Abstract
Children with Down syndrome (DS) show a spectrum of clinical anomalies, including cognitive impairment, cardiac malformations, and craniofacial dysmorphy. Moreover, hematologists have also noted that these children commonly show macrocytosis, abnormal platelet counts, and an increased incidence of transient myeloproliferative disease (TMD), acute megakaryocytic leukemia (AMKL), and acute lymphoid leukemia (ALL). In this review, we summarize the clinical manifestations and characteristics of these leukemias, provide an update on therapeutic strategies and patient outcomes, and discuss the most recent advances in DS-leukemia research. With the increased knowledge of the way in which trisomy 21 affects hematopoiesis and the specific genetic mutations that are found in DS-associated leukemias, we are well on our way toward designing improved strategies for treating both myeloid and lymphoid malignancies in this high-risk population.
Introduction
Down syndrome (DS), or constitutional trisomy 21, is the most common human aneuploidy, with an incidence of 1 in 700 births. Nearly 80 different clinical phenotypes have been identified in people with DS, including cognitive impairment, craniofacial dysmorphy, gastrointestinal tract abnormalities, congenital heart defects, endocrine abnormalities, neuropathology leading to dementia, and immunologic defects. With respect to the hematopoietic system, children with DS frequently show macrocytosis, abnormalities in platelet counts, and an increased prevalence of leukemia.1,2 The incidence of acute lymphoblastic leukemia (ALL; the most common leukemia in childhood) in children with DS is approximately 20-fold higher than in the general population, while the incidence of acute megakaryoblastic leukemia (AMKL) is 500-fold higher.2 Furthermore, it has been estimated that between 4% and 10% of infants with DS are born with transient myeloproliferative disease (TMD), a clonal disease that is characterized by immature megakaryoblasts in the fetal liver and peripheral blood.3,4 Although TMD spontaneously disappears in most cases, it is regarded as a preleukemic syndrome; approximately 20% of children diagnosed with TMD develop DS-AMKL within 4 years. The natural history of leukemia in children with DS suggests that trisomy 21 directly and functionally contributes to the malignant transformation of hematopoietic cells. It is important to note, however, that DS is not a classic genomic instability syndrome, as the overall risk of developing cancer, in particular solid tumors, is lower in these people.5 In line with these data, experiments with a mouse model of DS showed that trisomy for orthologs of about half of the genes on chromosome 21 led to a significant reduction in the number of adenomatous polyposis coli (multiple intestinal neoplasia [APC(min)]–mediated intestinal tumors.6 To better understand the impact of trisomy 21 on hematopoiesis, studies have been undertaken with human fetal liver cells as well as animal and cell-line models to determine the causative relationship between gene dosage imbalance and phenotypes of DS-associated leukemia. Before highlighting these research advances, we will review the manifestations of hematologic malignancies in people with DS.
Manifestations of leukemia in DS
TMD
The true frequency of TMD is unknown because it is quite likely that a significant proportion of these patients are not routinely diagnosed. As GATA1 mutations are uniformly associated with TMD7–10 and occur in utero,11 ongoing studies in Europe and North America combined screening for GATA1 mutations, and examination of neonatal blood smears will present a more precise picture of the true incidence of TMD. In one such recent study, Pine and colleagues examined DNA from Guthrie cards of 590 infants with DS and reported that GATA1 mutations (which result in expression of the GATA1s isoform; see “Mechanisms”) were detected in 3.8% of the infants.4 Moreover, they found that Hispanic newborns were 2.6 times more likely to have a mutant GATA1 gene than non-Hispanics. Thus, it is likely that the frequency of TMD is not higher than 5% of DS newborns.
TMD occasionally presents as hydrops fetalis, but is usually diagnosed during the first few weeks after birth. The neonate may be asymptomatic other than elevated blood count with hepatomegaly. Less commonly, infants with TMD may display jaundice and bleeding diatheses, respiratory distress coupled with ascites, pleural effusion, signs of heart failure, and skin infiltrates. Within the liver, there is megakaryocytic infiltration and liver fibrosis, likely caused by excess cytokines secreted from the megakaryoblasts. The full clinical syndrome may develop only at the second or third week of life. Laboratory tests are significant for either thrombocytosis or thrombocytopenia accompanied by elevated white blood cell count (WBC) with excess of blasts. The blood smear may show nucleated red cells, giant platelets and megakaryocyte fragments, and, most significantly, typical deeply basophilic blasts with blebs characteristic to megakaryocytic blasts. Flow cytometry reveals that the blasts are positive for CD34, CD33, CD41, CD61, glycophorin A, and quite often CD7 and CD36.12,13 The differential diagnosis includes leukoerythroblastic reaction associated with prematurity, sepsis, or asphyxia. However, the blasts of TMD usually persist for several weeks, and mutations in GATA1 are invariably found. When a typical TMD with GATA1 mutation is detected in an apparently normal baby, cytogenetic analysis to rule out mosaic DS is indicated.
Three large studies from the United States, Japan, and Europe have recently reported the natural course of TMD in 264 infants with DS.13–15 These studies confirmed the transient course of this disease that usually resolved spontaneously within the first 3 months of life. However, these studies revealed that the disease is not benign, as early death was reported in 15% to 20%. Because neither study prospectively screened all infants with DS for TMD, but rather included only those who were diagnosed with TMD, this relatively high mortality rate pertains to clinically evident TMD. Poor prognostic factors included high WBC count, prematurity, severe liver failure manifested by increasing jaundice and bleeding diatheses, and failure of spontaneous remission within the first 3 months (Table 1). Such patients benefit from low-dose cytarabine (ARA-C; 1 mg/kg per day for 7 days).14
Table 1.
Poor prognostic parameters that may necessitate treatment
| Parameters |
|---|
| Clinical |
| Prematurity |
| Ascites |
| Bleeding |
| Laboratory |
| White blood cells > 100 × 109/L |
| Severe coagulopathy |
| Progressive liver failure |
| No complete remission by 3 mo |
DS-AMKL
DS-AMKL is prevalent in children with DS aged 4 years or younger, and is most commonly seen by the age of 2 years. In contrast, myeloid leukemias in people with DS aged 4 years or older are usually negative for GATA1 mutations, and their prognosis does not differ from AML in patients without DS.16 It is unclear whether all DS-AMKLs are preceded by TMD, because many TMD cases are not diagnosed. A recent study suggests that the prognosis of AMKLs preceded by TMD is better than de novo DS-AMKL.14 DS-AMKL often presents with a myelodysplastic phase with thrombocytopenia. Like other AMKLs, bone marrow (BM) aspiration is often dry, and fibrosis is detected in BM biopsy. Unlike TMD, there is no clinical involvement of the liver. The immunophenotype of the blasts is generally similar to TMD, except that the percentage of CD34+ cells may be lower.12
DS-ALL
Patients with DS are also at a markedly increased risk for childhood ALL. Indeed, DS-ALL is more frequent than DS-AMKL.5 In most published multi-institutional ALL protocols, DS-ALL comprises approximately 1% to 3% of total patients.17–23
The age distribution and the immunophenotype are similar to “common ALL,” the B-cell precursor leukemia that occurs most commonly in young preschool-aged children.17,18,21,24 Trisomy or tetrasomy of human chromosome 21 (Hsa21) is the most common acquired somatic chromosomal abnormality in common ALL. Hence, it is tempting to speculate that constitutional and somatic trisomy 21 may facilitate leukemogenesis in a similar fashion, and therefore the study of DS-ALL may have direct implications for sporadic childhood ALL.
Clinically, DS-ALL is indistinguishable from other childhood common ALLs.17,18,21,25 The peak age is about 5 years, slightly older than the peak age of sporadic ALLs, possibly related to the absence of DS-ALL below the age of 1 year. The immunophenotype is typical for B-cell precursor ALL, namely positive for CD10, CD19, and CD79a. Unlike sporadic common ALL, many fall by National Cancer Institute (NCI) clinical criteria into a higher risk group.26
Outcomes
The prognosis of DS-AMKL is excellent on contemporary AML protocols, with an approximately 80% cure rate.27–29 DS-AMKL blasts have unusual sensitivity to multiple chemotherapeutic drugs.30 They are especially sensitive to cytarabine (ARA-C), possibly to the effect of the GATA1 mutations and trisomy 21 on the levels of cytarabine-metabolizing enzymes.31 Indeed, many patients may respond favorably to a simple regimen including low-dose cytarabine,32 although this is currently not the standard of care. Because toxic deaths, infections, and cardiac toxicity, not relapses, have been the major problems in treating DS-AMKL, new, less-intensive protocols have been initiated in the United States and Europe (Creutzig et al27; International Cooperative Pediatric AML Study Group Myeloid Leukemia DS 2006 [European Clinical Trials Database (EUDRACT) no. 2007-006219-22]; Children's Oncology Group: The Treatment of Down Syndrome Children with Acute Myeloid Leukemia [AML] and Myelodysplastic Syndrome [MDS] Under the Age of 4 Years [COG-AAML0431]; Low Dose Cytarabine in Treating Infants with Down Syndrome and Transient Myeloproliferative Disorder [COG-AAML0532]).
