Abstract
Parathyroid hormone-related protein (PTHrP) plays a primary role in the development of humoral hypercalcemia of malignancy seen in the majority of adult T-cell leukemia/lymphoma (ATLL) patients with human T-cell lymphotropic virus type-1 (HTLV-1) infection. HTLV-1 Tax has been shown to complex with ETS-1 and SP1 to transactivate the PTHrP P3 promoter. Previously, we established a SCID/bg mouse model of human ATL with RV-ATL cells and showed that PTHrP expression was independent of Tax. In this study, we report an inverse correlation of PTHrP with tax/rex mRNA in multiple HTLV-1-positive cell lines and RV-ATL cells. Stimulation of Jurkat T cells with PMA/ionomycin upregulated the PTHrP P3 promoter by a previously characterized Ets binding site and also induced protein/DNA complex formation identical to that observed in RV-ATL cells. Further, we provide evidence that cotransfection with Ets-1 and constitutively active Mek-1 in HTLV-1-negative transformed T cells with stimulation by PMA/ionomycin not only resulted in a robust induction of PTHrP P3 but also formed a complex with ETS-1/P3 EBS similar to that in ATLL cells. Our data demonstrate that transcriptional regulation of PTHrP in ATLL cells can be controlled by T-cell receptor signaling and the ETS and MAPK ERK pathway in a Tax-independent manner.
Keywords: parathyroid hormone-related protein (PTHrP), adult T-cell leukemia/lymphoma (ATLL), HTLV-1, transcriptional regulation, ETS-1, humoral hypercalcemia of malignancy (HHM)
Introduction
Adult T-cell leukemia/lymphoma (ATLL) is an aggressive and often fatal malignancy of mature helper T cells infected with human T-cell lymphotropic virus type-1 (HTLV-1)1 that develops in chronically infected individuals 20–50 years following initial infection. About 80% of ATL patients develop humoral hypercalcemia of malignancy (HHM). HHM is a life-threatening paraneoplastic syndrome seen in a wide variety of cancers2 in addition to ATLL. HHM develops in cancer patients with increased circulating parathyroid hormone-related protein (PTHrP) that stimulates parathyroid hormone-1 receptors (PTH1R) to induce osteoclastic bone resorption and increase calcium reabsorption in kidneys, resulting in hypercalcemia.3 In addition to its role in the induction of HHM, PTHrP has been shown to be involved in the regulation of cell proliferation and apoptosis in a wide variety of normal and neoplastic tissues.4–15
The human PTHrP gene is composed of nine exons spanning more than 15 kilobases of genomic DNA and 15 different transcripts can be generated by alternative splicing of the 5′ and 3′ exons. Nevertheless, all PTHrP transcripts share products of exons 5 and 6 that encode for the prepro region and the majority of the mature peptide. PTHrP transcription is regulated by three distinct promoters identified as P1, P2 and P3. P1 and P3 contain a typical TATA box,16–18 while P2 is a GC-rich promoter region.19 Previous evaluation of PTHrP alternative promoter usage by qualitative20,21 and quantitative reverse transcription (RT)-PCR22 revealed that P3-initiated transcripts were detectable at high levels in most tumors and cell lines examined, including the (HTLV-1)-positive cell line MT-2, whereas P1 or P2-derived transcripts were not detected or expressed only in a subset of tumors.
Tax is a 40-kDa nuclear-localizing phosphoprotein, encoded from open reading frame IV of HTLV-1 that induces transcription of viral and numerous cellular genes such as IL-2,23 IL-2 receptor α,24 c-fos,25 and c-myc26 through activation of cyclic AMP response element binding protein (CREB/ATF), NF-kappaB (NF-κB) and serum response factor (SRF). Rex is encoded from ORFIII and regulates viral gene expression by interacting with cis-acting elements called the Rex-responsive element (RxRE) present in the 3′ and 5′ long terminal repeat (LTR),27,28 which determines the level of expression of the virion components. Previous reports showed that Tax could transactivate PTHrP transcription in vitro.29–31 A physical interaction between Tax and transcription factor Ets-1 was reported, and it was shown that their coexpression enhanced PTHrP P3 transcriptional activation in an osteosarcoma cell line, OsA-CL.32 However, tax/rex mRNA expression is undetectable or present at very low levels in ATLL cells in vivo.33,34 These results suggest that stimuli other than HTLV-1 Tax are likely involved in PTHrP transcriptional regulation in ATLL cells. Furthermore, IL-2 upregulates PTHrP gene expression and secretion in ATLL cells via both transcriptional activation and increased mRNA stability.35,36 Still, little is known about the regulation of PTHrP gene expression and its transcriptional activation in ATLL cells in vivo.
Expression of Ets-1, a member of the Ets family of transcription factors, is limited to lymphoid tissues in adults,37 including mature T cells. Activation of Ets-1 occurs via Ras-mitogen activated protein kinase (MAPK) signaling involving Mek-1 and Erk-1/2 kinases. Activated Ets-1 was shown to transactivate PTHrP using an Ets-binding site in P3 in breast cancer cells,38–40 murine keratinocytes41 and HTLV-1-infected cells.30
We have previously reported that severe combined immunodeficient (SCID)/beige (bg) mice inoculated with ATLL cells develop solid lymphoma and HHM characterized by severe hypercalcemia, increased osteoclastic bone resorption, decreased bone mineral density and increased circulating PTHrP that correlated positively with plasma calcium concentrations.42 Xenografted ATLL cells (RV-ATL) expressed very high levels of PTHrP mRNA as compared to other HTLV-1 positive cell lines. In addition, HTLV-1 Tax protein and tax/rex mRNA were not detectable by Western blotting and RT-PCR in RV-ATL xenografted cells, but were expressed at high levels in other HTLV-1-infected cell lines. Altogether, our results demonstrated that HHM could develop due to an increase in circulating PTHrP concentrations independent of HTLV-1 Tax in ATLL.
In the present study, we compared PTHrP to tax/rex mRNA expression levels and determined the alternative PTHrP promoter usage in ATLL cells xenografted in SCID/bg mice and four HTLV-1-positive cell lines using real-time RT-PCR. We also characterized and demonstrated a constitutive protein/DNA complex formation on a composite region of the previously characterized Ets-binding site and a potential γ-interferon-activated site (GAS) located within the third promoter (P3) of the human PTHrP gene in HTLV-1-infected cells. Finally, we present evidence that transactivation of the PTHrP P3 promoter can occur in HTLV-1-negative transformed human T cells via activation of Ets-1 with an EBS after T-cell stimulation with combined phorbol-12-myristate-13-acetate (PMA) and ionomycin or coexpression of Ets-1 with a constitutively active MAPK Mek-1 mutant in a Tax-independent manner.
Materials and methods
Animals and inoculation
Homozygous C.B-17/IcrCrl-scid-bgBR (SCID/bg) immunodeficient mice were purchased from Charles River Laboratories (Wilmington, MA, USA) and maintained under specific pathogen-free conditions in the animal facility of the College of Veterinary Medicine at The Ohio State University (Columbus, OH, USA). Male mice aged 5 weeks were used as recipients, anesthetized with xylazine-ketamine, and injected intraperitonally with 2–4 × 107 RV-ATL cells suspended in RPMI 1640 medium. The source of the RV-ATL cell line was previously described.43 Approximately 1 × 107 RV-ATL cells were collected between 21 and 28 days postinoculation by peritoneal lavage.42
Cell lines
MT-2, SLB-1, Jurkat T and HUT-102 cells were cultured in RPMI 1640 media supplemented with 10% heat-inactivated (56°C, 30 min) FBS, l-glutamine (2 mm), penicillin (50 U/ml) and streptomycin (50 μg/ml) (Invitrogen, Carlsbad, CA, USA), at 37°C and 5% CO2. HT1-RV cells were maintained in Iscovés medium supplemented with 20% FBS, l-glutamine, penicillin and streptomycin.
