Abstract
Human Argonaute (Ago) proteins are essential components of the RNA-induced silencing complexes (RISCs). Argonaute 2 (Ago2) has a P-element-induced wimpy testis (PIWI) domain, which folds like RNase H and is responsible for target RNA cleavage in RNA interference1. Proteins such as Dicer, TRBP, MOV10, RHA, RCK/p54 and KIAA1093 associate with Ago proteins and participate in small RNA processing, RISC loading and localization of Ago proteins in the cytoplasmic messenger RNA processing bodies1,2. However, mechanisms that regulate RNA interference remain obscure. Here we report physical interactions between Ago2 and the α-(P4H-α(I)) and β-(P4H-β) subunits of the type I collagen prolyl-4-hydroxylase (C-P4H(I)). Mass spectrometric analysis identified hydroxylation of the endogenous Ago2 at proline 700. In vitro, both Ago2 and Ago4 seem to be more efficiently hydroxylated than Ago1 and Ago3 by recombinant human C-P4H(I). Importantly, human cells depleted of P4H-α(I) or P4H-β by short hairpin RNA and P4H-α(I) null mouse embryonic fibroblast cells showed reduced stability of Ago2 and impaired short interfering RNA programmed RISC activity. Furthermore, mutation of proline 700 to alanine also resulted in destabilization of Ago2, thus linking Ago2 P700 and hydroxylation at this residue to its stability regulation. These findings identify hydroxylation as a post-translational modification important for Ago2 stability and effective RNA interference.
To identify the protein network involved in regulating the RNA interference (RNAi) machinery, we established stable HeLa S3 cell lines expressing Flag–HA(haemagglutinin)-tagged human Ago1−4, respectively. Cytoplasmic extracts were immunoprecipitated with sequential anti-Flag and anti-HA antibody resins. Silver staining of a representative purification (Ago2) is shown in Fig. 1a. Mass spectrometric analysis of the purifications identified the known Ago-interacting proteins1, among which Dicer, KIAA1093 and eEF1A1 are present in all four Ago purifications, whereas TRBP and MOV10 are abundantly present in Ago3 and 4 purifications. In addition, unique peptides of P4H-α(I) and P4H-β, the α- and β-subunits of C-P4H(I) (EC 1.14.11.2), were found in sub-stoichiometric quantities in Ago2 purification (Fig. 1a). To confirm the P4H-α(I) interaction with Ago proteins, we performed co-immunoprecipitation experiments using cell lysates from 293ET cells co-transfected with Flag-tagged Ago 1−4 and HA-tagged P4H-α(I), and found interactions of P4H-α(I) with Ago2 and Ago4 (Fig. 1b). Co-immunoprecipitation using a polyclonal anti-P4H-β antibody co-precipitated endogenous Ago2 from 293ET cells (Fig. 1c), supporting the in vivo interaction between C-P4H(I) subunits with Ago2. That a small percentage of Ago2 protein was found to associate with P4H-β may reflect the dynamic nature of this enzyme–substrate interaction.
Figure 1. Human Ago2 associates with C-P4H(I) subunits.
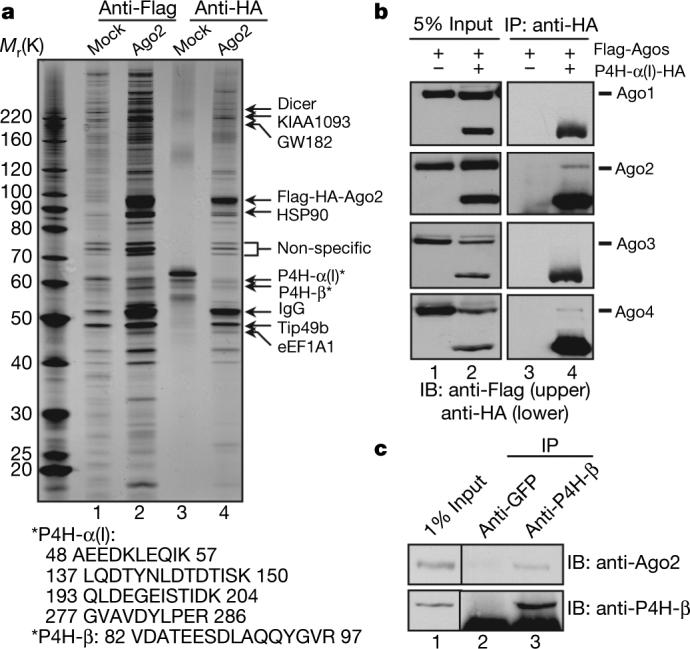
a, Tandem affinity purification was conducted on cytoplasmic extracts from Mock (lanes 1 and 3) and Flag–HA-tagged Ago2 stable HeLa S3 cells (lanes 2 and 4) with sequential anti-Flag and HA immunoprecipitation. The peptides for P4H-α(I) and P4H-β are shown below. b, Flag–Ago1−4 were co-transfected with empty vector (lanes 1 and 3) or P4H-α(I)-HA (lanes 2 and 4) in 293ET cells. The immunoprecipitates with anti-HA antibody were blotted with anti-Flag and HA antibodies, respectively. IP, immunoprecipitate; IB, immunoblot. c, 293ET cells were immunoprecipitated with anti-GFP or P4H-β antibodies and followed with western blotting with anti-P4H-β and Ago2 antibodies.