An interesting clinical question, currently under study in prospective clinical trials, is whether treatment of TMD by low-dose cytarabine could prevent DS-AMKL. A related question is whether treatment of clinically silent disease, identified by molecular detection of GATA1 mutations in patients who recovered from TMD, can prevent the future development of DS-AMKL.
Unlike DS-AMKL, which is uniquely sensitive to chemotherapy, the prognosis of DS-ALL is less favorable.17,18,21,24,25 Patients with DS may be especially sensitive to the toxic effects of methotrexate due to the excess activity of the folate transporter encoded by a gene on chromosome 21.33 Severe toxicity is also observed with anthracyclines and with the marked immunosuppressive effect of ALL therapy. Toxicity can be manifested by increased mucositis, infections, and death during intensive periods of chemotherapy. The reason why patients with DS-ALL do not have a more favorable outcome than patients without DS, in light of the increased sensitivity to methotrexate, is unclear. One possibility is that the gain of increased sensitivity of leukemic cells to chemotherapy is offset by an increased toxicity of normal cells. In DS-AMKL, the increased sensitivity to ARA-C is a consequence of GATA1 mutations,31 which are restricted to the leukemic population. In contrast, the increased sensitivity to chemotherapeutics in ALL is found in all cells with trisomy 21.
Importantly, marked reduction of chemotherapy may be a mistake in DS-ALL. A recent Children's Cancer Group study demonstrated a surprisingly good survival in children with DS-ALL treated by risk-adjusted intensive chemotherapy protocols. Event-free (56% vs 74%; P < .001) and disease-free (55% vs 73%; P < .001) survival at 10 years was significantly lower in the standard-risk DS-ALL population compared with ALL in the non-DS population, but not in the high-risk DS-ALL population (event-free survival, 62% vs 59%; P = .9; disease-free survival, 64% vs 59%; P = .9), and these differences persisted regardless of treatment era (early era [1983-1989] vs recent era [1989-1995]). Multivariate analysis revealed that presence of DS demonstrated an independent significant adverse prognostic effect for the standard-risk population, but not for the high-risk patients.21 These results suggest that intensification of therapy for patients with DS-ALL is needed to maintain outcome comparable with those of ALL in patients without DS. Similarly, a recent survey of 8 children with DS-ALL who underwent bone marrow transplantation reported that relapse and not treatment-related toxicity were the major causes for treatment failure. Indeed, the only surviving patients were those that were treated by myeloablative chemotherapy.34
To better understand the response to therapy, toxicity, and treatment, an international retrospective survey of the fate of children with DS-ALL treated on major cooperative ALL protocols has been initiated. Meanwhile, clinicians should be careful in dose reduction of patients with DS-ALL. Prospective clinical trials to incorporate potentially less toxic drugs, such as JAK2 inhibitors (see “Evolution of DS-ALL”), should be pursued by international cooperative childhood cancer treatment groups for this specific class of leukemia-prone children. Although JAK inhibitors have not yet been tested in DS-associated leukemias, animal studies are underway and future clinical trails are reasonable given the prevalence of JAK2 mutations and the promising clinical data from trials of JAK2 inhibitors in patients with myelofibrosis.
Mechanisms
DS-associated leukemias represent natural genetic models of “multistep pathogenesis” that are dependent on both time (initiating in fetuses and infants) and spatial (likely originating in fetal liver progenitors). In this section, the different experimental models used to understand each stage of this multistep leukemogenesis will be presented and discussed. In addition, we will highlight recent studies that shed light on the genetic basis of these diseases.
Oncogenic cooperation in DS-associated leukemia
Beyond trisomy 21 (discussed under “Impact of trisomy 21 on hematopoiesis”), what are the other genetic events that fuel the development of ALL or AMKL in children with DS? Recent studies have identified several cooperating mutations that are associated with evolution of DS-ALL or DS-AMKL. The most significant recent advances are the discoveries of mutations in JAK2 and JAK3 in DS-ALL and DS-AMKL, respectively.
Evolution of DS-AMKL.
The most significant event that shed light on the pathogenesis of DS-AMKL was the discovery of GATA1 mutations in TMD and DS-AMKL (reviewed in Vyas and Crispino35). GATA1 mutations in these disorders consist of short deletions, insertions, and point mutations clustered within exon 2 that uniformly lead to a block in expression of the full-length protein, but allow for expression of a shorter isoform, named GATA1s. Underscoring the requirement for trisomy 21 in DS-AMKL, people with germline GATA1 mutations analogous to those seen in TMD and DS-AMKL have no history of developing leukemia.36 Based on this observation and extensive work with GATA1 mutants by many research groups, we conclude that TMD and DS-AMKL require both trisomy 21 and a GATA1 mutation. However, it remains unclear whether these 2 alterations are sufficient to fuel TMD. Moreover, the specific secondary mutations that foster the evolution of TMD to DS-AMKL are still largely unknown, although a few candidate cooperating mutations have been identified. These include mutations in TP53 and FLT3 (Table 2).37–39
Table 2.
Summary of the genetic abnormalities identified in Down syndrome–associated leukemia
| Type of leukemia/genetic abnormalities | Reported cases | References |
|---|---|---|
| DS-ALL | ||
| Chromosomal | ||
| t(12;21)(p13;q22) | 17/322 | 24,25,44–46,49,99,100 |
| t(8;14)(q11;q32) | 4/209 | 44–46,49 |
| del(9p) | 15/186 | 44,46,49 |
| add(X) | 80/238 | 2,44–46,49 |
| Intragenic | ||
| JAK2 (ΔIREED, R683S, R683G, R683K) | 28/130 | 46,48,49 |
| PTPN11 (E76K) | 1/10 | 46,47 |
| RAS (G12D) | 1/10 | 46,47 |
| TMD | ||
| Intragenic | ||
| GATA1 | 73/75 | 7–10,41,43,102 |
| JAK3 | 5/40 | 37,41–43,102,104 |
| TP53 | 1/13 | 38,39,41 |
| DS-AMKL | ||
| Chromosomal | ||
| Trisomy 8 | 6/34 | 45,82,103 |
| Monosomy 7/complete or partial del(7q) | 7/34 | 45,82,103 |
| Intragenic | ||
| GATA1 | 58/65 | 8–10,41,101,102 |
| JAK3 | 7/53 | 37,40–42,102,104 |
| FLT3 | 2/35 | 37,40,41,104 |
| TP53 | 6/28 | 38,39,41 |
| JAK2 | 2/32 | 37,102,104 |
DS indicates Down syndrome; ALL, acute lymphoid leukemia; TMD, transient myeloproliferative disease; and AMKL, acute megakaryocytic leukemia.
The recent identification of activating mutations in tyrosine kinase genes in TMD and DS-AMKL specimens has provided exciting new insights into the evolution of DS-AMKL. First identified in 2 AMKL specimens and the CMK cell line,40 JAK3 mutations have been detected in a small but significant fraction of DS-leukemia samples (Table 2).37,41–43 Of these mutations, 7 are considered to be gain of function, as they lead to a cytokine-independent growth of Ba/F3 cells in vitro.37,40,43 Furthermore, JAK3 A572V, which was identified in the CMK cell line, induces a lethal biphenotypic hematopoietic disorder in recipient mice with features of megakaryoblastic leukemia.40 Three other JAK3 mutants have been proposed to be loss of function.42 Additional experiments are necessary to determine how these different JAK3 variants affect hematopoiesis and megakaryocyte development.
Evolution of DS-ALL.
The most common cytogenetic abnormality in DS-ALL is an extra chromosome X, observed in between 25% and 50% of patients17,44 (Table 2). The additional copy of chromosome X is usually present in hyperdiploid sporadic ALL, displaying additional copies of chromosomes 21, X, and 6, and occasionally chromosomes 10, 14, 17, and 18. However, the combination of trisomy 21 and extra chromosome X as a single cytogenetic abnormality seems typical of DS-ALL, and suggests a yet-unknown collaborating event between gene(s) on chromosomes 21 and X. Other cytogenetic lesions seen in ALL include t(8;14) and del(9p), which are significantly more frequent in DS-ALL than in their non-DS counterparts.44–46 Apart from chromosomal changes, activating mutations in PTPN11 and NRAS have also been found in a few patients with DS-ALL.47 Both of these mutations were found in specimens with a simple karyotype consisting of only constitutional trisomy 21.
