Abstract
Calcineurin/NFAT signaling is involved in multiple aspects of skeletal muscle development and disease. The myogenic basic helix-loop-helix transcription factors, MyoD, myogenin, Myf5, and MRF4 specify the myogenic lineage. Here we show that calcineurin/NFAT (nuclear factor of activated T cells) signaling is required for primary myogenesis by transcriptional cooperation with the basic helix-loop-helix transcription factor MyoD. Calcineurin/NFAT signaling is involved in myogenin expression in differentiating myoblasts, where the myogenic regulatory factor MyoD synergistically cooperates with NFATc2/c3 at the myogenin promoter. Using gel shift and chromatin immunoprecipitation assays, we identified two conserved NFAT binding sites in the myogenin promoter that were occupied by NFATc3 upon skeletal muscle differentiation. The transcriptional integration between NFATc3 and MyoD is crucial for primary myogenesis in vivo, as myogenin expression is weak in myod:nfatc3 double null embryos, whereas myogenin expression is unaffected in embryos with null mutations for either factor alone. Thus, the combined findings provide a novel transcriptional paradigm for the first steps of myogenesis, where a calcineurin/NFATc3 pathway regulates myogenin induction in cooperation with MyoD during myogenesis.
In vertebrates all trunk muscles originate from the dermomyotome, an epithelial sheet formed by the paraxial mesoderm that develops from the dorsal part of the epithelial somite and overlays the sclerotome. Vertebrate skeletal muscle develops through the fusion of a variable number of myoblasts, muscle precursors committed to the skeletal muscle lineage within the myotome, to form syncytial myofibers (1). Myf5 is the first myogenic regulatory protein expressed in the skeletal muscle lineage. In concert with Pax3, Myf5 activates a network of myogenic regulatory factors including MyoD, myogenin, and MRF4 (Myf-6) in the muscle precursors to initiate and maintain the expression of muscle-specific genes (for reviewed, see Ref. 2). Genetic studies indicate that both Myf5 and MRF4 act upstream of MyoD to specify the myogenic lineage (3, 4), whereas myogenin has a crucial role in the terminal differentiation of committed muscle cells (5, 6). Protein motifs conserved in MyoD and Myf5 are necessary to initiate the expression of a subset of genes critical for the myogenic program, including transcription of the myogenin gene (7, 8).
The second messenger calcium regulates many signaling pathways critical for skeletal muscle homeostasis. A number of studies demonstrate that the calcium/calmodulin-dependent protein phosphatase calcineurin plays a regulatory role in skeletal muscle adaptation and muscle regeneration by its ability to promote myotube differentiation (9, 10). Calcineurin dephosphorylates members of the nuclear factor of activated T cells (NFAT)2 transcription factor family, allowing NFAT to translocate to the nucleus where it cooperates with other transcription factors to induce transcription of target genes. Five NFAT genes have been identified, NFATc1-c4 and NFAT5 (11). Forced calcineurin activity provokes nuclear translocation of NFATc3 and differentiation of myoblasts. These in vitro results are consistent with the muscle phenotype of nfatc3 null mice, which display reduced muscle mass due to a decrease in the number of myofibers (10). Although the precise mechanisms by which this occurs remain unresolved, these findings suggest that NFATc3 may serve a specialized role in primary myogenesis.
In this study we provide mechanistic insights as to how calcineurin/NFAT signaling regulates primary myogenesis. We show that calcineurin/NFAT signaling induces myogenin expression in differentiating C2C12 cells by transcriptional cooperation with the basic helix-loop-helix (bHLH) transcription factor MyoD. Our data demonstrate that the myogenic regulatory factor MyoD cooperates with NFATc2/c3 to activate the myogenin promoter in differentiating myoblasts. We demonstrate that NFATc3 and MyoD both play a crucial role in somite differentiation as double null myod/nfactc3 embryos express very low amounts of myogenin transcripts, whereas myogenin expression in mice with single null mutations for either nfatc3 or myod is unaffected. Thus, a calcineurin/NFATc3 pathway plays a crucial role in the first steps of myogenesis by regulating myogenin induction in cooperation with MyoD.
EXPERIMENTAL PROCEDURES
Animals—Myod, nfatc3, and nfatc2 null mice were generously provided by Shahragim Tajbakhsh and Laurie Glimcher and were described previously (3, 12, 13). Double null myod: nfatc2 and myod:nfatc3 mice were generated by cross-breeding single knock out mice. All protocols were performed according to institutional guidelines and approved by local Animal Care and Use Committees.
Processing of Embryos and Detection of Transcripts—Embryos were processed for whole mount in situ hybridization (WM-ISH) (14). Riboprobes for myogenin and paraxis were as described previously (15). For comparative WM-ISH experiments, age-matched and litter-matched embryos were used with independent probe sets and litters, and the ISH reactions were stopped at the same time.