C-P4H catalyses proline hydroxylation of collagens in the X-Pro-Gly (X-P-G) triplets. Three isoforms (I, II, III) of the α-subunit have been identified3 and they all interact with the β-subunit to form active C-P4H α2β2 tetramers. However, these isoforms show differential tissue expression pattern and abundance. In addition to collagens, other proteins containing collagen-like sequences, such as pulmonary surfactant, apoproteins, complement protein C1q and the 18S form of acetylcholinesterase, are also hydroxylated by C-P4H (ref. 3). C-P4H also mediates hydroxylation of the X-Pro-Ala (X-P-A) motif found in the complement protein C1q and type IV collagen4. We identified both X-P-G and X-P-A triplets within the Ago proteins (Supplementary Fig. 1). Three X-P-G triplets (119−121, 522−524, 699−701) were found in Ago2. They were conserved among other human Ago proteins (Supplementary Fig. 1) and Ago proteins from Mus musculus, Drosophila melanogaster (Ago1) and Caenorhabditis elegans (Alg1), but not Arabidopsis thaliana and Schizosaccharomyces pombe. Ago4 contains two additional X-P-G motifs at its amino terminus, of which the R-P-G (18−21) motif is conserved in Ago1 and 3. In addition, Ago2 has seven X-P-A triplets; five of them are conserved in human Ago1, 3, 4 and Ago proteins from M. musculus and D. melanogaster.
To determine the hydroxylation site(s) in Ago2, endogenous Ago2 was immunoprecipitated from 293ET cells and analysed by tandem mass spectrometry (MS/MS). The analysis identified one hydroxylation site at Pro 700 (DYQP*GITFIVVQK) in Ago2. The hydroxylated peptides (Fig. 2a, upper panel) represented approximately 20% (7,900/39,900) of total Ago2 peptides detected (DYQPGITFIVVQK). However, this is probably an underestimation because the MS/MS pattern of this hydroxylated peptide is very similar to that of its unmodified peptide (Fig. 2a, lower panel), as would have been expected for this type of modification. In contrast, we did not detect peptides containing modified proline at other X-P-G sites such as Pro 120 or 523, suggesting that Ago2 hydroxylation at Pro 700 is likely to be specific. This analysis, however, does not exclude cell type and tissue- and/or developmental-stage-specific hydroxylation at other proline residues of the Ago proteins.
Figure 2. In vivo and in vitro hydroxylation of Ago2.
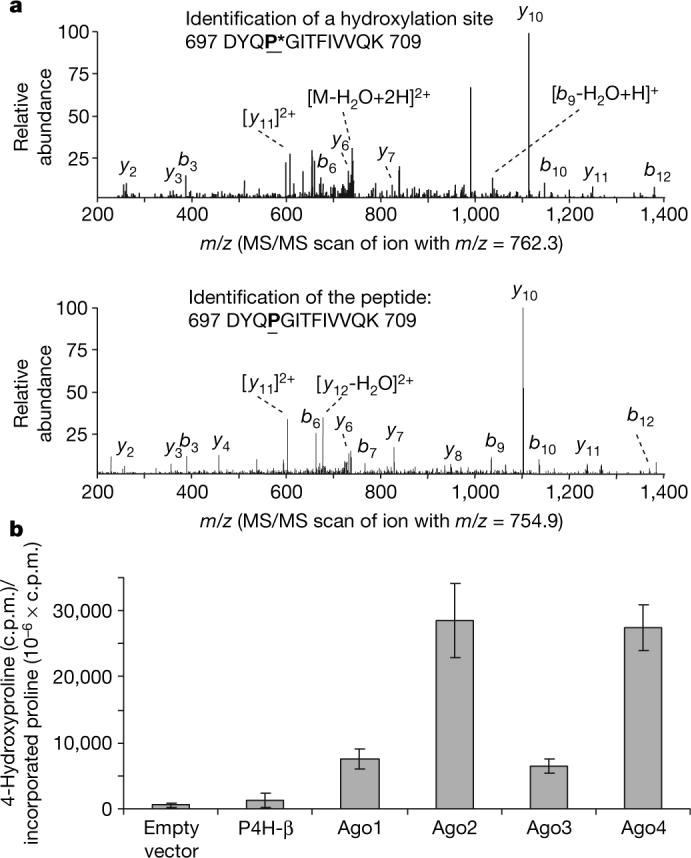
a, Endogenous Ago2 is hydroxylated at proline 700. Endogenous Ago2 was immunoprecipitated with a polyclonal anti-Ago2 antibody and analysed by LC–MS/MS. The MS/MS scanned hydroxylated peptides with ion m/z 762.3 (upper panel) and ion m/z 754.9 (lower panel). b, In vitro hydroxylation of Ago proteins. pcDNA3–Flag-HA-Ago1−4 were in vitro translated in the presence of L-[2,3,4,5-3H]proline. The in vitro translated lysates were incubated with purified recombinant C-P4H(I) enzyme and the reaction cofactors. The amount of 4-hydroxy[3H]proline formed in control, P4H-β and Ago 1−4 was determined as described in Methods. Data are presented as the mean ± standard deviation for three independent experiments.