Perhaps the most exciting recent breakthrough in understanding the pathogenesis of ALL in DS was the discovery that JAK2 mutations are present in approximately 20% of patients with DS-ALL.46,48,49 DS-ALL–associated mutations specifically affect Arg683 and surrounding residues within the JH2 pseudokinase domain. Interestingly, these mutations are distinct from those seen in people with myeloproliferative disorder (MPD) and reside in an exposed conserved region separate from V617F, leading to the hypothesis that the location of the JAK2 mutations might affect different downstream signaling events in a cell-dependent context. Of note, the R683G mutation was found in one patient with non-DS ALL who harbored an isochromosome 21q, underscoring the link between +21 and these novel mutations in JAK2.49 Moreover, all 3 studies found that the JAK2 mutations were not present in remission samples, confirming that the mutations are secondary events acquired by trisomic progenitors. Another important observation was that children with a JAK2 mutation were significantly younger (mean age, 4.5 vs 8.6 years; P < .001) at diagnosis.49 Similar confirmatory results have been obtained in other groups in the United States and in Europe,46,50 although the younger age has not been confirmed in the US study.
With respect to function, DS-ALL–associated JAK2 mutants immortalized primary mouse hematopoietic progenitors and caused constitutive JAK/STAT activation and cytokine-independent growth of Ba/F3 cells that was sensitive to pharmacologic inhibition of JAK/STAT signaling.46,48,49 Of note, although these ALL-associated mutations have not been found in people with MPD, they appear to affect myeloid progenitors in the same way as the V617F mutation. Expression of JAK2 R683 mutants enhanced the replating activity of primary BM cells in myeloid colony-forming assays.49 Hence, these results suggest that JAK2 mutations are a late acquired genetic event arising in already committed B-lymphoid progenitors conferring a proliferative advantage to these cells. Together, these results not only provide the first specific molecular lesions of DS-ALL, but also suggest that these leukemias are candidates for therapy with novel JAK2 inhibitors.
Impact of trisomy 21 on hematopoiesis
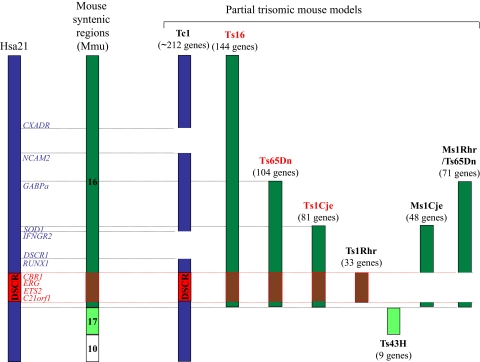
Hsa21 is approximately 33.7 Mb and contains 364 known genes, 31 antisense transcripts, and 5 different miRNAs (miR-99a, let-7c, miR-155, miR-125b-2, and miR-802). Genotype-phenotype correlations of partial trisomy in children suspected of having DS has led to the identification of the “Down Syndrome Critical Region” (DSCR), which lies between the DS21S17 marker and MX1 (Figure 1).51–53 Characterization of this region, centered on the 21q22 band, has narrowed the list of genes potentially implicated in the clinical phenotypes. However, the functional correlation between genes within this particular region and the majority of the clinical phenotypes is still controversial.54 For example, this region has been shown to be necessary but not sufficient for the craniofacial characteristics observed in mouse models of DS, underscoring the complexity of gene interactions in DS.55 Although no specific genes within this region have been unequivocally linked to the increased incidence of leukemia in DS, a few genes are strong candidates, including ERG, ETS2, and RUNX1 (discussed under “Candidate leukemia oncogenes encoded by chromosome 21”).
Figure 1.
Diagram of Hsa21 and the regions of trisomy in the various mouse models of DS. Human chromosome 21 and specific genes that may contribute to the development of leukemia in children with DS are shown on the left. Syntenic regions on Mmu16, and the mouse models in which they exist in 3 copies, are depicted on the right. Note that the Tc1 mouse is trisomic for the majority of human chromosome 21. Mouse models outlined in red are discussed under “Utility of animal models of DS.”
Several observations argue for a functional contribution of the trisomy 21 in hematologic malignancies. These include the high incidence of leukemia in patients with DS, the fact that TMD and DS-AMKL blasts always harbor trisomy 21 (even in children without DS), and that acquired trisomy or tetrasomy of chromosome 21 is frequently observed in blasts of different types of leukemia, including hyperdiploid ALL and de novo AML (reviewed in Vyas and Crispino35). In addition to these observations, recent data obtained from studies with fetal liver specimens collected from human embryos with DS and from research with animal models of DS further argue that trisomy 21 directly contributes to the leukemic process.
Effects of trisomy 21 on fetal hematopoiesis.
Given that TMD has its origins in a bipotential fetal liver progenitor and is restricted to people with DS (or to rare cases of acquired trisomy 21), it is likely that trisomy 21 directly affects the development of hematopoietic cells during gestation. Previous studies have shown that GATA1 mutations can develop in embryos as early as 21 weeks of gestation.11 To define the cellular context in which GATA1 mutations occur, 2 groups have recently studied hematopoiesis in trisomy 21 human fetal livers (FLs).56,57 They found that although trisomy 21 did not alter the proportion of CD34+CD38− cells, trisomy 21 FLs showed a 2- to 3-fold increase of megakaryocyte erythroid progenitors (MEPs), which appeared to increase over time (35% at 16 weeks to 65% at 18 weeks). Moreover, trisomy 21 FL gave rise to a significant increase in the number of burst-forming units–erythroid (BFU-E) and colony-forming units–megakaryocyte (CFU-Mk) colonies in methylcellulose assays and CFU–granulocyte, erythroid, macrophage, megakaryocyte (GEMM) colonies obtained from trisomic CD34+ progenitors have a self-renewal advantage as evidenced by their enhanced replating activity. In the process of analyzing the differentiation potential of trisomic progenitors, both studies observed a preferential expansion of megakaryocytes in liquid cultures of trisomic FLs. An expansion of the MEP population in trisomy 21 fetuses was also validated in vivo by transplantation assays: mice that received trisomy 21 FL progenitors showed an enhanced expression of the human isoforms of glycophorin A and CD41 compared with those that received euploid FL progenitors.56 These results reveal that there is a cell-autonomous enhanced proliferation of erythroid and megakaryoblastic cell types from trisomy 21 FLs. Notably, these differences in composition and growth of these cells are independent of GATA1 mutations, as none were detected in the fetuses under study.
Together, these studies strongly implicate trisomy 21 as a causative agent in aberrant megakaryopoiesis. The functional perturbations induced by trisomy 21 likely create a cellular context that is highly susceptible to additional transforming events such as mutagenesis of GATA1 in TMD. This FL cell-based assay provides a powerful way to identify the specific Hsa21 genes that participate in TMD. Preliminary quantitative reverse transcription–polymerase chain reaction (qRT-PCR) studies have shown that there are no significant differences in expression of ERG, ETS2, RUNX1, and SON, top-ranked candidate leukemia oncogenes, in trisomic versus euploid FLs.56,57 However, functional studies, such as knockdown of one or more of these candidates genes in FL progenitors followed by colony assays and transplantation experiments, are necessary to determine the requirements for these genes in leukemia. In addition, in vitro and in vivo analyses of trisomic FLs engineered to express GATA1s will also provide important new insights into how trisomy 21 affects hematopoiesis and drives leukemia.
Utility of animal models of DS.
Although research with human tissues has provided critical new insights into the role of trisomy 21 in hematopoiesis, animal models continue to provide a useful way to study the genetics of complex diseases. In mice, the orthologs of Hsa21 genes are spread among 3 chromosomes with 23.2 Mb synteny (representing 154 conserved genes) on mouse chromosome 16 (Mmu16), 1.1 Mb (with 9-15 genes) on Mmu17, and 2.3 Mb (with 24 genes) on Mmu10.58–61 Several mouse models of DS, each containing partial trisomy for various regions of Mmu16, have been used to study the effect of trisomy on development and organogenesis (Figure 1; Table 3).55,62,63
Table 3.