Cell Culture, Transfections, and Adenoviruses—Cell culture of C2C12 and COS7 cells was described previously (16). Transfections were performed in 48-well plates using FuGENE 6 (Roche Applied Science) and the dual luciferase system (Promega). HEK-293 cells were cultured and transfected by the modified calcium phosphate method as described (17). AdGFP was generated as described previously (18). AdVIVIT was generated by using VIVIT-eGFP (19) and the pAdTrack-CMV system (20). AdNFAT9mer-luc was generated via ligation of NFAT9mer-Luc in vector pDC511 and FLP mediated recombination with pBGHfrt (Microbix). Small interfering RNA (NFATc3: s70514, Ambion; NFATc2: s70506, Ambion) transfections were performed in 24-well plates using DharmaFECT3 (Dharmacon) 48 h after regular transfections. Luciferase activity was measured 48 h later using the dual luciferase system (Promega).
Supplemental Material—The supplemental material includes a supplemental figure with WM-ISH for paraxis in embryos with different genotypes and an “Expanded Methods” section providing details about expression vectors, real-time PCR detection methods, electromobility shift assays, chromatin immunoprecipitation assays, and coimmunoprecipitation assays.
Statistical Analysis—The results are presented as the means ± S.E. Statistical analyses were performed using INSTAT 3.0 software (GraphPad, San Diego, CA) and Student's t test or analysis of variance followed by Tukey's post-test when appropriate. Statistical significance was accepted at a p value < 0.05.
RESULTS
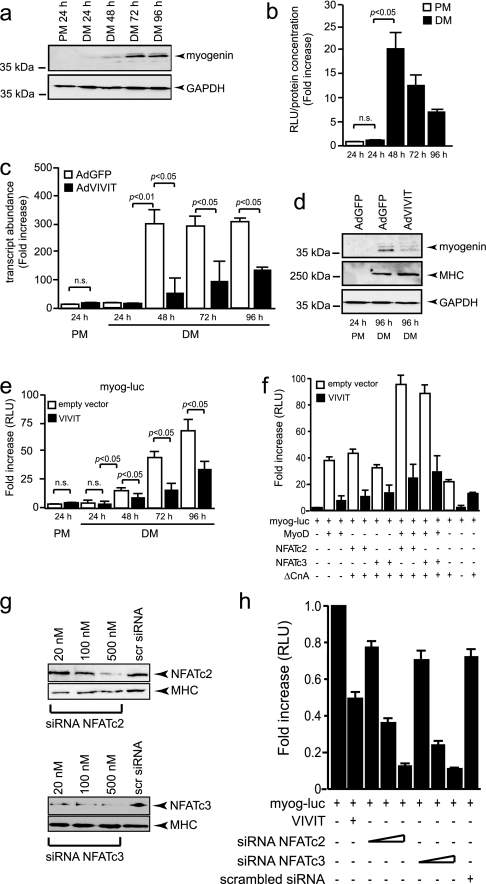
Calcineurin/NFAT Signaling Is Involved in Myogenin Expression—Genetic studies indicate that MyoD and Myf5 are necessary to specify the skeletal muscle lineage, whereas myogenin (Myog) has a critical role in the terminal differentiation of the specified muscle cells (5, 6). We first analyzed the timing of Myog induction in cultured C2C12 skeletal myoblasts upon differentiation to skeletal myotubes. Western blot analyses reveal a slight increase in Myog protein after 48 h of differentiation which is dramatically enhanced after 72 and 96 h of differentiation (Fig. 1a). Next, we induced myotube differentiation of myoblasts pre-infected with an adenovirus harboring 9 copies of the consensus NFAT-binding site from the interleukin-4 gene (9-mer) upstream of a minimal TATA box-driving luciferase (AdNFAT-luc) to track the activity of endogenous NFAT in skeletal myoblasts. In either proliferation medium (PM) or upon differentiation for 24 h, endogenous NFAT transcriptional activity was barely detectable (Fig. 1b). In contrast, NFAT activity is massively and transiently increased in C2C12 starting at 48 h of differentiation (Fig. 1b), a time point that corresponds with the induction of Myog (Fig. 1a).
FIGURE 1.
NFAT transcriptional activity is involved in myogenin expression. a, C2C12 myoblasts were cultured in PM or differentiation medium (DM) and analyzed for myogenin protein abundance using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as loading control. b, C2C12 myoblasts were infected with AdNFAT9mer-luc, grown in PM or DM, and luciferase induction-analyzed. Data are provided as the mean ± S.E. (n = 3) as -fold increase of the ratio relative light units/protein content in differentiation over proliferation. c and d, C2C12 cells were infected with either control AdGFP or AdVIVIT adenoviruses and cultured in PM or DM for indicated time periods, and myogenin transcript (c) or protein abundance (d) was analyzed by real-time PCR or Western blotting, respectively. The data in c represent the mean ± S.E. of two independent experiments. e, co-transfection assays in C2C12 cells culture in PM or DM using a 0.6-kb myog-luc vector in the absence (empty vector) or presence of VIVIT-eGFP expression vector demonstrates the involvement of endogenous NFAT transcriptional activity in induction of the myog-luc vector. n.s., not significant. f, co-transfection assays in C2C12 cells cultured 96 h in DM using a 0.6-kb myog-luc vector and the indicated expression vectors in the absence (empty vector) or presence of VIVIT-eGFP expression vector demonstrates a synergistic cooperation between MyoD and either NFATc2 or NFATc3. g, Western blot analysis to demonstrate efficient knockdown of NFATc2 (upper panel) or NFATc3 (lower panel) in C2C12 cells transfected with increasing doses of small interfering RNA specific for NFATc2 or NFATc3. Myosin heavy chain (MHC) was used as loading control. siRNA, small interfering RNA. h, transfection assay in C2C12 cells culture in DM using a 0.6-kb myog-luc vector in the presence of VIVIT-eGFP expression vector or increasing amounts of small interfering RNA directed against NFATc2 or NFATc3 (20, 100, and 500 nm) indicates that NFATc2 and NFATc3 participate to the activation of the myogenin reporter in culture. RLU, relative light units.