Human Ago2 was originally reported as GERp95 (Golgi endoplasmic reticulum protein 95 kDa)5. Biochemical analysis suggests that Ago2 is a cytoplasmically exposed, peripheral membrane protein that exists in a protease-resistant complex5. Although Ago2 does not have either signal peptides or transmembrane domains, the expression of the exogenously introduced Ago2 overlaps with that of the ER marker BiP in NRK and COS cells5. Endogenous Ago2 is largely cytoplasmically localized, but is also enriched where P4H-β signal is found in a percentage of U2OS cells. Partial overlap with P4H-β was also observed in HeLa and 293ET cells (Supplementary Fig. 2). Importantly, the in vitro hydroxylation assay demonstrated that only C-P4H(I) but not the cytoplasmically localized prolyl-4-hydroxylases (PHD 1−3) was able to mediate Ago protein hydroxylation in vitro (Fig. 2b and data not shown). All four Ago proteins are hydroxylated in vitro by recombinant human C-P4H(I); however, Ago2 and Ago4 seem to be hydroxylated to a greater extent (Fig. 2b), consistent with the co-immunoprecipitation data that P4H-α(I) associated with Ago2 and 4 more readily than with Ago1 and Ago3 (Fig. 1b).
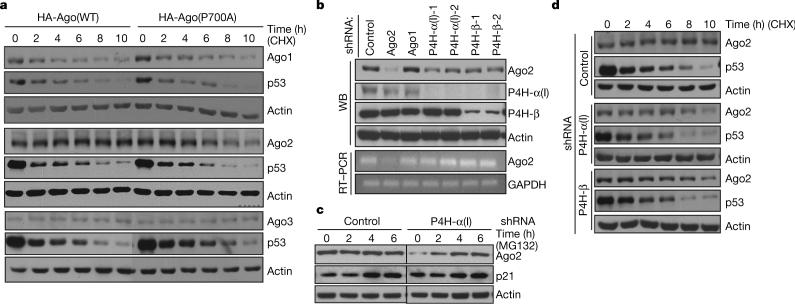
Hydroxylation of collagen and collagen-like proteins is essential for their functional triple-helical folding3. Inhibition of the C-P4H(I) enzyme generates unstable collagens, resulting in reduced collagen production6. In contrast, overexpression of P4H-α(I) is associated with excess collagen production7. To determine whether hydroxylation of proline at position 700 regulates Ago stability, we mutated the proline residue to alanine (P700A). As shown in Fig. 3a, Ago2 with the P700A mutation was less stable (t1/2 approximately 6 h) than the wild-type protein whose half-life was estimated to be greater than 10 h (Fig. 3a). In contrast, P700A mutation had no effect on the stability of Ago1 and Ago3, suggesting that proline 700 is important for Ago 2 stability.
Figure 3. Impaired hydroxylation downregulates Ago2 stability.
a, U2OS cells transfected with HA–Ago1, 2, 3 (wild type, WT) and their mutants (P700A) were treated with 50 μg ml−1 of cycloheximide (CHX) for the indicated times. Agos, p53 and actin were detected by western blot. The results shown are representative for three independent experiments. b, U2OS cells were transfected with indicated shRNAs followed by puromycin selection. Western blot with indicated antibodies and RT–PCR were performed. c, P4H-α(I)-depleted and control U2OS cells were treated with 25 μM of MG132 for the indicated times. Endogenous Ago2, p21 and actin were detected by western blot. d, U2OS cells were transfected with P4H-α or -β shRNA followed with puromycin selection. Cells were then treated as in a. Ago2, p53 and actin were monitored by western blot. The results are representative of three independent experiments.