Hematopoietic phenotypes of partially trisomic mouse models
| Strain/fetal hematopoiesis | Adult hematopoiesis | Transplantability | References |
|---|---|---|---|
| Ts16 | |||
| Thymic hypoplasia, delayed maturation of T lymphocytes, decreased B lymphocytes, and stem cells deficiency | Not studied because Ts16 embryos die in utero | FL donor cells: decreased repopulating ability; anemia | 68,105 |
| Ts65Dn | |||
| No significant differences in in vitro colony formation or number of megakaryocytes compared with euploid littermates | Macrocytosis, decreased red cell number, thrombocytosis, BM fibrosis, extramedullary hematopoiesis, increased CD34+ cells (increased apoptosis in vitro), increased GMP, and less quiescent LSK cells | BM donor cells: weaker long-term repopulating ability; macrocytosis | 66,67 |
| Ts1Cje | |||
| Fewer and smaller colonies. Normal number of HSCs, but impaired CFU-S | Macrocytosis, decreased red cell number and late erythroid differentiated stages, increased GMP | FL donor cells: no competitive reconstitution; BM donor cells: macrocytosis and anemia | 71 |
FL indicates fetal liver; BM, bone marrow; GMP, granulocyte macrophage progenitor; LSK, Lin−c-Kit+Sca1+; HSC, hematopoietic stem cell; and CFU-S, colony-forming unit–spleen.
The most widely used mouse model of partial trisomy 21 is the Ts65Dn strain, which harbors a constitutional translocation comprising the distal end of Mmu16 attached to the centromeric end of Mmu17.64 These mice are trisomic for 15.6 Mb and contain orthologs of 104 genes of Hsa21 (based on Gardiner et al59). Among the DS phenotypes, the animals show cognitive impairment, craniofacial dysmorphy, and cardiac abnormalities.64,65 With respect to the hematopoietic system, one report demonstrated that Ts65Dn CD34+ cells have a drastically reduced growth capacity in vitro compared with those of their littermates.66 More recently, we have shown that trisomy for 104 orthologs of Hsa21 in the Ts65Dn strain has profound effects on hematopoiesis.67 Ts65Dn mice develop a MPD characterized by thrombocytosis, megakaryocyte hyperplasia, dysplastic megakaryocytic morphology, and myelofibrosis, a feature commonly observed in human megakaryocytic leukemia. Flow cytometry and colony assays revealed the presence of extramedullary hematopoiesis with striking increases of CD41+, c-Kit+, and Ter119+ populations in the spleen. Trisomy was also found to affect the hematopoietic progenitor and stem cell compartments, as evidenced by a decreased proportion of Lin−c-Kit+Sca1− cells and increased proportion of Lin−c-Kit+Sca1+ (LSK) cells in trisomic BM, respectively. Within the myeloid progenitor fraction, Ts65Dn mice displayed an increase in granulocyte macrophage progenitors (GMPs), with a concomitant decline in MEPs. Furthermore, the LSK cells are less quiescent than those of their littermates. Transplantation studies revealed that although Ts65Dn mice have an increase in hematopoietic stem cells, only 2 of 5 recipients of Ts65Dn BM survived at 6 months after transplantation.67 Several hypotheses may explain this loss of repopulating ability of trisomic hematopoietic stem cells (HSCs), such as a decreased number of long-term (LT)–HSCs or loss of reconstitution functionality of the HSCs. Interestingly, the less-effective reconstitution of trisomic stem cells as well as the transplantable anemia were also observed in BM reconstitution experiments using FLs from mice with trisomy 16, which are trisomic for 144 orthologs of Hsa21 genes (Figure 1; Table 3).68,69
The Ts1Cje mouse model of DS was generated during a gene-targeting event for sod1 that resulted in translocation of the distal end of Mmu16 to Mmu12.70 These mice are trisomic for nearly two-thirds of the Ts65Dn region, with 10.3 Mb containing 81 orthologous genes (Figure 1; Table 3). Similar to the phenotype of Ts65Dn mice, Ts1Cje mice display macrocytosis and anemia, both of which are transplantable.71 Ts1Cje mice also show an increased proportion of GMPs relative to other myeloid progenitors. CFU–spleen (S) assays and competitive BM and FL reconstitution experiments further revealed that there was an impaired reconstitution ability of the Ts1Cje stem cells.
Although there are similarities in the phenotypes of the Ts65Dn and Ts1Cje models, there are striking differences. Most notably, thrombocytosis, extramedullary hematopoiesis, and myelofibrosis are prevalent in the Ts65Dn strain, but not observed in Ts1Cje mice. The most logical explanation for this discrepancy is that the gene (or genes) that elicits these hematopoietic phenotypes is among the 23 genes that are trisomic in Ts65Dn, but disomic in Ts1Cje mice. It is also possible that differential gene expression outside the trisomic region could alter the megakaryocytic phenotype. This latter explanation is especially important to consider because these 2 strains are in different mixed genetic backgrounds.
Taken together, these reports, which show that partial trisomy for orthologs of Hsa21 leads to a plethora of hematologic phenotypes in mice, strongly suggest that gene dosage directly affects hematopoiesis, but several important questions remain unresolved. For example, what are the effects of trisomy on murine FL progenitors, and why do the animals fail to develop TMD and DS-AMKL? Ts1Cje FL cells were found give rise to reduced numbers of myeloid colonies in vitro and to be functionally impaired in their ability to regenerate the hematopoietic system of recipient mice.71 In contrast, Ts65Dn FLs did not show dramatic differences in in vitro colony formation or megakaryocyte numbers compared with euploid littermates (G. Kirsammer and J.D.C., unpublished observations, June 2005). The absence of a similar phenotype between these 2 animal models and human trisomy 21 fetuses might be the consequence of the absence of key Hsa21 gene orthologs from the trisomic regions. Differences in gene dosage of microRNAs (miRNAs) encoded by Hsa21 but not within the trisomic Mmu16 region might also contribute to the lack of a profound fetal DS phenotype. For example, miR-125b-2, which has been implicated in DS-AMKL,72 (see “miRNAs encoded by Hsa21”) is not trisomic in either Ts65Dn or Ts1Cje mice.
Role of specific chromosome 21 genes in DS-associated leukemia
A key unanswered question is which genes are encoded by Hsa21 that promote aberrant hematopoiesis in humans with DS. In the past several years, multiple groups have pursued answers to this question by examining microarray data, gene sequencing, and functional studies.
Microarrays analyses and gene expression data.
Two microarray studies comparing DS-AMKL versus non-DS AMKL have recently been reported.73,74 These studies share 76 genes that discriminate between DS and non-DS AMKL. Among them, genes encoding erythroid markers, such as glycophorin A and CD36, appear to be significantly overexpressed in DS-AMKL, as confirmed by immunophenotypic analysis of blasts.12
Analysis of the gene expression data also revealed that there is an overall increase in expression of chromosome 21 genes in DS-AMKL, relative to non-DS AMKL.74 Indeed, 47 Hsa21 genes, including BACH1, SON, C21orf66, and GABPA, were found to contribute to the enrichment by gene set enrichment analysis (GSEA), but the distinction between the 2 types of AMKL was not driven by differences in expression of chromosome 21 genes. Similarly, Ge et al found that only 7 of 551 distinctive genes were differentially expressed and encoded by chromosome 21.73 This list included BACH1, CXADR, SYNJ1, and STCH. Of note, BACH1 and CXADR are the only 2 genes commonly overexpressed in DS-AMKL in both studies compared with non-DS AMKL. Differences in the 2 datasets might be explained by different thresholds for considering a given change in gene expression to be significant, or to differences in purity of the samples.
To our knowledge, no microarray studies directly comparing DS-ALL versus non-DS ALL have been reported. Because DS-ALL is very close clinically and cytogenetically to “common” ALL, it is possible that very few differences might exist. Nevertheless, it is interesting to note that global microarrays performed on B-ALL led to the identification of several chromosome 21 genes encoded in the hyperdiploid subgroup, which contains multiple acquired copies of chromosome 21, compared with BCR-ABL+, TEL-AML1+, E2A-PBX1+, or MLL+ B-ALL subtypes.75 Among these genes, SOD1, MX1, HMG14, and SUMO3 were designated as discriminating genes of the B-ALL hyperdiploid subtypes, and the STCH gene has been identified as predictors of relapse from hyperdiploid B-ALL samples.
Candidate leukemia oncogenes encoded by chromosome 21.