Real-time RT-PCR
Total RNA was extracted with TRIzol reagent (Invitrogen, San Diego, CA, USA). Quantitative evaluation of HTLV-1 tax/rex and PTHrP mRNA expression and alternative promoter usage was performed as previously described.22 RT reactions were carried out with 5 μg of total RNA according to the manufacturer's protocol using oligo(dT)12–18 and 50 U of Superscript II reverse transcriptase (Invitrogen) and were diluted to 50 μl after RT. cDNA standards were prepared by PCR amplification of cDNA from human lung carcinoma cells BEN (PTHrP and β2-microglobulin) or MT-2 cells (tax/rex standards) and calibrators ranging from 250 to 2.5 × 108 copies/μl were prepared by serial dilution. The following primers were used for tax/rex transcript (tax1.fw; 5′-ccgccgatcccaaagaaa-3′/tax1.rv; 5′-ccgaacatagtcccccaga-3′, exon 2/3, 144-bp amplicon). Data were normalized using the ratio of the target cDNA concentration to β2-microglobulin.
Preparation of nuclear extracts
Cells (0.5–1 × 107) were washed twice with cold phosphate-buffered saline and cell pellets were resuspended in 500–600 μl of hypotonic buffer A (10 mmol/l HEPES, pH 8.0, 10 mmol/l KCl, 1.5 mmol/l MgCl2, 0.1 mmol/l EDTA, 1 mmol/l dithiothreitol (DTT), 0.5 mmol/l phenylmethylsulfonyl fluoride (PMSF) and 0.065% Nonidet P-40) for 10 min at 4°C. Nuclei were isolated by microcentrifugation (800 rpm) for 4 min at 4°C. The nuclear pellet was suspended in 75–100 μl of extract buffer C (20 mmol/l HEPES, pH 8.0, 400 mmol/l NaCl, 1.5 mmol/l MgCl2, 1 mmol/l EDTA, 1 mmol/l DTT, 20% glycerol, 33 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 50 mmol/l sodium fluoride and 1.5 mmol/l sodium orthovanadate) and incubated for 30 min at 4°C with continuous gentle mixing. The mixture was micro-centrifuged (14 000 rpm) for 30 min at 4°C. Protein concentrations of nuclear extracts were determined using the Bradford assay (Pierce, Rockford, IL, USA).
Oligonucleotides and electrophoretic mobility shift assay
Double-stranded PTHrP P3 wild type (sense strand; 5′-gatctcaactttccggaagcaacca-3′), P3 GAS mutant (sense strand; 5′-gatctcaacttagcggaagcaacca-3′, GAS element is underlined), P3 EBS mutant (sense strand; 5′-gatctcaactttccgttagcaacca-3′, ETS-binding element is underlined) and P3 GAS/EBS mutant (sense strand; 5′-gatctcaacttagcgttagcaacca-3′) oligonucleotides were prepared by annealing and fill-in reaction using [α-32P] or nonradiolabeled deoxyadenosine triphosphate in the presence of the Klenow fragment of DNA polymerase I. Oligonucleotides containing known consensus for ETS family, STAT-1, STAT-3 and STAT-5 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were end-labeled using T4-polynucleotide kinase (Invitrogen) in the presence of [γ-32P]adenosine triphosphate. Nuclear extracts (7.5 μg of protein) were incubated in 18 μl total reaction volume, containing 10 mmol/l Tris-HCl, pH 7.5, 50 mmol/l NaCl, 1 mmol/l EDTA, 1 mmol/l DTT, 10% glycerol, 1 μg of poly(dI-dC) · poly(dI-dC) (Amersham Biosciences, Piscataway, NJ, USA) and 0.4 μl/ml bovine serum albumin for 15 min at room temperature. The reaction mixture was then incubated with the radiolabeled oligonucleotide (50 000 cpm) alone or with 25-fold excess of nonradiolabeled oligonucleotide for 15 min at RT. Samples were analyzed by electrophoresis using 4% nondenaturing polyacrylamide gels with 0.25× TBE buffer (22.3 mmol/l Tris, 22.2 mmol/l boric acid and 0.5 mmol/l EDTA). The gels were dried and analyzed by autoradiography.
Plasmids and site-directed mutagenesis
A human PTHrP P3-luciferase reporter gene construct, originally identified as the Bg/I–HindIII construct,44 was used in luciferase expression assays. It was derived from human P3 promoter PTHrP-CAT constructs19 and cloned into pGL-2 basic vector (Promega, Madison, WI, USA). Constructs bearing mutations in the GAS element, ETS (EBS) and SMAD-binding sites (SBS) were generated with the QuickChange™ Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) and the following primers (mutations indicated with italic letters); EBS-mutant (sense strand: 5′-gtggaatcaactttccgttagcaaccagcccaccag-3′), GAS-mutant (sense strand: 5′-gtgtgtggaatcaacttagcggaagcaaccagccc-3′), GAS/EBS-mutant (sense strand: 5′-gggtgtgtggaatcaacttagcgttagcaaccagcccaccagagg-3′), SBS-mutant (sense strand: 5′-gaggaggtagggaggtagatacatagctatgtatatatatgtggg-3). Sequences in mutant plasmids were confirmed with an Applied Biosystems automated 3700 DNA Analyzer. The pcDNA-3-based expression vectors of Ets-1 (pcEts-1) and constitutively active Mek-1 (pMek-1-DE) have been described previously45,46 and were a kind gift of Dr M Ostrowski, The Ohio State University. pβgal-control vector (BD Biosciences, San Jose, CA, USA) was used as an internal control to correct for transfection efficiency in all experiments.
T-cell transfection and stimulation
Approximately 2.5 × 106 Jurkat T cells were electrotransfected in a BioRad Gene Pulser II (250 V, 950 μF) with 5 μg of wild type or mutant (m1–m7) PTHrP P3-luc constructs. Cells were either left untreated or stimulated with 20 ng/ml PMA (Sigma, St Louis, MO, USA) and 2 μm ionomycin (Sigma) at 6 h post-transfection, followed by an 18-h incubation period prior to lysis for measurement of luciferase and β-galactosidase activities. For EMSA studies, Jurkat T cells were maintained at 5 × 105 cells/ml in RPMI supplemented with 10% FBS and were stimulated with PMA or ionomycin alone or in combination for a period ranging from 15 to 60 min prior to nuclear extract preparation. To investigate the effect of Ets-1 and Mek-1 on PTHrP transcriptional regulation, Jurkat T cells were cotransfected with 5 μg of wild type or EBS mutant (m2 and m7) P3-luc constructs in the presence or absence of expression plasmids pcEts1 (0–8 μg) or pcDNA-3 (10 μg), and/or pMek-1-DE (2.5 μg). Different groups were stimulated with PMA and ionomycin at 30 h post-transfection, followed by an 18 h incubation. In all cases, the plasmid pβgal-Control Vector (2 μg) was cotransfected and served as internal control to correct for transfection efficiency. Transfected cells were lysed after 48 h in 100 μl of passive lysis buffer (Promega). Luciferase activity was measured with the Luciferase Assay System (Promega) using 50 μl of lysate. Simultaneously, β-galactosidase activity was measured with the Luminescent β-Galactosidase Detection Kit II (BD Biosciences). Each reporter assay experiment was repeated at least three times.
Statistical analysis
Comparison between groups in luciferase expression assays was performed using the nonparametric Mann–Whitney U-test and Student's t-test (two-tailed) with P<0.05 considered significant. Statistical analysis was performed using Instat 3.01 (GraphPAD software).