To determine whether the reduced stability of the Ago2 (P700A) mutant was due to loss of hydroxylation, we first compared the steady-state levels of the endogenous Ago2 in the wild type versus cells in which the expression of the hydroxylase, C-P4H(I), was inhibited by RNAi. We found knockdown of P4H-α(I) or -β reduced the steady-state level of the endogenous Ago2 protein in U2OS (Fig. 3b, lanes 4−7) and HeLa cells (data not shown) without affecting its mRNA level (Fig. 3b), suggesting that C-P4H(I) may regulate Ago2 protein stability. Upon MG132 treatment, the reduced endogenous Ago2 protein caused by P4H-α(I) RNAi was restored to the level comparable to that of the control RNAi sample (Fig. 3c), suggesting that reduced Ago2 is likely to be due to degradation by the proteasome-mediated pathway.
We examined the protein turnover rate of the endogenous Ago2 with or without an intact C-P4H(I). In the control RNAi U2OS cells, we found endogenous Ago2 to be quite stable with very little turnover, even at 10 h post-cycloheximide (50 μg ml−1) treatment (Fig. 3d). In the cells where C-P4H(I) expression was inhibited by short hairpin RNA (shRNA), Ago2 became slightly destabilized at 4 h, and the steady-state level was further decreased at later time points (Fig. 3d). These results show that a reduction of the steady-state level of Ago2 is correlated with reduction of the hydroxylase C-P4H(I). Taken together, these findings suggest that hydroxylation of Ago2 at P700 regulates its stability.
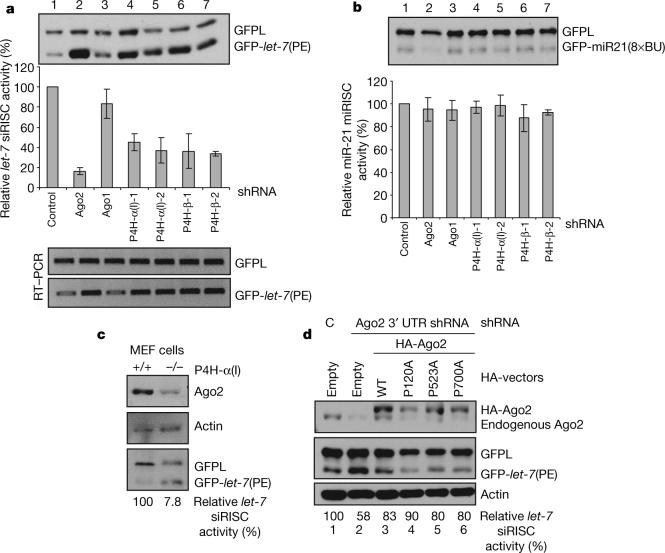
The observation that Ago2 hydroxylation regulates its stability predicts that the hydroxylase C-P4H(I) is likely to play a role in RNAi. To address this issue, we assayed the requirements of C-P4H(I) for short interfering (si)RNA- or miRNA-programmed RISC (si or miRISC) activity. We first constructed and validated GFP (green fluorescent protein) reporters containing perfect (PE) or bulged (BU) complementary sequences for miRNA let-7 or miR21 (Supplementary Fig. 3). We next established U2OS stable cell lines expressing long GFP (GFPL) and GFP–let-7 (PE), GFPL and GFP–miR21 (8 × BU), respectively. The siRISC or miRISC activities were measured by determining the ratio between the reporter GFP–let-7 (PE) or GFP–miR21 (8 × BU) and the GFPL control. As shown in Fig. 4a, inhibition of Ago2, not Ago1, reduced the siRISC activity to 20% of that with the control RNAi. Knockdown of P4H-α(I) and -β reduced let-7 siRISC activity to approximately 40% (Fig. 4a). Reverse transcription–PCR (RT–PCR) of GFPL and GFP–let-7 (PE) showed derepression of the GFP–let-7 (PE) reporter at the RNA level (Fig. 4a). Consistently, P4H-α(I) null mouse embryonic fibroblast (MEF) cells8 also showed reduced Ago2 protein level and let-7-guided siRISC activity compared with wild-type MEF cells (Fig. 4c). In contrast, miRISC activity was not altered upon C-P4H(I) knockdown as measured by the GFP–miR21 (8 × BU) reporter (Fig. 4b), although whether this observation is generally applicable to other miRNA-mediated translational inhibition remains to be determined. Hydroxylation appears to be dispensable for the catalytic activity of Ago2 as the Ago2 hydroxylation mutants were capable of restoring siRISC activity upon overexpression in cells in which the endogenous Ago2 expression was inhibited by an shRNA targeting the 3′ untranslated region of Ago2 (Fig. 4d). Furthermore, bacterially produced Ago2 binds single-stranded siRNAs and is able to cleave siRNA targets, suggesting that hydroxylation is not required for Ago2 siRNA binding and slicing activity because bacteria do not have C-P4H (ref. 9). Taken together, C-P4H(I) appears to play an important role in RNAi by regulating Ago2 stability.
