Discussion
We have demonstrated that diverse TEs are transiently activated in a coordinated fashion in the vegetative nuclei of pollen, and this leads to new somatic transpositions. Several independent lines of evidence demonstrate that TE reactivation in pollen is confined to the VN: (1) GUS staining patterns, (2) sperm and pollen transcriptome data, (3) the lack of inheritance of transposed elements, and (4) the reduced methylation of DNA isolated from whole pollen grains compared to purified sperm. This epigenetic reactivation is specific to TEs, and is co-incident with a substantial loss of heterochromatin (Figure 2)(McCormick, 1993). In pollen from Lilium, changes in histone H4 acetylation levels occur specifically in the VN (Janousek et al., 2000). In both Lilium and Arabidopsis histone H3 and H2A variants are specific to SCs (Ingouff et al., 2007; Okada et al., 2006; Talbert et al., 2002; Ueda and Tanaka, 1995), suggesting genome-wide reprogramming in pollen.
Reprogramming of TEs during plant development
Global reactivation of TEs in the VN likely depends on the down-regulation of genes required for both siRNA biogenesis and heterochromatin formation. Loss of the 24nt siRNA (rdr2/dcl3) pathway alone, at least in the plant body, does not result in transposition of AtMu1 (data not shown). Transposition only occurs in mutants that lose both heterochromatic histone modification and DNA methylation, such as ddm1(Miura et al., 2001; Singer et al., 2001). TEs may also be reactivated, to a lesser degree, in the developing seed (Figure 1). DDM1 is down regulated in the immature seed (Supplemental Figure 7) and the DNA methyltransferase MET1 is down regulated in some cells of the female gametophyte (FitzGerald et al., 2008). This could contribute to TE expression in the endosperm, which is reduced to just a few cells at the late stages of seed development and also does not contribute DNA to the embryo.
In addition to the normal developmental plan of Arabidopsis, we have recently found Athila 21nt siRNA in cultured suspension cells, in which some TEs are also reactivated(Tanurdzic et al., 2008). This suggests that the Athila 21nt siRNAs are specifically associated with active TEs, and potentially with dividing cells. TEs in both cultured cells and in pollen are also selectively hypomethylated (Tanurdzic et al., 2008). In pollen, the reduced AtMu1a DNA methylation in the VN is likely to result from an active process, as only one cell division separates it from the sperm generative cell, and asymmetric CHH methylation is lost from both strands (Figure 4). We propose that a DNA demethylase is active only in the VN, which acts to selectively remove DNA methylation from some TEs. Recently, a potential mechanism for demethylation has been described involving the siRNA binding protein ROS3, which appears to guide the DNA glycosylase ROS1 in demethylating its targets (Zheng et al., 2008). However, ROS3 is not highly up regulated in pollen, so another protein or mechanism is likely responsible.
The reactivation of TEs in the pollen VN likely explains somaclonal variation in doubled haploid plants generated by anther culture, a procedure that is widely used to aid plant breeding. For example, MITE TEs are activated in rice anther cultures (Kikuchi et al., 2003). Controlling the contribution of the pollen VN to the haploid plant embryo might reduce TE activity and somaclonal variation in doubled haploids. On an evolutionary scale, the seed-free tracheophytes (such as lycophytes and ferns), despite their highly elaborated gametophytic phase, do not have sperm companion cells comparable to those found in angiosperms. Fern genomes are large and have little differential methylation of transposons and genes (Rabinowicz et al., 2005), and therefore it is tempting to speculate that the evolution of the sperm-companion VN helped to promote TE silencing in angiosperms.
Germline mechanisms for revealing and silencing TEs
The reactivation of TEs in the pollen VN represents a cell type-specific epigenetic reprogramming that effectively “reveals” the presence of TEs in the genome. The pollen VN is terminally differentiated, however this terminal differentiation is likely not responsible for the programmed reactivation of TEs in pollen. For example, Arabidopsis TEs are not coordinately reactivated in terminally differentiated senescing leaf cells (Figure 1). Our data demonstrates that TE activation is completely under host (plant) control, as the genes responsible for defense against TEs are down regulated in the VN. This suggests that the TE reprogramming in the VN has evolved for a function.