Results
A strong inverse correlation between PTHrP and tax/rex mRNA expression was observed in HTLV-1-positive cell lines and RV-ATL cells
We compared PTHrP to HTLV-1 tax/rex mRNA expression in five different HTLV-1 positive cell lines using quantitative real-time RT-PCR. First, we examined PTHrP gene expression using primers to amplify products of exons 5 and 6, which are common to all PTHrP mRNA species regardless of their respective 5′ or 3′ alternative splicing. RV-ATL cells engrafted in SCID/bg mice showed the highest PTHrP mRNA expression among the HTLV-1 cell lines tested. Comparatively, PTHrP mRNA expression in MT-2, SLB-1, HT1-RV and Hut-102 was lower and corresponded to 15, 13, 6 and 1% of its expression in RV-ATL cells, respectively (Figure 1). Simultaneously, we also measured HTLV-1 tax/rex gene expression in the same samples using primers amplifying doubly spliced viral RNA encoding for the tax and rex gene products. We observed that tax/rex mRNA expression was greater in HT1-RV cells when compared to the other cell lines tested. Hut-102, MT-2 and SLB-1 cells expressed moderate levels of tax/rex mRNA representing 13, 8 and 7% of the expression observed in HT1-RV cells. The level of tax/rex gene expression in RV-ATL cells was at the limit of detection of our assay and corresponded to less than 1% of levels detected in HT1-RV cells. Final real-time PCR products were of expected size when separated on a 2% agarose gel by electrophoresis. In addition, the intensity of the bands correlated well with the quantitative data obtained from the LightCycler. The quantitative real-time RT-PCR results in cell lines examined confirmed our previous qualitative evaluation of PTHrP gene expression by standard RT-PCR (data not shown) using primers published previously.20 Ratios of PTHrP to tax/rex mRNA expression in cell lines examined were also determined. HT1-RV and Hut-102 cells had ratios (PTHrP:tax/rex) of 1:200 and 1:167, whereas the ratios in MT-2, SLB-1 and RV-ATL cells were 1:10, 1:7 and 89:1, respectively. Our data demonstrate a strong inverse correlation between both transcripts in HTLV-1-positive cells.
Figure 1.
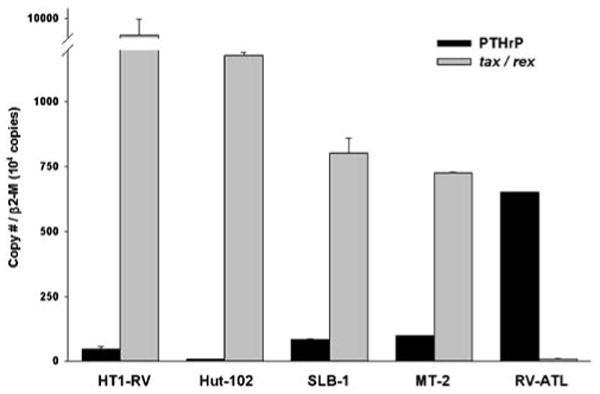
Comparison of PTHrP and HTLV-1 tax/rex mRNA expression in HT1-RV, Hut-102, SLB-1 and MT-2 with xenografted RV-ATL cells by quantitative real-time RT-PCR. Copy numbers for both genes transcripts were normalized using the ratio of target cDNA to β2-microglobulin (β2-M). Bars represent the mean ± s.d.
Examination of alternative PTHrP promoter usage in HTLV-1-positive cell lines and RV-ATL cells
Transcripts derived from the three identified promoters that regulate PTHrP gene transcription in HTLV-1-infected cell lines was evaluated using quantitative real-time RT-PCR. Primers used in the assay were designed to amplify specific regions of P1, P1/P2 and P3-derived spliced mRNA. Based on the assays, ratios of the different promoter-derived transcripts (P1:P2:P3) in RV-ATL, SLB-1, MT-2, Hut-102 and HT1-RV were 1:15:84, 1:11:88, 1:15:84, 6:78:16 and 1:63:36, respectively. Very low copy numbers of transcripts from P1 activation were detected in all HTLV-1-positive cell lines and RV-ATL cells when compared to copy numbers measured for P2 or P3-derived transcripts (Figure 2). Cells expressing high PTHrP mRNA levels (RV-ATL, SLB-1 and MT-2) had P2:P3 ratios ranging from 1:6 to 1:8. In contrast, P2:P3 ratios were 2:1 and 5:1 in HT1-RV and Hut-102 cells, which express low levels of PTHrP mRNA. Quantitative real-time RT-PCR data evaluating the alternative promoter usage correlated well with our previous observations using standard RT-PCR (data not shown). The results demonstrated that HTLV-1-positive cells expressing high levels of tax/rex mRNA have a P2:P3 ratio in favor of P2-derived transcripts in contrast to a predilection for usage of the P3 promoter in cell lines expressing modest to low levels of tax/rex mRNA.
Figure 2.
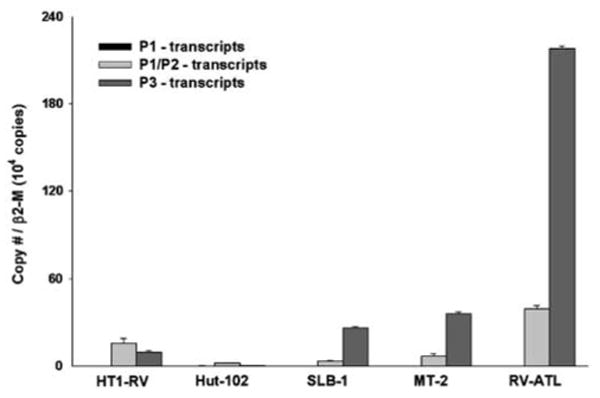
Comparative PTHrP alternative promoter usage in HT1-RV, Hut-102, SLB-1 and MT-2 with xenografted RV-ATL cells by quantitative real-time RT-PCR. Copy numbers for each transcript were normalized using the ratio of target cDNA to the house keeping gene, β2-microglobulin (β2-M). Bars represent the mean ± s.d.
Constitutive and Ets-specific binding to PTHrP P3 in HTLV-1-positive cell lines and RV-ATL cells
Constitutive formation of a distinct ETS-specific protein–DNA complex (C1) on the composite GAS/EBS regulatory element located within the third promoter (P3) of human PTHrP gene was revealed by EMSA using nuclear extracts from RV-ATL cells xenografted in SCID/bg mice (lane 1) (Figure 3a). A second protein–DNA complex (C2) was formed with a mutant oligonucleotide (lane 2) containing a disrupted EBS motif (GAS/ebs). Formation of the ETS-specific protein–DNA complex (C1) was markedly reduced in the reaction with a mutant oligonucleotide (lane 3) containing a disrupted GAS element (gas/EBS). Comparison of the complexes formed between the P3 oligonucleotide and other oligonucleotides with consensus sequences for Ets-1 (lane 4), STAT-1 (lane 5), STAT-3 (lane 6) and STAT-5 (lane 7) suggested that a protein–DNA complex formed on the Ets-1 oligonucleotide was identical to the C1 complex, while the C2 complex was the same as the protein–DNA complex associated with the STAT-5 oligonucleotide. Examination of protein–DNA complex formation on the P3 GAS/EBS element in other HTLV-1-positive cells and Jurkat T cells revealed constitutive formation of complex C1 that was fully abolished with the addition of 25-fold excess of nonradiolabeled P3 oligonucleotide (Figure 3b). Complexes with the highest intensity were observed in RV-ATL cells followed by MT-2, SLB-1 and Hut-102 cells, whereas the intensity of C1 complexes was lower in HT1-RV and Jurkat T-cells. Formation of C2 complexes with the GAS/ebs oligonucleotide was seen only in RV-ATL, MT-2 and SLB-1 cells. C1 complex formation was partially abolished when mutant oligonucleotide gas/EBS was tested. The oligonucleotide containing a mutation in both GAS and EBS elements (gas/ebs) fully prevented the formation of C1 and C2 complexes in all cell lines tested. Taken together, these results strongly supported that a constitutive EBS-specific DNA-binding complex (C1) formed with Ets transcription factor(s) in all HTLV-1-positive cell lines tested, and that a second complex (C2) containing STAT-5 could form when the EBS element was disrupted, suggesting that a factor(s) in the complex C2 had a lower affinity or was simply present at lower concentrations than Ets transcription factor(s) included in complex C1.
Figure 3.