Figure 4. Impaired C-P4H(I) reduced let-7 guided siRISC activity.
a, b, U2OS stable cell line expressing GFPL/GFP–let-7 (PE) (a) or GFPL/GFP–miR21(8 × BU) (b) were transfected with indicated shRNAs and selected by puromycin. The expression ratios between GFPL and GPF reporter were calculated and the intact (100%) si/miRISC activity was considered in control shRNA cells. The relative si/miRISC activities under knockdown conditions of the indicated genes were obtained by comparison with the control shRNA transfection. The standard deviations were obtained from three independent experiments. c, Control and P4H-α(I) null MEF cells were transfected with GFPL and GFP–let-7 (PE) plasmids. Ago2, actin and GFP expressions were detected by western blot. d, Ago2(P700A) is catalytically active. GFPL/GFP–let-7 (PE) stable cell line was co-transfected with control shRNA (lane 1) or shRNA targeting the 3′ untranslated region of Ago2 (lanes 2−6) together with empty vector (lanes 1 and 2) or HA–Ago2 constructs as indicated. GFP, Ago2 proteins were detected. let-7 guided siRISC activity was calculated as in a.
It has been reported that Ago2 is localized in cytoplasmic mRNA processing bodies (P-bodies)1 and is recruited to stress granules in response to oxidative stress or specific translational inhibitors10. Although the P-body does not appear to be required for RNAi11, it is important for RNA-related functions in controlling mRNA translation and degradation12. Thus, Ago2 P-body localization may play a yet-to-be-identified function in RNA-mediated gene regulation. Therefore, we investigated whether hydroxylation has any impact on Ago2 P-body and stress-granule localization. We found that depletion of the C-P4H(I) subunits by RNAi or genetic ablation of P4H-α(I) in MEF cells reduced Ago2 P-body localization marked by Dcp1a, which resides in P-bodies12-14 (Supplementary Fig. 4a). The reduced P-body localization of Ago2 might be a reflection of the reduced steady-state level of Ago2 protein when the expression of the hydroxylase is impaired. We also found that P700A mutation caused the mutant proteins to localize away from Dcp1a, compared with wild type and the other proline mutants of Ago2 (P120A and P523A) (Supplementary Fig. 4b). P700A also delocalized Ago1 from P-bodies. In contrast, P700A did not cause delocalization of Ago3 from P-bodies, but reduced Ago4 P-body localization (Supplementary Fig. 4b), suggesting that proline 700 may impact different Ago proteins differently. Affinity purification and co-immunoprecipitation from wild-type and mutant (P700A) Ago2 showed unchanged Dicer but reduced Dcp1a and KIAA1093 association with Ago2 (P700A) (Supplementary Fig. 4c, d). This finding suggests that Ago1 and 2 may be regulated similarly in their P-body localization. To ask whether hydroxylation has any effects on Ago2 stress-granule re-localization, we treated U2OS cells containing either intact or impaired C-P4H(I) with arsenite (Supplementary Fig. 5), translation inhibitor (hippuristanol) or heat shock (data not shown) and found no dramatic changes in Ago2 stress-granule re-localization.
Hydroxylation is an important post-translational modification, and plays important biological roles. C-P4H-mediated hydroxylation of collagen proteins is essential for their functional folding and stability3. Proline hydroxylation of the hypoxia-inducible factor (HIF) α-subunit by a separate P4H family causes recruitment of the von Hippel Lindau (VHL) ubiquitin ligase complex for HIF-α degradation15. We have shown that hydroxylation regulates Ago2 stability, suggesting a novel role for C-P4H. It will be important to investigate whether this modification is conserved in other species. For example, proline 700 is conserved in C. elegans Alg1, Alg2 and T23D8.7 proteins, which function in miRNA biosynthesis16. Additionally, the P4H-α subunit is rate-limiting in the formation of active C-P4H. Several pathways or stimuli regulate P4H-α(I) at transcriptional and post-transcriptional levels. For example, transforming growth factor (TGF)-β1 stimulates transcription of the P4H-α(I) gene17. Hypoxia upregulates the mRNA levels of P4H-α(I) and (II)18. At post-transcriptional level, the RNA-binding protein nucleolin has been demonstrated to bind to the 5′ and 3′ untranslated regions of P4H-α(I) mRNA and enhance P4H-α(I) translation efficiency under hypoxia19. Further efforts to delineate if the P4H-α regulators might also regulate RNAi machinery through regulation of hydroxylation of Ago proteins will provide additional insights into RNAi regulatory mechanisms.
