Abstract
The Arabidopsis thaliana mutant psbo1 contains a point mutation in the psbO-1 gene (At5g66570) leading to the loss of expression of the PsbO-1 protein and overexpression of the PsbO-2 protein (Murakami, R., Ifuku, K., Takabayashi, A., Shikanai, T., Endo, T., and Sato, F. (2002) FEBS Lett. 523, 138–142). Previous characterization of fluorescence induction and decay kinetics by our laboratory documented defects on both the oxidizing and reducing sides of Photosystem II. Additionally, anomalous flash oxygen yield patterns indicated that the mutant contains a defective oxygen-evolving complex that appears to exhibit anomalously long-lived S2 and S3 oxidation states (Liu, H., Frankel, L. K., and Bricker, T. M. (2007) Biochemistry 46, 7607–7613). In this study, we have documented that the S2 and S3 states in psbo1 thylakoids decay very slowly. The total flash oxygen yield of the psbo1 mutant was also significantly reduced, as was its stability. Incubation of psbo1 thylakoids at high NaCl concentrations did not increase the rate of S2 and S3 state decay. The oxygen-evolving complexes of the mutant did, however, exhibit somewhat enhanced stability following this treatment. Incubation with CaCl2 had a significantly more dramatic effect. Under this condition, both the S2 and S3 states of the mutant decayed at nearly the same rate as the wild type, and the total oxygen yield and its stability following CaCl2 treatment were indistinguishable from that of the wild type. These results strongly suggest that the principal defect in the psbo1 mutant is an inability to effectively utilize the calcium associated with Photosystem II. We hypothesize that the PsbO-2 protein cannot effectively sequester calcium at the oxygen-evolving site.
Photosystem II (PS II)2 functions as a light-driven, water-plastoquinone oxidoreductase. In higher plants and cyanobacteria at least six intrinsic proteins appear to be required for O2 evolution (1–3). These are CP47, CP43, D1, D2, and the α and β subunits of cytochrome b559. Deletion of these subunits uniformly results in the loss of PS II function and assembly (4). Additionally, in higher plants, three extrinsic proteins, with apparent molecular masses of 33 kDa (PsbO), 24 kDa (PsbP), and 16 kDa (PsbQ), are also required for maximal rates of O2 evolution at physiological inorganic cofactor concentrations. Of these three proteins, the PsbO protein appears to play a central role in the stabilization of the manganese cluster, is essential for efficient and stable O2 evolution, and is required, along with PsbP, for photoautotrophic growth and PS II assembly in higher plants propagated under normal growth conditions (5, 6).
In Arabidopsis thaliana, two genes that encode PsbO (psbO-1, At5g66570 and psbO-2, At3g50820) are normally expressed, yielding two different PsbO proteins (PsbO-1 and PsbO-2, respectively). A highly fluorescent mutant, psbo1, was recently identified in which a stop codon has been introduced in the psbO-1 gene by ethane methylsulfonate mutagenesis at amino acid residue 74 of the mature PsbO protein (Gln74 → Stop), which leads to the loss of this component (7). The mutant exhibits a lower variable fluorescence yield (FV/FM), lower rates of steady state O2 evolution, and retarded growth. It was demonstrated that PsbO-1 is the major isoform in the wild type under normal growth conditions and that in the psbo1 mutant the PsbO-2 protein is up-regulated in a semicompensatory manner. The mechanism that leads to the increased expression of the PsbO-2 protein in the psbo1 mutant is unclear at this time. Domain-swapping analysis followed by in vitro reconstitution experiments were interpreted as indicating that two amino acid differences between the PsbO-1 and PsbO-2 components, Val186 → Ser and Leu246 → Ile, could explain the functional differences between the two PsbO proteins (8). It was concluded that inherent functional defects of this component are responsible for the phenotype observed in the mutant psbo1. Further characterization by our laboratory (9) indicated that the psbo1 mutant possesses significant functional defects on both the reducing and oxidizing sides of the photosystem. During fluorescence induction, the psbo1 mutant exhibited an enhanced O-to-P transition. Additionally, the J-to-I transition accounted for a <2% rise of the total fluorescence yield, whereas in the wild type, this transition accounted for >30% of the total fluorescence yield. Analysis of the flash-induced fluorescence rise in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea indicated that in the wild type the ratio of PS IIα to PS IIβ reaction centers is ∼1.2, whereas in the mutant, the ratio is ∼0.3. Fluorescence decay kinetics in the absence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea indicated that electron transfer to QB was significantly altered in the mutant. Fluorescence decay kinetics in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea indicated that the charge recombination between QA– and the higher S states of the oxygen-evolving complex was retarded. Interestingly, flash oxygen yield analysis indicated that, after 5 min of dark incubation, a higher proportion of PS II reaction centers were in the S2 and S3 states. In the wild type, 6 ± 8% of the oxygen-evolving PS II reaction centers were in the higher S states, whereas in the mutant, this value was 28 ± 8%. These latter two observations appeared to indicate that the higher S states were more stable in the psbo1 mutant than in the wild type.