To determine whether NFAT transcriptional activity is required for the induction of Myog upon differentiation, myoblasts were infected with either a control adenovirus (AdGFP) or an adenovirus expressing a fusion between GFP and the high affinity peptide VIVIT, which specifically inhibits calcineurin-mediated activation of NFAT (19). Cells were allowed to differentiate into myotubes, and Myog transcript and protein abundance were documented. AdGFP-infected myoblasts displayed a dramatic induction of Myog transcripts after 48 h of differentiation (Fig. 1c), which mirrors the induction of Myog protein expression (Fig. 1a). In contrast, AdVIVIT-infected cells express substantially lower amounts of Myog transcripts at every time point analyzed (Fig. 1c). The inhibition of Myog induction by VIVIT-mediated inhibition of NFAT transcriptional activity was confirmed at the protein level (Fig. 1d). Under these conditions, myosin heavy chain induction was not affected by AdVIVIT infection, suggesting a specific effect of calcineurin/NFAT signaling on Myog induction (Fig. 1d).
To determine whether endogenous calcineurin/NFAT signaling can activate the myog promoter, a DNA fragment extending from +73 to –522 bp relative to the transcription initiation site of the mouse myog gene was fused to a luciferase reporter (myog-luc) and transfected in myoblasts, and cells were allowed to differentiate to myotubes. Transcriptional activity of the myog promoter was very low in myoblasts. Myog-luc was strongly activated after 48 h of differentiation, mimicking the temporal activation profile of endogenous myog transcripts (Fig. 1c), with a maximum at 96 h (72 ± 6-fold increase; Fig. 1e). In contrast, when myog-luc was co-transfected with an expression vector expressing a fusion between GFP and the high affinity peptide VIVIT, activation of the myog promoter construct was significantly abrogated at each time point analyzed (Fig. 1e).
Next, to analyze whether forced overexpression of calcineurin/NFAT and MyoD could activate myog-luc, we performed transient co-transfection assays (Fig. 1f). Luciferase activity was induced in C2C12 cells cultured 96 h in differentiating medium and transfected with MyoD, NFATc2, or NFATc3. Combined transfections of MyoD with either NFATc2 or NFATc3 were required to get a maximal induction of luciferase activity compared with C2C12 cells transfected with myog-luc alone. The induction of luciferase activity was partially abrogated when the expression vector VIVIT was cotransfected. These results indicate the existence of synergistic cooperation between MyoD and NFATc2 or NFATc3 to activate the myogenin reporter in transfected differentiating C2C12 cells (Fig. 1f).
To test whether endogenous NFATc2 and/or NFATc3 cooperate to activate myogenin expression, differentiating C2C12 cells were cotransfected with the myog-luc reporter and small interfering RNAs directed against either NFATc2 or NFATc3 (Fig. 1g). This transfection assay resulted in a dose-dependent reduction of luciferase activity (Fig. 1h), suggesting that both endogenous NFATc2 and NFATc3 are able to participate in a transactivation complex activating myogenin expression in differentiating myoblasts. Conclusively, these findings demonstrate that NFAT activity is induced upon differentiation of myoblasts to myotubes and that the two isoforms NFATc2 and NFATc3 are involved in myogenin expression in culture.
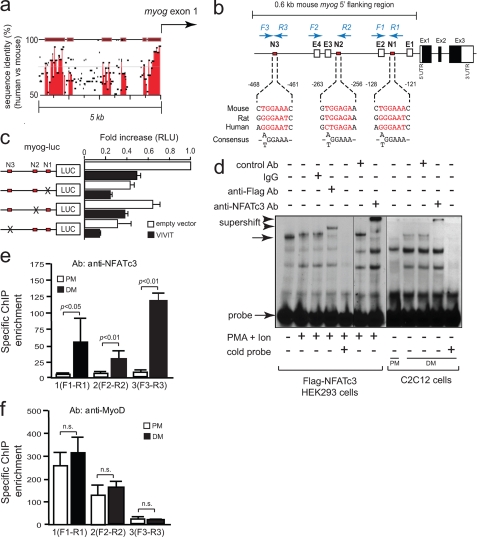
NFATc3 Directly Activates the Myog Promoter—To define the mechanisms behind the regulation of Myog by calcineurin/NFAT signaling, we searched for enhancers that might regulate myog transcription in vivo. Comparison of genomic sequences across species using rVISTA revealed several regions with conservation between human and mouse within a 5.0-kb 5′-flanking genomic region of myog (Fig. 2a). Within the most proximal 0.6-kb part of murine myog, 3 potential NFAT consensus binding sites ((T/A)GGAAA) were identified which we designated N1, N2, and N3 (Fig. 2b). By transfection assays we observed that myogenin reporter activity was mainly altered when myog-N1 or myog-N3 binding sites were mutated (Fig. 2c), suggesting that among the three NFAT binding sites, myog-N1 and myog-N3 could be considered as the most relevant NFAT binding sites within the most proximal part of the myogenin promoter.