METHODS SUMMARY
Tandem affinity purification, in vivo hydroxylation and mass spectrometry
The tandem affinity purification was performed as described previously20. Briefly, HeLa S3 stable cell lines expressing Flag–HA-tagged Ago proteins were cultured in 32 litres. Cytoplasmic extracts were immunoprecipitated with an anti-Flag M2 antibody (Sigma) followed by anti-HA antibody resins (Santa Cruz) with buffer A (20 mM Tris, pH 8.0, 100 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 0.1% Tween-20, 10 mM 2-mercaptoethanol and 0.25 mM phenylmethylsulphonyl fluoride (PSMF)). HA immunoprecipitation elution 1 (50 μl) was analysed by mass spectrometry using an LTQ-FT hybrid mass spectrometer (Thermo Finnigan). To identify the hydroxylation site(s) of endogenous Ago2, immunoprecipitation was performed using an anti-Ago2 polyclonal antibody (Upstate) from 293ET cells (see immunoprecipitation condition and buffer in Methods). The Ago2-containing band was cut from SDS–polyacrylamide gel electrophoresis gel, fully trypsinized and analysed by reverse-phase liquid chromatography–tandem mass spectrometry (LC–MS/MS) as previously reported21. An MS/MS scan of the precursor ion m/z 762.3, in which it was fragmented into multiple labelled product ions (b and y ions), identified a peptide according to the mass shift (+16 Da) caused by hydroxylation. An MS/MS scan of the precursor ion m/z 754.9 indicated the peptide without modification.
METHODS
Plasmid constructions, antibodies and chemicals
Human Ago4 (KIAA1567), KIAA1093, KIAA1582 and GW182 (KIAA1460) cDNAs were obtained from the Kazusa DNA Research Institute (Japan), the others were amplified by RT–PCR. For tandem affinity purification, pOZ-FH-N-Ago1−4 were constructed by inserting the open reading frames into the XhoI/NotI S sites of pOZ-Flag-HA-N vector. For pEGFPC1-Agos, SalI/BamHI and SalI/SmaI were chosen for Ago1, 2, 3 and 4 respectively. To generate amino-terminal Flag-tagged Agos, Ago1, 2, 3 and 4 were cloned into HindIII/NotI and NotI/SalI sites of 3 × Flag-CMV-7.1. To express Flag–HA-tagged Agos in pcDNA3 vector, the coding regions and Flag–HA (FH) tags were PCR-amplified from pOZ-FH-N-Agos and cloned into EcoRI/NotI (Ago1, 2 and 3) and KpnI/NotI (Ago4) sites of pcDNA3 vector. For HA-tagged vectors, KIAA1093 was cloned into BamHI/XbaI sites of pcDNA–C-HA. Point mutations were introduced following standard mutagenesis procedures. Polyclonal anti-P4H-α(I) was generated (Innovagen) against purified recombinant P4H-α(I) peptide-substrate-binding domain22. Polyclonal anti-P4H-β antibody, polyclonal anti-TIA-1, monoclonal anti-CDC25a, monoclonal anti-GFP(B2) and monoclonal anti-p53 antibodies were purchased from Santa Cruz. Rabbit polyclonal anti-Ago2 antibody was obtained from Upstate, and polyclonal anti-Dcp1a antibody was a gift from J. Lykke-Andersen. Monoclonal antibodies for Ago2 (Novus Biochemicals and Wako Chemicals), Dicer (Abcam), p21 (Calbiochem), HA (Covance) and Flag M2 (Sigma) were also used. Cycloheximide (Sigma), Z-Leu-Leu-Leu-al (MG132) (Sigma), arsenite (Riedel de Haen) and hippuristanol (a gift from the P. Sharp laboratory) were obtained.
In vitro hydroxylation
pcDNA3–Flag–HA–Ago1−4 were translated in vitro in the presence of radioactively labelled proline, L-[2,3,4,5-3H]proline (85 mCi mmol−1) in rabbit reticulocyte lysate using TNT® Quick-coupled transcription/translation system (Promega). The empty vector and pcDNA–P4H-β were used as negative controls. The in vitro translated lysates were aliquoted into two portions for C-P4H(I) hydroxylation reaction in which 0.33 pmol of recombinant C-P4H(I) purified from insect cells23 or purified HIF PHD (1−3)24 was added. The enzyme was omitted from the parallel tubes that served as negative controls. The reactions were dialysed extensively to remove any remaining free radioactive proline. The amount of 4-hydroxy[3H]proline formed in the substrate was determined by a specific radiochemical procedure as previously described25.