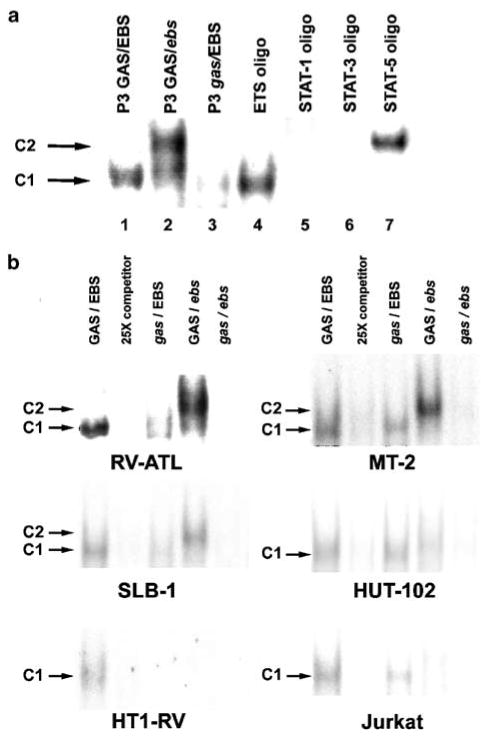
Constitutive activation of Ets factor(s) in HTLV-1-positive and Jurkat T cells. (a) Nuclear extracts of xenografted RV-ATL cells formed two protein/DNA complexes (C1 and C2) when subjected to EMSA using oligoprobes to PTHrP P3 wild-type GAS/EBS (lane 1), P3 GAS/EBS mutant (italic) oligos (lanes 2–3) and ETS-1, STAT1, STAT3 and STAT5 consensus sequences (lanes 4–7) as a probes. (b) Nuclear extracts from RV-ATL, MT-2, SLB-1, Hut-102, HT1-RV and Jurkat T cells were subjected to EMSA with PTHrP GAS/EBS wild type and mutant oligoprobes. 25-fold excess of unlabeled wild-type probe was used as competitor.
Stimulation of Jurkat T cells with PMA and ionomycin induces transcriptional activation of PTHrP P3 through an Ets-binding site
HTLV-1 negative Jurkat T cells were transfected with a PTHrP luciferase reporter construct under the control of human PTHrP promoter P3 and stimulated with PMA and ionomycin. There was a 5.5-fold induction of luciferase activity with wild-type PTHrP P3-luciferase construct in Jurkat T cells in response to PMA and ionomycin stimulation (Figure 4a). A 2.5-fold increase in luciferase activity was observed in cells stimulated with either ionomycin or PMA. Luciferase activity of a series of mutant PTHrP P3-luciferase constructs (m1–m7) carrying point mutations that disrupted the GAS, EBS and SBS elements individually or in combination was compared to that of the wild-type construct (Figure 4b). Lysates from Jurkat T cells transfected with mutants containing a disrupted EBS (m2, m5, m6 and m7) and costimulated with PMA and ionomycin resulted in a 65–73% decreased luciferase activity compared to the wild-type construct. Stimulation with PMA/ionomycin with the plasmids containing mutations resulted in a 78–83% decrease in luciferase activity. Interestingly, transfection with mutants containing disruption of the GAS and/or SBS (m1, m3 and m6) resulted in reduced luciferase activity, but was not significantly different from wild type demonstrating the central role played by the EBS element in the transcriptional activation of PTHrP P3 in response to cell signaling downstream of the T-cell receptor in transformed human T cells.
Figure 4.
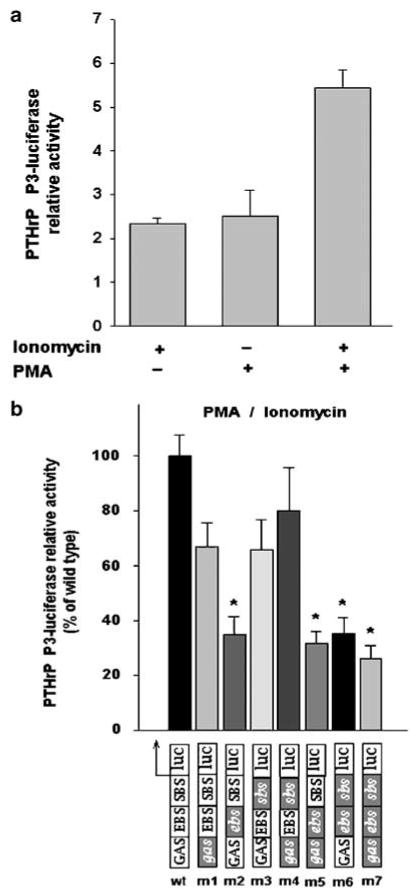
Relative luciferase activity in Jurkat T cells transfected with PTHrP P3-luciferase constructs stimulated with PMA or ionomycin or PMA/ionomycin. (a) Wild-type PTHrP P3-luciferase activity relative to unstimulated cells. (b) Mutants (m1–m7) PTHrP P3-luciferase activity relative to wild-type construct. Mutations indicated by gray boxes and italic text. Bars represent the mean ± s.d. of three independent experiments. (∗) indicates significant differences between mutants and wild type (P<0.05).
Combined PMA and ionomycin stimulation of Jurkat T cells induces a major protein–DNA complex containing Ets transcription factor(s)
EMSAs were performed with nuclear extracts from Jurkat T cells activated with PMA and or ionomycin for 15–60 min to test whether the complex (C1) containing Ets factor(s) detected in HTLV-1-positive cell lines was inducible by signaling downstream of the T-cell receptor (TCR). Combined stimulation by PMA and ionomycin induced the formation of a protein–DNA complex (Figure 5). Intensity of C1 complexes was greater with extracts from PMA/ionomycin-stimulated cells when compared to controls, and was most remarkable in samples from cells stimulated for 30 and 60 min. The protein–DNA complex (C1) induced by stimulation was identical to the complex (C1) observed previously in other HTLV-1-positive cell lines and RV-ATL cells xenografted in SCID/bg mice (Figure 3a). A modest induction of the protein–DNA complex C1 was also present with extracts from Jurkat T cells stimulated with PMA alone. Stimulation by ionomycin alone did not induce more protein–DNA complex formation when compared to controls. In all cases, the C1 complex was moderately reduced when the oligonucleotide contained a mutation in the GAS element (gas/EBS), almost completely abrogated with the mutant oligonucleotide for EBS (GAS/ebs), and fully prevented when the double-mutant oligonucleotide (gas/ebs) was tested.
Figure 5.
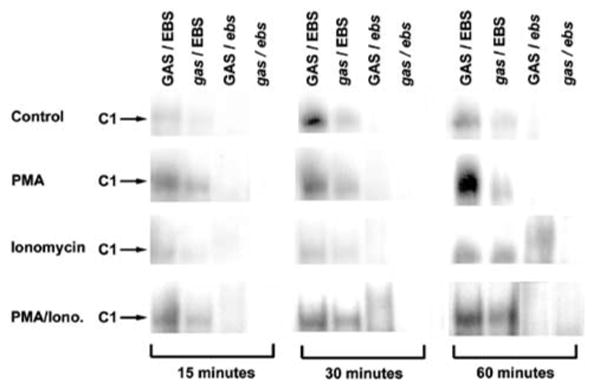
Activation of Ets factor(s) in Jurkat T cells stimulated with PMA, ionomycin and PMA/ionomycin. Induction of protein/DNA complex (C1) as detected by EMSA in Jurkat T-cells stimulated by PMA, ionomycin or PMA/ionomycin for 15–60 min prior to nuclear extraction. Complex formed between Ets factor(s) and PTHrP P3 GAS/EBS induced by stimulation was identical to complex C1 observed in all HTLV-1-positive cells (Figure 3). Mutations are indicated in italics.