FIGURE 2.
Presence of functional NFAT consensus binding sites in the myogenin promoter. a, comparison of the myog genomic regions between mouse and human. Percentage conservation of a 5′ 5.0-kb genomic region upstream myogenin first exon is shown. b, schematic presentation of 0.6-kb 5′-flanking region in mouse myog and location of NFAT binding sites (N1, N2, N3) and E-boxes (E1, E2, E3, E4). Primers yielding chromatin immunoprecipitation amplicons are indicated in blue. c, C2C12 cells were transiently transfected with the 0.6-kb myog-luc vector and the same construct containing site-directed mutations for the myog-N1, myog-N2, or myog-N3 NFAT binding sites in the absence (empty vector) or presence of VIVIT-eGFP expression vector. Cells were cultured 96 h in DM and assayed for luciferase activity. RLU, relative light units. d, nuclear extracts from HEK293 cells transfected with FLAG-tagged NFATc3 either unstimulated or stimulated with phorbol ester plus calcium ionophore (PMA + Ion) for 4 h and nuclear extracts from C2C12 cells grown in PM or in DM were analyzed by electrophoretic mobility shift assay. Gel shift assays were performed using an oligonucleotide probe of the NFAT-like site myog-N1 from the myog promoter in the presence or absence of 0.8 μg of the indicated antibodies. Ab, antibody. A 20-fold molar excess of unlabeled myog-N1 oligonucleotide was added to the binding reaction mixtures to determine the specific binding. Complexes are indicated by arrows, and retarded complexes are indicated by arrowheads. e and f, chromatin immunoprecipitation (ChIP) assays were performed on C2C12 cells grown in PM or DM with antibodies for NFATc3 (e) or MyoD (f). Bars represent -fold enrichment of amplicons with indicated primer sets (b), normalized to input controls obtained with primers spanning a non-coding genomic region 3′ of the myog gene. n.s., not significant.
To test the functionality of the sequences identified as NFAT binding elements, we performed an electrophoretic mobility shift assay with nuclear extracts of FLAG-tagged NFATc3-transfected HEK293 cells stimulated or not with phorbol ester plus calcium ionophore and using radiolabeled DNA probes of myog-N1, myog-N2, and myog-N3 sites (Fig. 2d). NFATc3 failed to bind myog-N2 and only weakly bound the myog-N3 probe in FLAG-NFATc3-transfected HEK293 cells (data not shown). In contrast, FLAG-tagged NFATc3 specifically bound the NFAT-like sequence N1 from the myog promoter, as FLAG-NFATc3 was specifically supershifted with the anti-FLAG antibody. A specific complex was also supershifted by a specific anti-NFATc3 antibody using nuclear extracts of differentiating C2C12 cells, indicating that endogenous NFATc3 also bound myog-N1. This specific complex was supershifted by an anti-NFATc3 antibody in a similar manner to that observed with nuclear extracts from HEK293 cells (Fig. 2d). Further evidence on the specificity of FLAG and endogenous NFATc3 binding was provided by competition experiments using a 20-fold molar excess of homologous oligonucleotide (Fig. 2d).
To further confirm the binding of NFATc3 to the promoter of myog, chromatin immunoprecipitation was carried out. C2C12 myoblasts were either maintained in PM or were allowed to differentiate for 48 h; the resultant nuclear fractions were immunoprecipitated using specific antibodies to NFATc3 or MyoD, and associated DNA was purified (Fig. 2e). Using specific primers to the myog promoter flanking the N1, N2, or N3 sites by real time PCR, all three PCR amplicons were observed to be significantly enriched in differentiated C2C12 cells compared with undifferentiated myoblasts (Fig. 2e). This association was specific for NFATc3 as enrichment of PCR products was not obtained when using beads alone or when using primers to an unrelated promoter, such as myoglobin (data not shown).
Conversely, chromatin immunoprecipitation analysis carried out with an antibody against MyoD demonstrated that MyoD was constitutively bound on the myog promoter in both proliferating and differentiating C2C12 cells (Fig. 2f). MyoD binding occurred roughly as efficiently to the E-boxes adjacent to N1 and N2, suggesting that E-boxes located in these regions are involved in myog gene activation. Taken together, these results indicate that calcineurin/NFAT signaling regulates the myog gene by direct transcriptional activation and unambiguously show the presence of endogenous MyoD and NFATc3 on the proximal myog promoter in vivo, supporting the idea that NFATc3 and the bHLH transcription factor MyoD may cooperate to activate myog transcription.