Endogenous miRNA-guided si/miRISC activity assay, shRNAs and RT–PCR
To generate the reporter system for miRNA-guided siRISC and miRISC activity reading, pcDNA3–GFP was first cloned by inserting the GFP coding region into BamHI/NotI sites of pcDNA3. Subsequently, one copy of perfect complementary sequence of miRNA let-7 (5′-AACTATACAACCTACTACCTCA-3′) or eight copies of bulged complementary sequence of miR21 (5′-TCAACATCAGAGAGATAAGCTA-3′) were cloned into the NotI/XhoI sites of pcDNA3–GFP to form pcDNA3–GFP–let-7 (PE) and pcDNA3–GFP–miR-21 (8 × BU). pcDNA3–GFPL was obtained by removing the stop codon from pcDNA3–GFP and produces a mutant long GFP (GFPL) (267 amino acids compared with its original form of 239 amino acids) serving as a control. pcDNA3–GFPL was co-transfected with pcDNA3–GFP–let-7 (PE) or pcDNA3–GFP–miR-21 (8 × BU) into U2OS cells followed by 2 mg ml−1 G418 selection for two weeks. The resistant colonies were propagated into a stable cell line. shRNAs used in this study were all cloned into XhoI/BamHI sites of pBabe-U6 vector, which also expresses a puromycin-resistant gene26. The sequences of shRNAs (passenger strand, 5′ to 3′) were as follows: Ago1 (GATGAAGAATGCCAGCTACAA), Ago2 (ORF) (GGCACAGCCAGTAATCGAGTT), Ago2 (3′ untranslated region) (GCTACACTCAGACCAACAGAT), P4H-α(I)-1 (GTATTATTCGCTTCCATGATA), P4H-α(I)-2 (GGCTAACTAGTACAGCGACAA), P4H-β-1 (GCCGACAGGACGGTCATTGAT), P4H-β-2 (GATGAACTGTAATACGCAAAG) and control (GAACGTCATCAAGCTGATCTA). For si/miRISC activity assay, 2 μg of corresponding shRNAs were transfected into 5 × 105 cells of the GFP–reporter stable cell lines in six-well plates, the cells were transferred to 10-cm plate after 24 h and selected with 2 μg ml−1 of puromycin for 36 h. For RT–PCR, total RNAs were extracted using Trizol reagents (Invitrogen) from either GFP reporter cell lines, or U2OS cells transfected with corresponding shRNAs. Primers used for RT–PCR as follows: common forward primer for GFPL and GFP–let-7 (PE): GAACGGCATCAAGGTGAACTT, specific reverse primers for GFPL: TAGCGTAATCTGGAACATCGTATGGGT, and GFP–let-7 (PE): GACGACCTCGAGTGAGGTAGTAGGTTGTATA, GAPDH: forward GAAGGTGAAGGTCGGAGTC and reverse GAAGATGGTGATGGGATTTC, Ago2: forward GATCGGCAAGAAGAGATTA and reverse CGTCTTGCCGGCCAGGATGAC.
Immunoprecipitation and immunofluorescence
Three × Flag-Ago constructs and HA–P4H-α(I) as well as 3 × Flag–wtAgo2, Ago2(P700A) and KIAA1093-HA were co-transfected into 293ET cells. Cells were lysed 48 h after transfection in lysis buffer containing 300 mM NaCl, 50 mM TrisHCl, 5 mM EDTA, 0.1% NP-40, 0.5 mM NaF, 0.5 mM Na3VO4 and 1 × proteinase inhibitor (Roche). The lysates were incubated with anti-HA resins (Santa Cruz) for 3 h and washed five times with washing buffer (lysis buffer with 150 mM NaCl). The immunoprecipitated products were separated by SDS–polyacrylamide gel electrophoresis and blotted with anti-HA and anti-Flag antibodies, respectively. To assay P-body localization of Ago2, U2OS cells were first transfected with shP4H-α(I)-2 and -β-2 followed by puromycin selection for 36 h. The selected cells were seeded on coverslips for immunofluorescence. Subsequently, the cells were fixed with 3.7% paraformaldehyde in PBS for 30 min followed by permeabilization by 0.2% Triton X-100. The samples were then incubated with either anti-HA or Ago2 monoclonal antibodies at 1:500 dilutions simultaneously with anti-Dcp1a polyclonal antibody at 1:1,000 dilutions. Secondary antibodies against mouse immunoglobulin G conjugated with Alexa Fluor 488, rabbit IgG with Alexa 594 and Hoechst 33258 (Molecular Probes) were applied correspondingly after three washes with 0.1% Tween-20 in PBS.