Cooperative transcriptional activation of PTHrP P3 in Jurkat T cells by Mek-1 and Ets-1 mediated through an Ets-binding site
To test the effect of Ets-1 overexpression and its activation by MAPK Mek-1 on PTHrP P3 transcriptional activation, Jurkat T-cells were cotransfected with constructs expressing Ets-1 and constitutively active Mek-1 mutant (Mek-1-DE) in addition to the PTHrP P3-luc construct. Co-transfection of Jurkat T cells with 2, 4 and 8 μg of construct expressing Ets-1 with wild-type PTHrP P3-luc resulted in a dose-dependent increase in luciferase activity ranging from 2.5- to 7-fold (Figure 6a) relative to control. Stimulation with PMA and ionomycin of cells cotransfected with pcETS-1 (8 μg) and PTHrP P3-luc constructs increased luciferase activity by 10-fold in comparison to controls. Lysates from cells cotransfected with Mek-1-DE construct in addition to 2, 4 or 8 μg of pcETS-1 had a dose-dependent 11- to 40-fold increase in luciferase activity when compared to controls. When the Mek-1-DE construct was cotransfected in the absence of pcETS-1, luciferase activity in cell lysates was 6.5-fold higher than in control. Costimulation of Jurkat T cells with PMA and ionomycin without addition of pcETS-1 and Mek-1-DE resulted in a fivefold induction of PTHrP P3-luciferase activation over controls. Two mutants of the PTHrP P3-luc construct with a single mutation in EBS (m2) or a triple mutation in GAS, EBS and SBS (m7) were tested for luciferase activity simultaneously with the wild-type construct in all experimental groups. A significant decrease in luciferase activity relative to wild type was observed in Jurkat T cells transfected with either mutant in all 10 experimental groups (Figure 6b). Reduction of luciferase activity relative to wild type ranged from 62 to 78% with the m2 PTHrP P3-luc construct and from 62 to 82% with the m7 PTHrP P3-luc construct. No significant difference in luciferase activity was observed between both mutant constructs in any of the experimental groups. These results support a cooperative effect of MAPK Mek-1 with the downstream transcription factor Ets-1 in the transcriptional activation of PTHrP P3 and suggest events occurred through a specific Ets-binding site.
Figure 6.
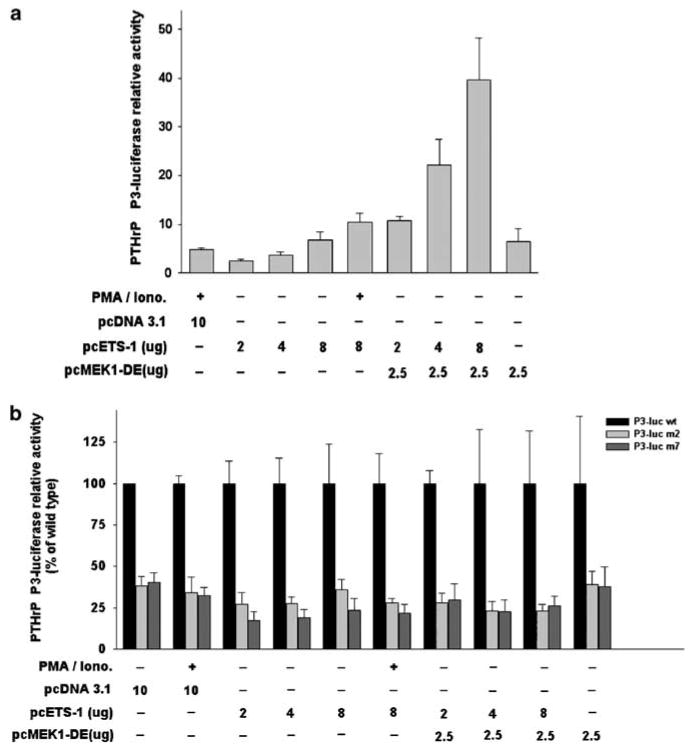
Relative luciferase activity in Jurkat T cells cotransfected with PTHrP P3-luciferase constructs, Ets-1 and/or constitutively active Mek-1 expression vectors. (a) Wild-type PTHrP P3-luciferase activity relative to unstimulated cells transfected with 10 μg of pcDNA3.1 (empty vector). (b) Mutants (m2 and m7) PTHrP P3-luciferase activity relative to wild-type construct. Quantity of expression vector indicated in (μg). Bars represent the mean ± s.d. of three independent experiments. (∗) indicates significant differences between mutants and wild type (P<0.05).
Co-expression of Ets-1 with constitutively active Mek-1 in Jurkat T cells induces a protein–DNA complex involving Ets-1 and PTHrP P3 Ets-binding site
Cotransfection with constructs expressing Ets-1 and constitutively active Mek-1 mutant was performed in Jurkat T cells to evaluate by EMSA the effect of Mek-1/Erk1/Ets-1 activation on the formation of the protein–DNA complex involving PTHrP P3 containing the Ets-binding site. The intensity of the protein–DNA complex (C1) formation with PTHrP wild-type oligonucleotide (GAS/EBS) was remarkably increased with nuclear extracts from Jurkat T cells cotransfected with increasing doses of pcETS-1 and Mek-1-DE constructs (Figure 7, bottom panel) in comparison to control cells (top panel). Extracts from cells transfected with 2–8 μg of the construct expressing Ets-1 had a moderate increase in the intensity of complex C1 formation (middle panel) when compared to control. A small to moderate increase was also observed with nuclear extracts from cells stimulated with PMA/ionomycin or transfected with construct Mek-1-DE alone (top panel). In all cases, the protein–DNA complex (C1) formed with PTHrP oligonucleotide GAS/EBS was identical to the one detected with extracts from RV-ATL cells xenografted in SCID/bg mice (top panel). Mutation of GAS and EBS in the PTHrP P3 oligonucleotide (gas/ebs) fully prevented the formation of protein–DNA complex in all experimental groups as well as the RV-ATL cells. These results confirmed the involvement of Ets-1 in the formation of a protein–DNA complex with the PTHrP P3 promoter containing a specific Ets-binding site in T cells. It also emphasized the important role of Mek-1 activation in the induction of Ets-1-PTHrP P3-EBS complex formation.
Figure 7.
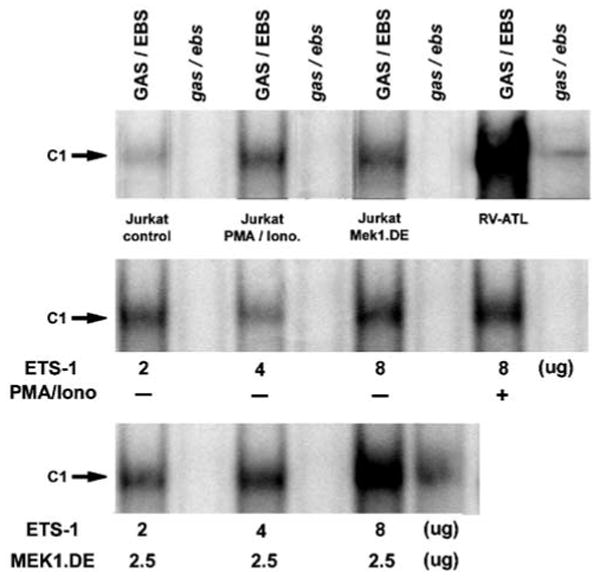
Activation of Ets-1 in Jurkat T cells cotransfected with vectors expressing Ets-1 and constitutively active Mek-1. Induction of protein/DNA complex (C1) as detected by EMSA in Jurkat T-cells cotransfected with pcETS-1 and/or Mek-1.DE construct in the presence or absence of PMA/ionomycin. The complex formed between Ets-1 and PTHrP P3 GAS/EBS was inducible by coexpression of Ets-1 and constitutively active Mek-1 as shown by EMSA. It was identical to complex C1 observed in RV-ATL cells. Mutations are indicated in italics.
Discussion
Although reports have attributed a central role to Tax in upregulation of PTHrP gene expression in ATLL cells based upon results that PTHrP promoter constructs could be transactivated by HTLV-1 Tax by in vitro assays,29–32 it remains unclear whether Tax is involved in the regulation of PTHrP gene expression in vivo since tax/rex mRNA expression is generally very low or undetectable in ATLL cells. We previously showed that immunodeficient mice inoculated with ATLL cells developed solid lymphoma and HHM as occurs in human ATLL patients independent of Tax expression. Here, we compared tax/rex and PTHrP mRNA levels in HTLV-1-positive cell lines and xenografted RV-ATL cells by real-time RT-PCR. Our results demonstrated a marked inverse relationship between PTHrP and tax/rex mRNA copy numbers in HTLV-1-infected cells.