Thus, the question remains: Why is epigenetic reactivation of TEs needed in the VN of pollen? The answer may be that the relative position of the VN next to the SCs is important for the silencing of TEs in the next generation. In animals, epigenetic marks are first lost, and then robustly reset in the germline each generation, resulting in TE expression in sperm and their progenitor cells(Calvi and Gelbart, 1994; Dupressoir and Heidmann, 1996; Ostertag et al., 2002; Pasyukova et al., 1997). Re-methylation and silencing depends on piRNA (Carmell et al., 2007). In plants, rather than reprogramming the sperm, the VN unexpectedly acts as the location of epigenetic reactivation of TEs. This reactivation potentially signals TE silencing in the neighboring sperm via Athila 21nt siRNAs, which accumulate at very high levels in mature SCs, and act either in the mature pollen grain (see model, Figure 7) or just prior to fertilization, as the SCs pass by the degenerating VN in the pollen tube prior to entering the ovule. Interestingly, SCs in Arabidopsis mature pollen are stalled in S-phase (Friedman, 1999), the phase of the cell cycle when epigenetic marks (such as histone modification) are established via siRNAs (Kloc et al., 2008). The result of TE reactivation in the VN may thus be silencing of Athila TEs in the sperm, at both the transcriptional and the post-transcriptional level, reminiscent of viral siRNA (and Athila’s presumed evolutionary origin as a retrovirus)(Dunoyer et al., 2007; Mathieu and Bender, 2004).
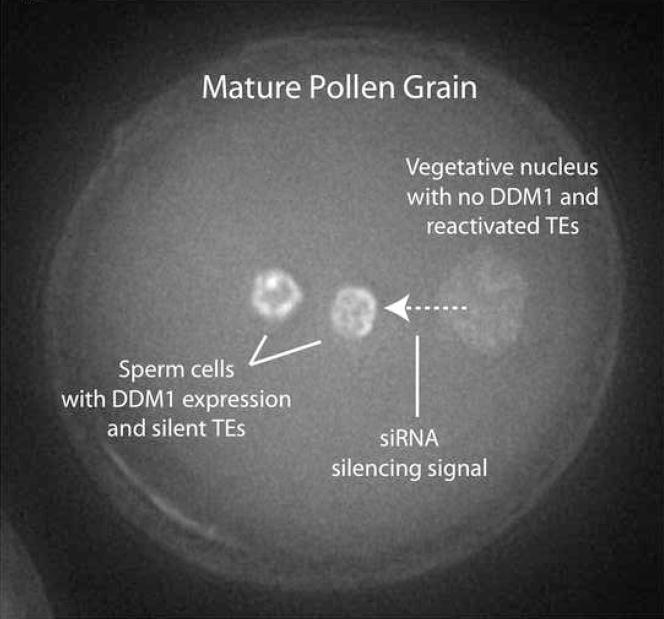
Figure 7. Model of pollen TE reactivation and potential reprogramming.
The pollen VN and SCs (stained here with DAPI) differ in DDM1 localization, TE expression, mobilization and DNA methylation. Reactivated Athila retrotransposons from the VN produce 21nt siRNAs, which accumulate in the SCs. Thus Athila retrotransposons may be “revealed” by epigenetic reprogramming of the VN specifically to target and reinforce their silencing in the SCs.
Along with germline reprogramming of TEs, movement of small RNA signals into germ cells may be highly conserved. In tobacco and in Arabidopsis cytoplasmic connections between SCs and the VN have previously been observed (McCormick, 2004; Yu et al., 1989) and might provide a channel for 21nt siRNA. In support of this idea, genes required for trans-acting siRNA biogenesis (RDR6, DCL1 and DCL4) are some of the few RNAi genes expressed in pollen and are required for cell-to-cell siRNA movement elsewhere in the plant (Dunoyer et al., 2005; Himber et al., 2003). In Drosophila, somatic follicular cells adjacent to female germline stem cells are the site of Gypsy inhibition via Flamenco and piRNA(Brennecke et al., 2007; Pelisson et al., 1994; Sarot et al., 2004). Furthermore, piRNA-mediated repression of hybrid dysgenesis by non-LTR I retrotransposons depends on expression of full-length autonomous elements in nurse cell nuclei adjacent to the oocyte (Chambeyron et al., 2008)(Brennecke et al., 2008). Finally, in protists, communication between the old somatic macronucleus and germinal micronucleus leads to elimination of TE-related sequences from new macronuclei, and depends on siRNA, albeit within the same cell(Meyer and Chalker, 2007). Thus the accumulation of siRNA in germ cells via TE reprogramming in companion cells may be a conserved mechanism for TE silencing in eukaryotes.