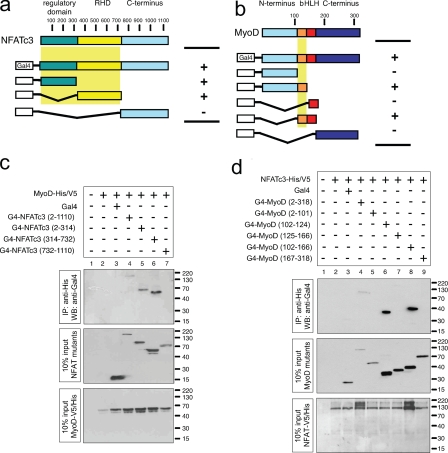
NFATc3 Physically Interacts with MyoD—Given that calcineurin/NFAT signaling is involved in Myog induction and MyoD is also capable of directly activating Myog expression, we tested whether NFATc3 may physically associate with MyoD to cooperatively activate Myog expression. To map the MyoD binding site(s) on NFATc3, a panel of NFATc3 deletion mutants was used in coimmunoprecipitation assays (Fig. 3a). Next, a C-terminal His/V5-tagged form of MyoD and Gal4-NFATc3 deletion constructs were coexpressed in COS7 cells and MyoD-immunoprecipitated with Ni-NTA beads, specific for the C-terminal His tag on MyoD. The presence of Gal4-NFATc3 deletion mutants was detected by immunoblotting against Gal4. Both the N-terminal regulatory domain of NFATc3 (residues 2–314) and the DNA binding Rel homology domain on NFATc3 (residues 314–732) interacted with MyoD (Fig. 3c, lanes 4–6). NFATc3 (732–1110) did not interact with MyoD (Fig. 3c, lane 7). We conclude that two separate domains located between residues 2–314 harboring the N-terminal regulatory domain and residues 314–732, which includes the Rel homology domain domain, respectively, are sufficient for MyoD binding.
FIGURE 3.
Mapping of NFATc3 and MyoD interaction. a and b, schematic overview of Gal4-NFATc3 (a) and Gal4-MyoD (b) deletion constructs and their ability to bind MyoD or NFATc3, respectively. RHD, Rel homology domains. c, protein extracts from COS7 cells transfected with MyoD-V5/His, empty vector (Gal4), and/or Gal4-NFATc3 deletion constructs were immunoprecipitated (IP) with Ni-NTA beads and subjected to Western blotting (WB) using an anti-Gal4 antibody. d, protein extracts from COS7 cells transfected with NFATc3-V5/His, empty vector (Gal4), and/or Gal4-MyoD deletion constructs were immunoprecipitated with Ni-NTA beads and subjected to Western blotting using an anti-Gal4 antibody.
Next, to map the NFATc3 binding site on MyoD, coimmunoprecipitation assays were also performed by using extracts from COS7 cells overexpressing epitope-tagged derivatives of MyoD. To this end a series of MyoD deletion mutants coupled to Gal4 (Fig. 3b) were coexpressed with a C-terminal His/V5-tagged full-length NFATc3 and an activated mutant of CnA. NFATc3-His/V5 was immunoprecipitated with Ni-NTA beads, and interacting MyoD mutants were identified with an antibody against Gal4. Deletion of N-terminal sequences up to amino acid 101 had no effect of MyoD binding to NFATc3 (Fig. 3d, lane 8). Likewise, deletion of C-terminal sequences from amino acid 167 to 318 had no noticeable effect on binding of MyoD to NFATc3 (Fig. 3d, lanes 6 and 8), indicating that interaction with NFATc3 centered on the bHLH region. Removal of residues between amino acids 125 and 166, the basic domain, led to a complete loss of MyoD binding (Fig. 3d, lanes 5, 7, and 9). In conclusion, these findings confirm that NFATc3 and MyoD physically interact and that residues 102–124 on MyoD, the basic domain of this class B bHLH transcript factor, and two separate domains on NFATc3 are required for this interaction.
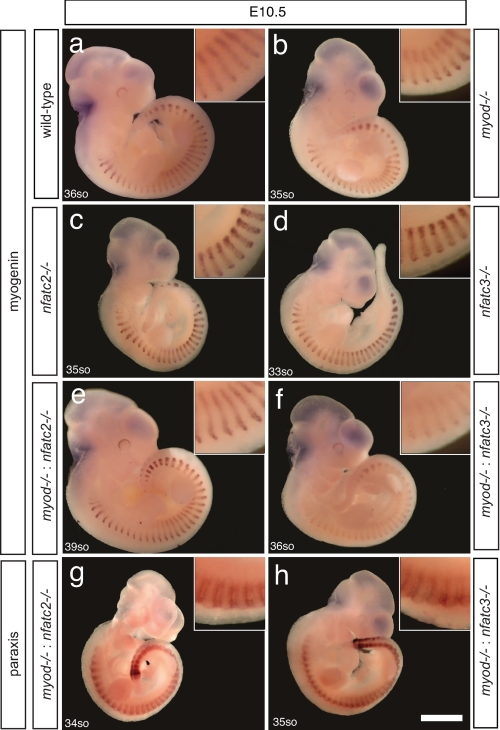
NFATc3 and MyoD Cooperatively Induce Myogenin Expression in Somites—The myogenic regulatory factors MyoD, Myog, Myf5, and MRF4 (Myf-6) regulate myogenesis in the developing embryo. Myog and MyoD are expressed in the myotome at E8.5 and E10.0, respectively (21). To analyze the implications of the uncovered transcriptional interaction between MyoD and NFATc3, we analyzed embryos deficient for myod, nfatc3, nfatc2 or combinations thereof for Myog transcript expression at E10.5. To this end we performed whole-mount in situ hybridization for myog in wild-type (Fig. 4a), myod null (Fig. 4b), nfatc2 null (Fig. 4c), and nfatc3 null (Fig. 4d) mice. Mutant embryos for myod, nfatc2, and nfatc3 displayed a level of myog expression in developing somites to approximately the same extent as in wild-type embryos (Fig. 4, a–d). Next, we analyzed myod:nfatc3 and myod:nfatc2 double null embryos. Remarkably, myog transcripts were expressed at very low amount in somites of myod: nfatc3 double null embryos. As myod:nfatc3 double null embryos die in utero between E12 and E15, the adult muscle phenotype cannot be analyzed. In contrast, myod: nfatc2 double null embryos displayed myog transcript levels to the same extent as somites from wild-type embryos, and no drastic muscle phenotype was observed in adult muscles. These data demonstrate the specificity of the interaction between MyoD and the NFATc3 isoform during myogenesis (Fig. 4, e and f).