In this study, we performed a detailed analysis of the S2 and S3 state lifetimes in psbo1 and wild type thylakoid membranes. We also examined the total flash oxygen yield and its stability during incubation at room temperature. Our findings indicate that both the S2 and S3 states decay extremely slowly in psbo1. The oxygen-evolving complex in the psbo1 mutant is also quite unstable and rapidly loses its ability to evolve oxygen. Incubation of psbo1 thylakoids at a high NaCl concentration did not increase the rate of S2 and S3 state decay or the total oxygen yield, but the stability of the oxygen-evolving complex was somewhat enhanced. Incubation with a moderate concentration of CaCl2 had a significantly more dramatic effect. Under this condition, the S2 and S3 states of the mutant decayed nearly as rapidly as in the wild type. Additionally, the total flash oxygen yield and its stability in psbo1 following CaCl2 treatment were indistinguishable from the wild type. Steady state oxygen evolution experiments in large measure support these findings. These results strongly suggest that the principal defect in the psbo1 mutant is an inability to effectively utilize the calcium cofactor associated with PS II. We hypothesize that the Arabidopsis PsbO-2 protein cannot effectively sequester this ion in the vicinity of the oxygen-evolving site.
EXPERIMENTAL PROCEDURES
Plant Materials—Seeds of wild type A. thaliana (var. Landsberg erecta) and the mutant psbo1 were germinated on potting mixture and grown at 19–22 °C under 50–80 μmol photons m–2 s–1 white light with an 8-h light/16-h dark diurnal cycle. This suppressed flowering and maximized the yield of leaf material. Only leaves of mature plants were used in these experiments.
Flash Oxygen Yield Experiments—Thylakoids were isolated by grinding leaves in a blender with a buffer containing 0.45 m sorbitol, 10 mm EDTA, 0.1% bovine serum albumin, 1% polyvinylpyrrolidone, and 20 mm Tricine-NaOH, pH 8.4. The homogenate was filtered through two layers of cheesecloth and one layer of Miracloth (Calbiochemical Co.), and the thylakoids were pelleted by centrifugation at 4 °C at 1500 × g. The membranes were then washed twice with 0.3 m sorbitol, 5 mm MgCl2, and 20 mm Tricine-NaOH, pH 7.6. The membranes were finally pelleted and applied to the platinum electrode as a thin paste. For treatment with 150 mm NaCl or 20 mm CaCl2, these salts were included in the wash buffer. In the case of CaCl2 treatment, the washed membranes were resuspended in 1 ml of wash buffer plus 20 mm CaCl2 and brought to 50 μm ionomycin (a calcium ionophore). After incubation for 5 min at 4 °C, the treated thylakoids were pelleted and applied to the platinum electrode as described above. The flash oxygen yield measurements were performed on a bare platinum electrode (Artesian Scientific Co., Urbana, IL). The samples were incubated briefly (30 s to 1 min) under fluorescent room light (∼5 μmol photons m–2 s–1) prior to the onset of the experiment to allow the equilibration of the S states and then dark-incubated for only 20 s (during the electrode polarization). The electrode was polarized at 0.73 V for 20 s, and a series of 16 saturating flashes was supplied by an integrated, computer-controlled xenon flash lamp (20 μs). The polarization was turned off, and the same sample was then incubated in the dark at room temperature for various lengths of time followed by additional flash series. The data were analyzed using a four-state, homogeneous model (10). Five- and six-state models that incorporated either an S–1 state or S–1 and S–2 states, respectively, uniformly failed to fit the data acquired from either wild type or mutant thylakoids. Prior to fitting, the raw oxygen yield data were normalized to the average oxygen yield obtained on flashes 13–16. It is important to note that standard methods for measuring S state lifetimes (11, 12) could not be used in this study. The fact that the oxygen evolution of the mutant thylakoid membranes is very unstable in the absence of added calcium (see below), coupled with the fact that the S2 and S3 states are quite persistent during dark incubation (9), precludes the use of the long dark incubation periods normally used in such experiments. We consider our measurements to be semiquantitative and satisfactory for our comparative studies (but not sufficiently accurate for more detailed bioenergetic studies, such as those performed in Ref. 11).