Measurement of PTHrP alternative promoter usage in the HTLV-1-positive cells revealed that P1 was minimally used in all HTLV-1-positive cell lines. This observation is consistent with other reports showing that PTHrP P1-derived transcripts were abundant in normal keratinocytes and squamous cell carcinomas, and were infrequently expressed in other tissues.19,21 P3 promoter usage was favored over P2 in HTLV-1-infected cells (RV-ATL, MT-2 and SLB-1) expressing the highest PTHrP and lowest tax/rex mRNA copy numbers whereas the converse was observed in cells (Hut-102 and HT1-RV) expressing low PTHrP and high tax/rex mRNA. The latter finding may be explained by a variety of simple or combined mechanistic actions of Tax or Rex on transcriptional or post-transcriptional regulation of PTHrP gene expression. First, Tax can activate NF-κB signaling through various mechanisms,47 and we identified two possible NF-κB DNA binding sites in the GC-rich PTHrP P2 promoter, one of which was able to bind NF-κB subunit p50 in RV-ATL cells (data not shown). Thus, elevated Tax expression in HTLV-1-infected cells might be associated with P2 transactivation via NF-κB, therefore leading to an increased ratio of P2 over P3-transcripts. Second, Tax can physically bind to transcription factors Ets-1 and Sp-1 to form a ternary complex,32 but little is known regarding the affinity and transactivation potential of this complex relative to other Ets factor containing complexes. Thus, it is conceivable that the Ets-1/Sp1/Tax complex has higher affinity to the PTHrP P3 EBS but lower than other complexes that do not contain Tax leading to a lower P3 transcriptional activation when Tax is expressed at high levels.
ATLL cells typically exhibit the phenotype and cell signaling of activated T-cells. The MAPK signaling pathway can also be induced through active protein kinase C (PKC) and Sos following TCR engagement and IL-2 receptor activation. First, we examined by EMSA the formation of a protein/DNA complex using oligonucleotides containing the EBS PTHrP P3. We observed a constitutive and specific complex formation between an Ets transcription factor and P3 oligonucleotides in all HTLV-1 cell lines tested and the RV-ATL cells. The intensity of the complex correlated well with copy numbers of P3-derived mRNA transcripts in the cells. The complex formed with P3 EBS in RV-ATL cells was identical to that with an oligonucleotide containing an Ets-1 consensus sequence. Then, we mimicked cell signaling downstream of TCR activation by stimulating Jurkat T cells with PMA and ionomycin and observed a five- to six-fold induction of luciferase activity regulated by human PTHrP P3. Most of the transcriptional effect was attributable to an intact EBS located upstream of the TATA-box as demonstrated by a significant decrease in luciferase activity in Jurkat T cells transfected with constructs bearing a disrupted EBS. Mutation of the GAS and/or SBS did not significantly influence the luciferase activity in comparison to the wild-type construct. In addition, EMSA results showed that stimulation of Jurtkat T-cells with PMA and ionomycin induced complex formation on PTHrP P3 EBS, which was identical to that seen in HTLV-1-infected cells, including the RV-ATL cells. These results were consistent with previous reports that indicated upregulation of PTHrP gene expression by phorbol esters48–50 and PKC activation,51 and this effect was mediated by an Ets family transcription factor. The effect of ionomycin and PMA were additive on the induction of luciferase activity as would be expected by the synergistic activation of PKC by these agents.52
The Raf/Mek-1/ERK1/2 pathway is activated by PKC upon T-cell activation and has been involved in PTHrP P3 transactivation in breast cancer cells.51 Cotransfection of Jurkat T cells with constitutive Mek-1 mutant and Ets-1 expression vectors resulted in up to a 40-fold induction of PTHrP P3-derived luciferase activity, and the induction was dramatically reduced by using P3 constructs bearing an EBS mutation whereas mutations in GAS and SBS did not affect the luciferase activity. Moreover, the protein/DNA complex formation observed by EMSA in all HTLV-1-positive cells was inducible by overexpression of active Mek-1 and Ets-1 as demonstrated in cotransfection experiments. Overall, our results showed that endogenous Ets-1 expression in Jurkat T cells as well as Mek-1 activity were insufficient to fully transactivate PTHrP P3, but this baseline transcription level could be synergistically enhanced by PMA and ionomycin stimulation in the presence of exogenous Ets-1.
These results support a model that chronically activated T-cells will have constitutive activation of the Ras/Mek-1/ERK1/2 coupled to high Ets-1 expression levels in order to upregulate PTHrP expression in ATLL cells. Other evidence to support this model comes from reports showing that NF-κB,53 AP-154 and Jak/STAT55 are constitutively active in primary ATLL cells in a Tax-independent manner and likely due to a cell signaling deregulation. Interestingly, all these signaling pathways are typically upregulated in activated T-cells. However, both Ets-1 expression and Ras/Mek-1/ERK1/2 activity have not been evaluated in ATLL. We evaluated the status of ERK-1/2 in HTLV-1-infected cell lines and RV-ATL cells and observed that a constitutive activation in ERK-1/2 was present in all cells examined (data not shown). Nevertheless, further investigation is necessary to correlate Ras/Mek-1/ERK1/2 pathway and Ets-1 expression with the level of PTHrP mRNA expression regulated by P3 in a subset of primary ATLL cells to fully validate the model.
In conclusion, our findings support the concept that PTHrP gene transcription upregulation in ATLL cells is attributable to transactivation of the third promoter (P3) via Ets-1 binding or other factors of the Ets family to a specific EBS response element located within the promoter and those events can occur upon activation of cell signaling downstream of the TCR independently of HTLV-1 Tax expression. This conclusion challenges the traditionally accepted paradigm that increased PTHrP expression and HHM in ATLL is due to Tax. Finally, further investigation is required to understand the underlying mechanisms involved in PTHrP P2 regulation in ATLL cells.
Acknowledgments
This work was supported by the National Cancer Institute (CA100730 and CA77911) and the National Center for Research Resources (RR00168). VR and MVPN were supported by the Barber Trust Fund from the College of Veterinary Medicine, The Ohio State University.