FIGURE 4.
NFATc3 and MyoD are required for myogenin expression in myogenesis. Representative myogenin WM-ISH of wild type (a), myod null (b), nfatc2 null (c), nfatc3 null (d), myod:nfatc2 double null (e), and myod:nfatc3 double null (f) E10.5 embryos. Paraxis WM-ISH in myod:nfatc2 double null (g) and myod:nfatc3 double null (h) E10.5 embryos is shown. Five to ten embryos of each genotype were obtained and analyzed.
Paraxis, a member of the Twist subfamily of bHLH transcription factors, has been shown to regulate morphogenetic events during somitogenesis, including the transition of cells from mesenchyme to epithelium and maintaining anterior/posterior polarity (22). To exclude the possibility that the observed Myog phenotype derived from general somite dysmorphogenesis or premature embryonic death before E10.5, we performed whole-mount in situ hybridization for paraxis on myod:nfatc2 and myod:nfatc3 double null embryos (Fig. 4, g and h). Paraxis expression in myod:nfatc3 and myod:nfatc2 double null embryos was unchanged, which relieves concerns about the specificity of the observed down-regulation of Myog in myod:nfatc3 double null mice. Collectively, these data confirm that Myog induction during primary myogenesis in vivo is established by cooperative interaction between MyoD and calcineurin/NFATc3 signaling.
DISCUSSION
Calcineurin/NFAT Signaling Regulates Skeletal Muscle Development, Adaptation, and Regeneration—In the embryo initial muscle development is controlled by the myogenic regulatory factors Myf5, MyoD, myogenin, and MRF4 (Myf-6) (2) in conjunction with the MEF2 MADS-box family of transcription factors (23). Notably, of the single knock-out mice for the myogenic regulatory factors, only myog-null mice exhibit severe skeletal muscle deficiencies, thereby demonstrating its unique stance in embryonic muscle differentiation and suggesting the absence of redundant or compensatory mechanisms to substitute its function in vivo (5, 6, 24).
Reminiscent to the myogenic factors, the calcineurin/NFAT pathway represents another pathway involved in skeletal muscle differentiation and muscle regeneration. Calcineurin activity affects myogenic differentiation of cultured myoblasts in vitro (16), whereas during pupal development in Drosophila, mutation of canB2, which encodes a regulatory subunit of calcineurin, provokes severe defects in the organization of indirect flight muscles (25). Calcineurin/NFAT also regulates postnatal skeletal muscle hypertrophy and fiber-type switching. Transgenic mice expressing an activated calcineurin mutant in skeletal muscle exhibit an increase in type I fibers (26), whereas a constitutively active NFAT mutant stimulates the MyHC (myosin heavy chain) slow promoter in adult fast muscles (27). Conversely, calcineurin-deficient mice have a reduced oxidative slow muscle fiber-type profile (28), providing evidence that calcineurin/NFAT signaling acts as a nerve activity sensor and controls activity-dependent myosin switching in adult skeletal muscle. In response to injury, quiescent satellite cells become activated and migrate to the site of injury where they proliferate, differentiate, and fuse to form new myofibers. Calcineurin/NFAT signaling is required for muscle precursor cell differentiation and the regenerative capacity of postnatal skeletal muscle (29).
Rather than regulating muscle differentiation in parallel, here we demonstrate that MyoD and the calcineurin/NFAT pathway converge at the transcriptional level to initiate embryonic muscle differentiation by coactivating the myog gene with implications for proper differentiation of somite derivatives in vivo. Although at least three NFAT isoforms are expressed in skeletal muscle (30), the combined findings from single NFAT null mice now provide evidence that the individual NFAT isoforms have a unique role in skeletal muscle development. Although both NFATc2 and NFATc3 were able to synergistically activate Myog reporters with MyoD in C2C12 myoblasts, the muscle phenotype of nfatc2 null mice is distinct from nfatc3 mutant mice, where double myod:nfatc2 genetic deletion has no influence on myogenin expression, indicating the limitation of the in vitro experimental system studied. NFATc2 has been shown to participate in myofiber and myoblast fusion, leading to the growth of multinucleated muscle tubes (9). Collectively, the combined observations point to isoform-specific and temporally distinct contributions of NFAT transcription factors to skeletal muscle development.