Supplementary Material
Acknowledgements
We thank C. C. Mello, A. Grishok and P. A. Sharp for reading this manuscript and discussion. We thank E. Lehtimäki and R. Juntunen for in vitro hydroxylation assays, J. Lykke-Andersen and N. Kedersha for polyclonal anti-Dcp1a antibody and anti-Tia1 antibody, and the personnel of the Biocenter Oulu Transgenic Animal Core Facility and the University of Oulu Laboratory Animal Centre for technical assistance. We also thank the Kazusa DNA Research Institute (Japan) for providing the KIAA clones with numbers 1567, 1093, 1582 and 1460 and N. R. Wall for pOZ-FH-Ago1 plasmid. H.H.Q. was supported by a Canadian Institute of Health Research postdoctoral fellowship. This work was supported by grants from the Health Science Council of the Academy of Finland (202469) and the S. Jusélius foundation to J.M. and National Institutes of Health grants to J.P. (AG025688) and Y.S. (GM53874).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 3.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Kivirikko KI, Myllylaä R, Pihlajaniemi T. In: Post Translational Modification of Proteins. Harding JJ, Crabbe MJC, editors. CRC Press; 1992. pp. 1–51. [Google Scholar]
- 5.Cikaluk DE, et al. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol. Biol. Cell. 1999;10:3357–3372. doi: 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocnik EF, Chan BM, Pickering JG. Evidence for a role of collagen synthesis in arterial smooth muscle cell migration. J. Clin. Invest. 1998;101:1889–1898. doi: 10.1172/JCI1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.John DC, et al. Expression of an engineered form of recombinant procollagen in mouse milk. Nature Biotechnol. 1999;17:385–389. doi: 10.1038/7945. [DOI] [PubMed] [Google Scholar]
- 8.Holster T, et al. Loss of assembly of the main basement membrane collagen, type IV, but not fibril-forming collagens and embryonic death in collagen prolyl 4-hydroxylase I null mice. J. Biol. Chem. 2007;282:2512–2519. doi: 10.1074/jbc.M606608200. [DOI] [PubMed] [Google Scholar]
- 9.Rivas FV, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 10.Leung AKLCJ, Sharp PA. Quatitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl Acad. Sci. USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nature Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nature Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 15.Kaelin WG., Jr. The von Hippel-Lindau protein, HIF hydroxylation, and oxygen sensing. Biochem. Biophys. Res. Commun. 2005;338:627–638. doi: 10.1016/j.bbrc.2005.08.165. [DOI] [PubMed] [Google Scholar]
- 16.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, et al. Human prolyl-4-hydroxylase α(I) transcription is mediated by upstream stimulatory factors. J. Biol. Chem. 2006;281:10849–10855. doi: 10.1074/jbc.M511237200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofbauer KH, et al. Oxygen tension regulates the expression of a group of procollagen hydroxylases. Eur. J. Biochem. 2003;270:4515–4522. doi: 10.1046/j.1432-1033.2003.03846.x. [DOI] [PubMed] [Google Scholar]
- 19.Fahling M, et al. Translational control of collagen prolyl 4-hydroxylase-α(I) gene expression under hypoxia. J. Biol. Chem. 2006;281:26089–26101. doi: 10.1074/jbc.M604939200. [DOI] [PubMed] [Google Scholar]
- 20.Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- 21.Peng J, Gygi SP. Proteomics: the move to mixtures. J. Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 22.Hieta R, et al. The peptide-substrate-binding domain of human collagen prolyl 4-hydroxylases. Backbone assignments, secondary structure, and binding of proline-rich peptides. J. Biol. Chem. 2003;278:34966–34974. doi: 10.1074/jbc.M303624200. [DOI] [PubMed] [Google Scholar]
- 23.Vuori K, et al. Characterization of the human prolyl 4-hydroxylase tetramer and its multifunctional protein disulfide-isomerase subunit synthesized in a baculovirus expression system. Proc. Natl. Acad. Sci. USA. 1992;89:7467–7470. doi: 10.1073/pnas.89.16.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirsilä M, et al. Effect of desferrioxamine and metals on the hydroxylases in the oxygen sensing pathway. FASEB J. 2005;19:1308–1310. doi: 10.1096/fj.04-3399fje. [DOI] [PubMed] [Google Scholar]
- 25.Berg RA. Determination of 3- and 4-hydroxyproline. Methods Enzymol. 1982;82:372–398. doi: 10.1016/0076-6879(82)82074-0. [DOI] [PubMed] [Google Scholar]
- 26.Liu G, Xia T, Chen X. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J. Biol. Chem. 2003;278:17557–17565. doi: 10.1074/jbc.M210696200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.