Of the genes on chromosome 21, several are compelling candidate leukemia oncogenes. Of these, 4 such candidates are AML1/RUNX1, which encodes the heterodimeric partner of CBFβ, and the 3 ETS transcription factors encoded by genes localized on chromosome 21 (ERG, ETS2, and GABPA). Several lines of evidence suggest that RUNX1 is involved in the megakaryocytic leukemia of DS, including the observation that mutations in the DNA-binding domain of RUNX1 have been identified in 5% of sporadic leukemia and in myeloid malignancies with acquired trisomy 21,76,77 and that RUNX1 is located on Hsa21 and cooperates with GATA1 during megakaryocyte differentiation.78 However, given that loss-of-function mutations in RUNX1 are associated with leukemia, it is unclear how 3 copies would promote tumorigenesis in DS.79 One possibility is that cells with trisomy 21 may express different relative levels of RUNX1 isoforms, which in turn may affect tumor development.80 Indeed, differential expression of RUNX1 isoforms was observed in human AMKL samples, although the total level of RUNX1 expression was lower in DS-AMKL compared with non-DS AMKL.74 In contrast, trisomy for Runx1 was found to not be required for the development of MPD in Ts65Dn mice.67 Moreover, the hematopoietic phenotype of Ts16 fetuses was not correlated with an increased expression of Runx1 or an altered ratio of its isoforms.69
Other potential candidates include the ETS transcription factors ERG, ETS2, and GABPα, which are all expressed and functionally implicated in megakaryocytes differentiation.
Dysregulation of the ERG proto-oncogene has been observed in many types of cancer. In hematologic malignancies, rare cases of AML harbored a chromosomal translocation t(16;21) leading to the expression of FUS(TLS)-ERG fusion gene,81,82 and overexpression of ERG has been found in AML samples with normal83 or complex acquired karyotypes involving Hsa21.84 Overexpression of ERG in K562 cells was found to induce a switch in differentiation toward the megakaryocytic lineage and to increase expression of the early megakaryocytic markers, including CD41 and CD61.85 Interestingly, homozygous mutant Erg mice died in utero, due to a defect in definitive hematopoiesis, while heterozygous Erg mutant mice showed thrombocytopenia without abnormalities in the number of BM megakaryocytes, confirming a role of ERG in late stages of megakaryopoiesis.86 Unpublished observations from our laboratories argue for a role of ERG in hematopoietic disorders. We observed that overexpression of ERG in mouse primary FL progenitors cells leads to a dramatic increase in megakaryocytic differentiation in liquid culture, an enhanced replating ability of wild-type and Gata1-deficient hematopoietic progenitors, and AMKL in transplant recipients (M. Stankiewicz and J.D.C., unpublished results, July 2007; S.I., unpublished observations, July 2007). These studies strongly implicate increased expression of ERG in megakaryocytic leukemia.
Overexpression of ETS2 has also been noted in various cancers, including AML.84 Furthermore, ETS2 transcripts are elevated in AMKL (both DS or non-DS) compared with DS-AML.87 These observations and its implication in regulation of megakaryocytic genes argue for a causative role of ETS2 in TMD or DS-AMKL. Whereas Ets2-deficient mice die before embryonic hematopoiesis could be studied,88 ETS2-transgenic mice develop lymphoid compartment abnormalities similar to that observed in people with DS.89 Like ERG, overexpression of ETS2 was also found to promote to a switch in differentiation of K562 cells from erythroid to megakaryocytic fate.87
Although the ETS family member GABPA has not been implicated as an oncogene, it is expressed in the megakaryocyte lineage, where it has been implicated in early stages of megakaryocytic maturation.90 Recent studies have shown that GABPα directly affects the cell cycle by regulating the expression of genes required of DNA synthesis and degradation of cell-cycle inhibitors.91 Of Hsa21 genes, GABPA was one of the few whose expression is elevated in DS-AML versus non-DS AMKL.74
Finally, ERG and/or GABPA may be involved in the pathogenesis of DS-ALL, as ERG is expressed in a pre–B-cell leukemia cell line but not in mature B-cell leukemia cells,85 overexpression of ERG is associated with adverse outcome in adults with T-ALL,92 and GABPα appears to be a critical regulator of B-lymphocyte development.93 Moreover, deletions encompassing ERG, which lead to expression of an N-terminal truncated protein that retains the ETS domain, have recently been found in pediatric ALL.94
miRNAs encoded by Hsa21: potential players in leukemic transformation?
Hsa21 is known to encode 5 miRNAs. Overexpression of several of these has been noted in brain and heart tissues of people with DS,95 and a subset of these miRNAs has been implicated in normal and pathologic hematopoiesis. For example, miR-99a is up-regulated during megakaryocytic differentiation of CD34+ cells, whereas miR-155 and let-7c are down-regulated.96 Notably, miR-155 has been linked to myeloproliferative and B-lymphoproliferative disorders.97,98 Recent studies have implicated miR-125b-2, which is overexpressed in TMD and AMKL samples compared with normal megakaryocytes, in the megakaryocytic leukemia of DS.72
Taken together, these experimental observations provide some clues about how chromosome 21 genes may act to deregulate the hematopoietic system. Although the list of genes presented here is not exclusive, experimental data support the hypothesis that increased gene dosage of Hsa21 ETS transcription factors facilitates abnormal megakaryopoiesis. Notably, these 3 ETS genes are all contained within the trisomic region of Ts65Dn mice, which develop a megakaryocytic MPD. Nevertheless, it is likely that other Hsa21 genes (including miRNA-encoding genes) are also required to drive the full spectrum of hematologic disorders in children with DS.
Summary and perspective
It is now well accepted that multiple genetic events, including trisomy 21 and GATA1 mutations, cooperate in TMD and DS-AMKL.35 Our model for the development of lymphoid leukemias in DS is depicted in Figure 2. The common links for both forms of leukemia are the presence of trisomy 21 and mutations in JAK tyrosine kinases. Although we believe that trisomy 21 is an essential component of these malignancies, the specific genes whose dysregulation is directly involved in tumorigenesis remain unknown. There are still many other important unanswered questions. Are the same Hsa21 genes responsible for the increased incidence of both ALL and AMKL in children with DS? Do both forms of leukemia share a common aberrant trisomy 21 stem cell, which then gives rise to ALL or AMKL depending on the nature of the secondary mutations? What is the identity of the cell that acquires the secondary mutation in these leukemias? Finally, we recognize that other genetic abnormalities, not yet identified, are necessary for the development of both types of DS-associated leukemia. We predict that these likely include mutations that induce hyperactivation of the STAT transcription factors (in patients who lack JAK2 or JAK3 mutations) and/or mutations or other genetic abnormalities in a B-cell transcription factor in DS-ALL.
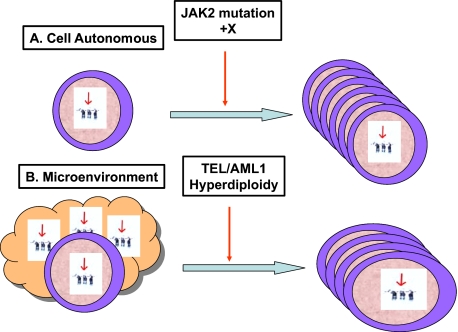
Figure 2.
Hypotheses to explain the pathogenesis of ALL in DS. (A) Constitutional trisomy 21 may enhance proliferation or survival of preleukemic lymphoid progenitors. An additional specific genetic event, such as a JAK2 mutation or additional chromosome X, cooperates with trisomy 21 to induce leukemia. This model is similar to the cooperation with GATA1 mutations in DS myeloid disorders. (B) The unique microenvironment in children with DS, caused by the presence of trisomy 21 in all body cells, results in an increased risk for usual childhood leukemogenic genetic events, such as TEL/AML1 and hyperdiploidy, and an increase in the common subtypes of childhood ALL. These 2 hypotheses are not mutually exclusive. Note that the latter model is speculative, but is presented as a hypothetical possibility to stimulate discussion and research in this area.
Acknowledgments
The authors thank the members of the Crispino and Izraeli laboratories as well as our international collaborators.
This review was supported by the US-Israel Binational Science Foundation (Jerusalem, Israel; J.D.C., S.I.), Israeli Science Foundation (Jerusalem, Israel; S.I.), the National Cancer Institute (Bethesda, MD; 1R01CA120772-01A2 to S.I. and 2R01CA101774 to J.D.C.), and the Samuel Waxman Cancer Research Foundation (New York, NY; J.D.C. and S.I.). J.D.C. is a scholar of the Leukemia & Lymphoma Society (White Plains, NY). S.M. acknowledges support from the Fondation pour la Recherche Medicale (FRM; Paris, France).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.M., S.I., and J.D.C. all contributed to the writing of this article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John D. Crispino, Division of Hematology/Oncology, Northwestern University, 303 East Superior St, Lurie 5-113, Chicago, IL 60611; e-mail: j-crispino@northwestern.edu; or Shai Izraeli, Department of Pediatric Hematology and Oncology and the Cancer Research Center, Sheba Medical Center, Tel Hashomer, Ramat Gan, 52621, Israel; e-mail: shai.izraeli@sheba.health.gov.il.