Experimental Procedures
Plant Material
Mature pollen was collected from Columbia reference strain plants using vacuum filtration (Johnson-Brousseau and McCormick, 2004). DNA and RNA were isolated from a ddm1-2 plant in the Columbia reference background. SCs were purified as in Borges et al. (Borges et al., 2008) and subjected to RT-PCR and microscopy analysis to ensure purity.
Small RNA Northern blots
Total RNA was extracted using Trizol reagent (Invitrogen). Small RNAs were enriched using the FlashPage Fractionator (Ambion). Gel electrophoresis and Northern blotting were performed as in Lippman et al.(Lippman et al., 2003). The size standard corresponds to the Decade RNA ladder (Ambion). For information on Northern probes, see Supplemental Data.
Analysis of microarray expression patterns
Affymetrix Athi1 microarray expression results from Schmid et al(Schmid et al., 2005) are available from http://www.weigelworld.org/resources/microarray/AtGenExpress/. This data was normalized for each ORF across experiments using gcRMA. For identification of TEs on the Ath1 array, see Supplemental Data. Figure 1 was generated using TIGR’s MultiExperiment Viewer.
GUS and DAPI staining
Enhancer and gene trap lines are described in Sundaresan et al(Sundaresan et al., 1995). GUS staining was performed as in Sundaresan et al(Sundaresan et al., 1995). The search for GUS staining in developing ovules was performed as in Galli et al(Galli et al., 2003). Additional information on GUS staining is available as Supplemental Data.
Numerical floral stages correspond to those described by Smyth et al(Smyth et al., 1990). Pollen was DAPI stained by incubating mature pollen in a 1 μg/ml DAPI solution in 15% sucrose for 5 minutes at room temperature. Pollen was washed twice in 15% sucrose.
Transposable element display
Transposable element display was performed as described in Casa et al(Casa et al., 2004). Primers used in this analysis are listed in Supplemental Table 1. For the transposable element display analysis shown in Figure 3, a stringent PCR amplification approach was used to detect only the AtMu1a and AtMu1b sub-family of AtMu1 TEs, using non-degenerate TE-specific PCR primers (Supplemental Table 1).
New polymorphism bands from the transposable element display were excised as in Casa et al(Casa et al., 2004). The gel in Figure 3A was not dried prior to imaging so PCR products could be easily excised. Excision products were diluted, re-amplified, TOPO-TA cloned into the pCRII vector (Invitrogen) and sequenced.
Bisulfite sequencing
DNA was bisulfite converted using the EpiTech Bisulfite conversion kit (Qiagen). PCR primers used for the amplification are listed in Supplemental Table 1. PCR products were TOPO-TA cloned into the pCRII vector (Invitrogen) and sequenced. Sequencing results were visualized using the KisMeth software (Gruntman et al., 2008).
Transgenic construction, transformation and microscopy analysis
The DDM1 promoter, 5′UTR and coding region (including introns) were moved from the pDONR-gDDM1 vector (T. Stokes and E. Richards, Washington University, St. Louis) to the pMDC107 GFP fusion binary vector using the Gateway recombination system (Invitrogen). Plants heterozygous for the ddm1-2 mutation were transformed and their seeds were selected on 50 μg/μl hygromycin. T1 and T2 generation transgenic individuals were genotyped for the ddm1-2 mutation, and homozygous mutants were used to assay transgene complementation (Supplemental Figure 8). Transformation yielded nine independent insertion events, of which six showed sperm-specific protein accumulation (Figure 6). Pollen grains were stained with DAPI by incubating mature pollen in a 1 μg/ml DAPI solution in 15% sucrose for 5 minutes at room temperature, and subsequently imaged using a restoration microscope system (DeltaVision RT, Applied Precision) equipped with an inverted microscope (Olympus IX-70), and a 100X oil immersion objective lens. Data was processed using the SoftWoRx 3.5.1 software (Applied Precision). Images in Figure 6 represent the flattened composite of ten Z-stacks separated at a distance of 0.5 μm.