References
- 1.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broadus AE, Mangin M, Ikeda K, Insogna KL, Weir EC, Burtis WJ, et al. Humoral hypercalcemia of cancer. Identification of a novel parathyroid hormone-like peptide. N Engl J Med. 1988;319:556–563. doi: 10.1056/NEJM198809013190906. [DOI] [PubMed] [Google Scholar]
- 3.Grill V, Martin TJ. Hypercalcemia of malignancy. Rev Endocr Metab Disord. 2000;1:253–263. doi: 10.1023/a:1026597816193. [DOI] [PubMed] [Google Scholar]
- 4.Inoue D, Matsumoto T, Ogata E, Ikeda K. 22-Oxacalcitriol, a noncalcemic analogue of calcitriol, suppresses both cell proliferation and parathyroid hormone-related peptide gene expression in human T cell lymphotrophic virus, type I-infected T cells. J Biol Chem. 1993;268:16730–16736. [PubMed] [Google Scholar]
- 5.Wake A, Tanaka Y, Nakatsuka K, Misago M, Oda S, Morimoto I, et al. Calcium-dependent homotypic adhesion through leukocyte function- associated antigen-1/intracellular adhesion molecule-1 induces interleukin-1 and parathyroid hormone-related protein production on adult T-cell leukemia cells in vitro. Blood. 1995;86:2257–2267. [PubMed] [Google Scholar]
- 6.Li X, Drucker DJ. Growth factor-like properties of parathyroid hormone-related peptide in transfected rodent cell line. Cancer Res. 1993;53:2980–2986. [PubMed] [Google Scholar]
- 7.Kaiser SM, Laneuville P, Bernier SM, Rhim JS, Kremer R, Goltzman D. Enhanced growth of a human keratinocyte cell line induced by antisense RNA for parathyroid hormone-related peptide. J Biol Chem. 1992;267:13623–13628. [PubMed] [Google Scholar]
- 8.Whitfield JF, Isaacs RJ, Jouishomme H, MacLean S, Chakravarthy BR, Morley P, et al. C-terminal fragment of parathyroid hormone-related protein, PTHrP-(107–111), stimulates membrane-associated protein kinase C activity and modulates the proliferation of human and murine skin keratinocytes. J Cell Physiol. 1996;166:1–11. doi: 10.1002/(SICI)1097-4652(199601)166:1<1::AID-JCP1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 9.Henderson JE, Amizuka N, Warshawsky H, Biasotto D, Lanske BM, Goltzman D, et al. Nucleolar localization of parathyroid hormone-related peptide enhances survival of chondrocytes under conditions that promote apoptotic cell death. Mol Cell Biol. 1995;15:4064–4075. doi: 10.1128/mcb.15.8.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luparello C, Burtis WJ, Raue F, Birch MA, Gallagher JA. Parathyroid hormone-related peptide and 8701-BC breast cancer cell growth and invasion in vitro: evidence for growth-inhibiting and invasion-promoting effects. Mol Cell Endocrinol. 1995;111:225–232. doi: 10.1016/0303-7207(95)03577-t. [DOI] [PubMed] [Google Scholar]
- 11.Rabbani SA, Gladu J, Liu B, Goltzman D. Regulation in vivo of the growth of Leydig cell tumors by antisense ribonucleic acid for parathyroid hormone-related peptide. Endocrinology. 1995;136:5416–5422. doi: 10.1210/endo.136.12.7588290. [DOI] [PubMed] [Google Scholar]
- 12.Iwamura M, Abrahamsson PA, Foss KA, Wu G, Cockett AT, Deftos LJ. Parathyroid hormone-related protein: a potential autocrine growth regulator in human prostate cancer cell lines. Urology. 1994;43:675–679. doi: 10.1016/0090-4295(94)90183-x. [DOI] [PubMed] [Google Scholar]
- 13.Burton PB, Moniz C, Knight DE. Parathyroid hormone related peptide can function as an autocrine growth factor in human renal cell carcinoma. Biochem Biophys Res Commun. 1990;167:1134–1138. doi: 10.1016/0006-291x(90)90641-y. [DOI] [PubMed] [Google Scholar]
- 14.Falzon M, Du P. Enhanced growth of MCF-7 breast cancer cells overexpressing parathyroid hormone-related peptide. Endocrinology. 2000;141:1882–1892. doi: 10.1210/endo.141.5.7470. In Process Citation. [DOI] [PubMed] [Google Scholar]
- 15.Dougherty KM, Blomme EA, Koh AJ, Henderson JE, Pienta KJ, Rosol TJ, et al. Parathyroid hormone-related protein as a growth regulator of prostate carcinoma. Cancer Res. 1999;59:6015–6022. [PubMed] [Google Scholar]
- 16.Mangin M, Ikeda K, Dreyer BE, Broadus AE. Identification of an up-stream promoter of the human parathyroid hormone-related peptide gene. Mol Endocrinol. 1990;4:851–858. doi: 10.1210/mend-4-6-851. [DOI] [PubMed] [Google Scholar]
- 17.Campos RV, Wang C, Drucker DJ. Regulation of parathyroid hormone-related peptide (PTHrP) gene transcription. Mol Endocrinol. 1992;6:1642–1652. doi: 10.1210/mend.6.10.1280327. [DOI] [PubMed] [Google Scholar]
- 18.Suva LJ, Mather KA, Gillespie MT, Webb GC, Ng KW, Winslow GA, et al. Structure of the 5′ flanking region of the gene encoding human parathyroid-hormone-related protein (PTHrP) Gene. 1989;77:95–105. doi: 10.1016/0378-1119(89)90363-6. [DOI] [PubMed] [Google Scholar]
- 19.Vasavada RC, Wysolmerski JJ, Broadus AE, Philbrick WM. Identification and characterization of a GC-rich promoter of the human parathyroid hormone-related peptide gene. Mol Endocrinol. 1993;7:273–282. doi: 10.1210/mend.7.2.8469240. [DOI] [PubMed] [Google Scholar]
- 20.Southby J, O'Keeffe LM, Martin TJ, Gillespie MT. Alternative promoter usage and mRNA splicing pathways for parathyroid hormone-related protein in normal tissues and tumours. Br J Cancer. 1995;72:702–707. doi: 10.1038/bjc.1995.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Southby J, Murphy LM, Martin TJ, Gillespie MT. Cell-specific and regulator-induced promoter usage and messenger ribonucleic acid splicing for parathyroid hormone-related protein. Endocrinology. 1996;137:1349–1357. doi: 10.1210/endo.137.4.8625910. [DOI] [PubMed] [Google Scholar]
- 22.Richard V, Luchin A, Brena RM, Plass C, Rosol TJ. Quantitative evaluation of alternative promoter usage and 3′ splice variants for parathyroid hormone-related protein by real-time reverse transcription-PCR. Clin Chem. 2003;49:1398–1402. doi: 10.1373/49.8.1398. [DOI] [PubMed] [Google Scholar]
- 23.Good L, Maggirwar SB, Sun SC. Activation of the IL-2 gene promoter by HTLV-I tax involves induction of NF-AT complexes bound to the CD28-responsive element. EMBO J. 1996;15:3744–3750. [PMC free article] [PubMed] [Google Scholar]
- 24.Ballard DW, Bohnlein E, Lowenthal JW, Wano Y, Franza BR, Greene WC. HTLV-I tax induces cellular proteins that activate the kappa B element in the IL-2 receptor alpha gene. Science. 1988;241:1652–1655. doi: 10.1126/science.241.4873.1652. [DOI] [PubMed] [Google Scholar]
- 25.Ohtani K, Tsujimoto A, Tsukahara T, Numata N, Miura S, Sugamura K, et al. Molecular mechanisms of promoter regulation of the gp34 gene that is trans-activated by an oncoprotein Tax of human T cell leukemia virus type I. J Biol Chem. 1998;273:14119–14129. doi: 10.1074/jbc.273.23.14119. [DOI] [PubMed] [Google Scholar]
- 26.Duyao MP, Kessler DJ, Spicer DB, Sonenshein GE. Transactivation of the c-myc gene by HTLV-1 tax is mediated by NFkB. Curr Top Microbiol Immunol. 1992;182:421–424. doi: 10.1007/978-3-642-77633-5_53. [DOI] [PubMed] [Google Scholar]