Calcineurin/NFAT-dependent, Organ-specific Responses by Transcriptional Synergy—The present study provides for the first time evidence of a combinatorial NFAT/MyoD transcriptional pathway controlling gene expression in myogenic cells. Our observations are reminiscent of the cooperative transcriptional integration between the bHLH transcription factor MyoD and members of the MEF2 family of transcription factors, which also plays an essential role in gene activation during muscle differentiation (23). Interestingly, MEF2 proteins also directly interact with NFAT transcription factors. In T-lymphocytes, NFATc2 interacts directly with MEF2D in a synergistic transcriptional complex to activate the Nur77 gene (31), whereas a combinatorial MEF2/NFAT regulatory transcriptional pathway controls gene expression in cardiac muscle cells (18).
The HLH transcription factor family has been classified based upon tissue distribution, dimerization capabilities, and DNA-binding specificities (32). Class B HLH proteins, which include the myogenic bHLH proteins MyoD, Myf5, myogenin, and MRF4, show a tissue restricted pattern of expression and are required for vital developmental processes, including hematopoiesis, cardiogenesis, myogenesis, and neurogenesis (32).
In contrast to the restricted tissue distribution of class B HLH proteins, NFAT proteins are expressed ubiquitously yet individual or combinatorial loss of NFAT isoforms in mice reveals highly specific defects in cardiovascular, myogenic, neuronal, or immune cell lineages (10, 12, 33–35). Consequently, one vexing question in NFAT biology relates to how the ubiquitously expressed NFAT factors can induce organ and cell type-selective responses despite their ubiquitous expression pattern. We now demonstrate that during myogenesis this specificity results from the transcriptional synergy between one single NFAT isoform and myogenic lineage-restricted bHLH transcription factor MyoD. This involvement is specific to one NFAT isoform, NFATc3, as double null myod:nfatc2 mice displayed no defect in myogenin expression at this stage of myogenesis. It is tempting to speculate that similar transcriptional cooperation may also exist between NFAT transcription factors and other members of the class B HLH transcriptional regulators to specify gene expression in distinct developmental processes and other organs. Future studies will be required to assess whether and to what extent the transcriptional cooperation between HLH proteins and the calcineurin/NFAT pathway also impinges on postnatal skeletal muscle adaptation, other developmental processes, and in disease.
Supplementary Material
Acknowledgments
We thank Laurie Glimcher, Shahragim Tajbakhsh, Eric Olson, and Paul Hogan for providing mouse models and reagents. We are grateful to Robert Kelly, Michael Schneider, and Jeffery Molkentin for helpful discussions.
This work was supported by Netherlands Organization for Health Research and Development Grants 912-04-054 and 912-04-017 and VIDI Award 917-863-72, Netherlands Heart Foundation Program Grant NHS2007B167, and European Union Contract LSHM-CT-2005-018833/EUGeneHeart (to L. J. D. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure and “Expanded Methods.”
Footnotes
The abbreviations used are: NFAT, nuclear factor of activated T-cells; bHLH, basic helix-loop-helix (HLH); DM, differentiation medium; GFP, green fluorescent protein; Myog, myogenin; PM, proliferation medium; WM-ISH, whole mount in situ hybridization; kb, kilobase.
References
- 1.Schnorrer, F., and Dickson, B. J. (2004) Dev. Cell 7 9–20 [DOI] [PubMed] [Google Scholar]
- 2.Buckingham, M. (1996) Biochem. Soc. Trans. 24 506–509 [DOI] [PubMed] [Google Scholar]
- 3.Rudnicki, M. A., Schnegelsberg, P. N., Stead, R. H., Braun, T., Arnold, H. H., and Jaenisch, R. (1993) Cell 75 1351–1359 [DOI] [PubMed] [Google Scholar]
- 4.Kassar-Duchossoy, L., Gayraud-Morel, B., Gomes, D., Rocancourt, D., Buckingham, M., Shinin, V., and Tajbakhsh, S. (2004) Nature 431 466–471 [DOI] [PubMed] [Google Scholar]
- 5.Hasty, P., Bradley, A., Morris, J. H., Edmondson, D. G., Venuti, J. M., Olson, E. N., and Klein, W. H. (1993) Nature 364 501–506 [DOI] [PubMed] [Google Scholar]
- 6.Nabeshima, Y., Hanaoka, K., Hayasaka, M., Esumi, E., Li, S., and Nonaka, I. (1993) Nature 364 532–535 [DOI] [PubMed] [Google Scholar]