Small RNA cloning and sequencing
Small RNAs of 19–28nt were size selected by denaturing 15% PAGE, and cloned as in (Brennecke et al., 2007). Additional details regarding the cloning of small RNAs are found in Supplementary Data. The small RNA libraries were sequenced on Illumina 1G sequencer. The total number of sequences perfectly matching the Arabidopsis genome were: wt inflorescence 4,158,848 (2,286,133 unique); wt pollen 1,034,665 (437,984 unique); ddm1 inflorescence 4,098,772 (1,637,771 unique); wt sperm 760,651 (429,972 unique).
For Figure 5B, the distributions of siRNA sizes in each library relative to Athila, AtMu1, and AtGP1 were calculated by summing the sequencing frequencies of all small RNAs of each size (20–26nt) that perfectly matched instances of the TE in the genome, then dividing that number by the total number of small RNAs in that size class that matched the genome at large. These values are expressed as library size normalized totals for each library to facilitate inter-library comparison.
For Figure 5C, siRNA sequencing frequencies for 100 base windows along all five chromosomes were calculated by summing the per-library sequencing frequencies of 21 and 24nt siRNAs matching with 100% identity within the window by the number of times they matched the genome as a whole. Because the results of these calculations are on different scales, based on the depth of each sequence library, a layer of per-library normalization based on the sum values of all small RNAs matching the genome was implemented. The results were converted into UCSC Wiggle format and displayed using the Generic Genome Browser visualization tool.
Supplementary Material
Acknowledgments
We thank J. Simorowski, M. Galli, T. Nawy, M. Bayer and S. Hearn, for assistance with GUS staining and microscopy, and T. Stokes, E. Richards, Y. Fang, D. Spector, D. Lisch and R. Schwab for providing tools and stocks. We additionally thank Dennis Stevenson (New York Botanical Garden) for discussions. R.K.S. is a recipient of a NIH postdoctoral fellowship (F32 CA125977). This work was supported by a grant from the NIH (R01 GM067014) to R.M., and grants PPCDT/BIA-BCM/61270/2004, PTDC/AGR-GPL/70592/2006 from Fundacão para a Ciência e a Tecnologia (FCT) to J.D.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijo JA. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 2003;133:713–725. doi: 10.1104/pp.103.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JD, Feijo JA. How many genes are needed to make a pollen tube? Lessons from transcriptomics. Ann Bot (Lond) 2007;100:1117–1123. doi: 10.1093/aob/mcm208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijo JA, Becker JD. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 2008;148:1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi BR, Gelbart WM. The basis for germline specificity of the hobo transposable element in Drosophila melanogaster. EMBO J. 1994;13:1636–1644. doi: 10.1002/j.1460-2075.1994.tb06427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Casa AM, Nagel A, Wessler SR. MITE display. Methods Mol Biol. 2004;260:175–188. doi: 10.1385/1-59259-755-6:175. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci U S A. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- Dupressoir A, Heidmann T. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol Cell Biol. 1996;16:4495–4503. doi: 10.1128/mcb.16.8.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady C, Lindsey K, Twell D. The Significance of Microspore Division and Division Symmetry for Vegetative Cell-Specific Transcription and Generative Cell Differentiation. Plant Cell. 1995;7:65–74. doi: 10.1105/tpc.7.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel ML, Holmes-Davis R, McCormick S. Green sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiol. 2005;138:2124–2133. doi: 10.1104/pp.104.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald J, Luo M, Chaudhury A, Berger F. DNA methylation causes predominant maternal controls of plant embryo growth. PLoS ONE. 2008;3:e2298. doi: 10.1371/journal.pone.0002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman WE. Expression of the cell cycle in sperm of Arabidopsis: implications for understanding patterns of gametogenesis and fertilization in plants and other eukaryotes. Development. 1999;126:1065–1075. doi: 10.1242/dev.126.5.1065. [DOI] [PubMed] [Google Scholar]