- 27.Toyoshima H, Itoh M, Inoue J, Seiki M, Takaku F, Yoshida M. Secondary structure of the human T-cell leukemia virus type 1 rex-responsive element is essential for rex regulation of RNA processing and transport of unspliced RNAs. J Virol. 1990;64:2825–2832. doi: 10.1128/jvi.64.6.2825-2832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Askjaer P, Kjems J. Mapping of multiple RNA binding sites of human T-cell lymphotropic virus type I rex protein within 5′- and 3′-Rex response elements. J Biol Chem. 1998;273:11463–11471. doi: 10.1074/jbc.273.19.11463. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe T, Yamaguchi K, Takatsuki K, Osame M, Yoshida M. Constitutive expression of parathyroid hormone-related protein gene in human T cell leukemia virus type 1 (HTLV-1) carriers and adult T cell leukemia patients that can be trans-activated by HTLV-1 tax gene. J Exp Med. 1990;172:759–765. doi: 10.1084/jem.172.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dittmer J, Gitlin SD, Reid RL, Brady JN. Transactivation of the P2 promoter of parathyroid hormone-related protein by human T-cell lymphotropic virus type I Tax1: evidence for the involvement of transcription factor Ets1. J Virol. 1993;67:6087–6095. doi: 10.1128/jvi.67.10.6087-6095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ejima E, Rosenblatt JD, Massari M, Quan E, Stephens D, Rosen CA, et al. Cell-type-specific transactivation of the parathyroid hormone-related protein gene promoter by the human T-cell leukemia virus type I (HTLV-I) tax and HTLV-II tax proteins. Blood. 1993;81:1017–1024. [PubMed] [Google Scholar]
- 32.Dittmer J, Pise-Masison CA, Clemens KE, Choi KS, Brady JN. Interaction of human T-cell lymphotropic virus type I Tax, Ets1, and Sp1 in transactivation of the PTHrP P2 promoter. J Biol Chem. 1997;272:4953–4958. doi: 10.1074/jbc.272.8.4953. [DOI] [PubMed] [Google Scholar]
- 33.Franchini G, Wong-Staal F, Gallo RC. Human T-cell leukemia virus (HTLV-I) transcripts in fresh and cultured cells of patients with adult T-cell leukemia. Proc Natl Acad Sci USA. 1984;81:6207–6211. doi: 10.1073/pnas.81.19.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinoshita T, Shimoyama M, Tobinai K, Ito M, Ito S, Ikeda S, et al. Detection of mRNA for the tax1/rex1 gene of human T-cell leukemia virus type I in fresh peripheral blood mononuclear cells of adult T-cell leukemia patients and viral carriers by using the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:5620–5624. doi: 10.1073/pnas.86.14.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikeda K, Okazaki R, Inoue D, Ohno H, Ogata E, Matsumoto T. Interleukin-2 increases production and secretion of parathyroid hormone-related peptide by human T cell leukemia virus type I-infected T cells: possible role in hypercalcemia associated with adult T cell leukemia. Endocrinology. 1993;132:2551–2556. doi: 10.1210/endo.132.6.8099324. [DOI] [PubMed] [Google Scholar]
- 36.Gotoh A, Kitazawa S, Mizuno Y, Takenaka A, Arakawa S, Matsumoto O, et al. Common expression of parathyroid hormone-related protein and no correlation of calcium level in renal cell carcinomas. Cancer. 1993;71:2803–2806. doi: 10.1002/1097-0142(19930501)71:9<2803::aid-cncr2820710919>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 37.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 38.Cataisson C, Gordon J, Roussiere M, Abdalkhani A, Lindemann R, Dittmer J, et al. Ets-1 activates parathyroid hormone-related protein gene expression in tumorigenic breast epithelial cells. Mol Cell Endocrinol. 2003;204:155–168. doi: 10.1016/s0303-7207(02)00298-8. [DOI] [PubMed] [Google Scholar]
- 39.Lindemann RK, Ballschmieter P, Nordheim A, Dittmer J. Transforming growth factor beta regulates parathyroid hormone-related protein expression in MDA-MB-231 breast cancer cells through a novel Smad/Ets synergism. J Biol Chem. 2001;276:46661–46670. doi: 10.1074/jbc.M105816200. [DOI] [PubMed] [Google Scholar]
- 40.Lindemann RK, Braig M, Ballschmieter P, Guise TA, Nordheim A, Dittmer J. Protein kinase Calpha regulates Ets1 transcriptional activity in invasive breast cancer cells. Int J Oncol. 2003;22:799–805. [PubMed] [Google Scholar]
- 41.Foley J, Wysolmerski JJ, Missero C, King CS, Philbrick WM. Regulation of parathyroid hormone-related protein gene expression in murine keratinocytes by E1A isoforms: a role for basal promoter and Ets- 1 site. Mol Cell Endocrinol. 1999;156:13–23. doi: 10.1016/s0303-7207(99)00151-3. [DOI] [PubMed] [Google Scholar]
- 42.Richard V, Lairmore MD, Green PL, Feuer G, Erbe RS, Albrecht B, et al. Humoral hypercalcemia of malignancy: severe combined immunodeficient/beige mouse model of adult T-cell lymphoma independent of human T-cell lymphotropic virus type-1 tax expression. Am J Pathol. 2001;158:2219–2228. doi: 10.1016/S0002-9440(10)64694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feuer G, Zack JA, Harrington WJ, Jr, Valderama R, Rosenblatt JD, Wachsman W, et al. Establishment of human T-cell leukemia virus type I T-cell lymphomas in severe combined immunodeficient mice. Blood. 1993;82:722–731. [PubMed] [Google Scholar]
- 44.Cataisson C, Gordon J, Roussiere M, Abdalkhani A, Lindemann R, Dittmer J, et al. Ets-1 activates parathyroid hormone-related protein gene expression in tumorigenic breast epithelial cells. Mol Cell Endocrinol. 2003;204:155–168. doi: 10.1016/s0303-7207(02)00298-8. [DOI] [PubMed] [Google Scholar]
- 45.Lindemann RK, Ballschmieter P, Nordheim A, Dittmer J. Transforming growth factor beta regulates parathyroid hormone-related protein expression in MDA-MB-231 breast cancer cells through a novel Smad/Ets synergism. J Biol Chem. 2001;276:46661–46670. doi: 10.1074/jbc.M105816200. [DOI] [PubMed] [Google Scholar]
- 46.Huang W, Erikson RL. Constitutive activation of Mek1 by mutation of serine phosphorylation sites. Proc Natl Acad Sci USA. 1994;91:8960–8963. doi: 10.1073/pnas.91.19.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li XH, Gaynor RB. Mechanisms of NF-kappaB activation by the HTLV type 1 tax protein. AIDS Res Hum Retroviruses. 2000;16:1583–1590. doi: 10.1089/08892220050192994. [DOI] [PubMed] [Google Scholar]
- 48.Ferrari SL, Rizzoli R, Bonjour JP. Effects of epidermal growth factor on parathyroid hormone-related protein production by mammary epithelial cells. J Bone Miner Res. 1994;9:639–644. doi: 10.1002/jbmr.5650090508. [DOI] [PubMed] [Google Scholar]
- 49.Okano K, Pirola CJ, Wang HM, Forrester JS, Fagin JA, Clemens TL. Involvement of cell cycle and mitogen-activated pathways in induction of parathyroid hormone-related protein gene expression in rat aortic smooth muscle cells. Endocrinology. 1995;136:1782–1789. doi: 10.1210/endo.136.4.7895691. [DOI] [PubMed] [Google Scholar]
- 50.Rodan SB, Wesolowski G, Ianacone J, Thiede MA, Rodan GA. Production of parathyroid hormone-like peptide in a human osteosarcoma cell line: stimulation by phorbol esters and epidermal growth factor. J Endocrinol. 1989;122:219–227. doi: 10.1677/joe.0.1220219. [DOI] [PubMed] [Google Scholar]
- 51.Lindemann RK, Braig M, Hauser CA, Nordheim A, Dittmer J. Ets2 and protein kinase C epsilon are important regulators of parathyroid hormone-related protein expression in MCF-7 breast cancer cells. Biochem J. 2003;372(Part 3):787–797. doi: 10.1042/BJ20030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chatila T, Silverman L, Miller R, Geha R. Mechanisms of T cell activation by the calcium ionophore ionomycin. J Immunol. 1989;143:1283–1289. [PubMed] [Google Scholar]
- 53.Mori N, Fujii M, Ikeda S, Yamada Y, Tomonaga M, Ballard DW, et al. Constitutive activation of NF-kappaB in primary adult T-cell leukemia cells. Blood. 1999;93:2360–2368. [PubMed] [Google Scholar]
- 54.Mori N, Fujii M, Iwai K, Ikeda S, Yamasaki Y, Hata T, et al. Constitutive activation of transcription factor AP-1 in primary adult T-cell leukemia cells. Blood. 2000;95:3915–3921. [PubMed] [Google Scholar]
- 55.Takemoto S, Mulloy JC, Cereseto A, Migone TS, Patel BK, Matsuoka M, et al. Proliferation of adult T cell leukemia/lymphoma cells is associated with the constitutive activation of JAK/STAT proteins. Proc Natl Acad Sci USA. 1997;94:13897–13902. doi: 10.1073/pnas.94.25.13897. [DOI] [PMC free article] [PubMed] [Google Scholar]