References
- 1.Roizen NJ, Amarose AP. Hematologic abnormalities in children with Down syndrome. Am J Med Genet. 1993;46:510–512. doi: 10.1002/ajmg.1320460509. [DOI] [PubMed] [Google Scholar]
- 2.Lange B. The management of neoplastic disorders of hematopoiesis in children with Down's syndrome. Br J Hematol. 2000;110:512–524. doi: 10.1046/j.1365-2141.2000.02027.x. [DOI] [PubMed] [Google Scholar]
- 3.Zipursky A. Transient leukemia: a benign form of leukemia in newborn infants with trisomy 21. Br J Haematol. 2003;120:930–938. doi: 10.1046/j.1365-2141.2003.04229.x. [DOI] [PubMed] [Google Scholar]
- 4.Pine SR, Guo Q, Yin C, Jayabose S, Druschel CM, Sandoval C. Incidence and clinical implications of GATA1 mutations in newborns with Down syndrome. Blood. 2007;110:2128–2131. doi: 10.1182/blood-2007-01-069542. [DOI] [PubMed] [Google Scholar]
- 5.Hasle H. Pattern of malignant disorders in individuals with Down's syndrome. Lancet Oncol. 2001;2:429–436. doi: 10.1016/S1470-2045(00)00435-6. [DOI] [PubMed] [Google Scholar]
- 6.Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Trisomy represses Apc(Min)-mediated tumors in mouse models of Down's syndrome. Nature. 2008;451:73–75. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- 7.Mundschau G, Gurbuxani S, Gamis AS, Greene ME, Arceci RJ, Crispino JD. Mutagenesis of GATA1 is an initiating event in Down syndrome leukemogenesis. Blood. 2003;101:4298–4300. doi: 10.1182/blood-2002-12-3904. [DOI] [PubMed] [Google Scholar]
- 8.Rainis L, Bercovich D, Strehl S, et al. Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood. 2003;102:981–986. doi: 10.1182/blood-2002-11-3599. [DOI] [PubMed] [Google Scholar]
- 9.Hitzler JK, Cheung J, Li Y, Scherer SW, Zipursky A. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood. 2003;101:4301–4304. doi: 10.1182/blood-2003-01-0013. [DOI] [PubMed] [Google Scholar]
- 10.Xu G, Nagano M, Kanezaki R, et al. Frequent mutations in the GATA-1 gene in the transient myeloproliferative disorder of Down syndrome. Blood. 2003;102:2960–2968. doi: 10.1182/blood-2003-02-0390. [DOI] [PubMed] [Google Scholar]
- 11.Taub JW, Mundschau G, Ge Y, et al. Prenatal origin of GATA1 mutations may be an initiating step in the development of megakaryocytic leukemia in Down syndrome. Blood. 2004;104:1588–1589. doi: 10.1182/blood-2004-04-1563. [DOI] [PubMed] [Google Scholar]
- 12.Langebrake C, Creutzig U, Reinhardt D. Immunophenotype of Down syndrome acute myeloid leukemia and transient myeloproliferative disease differs significantly from other diseases with morphologically identical or similar blasts. Klin Padiatr. 2005;217:126–134. doi: 10.1055/s-2005-836510. [DOI] [PubMed] [Google Scholar]
- 13.Massey GV, Zipursky A, Chang MN, et al. A prospective study of the natural history of transient leukemia (TL) in neonates with Down syndrome (DS): Children's Oncology Group (COG) study POG-9481. Blood. 2006;107:4606–4613. doi: 10.1182/blood-2005-06-2448. [DOI] [PubMed] [Google Scholar]
- 14.Klusmann JH, Creutzig U, Zimmermann M, et al. Treatment and prognostic impact of transient leukemia in neonates with Down syndrome. Blood. 2008;111:2991–2998. doi: 10.1182/blood-2007-10-118810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muramatsu H, Kato K, Watanabe N, et al. Risk factors for early death in neonates with Down syndrome and transient leukemia. Br J Haematol. 2008;142:610–615. doi: 10.1111/j.1365-2141.2008.07231.x. [DOI] [PubMed] [Google Scholar]
- 16.Hasle H, Abrahamsson J, Arola M, et al. Myeloid leukemia in children 4 years or older with Down syndrome often lacks GATA1 mutation and cytogenetics and risk of relapse are more akin to sporadic AML. Leukemia. 2008;22:1428–1430. doi: 10.1038/sj.leu.2405060. [DOI] [PubMed] [Google Scholar]
- 17.Pui CH, Raimondi SC, Borowitz MJ, et al. Immunophenotypes and karyotypes of leukemic cells in children with Down syndrome and acute lymphoblastic leukemia. J Clin Oncol. 1993;11:1361–1367. doi: 10.1200/JCO.1993.11.7.1361. [DOI] [PubMed] [Google Scholar]
- 18.Dordelmann M, Schrappe M, Reiter A, et al. Down's syndrome in childhood acute lymphoblastic leukemia: clinical characteristics and treatment outcome in 4 consecutive BFM trials. Berlin-Frankfurt-Munster Group. Leukemia. 1998;12:645–651. doi: 10.1038/sj.leu.2400989. [DOI] [PubMed] [Google Scholar]
- 19.Chessells JM, Harrison G, Richards SM, et al. Down's syndrome and acute lymphoblastic leukemia: clinical features and response to treatment. Arch Dis Child. 2001;85:321–325. doi: 10.1136/adc.85.4.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravindranath Y. Down syndrome and leukemia: new insights into the epidemiology, pathogenesis, and treatment. Pediatr Blood Cancer. 2005;44:1–7. doi: 10.1002/pbc.20242. [DOI] [PubMed] [Google Scholar]
- 21.Whitlock JA, Sather HN, Gaynon P, et al. Clinical characteristics and outcome of children with Down syndrome and acute lymphoblastic leukemia: a Children's Cancer Group study. Blood. 2005;106:4043–4049. doi: 10.1182/blood-2003-10-3446. [DOI] [PubMed] [Google Scholar]
- 22.Ross JA, Spector LG, Robison LL, Olshan AF. Epidemiology of leukemia in children with Down syndrome. Pediatr Blood Cancer. 2005;44:8–12. doi: 10.1002/pbc.20165. [DOI] [PubMed] [Google Scholar]
- 23.Zeller B, Gustafsson G, Forestier E, et al. Acute leukemia in children with Down syndrome: a population-based Nordic study. Br J Haematol. 2005;128:797–804. doi: 10.1111/j.1365-2141.2005.05398.x. [DOI] [PubMed] [Google Scholar]
- 24.Arico M, Ziino O, Valsecchi MG, et al. Acute lymphoblastic leukemia and Down syndrome: presenting features and treatment outcome in the experience of the Italian Association of Pediatric Hematology and Oncology (AIEOP). Cancer. 2008;113:515–521. doi: 10.1002/cncr.23587. [DOI] [PubMed] [Google Scholar]
- 25.Bassal M, La MK, Whitlock JA, et al. Lymphoblast biology and outcome among children with Down syndrome and ALL treated on CCG-1952. Pediatr Blood Cancer. 2005;44:21–28. doi: 10.1002/pbc.20193. [DOI] [PubMed] [Google Scholar]
- 26.Whitlock JA. Down syndrome and acute lymphoblastic leukemia. Br J Haematol. 2006;135:595–602. doi: 10.1111/j.1365-2141.2006.06337.x. [DOI] [PubMed] [Google Scholar]
- 27.Creutzig U, Reinhardt D, Diekamp S, Dworzak M, Stary J, Zimmermann M. AML patients with Down syndrome have a high cure rate with AML-BFM therapy with reduced dose intensity. Leukemia. 2005;19:1355–1360. doi: 10.1038/sj.leu.2403814. [DOI] [PubMed] [Google Scholar]
- 28.Rao A, Hills RK, Stiller C, et al. Treatment for myeloid leukemia of Down syndrome: population-based experience in the UK and results from the Medical Research Council AML 10 and AML 12 trials. Br J Haematol. 2006;132:576–583. doi: 10.1111/j.1365-2141.2005.05906.x. [DOI] [PubMed] [Google Scholar]
- 29.Gamis AS, Woods WG, Alonzo TA, et al. Increased age at diagnosis has a significantly negative effect on outcome in children with Down syndrome and acute myeloid leukemia: a report from the Children's Cancer Group Study 2891. J Clin Oncol. 2003;21:3415–3422. doi: 10.1200/JCO.2003.08.060. [DOI] [PubMed] [Google Scholar]
- 30.Zwaan CM, Kaspers GJ, Pieters R, et al. Different drug sensitivity profiles of acute myeloid and lymphoblastic leukemia and normal peripheral blood mononuclear cells in children with and without Down syndrome. Blood. 2002;99:245–251. doi: 10.1182/blood.v99.1.245. [DOI] [PubMed] [Google Scholar]
- 31.Ge Y, Stout ML, Tatman DA, et al. GATA1, cytidine deaminase, and the high cure rate of Down syndrome children with acute megakaryocytic leukemia. J Natl Cancer Inst. 2005;97:226–231. doi: 10.1093/jnci/dji026. [DOI] [PubMed] [Google Scholar]