- Galli M, Theriault A, Liu D, Crawford NM. Expression of the Arabidopsis transposable element Tag1 is targeted to developing gametophytes. Genetics. 2003;165:2093–2105. doi: 10.1093/genetics/165.4.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM. Anther development: basic principles and practical applications. Plant Cell. 1993;5:1217–1229. doi: 10.1105/tpc.5.10.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen R, Sachidanandam R. Kismeth: Analyzer of plant methylation states though bisulfite sequencing. BMC Bioinformatics. 2008;9:371. doi: 10.1186/1471-2105-9-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol. 2007;17:1032–1037. doi: 10.1016/j.cub.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Janousek B, Zluvova J, Vyskot B. Histone H4 acetylation and DNA methylation dynamics during pollen development. Protoplasma. 2000;211:116–122. [Google Scholar]
- Johnson-Brousseau SA, McCormick S. A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. The Plant Journal. 2004;39:761–775. doi: 10.1111/j.1365-313X.2004.02147.x. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Terauchi K, Wada M, Hirano H. The plant MITE mPing is mobilized in anther culture. Nature. 2003;421:167–170. doi: 10.1038/nature01218. [DOI] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. Mutator transposons. Trends Plant Sci. 2002;7:498–504. doi: 10.1016/s1360-1385(02)02347-6. [DOI] [PubMed] [Google Scholar]
- Mathieu O, Bender J. RNA-directed DNA methylation. J Cell Sci. 2004;117:4881–4888. doi: 10.1242/jcs.01479. [DOI] [PubMed] [Google Scholar]
- May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005;1:e79. doi: 10.1371/journal.pgen.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. Male Gametophyte Development. Plant Cell. 1993;5:1265–1275. doi: 10.1105/tpc.5.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. Control of male gametophyte development. Plant Cell. 2004;16(Suppl):S142–153. doi: 10.1105/tpc.016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Chalker DL. In: Epigenetics of Ciliates. Allis Epigenetics DC, Jenuwein T, Reinberg D, Caparros M-, editors. Cold Spring Harbor Laboratory Press; 2007. pp. 127–150. [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, Belo A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang GL, Meyers BC. An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol. 2007;25:473–477. doi: 10.1038/nbt1291. [DOI] [PubMed] [Google Scholar]
- Okada T, Singh MB, Bhalla PL. Histone H3 variants in male gametic cells of lily and H3 methylation in mature pollen. Plant Mol Biol. 2006;62:503–512. doi: 10.1007/s11103-006-9036-8. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, Kazazian HH., Jr A mouse model of human L1 retrotransposition. Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- Pasyukova E, Nuzhdin S, Li W, Flavell AJ. Germ line transposition of the copia retrotransposon in Drosophila melanogaster is restricted to males by tissue-specific control of copia RNA levels. Mol Gen Genet. 1997;255:115–124. doi: 10.1007/s004380050479. [DOI] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud’homme N, Smith PA, Bucheton A, Corces VG. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. Embo J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 2005;138:744–756. doi: 10.1104/pp.104.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Citek R, Budiman MA, Nunberg A, Bedell JA, Lakey N, O’Shaughnessy AL, Nascimento LU, McCombie WR, Martienssen RA. Differential methylation of genes and repeats in land plants. Genome Res. 2005;15:1431–1440. doi: 10.1101/gr.4100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizada MN, Benito MI, Walbot V. The MuDR transposon terminal inverted repeat contains a complex plant promoter directing distinct somatic and germinal programs. Plant J. 2001;25:79–91. doi: 10.1046/j.1365-313x.2001.00939.x. [DOI] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Singer T, Yordan C, Martienssen RA. Robertson’s Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1) Genes Dev. 2001;15:591–602. doi: 10.1101/gad.193701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanurdzic M, Vaughn MW, Jiang H, Lee T-J, Slotkin RK, Sosinski B, Thompson WF, Doerge RW, Martienssen RA. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 2008 doi: 10.1371/journal.pbio.0060302. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Tanaka I. The appearance of male gamete-specific histones gH2B and gH3 during pollen development in Lilium longiflorum. Dev Biol. 1995;169:210–217. doi: 10.1006/dbio.1995.1138. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 2008;148:1201–1211. doi: 10.1104/pp.108.126375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Hu S, Zhu C. Ultrastructure of sperm cells of the male germ unit in pollen tubes of Nicotiana tabacum. Protoplasma. 1989;152:29–36. [Google Scholar]
- Zheng X, Pontes O, Zhu J, Miki D, Zhang F, Li WX, Iida K, Kapoor A, Pikaard CS, Zhu JK. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.