- 7.de la Serna, I. L., Ohkawa, Y., Berkes, C. A., Bergstrom, D. A., Dacwag, C. S., Tapscott, S. J., and Imbalzano, A. N. (2005) Mol. Cell. Biol. 25 3997–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edmondson, D. G., Cheng, T. C., Cserjesi, P., Chakraborty, T., and Olson, E. N. (1992) Mol. Cell. Biol. 12 3665–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horsley, V., Jansen, K. M., Mills, S. T., and Pavlath, G. K. (2003) Cell 113 483–494 [DOI] [PubMed] [Google Scholar]
- 10.Kegley, K. M., Gephart, J., Warren, G. L., and Pavlath, G. K. (2001) Dev. Biol. 232 115–126 [DOI] [PubMed] [Google Scholar]
- 11.Rao, A., Luo, C., and Hogan, P. G. (1997) Annu. Rev. Immunol. 15 707–747 [DOI] [PubMed] [Google Scholar]
- 12.Hodge, M. R., Ranger, A. M., Charles de la Brousse, F., Hoey, T., Grusby, M. J., and Glimcher, L. H. (1996) Immunity 4 397–405 [DOI] [PubMed] [Google Scholar]
- 13.Oukka, M., Ho, I. C., de la Brousse, F. C., Hoey, T., Grusby, M. J., and Glimcher, L. H. (1998) Immunity 9 295–304 [DOI] [PubMed] [Google Scholar]
- 14.Roelen, B. A., de Graaff, W., Forlani, S., and Deschamps, J. (2002) Mech. Dev. 119 81–90 [DOI] [PubMed] [Google Scholar]
- 15.Wright, W. E., Sassoon, D. A., and Lin, V. K. (1989) Cell 56 607–617 [DOI] [PubMed] [Google Scholar]
- 16.Delling, U., Tureckova, J., Lim, H. W., De Windt, L. J., Rotwein, P., and Molkentin, J. D. (2000) Mol. Cell. Biol. 20 6600–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez, A., Martinez-Martinez, S., Lopez-Maderuelo, M. D., Ortega-Perez, I., and Redondo, J. M. (2005) J. Biol. Chem. 280 9980–9984 [DOI] [PubMed] [Google Scholar]
- 18.van Oort, R. J., van Rooij, E., Bourajjaj, M., Schimmel, J., Jansen, M. A., van der Nagel, R., Doevendans, P. A., Schneider, M. D., van Echteld, C. J., and De Windt, L. J. (2006) Circulation 114 298–308 [DOI] [PubMed] [Google Scholar]
- 19.Aramburu, J., Yaffe, M. B., Lopez-Rodriguez, C., Cantley, L. C., Hogan, P. G., and Rao, A. (1999) Science 285 2129–2133 [DOI] [PubMed] [Google Scholar]
- 20.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W., and Vogelstein, B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sassoon, D., Lyons, G., Wright, W. E., Lin, V., Lassar, A., Weintraub, H., and Buckingham, M. (1989) Nature 341 303–307 [DOI] [PubMed] [Google Scholar]
- 22.Johnson, J., Rhee, J., Parsons, S. M., Brown, D., Olson, E. N., and Rawls, A. (2001) Dev. Biol. 229 176–187 [DOI] [PubMed] [Google Scholar]
- 23.Molkentin, J. D., and Olson, E. N. (1996) Curr. Opin. Genet. Dev. 6 445–453 [DOI] [PubMed] [Google Scholar]
- 24.Venuti, J. M., Morris, J. H., Vivian, J. L., Olson, E. N., and Klein, W. H. (1995) J. Cell Biol. 128 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gajewski, K., Wang, J., Molkentin, J. D., Chen, E. H., Olson, E. N., and Schulz, R. A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1040–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu, H., Rothermel, B., Kanatous, S., Rosenberg, P., Naya, F. J., Shelton, J. M., Hutcheson, K. A., DiMaio, J. M., Olson, E. N., Bassel-Duby, R., and Williams, R. S. (2001) EMBO J. 20 6414–6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCullagh, K. J., Calabria, E., Pallafacchina, G., Ciciliot, S., Serrano, A. L., Argentini, C., Kalhovde, J. M., Lomo, T., and Schiaffino, S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10590–10595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons, S. A., Wilkins, B. J., Bueno, O. F., and Molkentin, J. D. (2003) Mol. Cell. Biol. 23 4331–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakuma, K., Nishikawa, J., Nakao, R., Watanabe, K., Totsuka, T., Nakano, H., Sano, M., and Yasuhara, M. (2003) Acta Neuropathol (Berl.) 105 271–280 [DOI] [PubMed] [Google Scholar]
- 30.Bourajjaj, M., Armand, A. S., da Costa Martins, P. A., Weijts, B., van der Nagel, R., Heeneman, S., Wehrens, X. H., and De Windt, L. J. (2008) J. Biol. Chem. 283 22295–22303 [DOI] [PubMed] [Google Scholar]
- 31.Youn, H. D., Chatila, T. A., and Liu, J. O. (2000) EMBO J. 19 4323–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massari, M. E., and Murre, C. (2000) Mol. Cell. Biol. 20 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de la Pompa, J. L., Timmerman, L. A., Takimoto, H., Yoshida, H., Elia, A. J., Samper, E., Potter, J., Wakeham, A., Marengere, L., Langille, B. L., Crabtree, G. R., and Mak, T. W. (1998) Nature 392 182–186 [DOI] [PubMed] [Google Scholar]
- 34.Graef, I. A., Chen, F., Chen, L., Kuo, A., and Crabtree, G. R. (2001) Cell 105 863–875 [DOI] [PubMed] [Google Scholar]
- 35.Graef, I. A., Wang, F., Charron, F., Chen, L., Neilson, J., Tessier-Lavigne, M., and Crabtree, G. R. (2003) Cell 113 657–670 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.