- 32.Al-Ahmari A, Shah N, Sung L, Zipursky A, Hitzler J. Long-term results of an ultra low-dose cytarabine-based regimen for the treatment of acute megakaryoblastic leukemia in children with Down syndrome. Br J Haematol. 2006;133:646–648. doi: 10.1111/j.1365-2141.2006.06097.x. [DOI] [PubMed] [Google Scholar]
- 33.Matherly LH, Taub JW. Methotrexate pharmacology and resistance in childhood acute lymphoblastic leukemia. Leuk Lymphoma. 1996;21:359–368. doi: 10.3109/10428199609093433. [DOI] [PubMed] [Google Scholar]
- 34.Meissner B, Borkhardt A, Dilloo D, et al. Relapse, not regimen-related toxicity, was the major cause of treatment failure in 11 children with Down syndrome undergoing hematopoietic stem cell transplantation for acute leukemia. Bone Marrow Transplant. 2007;40:945–949. doi: 10.1038/sj.bmt.1705844. [DOI] [PubMed] [Google Scholar]
- 35.Vyas P, Crispino JD. Molecular insights into Down syndrome-associated leukemia. Curr Opin Pediatr. 2007;19:9–14. doi: 10.1097/MOP.0b013e328013e7b2. [DOI] [PubMed] [Google Scholar]
- 36.Hollanda LM, Lima CS, Cunha AF, et al. An inherited mutation leading to production of only the short isoform of GATA-1 is associated with impaired erythropoiesis. Nat Genet. 2006;38:807–812. doi: 10.1038/ng1825. [DOI] [PubMed] [Google Scholar]
- 37.Malinge S, Ragu C, Della-Valle V, et al. Activating mutations in human acute megakaryoblastic leukemia. Blood. 2008;112:4220–4226. doi: 10.1182/blood-2008-01-136366. [DOI] [PubMed] [Google Scholar]
- 38.Malkin D, Brown EJ, Zipursky A. The role of p53 in megakaryocytic differentiation and the megakaryocytic leukemias of Down syndrome. Cancer Genet Cytogenet. 2000;116:1–5. doi: 10.1016/s0165-4608(99)00072-2. [DOI] [PubMed] [Google Scholar]
- 39.Hirose Y, Kudo K, Kiyoi H, Hayashi Y, Naoe T, Kojima S. Comprehensive analysis of gene alterations in acute megakaryoblastic leukemia of Down's syndrome. Leukemia. 2003;17:2250–2252. doi: 10.1038/sj.leu.2403121. [DOI] [PubMed] [Google Scholar]
- 40.Walters DK, Mercher T, Gu TL, et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 2006;10:65–75. doi: 10.1016/j.ccr.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Kiyoi H, Yamaji S, Kojima S, Naoe T. JAK3 mutations occur in acute megakaryoblastic leukemia both in Down syndrome children and non-Down syndrome adults. Leukemia. 2007;21:574–576. doi: 10.1038/sj.leu.2404527. [DOI] [PubMed] [Google Scholar]
- 42.De Vita S, Mulligan C, McElwaine S, et al. Loss-of-function JAK3 mutations in TMD and AMKL of Down syndrome. Br J Haematol. 2007;137:337–341. doi: 10.1111/j.1365-2141.2007.06574.x. [DOI] [PubMed] [Google Scholar]
- 43.Sato T, Toki T, Kanezaki R, et al. Functional analysis of JAK3 mutations in transient myeloproliferative disorder and acute megakaryoblastic leukemia accompanying Down syndrome. Br J Haematol. 2008;141:681–688. doi: 10.1111/j.1365-2141.2008.07081.x. [DOI] [PubMed] [Google Scholar]
- 44.Forestier E, Izraeli S, Beverloo B, et al. Cytogenetic features of acute lymphoblastic and myeloid leukemias in pediatric patients with Down syndrome: an iBFM-SG study. Blood. 2008;111:1575–1583. doi: 10.1182/blood-2007-09-114231. [DOI] [PubMed] [Google Scholar]
- 45.Lo KC, Chalker J, Strehl S, et al. Array comparative genome hybridization analysis of acute lymphoblastic leukemia and acute megakaryoblastic leukemia in patients with Down syndrome. Br J Haematol. 2008;142:934–945. doi: 10.1111/j.1365-2141.2008.07280.x. [DOI] [PubMed] [Google Scholar]
- 46.Kearney L, Gonzalez De Castro D, Yeung J, et al. A specific JAK2 mutation (JAK2R683) and multiple gene deletions in Down syndrome acute lymphoblastic leukemia. Blood. 2009;113:646–648. doi: 10.1182/blood-2008-08-170928. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto T, Isomura M, Xu Y, et al. PTPN11, RAS and FLT3 mutations in childhood acute lymphoblastic leukemia. Leuk Res. 2006;30:1085–1089. doi: 10.1016/j.leukres.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Malinge S, Ben-Abdelali R, Settegrana C, et al. Novel activating JAK2 mutation in a patient with Down syndrome and B-cell precursor acute lymphoblastic leukemia. Blood. 2007;109:2202–2204. doi: 10.1182/blood-2006-09-045963. [DOI] [PubMed] [Google Scholar]
- 49.Bercovich D, Ganmore I, Scott LM, et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet. 2008;372:1484–1492. doi: 10.1016/S0140-6736(08)61341-0. [DOI] [PubMed] [Google Scholar]
- 50.Gaikwad AS, Rye CL, Devidas M, et al. Prevalence and clinical correlates of JAK2 mutations in pediatric Down syndrome acute lymphoblastic leukemia [abstract]. Blood. 2008;112 doi: 10.1111/j.1365-2141.2008.07552.x. Abstract no. 1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delabar JM, Theophile D, Rahmani Z, et al. Molecular mapping of 24 features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- 52.Korenberg JR, Chen XN, Schipper R, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci U S A. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barlow GM, Chen XN, Shi ZY, et al. Down syndrome congenital heart disease: a narrowed region and a candidate gene. Genet Med. 2001;3:91–101. doi: 10.1097/00125817-200103000-00002. [DOI] [PubMed] [Google Scholar]
- 54.Lyle R, Bena F, Gagos S, et al. Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur J Hum Genet. 2008 doi: 10.1038/ejhg.2008.214. prepublished online November 12, 2008, as DOI: 10.1038/ejhg.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olson LE, Richtsmeier JT, Leszl J, Reeves RH. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science. 2004;306:687–690. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chou ST, Opalinska JB, Yao Y, et al. Trisomy 21 enhances human fetal erythro-megakaryocytic development. Blood. 2008;112:4503–4506. doi: 10.1182/blood-2008-05-157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tunstall-Pedoe O, Roy A, Karadimitris A, et al. Abnormalities in the myeloid progenitor compartment in Down syndrome fetal liver precede acquisition of GATA1 mutations. Blood. 2008;112:4507–4511. doi: 10.1182/blood-2008-04-152967. [DOI] [PubMed] [Google Scholar]
- 58.Wiltshire T, Pletcher M, Cole SE, et al. Perfect conserved linkage across the entire mouse chromosome 10 region homologous to human chromosome 21. Genome Res. 1999;9:1214–1222. doi: 10.1101/gr.9.12.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gardiner K, Fortna A, Bechtel L, Davisson MT. Mouse models of Down syndrome: how useful can they be? Comparison of the gene content of human chromosome 21 with orthologous mouse genomic regions. Gene. 2003;318:137–147. doi: 10.1016/s0378-1119(03)00769-8. [DOI] [PubMed] [Google Scholar]
- 60.Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- 61.Vacik T, Ort M, Gregorova S, et al. Segmental trisomy of chromosome 17: a mouse model of human aneuploidy syndromes. Proc Natl Acad Sci U S A. 2005;102:4500–4505. doi: 10.1073/pnas.0500802102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moore CS, Roper RJ. The power of comparative and developmental studies for mouse models of Down syndrome. Mamm Genome. 2007;18:431–443. doi: 10.1007/s00335-007-9030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Doherty A, Ruf S, Mulligan C, et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309:2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reeves RH, Irving NG, Moran TH, et al. A mouse model for Down syndrome exhibits learning and behavior deficits. Nat Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 65.Moore CS. Postnatal lethality and cardiac anomalies in the Ts65Dn Down syndrome mouse model. Mamm Genome. 2006;17:1005–1012. doi: 10.1007/s00335-006-0032-8. [DOI] [PubMed] [Google Scholar]
- 66.Jablonska B, Ford D, Trisler D, Pessac B. The growth capacity of bone marrow CD34 positive cells in culture is drastically reduced in a murine model of Down syndrome. C R Biol. 2006;329:726–732. doi: 10.1016/j.crvi.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Kirsammer G, Jilani S, Liu H, et al. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood. 2008;111:767–775. doi: 10.1182/blood-2007-04-085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herbst EW, Winking H. Adoptive transfer of the hematopoietic system of trisomic mice with limited life span: stem cells from 6 different trisomies are capable of survival. Dev Genet. 1991;12:415–422. doi: 10.1002/dvg.1020120606. [DOI] [PubMed] [Google Scholar]
- 69.Gjertson C, Sturm KS, Berger CN. Hematopoietic deficiencies and core binding factor expression in murine Ts16, an animal model for Down syndrome. Clin Immunol. 1999;91:50–60. doi: 10.1006/clim.1998.4685. [DOI] [PubMed] [Google Scholar]
- 70.Sago H, Carlson EJ, Smith DJ, et al. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci U S A. 1998;95:6256–6261. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carmichael CL, Majewski IJ, Alexander WS, et al. Hematopoietic defects in the Ts1Cje mouse model of Down syndrome. Blood. 2009;113:1929–1937. doi: 10.1182/blood-2008-06-161422. [DOI] [PubMed] [Google Scholar]
- 72.Klusmann JH. Chromosome 21-encoded miR-125b and its role in the development of myeloid leukemia in children with Down syndrome [abstract]. Blood. 2007;110 Abstract no. 716. [Google Scholar]
- 73.Ge Y, Dombkowski AA, LaFiura KM, et al. Differential gene expression, GATA1 target genes, and the chemotherapy sensitivity of Down syndrome megakaryocytic leukemia. Blood. 2006;107:1570–1581. doi: 10.1182/blood-2005-06-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bourquin JP, Subramanian A, Langebrake C, et al. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc Natl Acad Sci U S A. 2006;103:3339–3344. doi: 10.1073/pnas.0511150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 76.Osato M, Asou N, Abdalla E, et al. Biallelic and heterozygous point mutations in the runt domain of the AML1 gene associated with myeloblastic leukemias. Blood. 1999;93:1817–1824. [PubMed] [Google Scholar]
- 77.Preudhomme C, Warot-Loze D, Roumier C, et al. High incidence of biallelic point mutations in the Runt domain of the AML1/PEBP2 alpha B gene in Mo acute myeloid leukemia and in myeloid malignancies with acquired trisomy 21. Blood. 2000;96:2862–2869. [PubMed] [Google Scholar]
- 78.Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- 79.Izraeli S. Leukemia: a developmental perspective. Br J Haematol. 2004;126:3–10. doi: 10.1111/j.1365-2141.2004.04986.x. [DOI] [PubMed] [Google Scholar]
- 80.Levanon D, Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- 81.Ichikawa H, Shimizu K, Hayashi Y, Ohki M. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 1994;54:2865–2868. [PubMed] [Google Scholar]
- 82.Dastugue N, Lafage-Pochitaloff M, Pages MP, et al. Cytogenetic profile of childhood and adult megakaryoblastic leukemia (M7): a study of the Groupe Francais de Cytogenetique Hematologique (GFCH). Blood. 2002;100:618–626. doi: 10.1182/blood-2001-12-0241. [DOI] [PubMed] [Google Scholar]
- 83.Marcucci G, Baldus CD, Ruppert AS, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:9234–9242. doi: 10.1200/JCO.2005.03.6137. [DOI] [PubMed] [Google Scholar]
- 84.Baldus CD, Liyanarachchi S, Mrozek K, et al. Acute myeloid leukemia with complex karyotypes and abnormal chromosome 21: amplification discloses overexpression of APP, ETS2, and ERG genes. Proc Natl Acad Sci U S A. 2004;101:3915–3920. doi: 10.1073/pnas.0400272101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rainis L, Toki T, Pimanda JE, et al. The proto-oncogene ERG in megakaryoblastic leukemias. Cancer Res. 2005;65:7596–7602. doi: 10.1158/0008-5472.CAN-05-0147. [DOI] [PubMed] [Google Scholar]
- 86.Loughran SJ, Kruse EA, Hacking DF, et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- 87.Ge Y, LaFiura KM, Dombkowski AA, et al. The role of the proto-oncogene ETS2 in acute megakaryocytic leukemia biology and therapy. Leukemia. 2008;22:521–529. doi: 10.1038/sj.leu.2405066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamamoto H, Flannery ML, Kupriyanov S, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wolvetang EJ, Wilson TJ, Sanij E, et al. ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum Mol Genet. 2003;12:247–255. doi: 10.1093/hmg/ddg015. [DOI] [PubMed] [Google Scholar]
- 90.Pang L, Xue HH, Szalai G, et al. Maturation stage-specific regulation of megakaryopoiesis by pointed-domain Ets proteins. Blood. 2006;108:2198–2206. doi: 10.1182/blood-2006-04-019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang ZF, Mott S, Rosmarin AG. The Ets transcription factor GABP is required for cell-cycle progression. Nat Cell Biol. 2007;9:339–346. doi: 10.1038/ncb1548. [DOI] [PubMed] [Google Scholar]
- 92.Baldus CD, Burmeister T, Martus P, et al. High expression of the ETS transcription factor ERG predicts adverse outcome in acute T-lymphoblastic leukemia in adults. J Clin Oncol. 2006;24:4714–4720. doi: 10.1200/JCO.2006.06.1580. [DOI] [PubMed] [Google Scholar]
- 93.Xue HH, Bollenbacher-Reilley J, Wu Z, et al. The transcription factor GABP is a critical regulator of B lymphocyte development. Immunity. 2007;26:421–431. doi: 10.1016/j.immuni.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 94.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 95.Kuhn DE, Nuovo GJ, Martin MM, et al. Human chromosome 21-derived miRNAs are overexpressed in down syndrome brains and hearts. Biochem Biophys Res Commun. 2008;370:473–477. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Garzon R, Pichiorri F, Palumbo T, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O'Connell RM, Rao DS, Chaudhuri AA, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 99.Lanza C, Volpe G, Basso G, et al. The common TEL/AML1 rearrangement does not represent a frequent event in acute lymphoblastic leukemia occurring in children with Down syndrome. Leukemia. 1997;11:820–821. doi: 10.1038/sj.leu.2400651. [DOI] [PubMed] [Google Scholar]
- 100.Steiner M, Attarbaschi A, Konig M, et al. Equal frequency of TEL/AML1 rearrangements in children with acute lymphoblastic leukemia with and without Down syndrome. Pediatr Hematol Oncol. 2005;22:229–234. doi: 10.1080/08880010590921603. [DOI] [PubMed] [Google Scholar]
- 101.Wechsler J, Greene M, McDevitt MA, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 102.Norton A, Fisher C, Liu H, et al. Analysis of JAK3, JAK2, and C-MPL mutations in transient myeloproliferative disorder and myeloid leukemia of Down syndrome blasts in children with Down syndrome. Blood. 2007;110:1077–1079. doi: 10.1182/blood-2007-03-080374. [DOI] [PubMed] [Google Scholar]
- 103.Hama A, Yagasaki H, Takahashi Y, et al. Acute megakaryoblastic leukemia (AMKL) in children: a comparison of AMKL with and without Down syndrome. Br J Haematol. 2008;140:552–561. doi: 10.1111/j.1365-2141.2007.06971.x. [DOI] [PubMed] [Google Scholar]
- 104.Klusmann JH, Reinhardt D, Hasle H, et al. Janus kinase mutations in the development of acute megakaryoblastic leukemia in children with and without Down's syndrome. Leukemia. 2007;21:1584–1587. doi: 10.1038/sj.leu.2404694. [DOI] [PubMed] [Google Scholar]
- 105.Epstein CJ, Hofmeister BG, Yee D, et al. Stem cell deficiencies and thymic abnormalities in fetal mouse trisomy 16. J Exp Med. 1985;162:695–712. doi: 10.1084/jem.162.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]