Steady State Oxygen Evolution—For these experiments, thylakoids were isolated as described above and resuspended in a small volume of wash buffer. Chlorophyll was measured by the method of Arnon (13). Oxygen evolution was measured polarographically in a Hansatech oxygen electrode at 22 °C in the wash buffer supplemented with 0.5 mm potassium ferricyanide, 0.5 mm dichlorobenzoquinone, and 10 mm ammonium chloride. The chlorophyll concentration was 10 μg/ml, and the light intensity was 2000 μmol photons m–2 s–1. In some experiments, 150 mm NaCl or 20 mm CaCl2 was added directly to the reaction mixture. In the CaCl2 experiments, the thylakoid membranes were pretreated with 50 μm ionomycin for 5 min in the dark.
RESULTS AND DISCUSSION
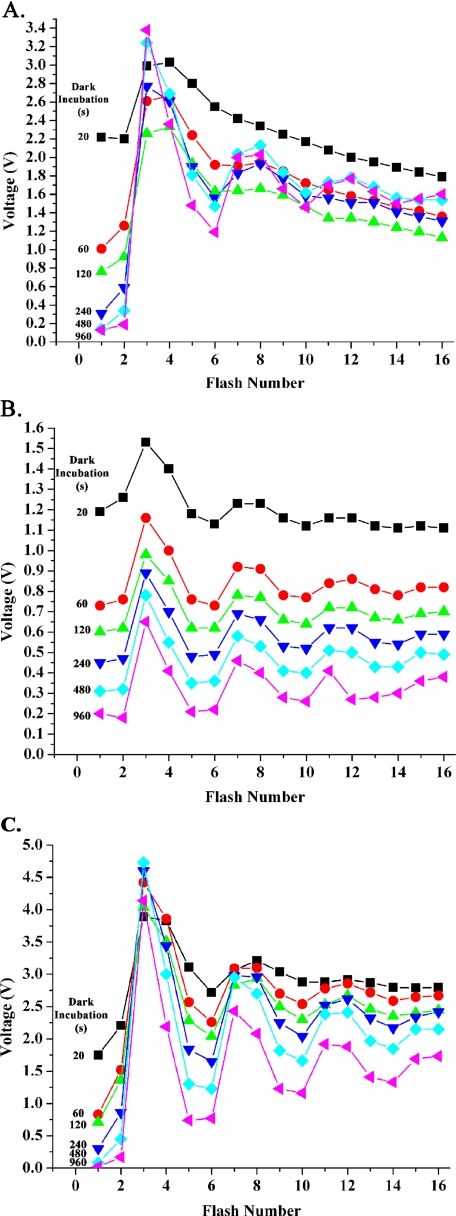
Fig. 1 shows examples of typical flash oxygen yield experiments performed for the wild type (Fig. 1A) and psbo1 mutant (Fig. 1B) under control cofactor conditions (10 mm chloride with no added calcium). Before the onset of these experiments, the samples were incubated in room light (5 μmol photons m–2 s–1) to allow mixing of the S states. The first flash series was performed after 20 s of dark incubation, which was required to allow polarization of the electrode. As expected, both wild type and mutant thylakoids exhibited large oxygen yields on the first and second flashes, indicating that a large proportion of PS II reaction centers (42–45%) were in the S2 + S3 states. Additional flash series were then performed after increasing intervals of dark incubation of the same sample. In the wild type (Fig. 1A), the oxygen yield observed on the first and second flashes decreased rapidly with increasing lengths of dark incubation. This indicates that the proportion of PS II reaction centers in the S2 and S3 states decreased rapidly with increasing length of dark incubation. After 960 s of dark incubation, <3% of the reaction centers were in the S2 + S3 states. In the mutant, a significantly different pattern was observed. Whereas the overall flash oxygen yield dropped substantially during the course of the experiment (see below), the proportion of reaction centers in the S2 + S3 states remained quite high (18%), even at the longest period of dark incubation. This indicates that the S2 and S3 states in the mutant are very long-lived, confirming our earlier observations (9).
FIGURE 1.
Raw data illustrating typical flash oxygen yield curves obtained from the wild type (A), the psbo1 mutant (B), and the psbo1 mutant + 20 mm CaCl2 + 50 μm ionomycin (C). A single sample was used to collect the data in each graph. The length of dark incubation between each flash series is shown. In A and B, the samples contained 10 mm chloride with no added calcium. In C, the sample contained 20 mm CaCl2 + 50 μm ionomycin. The dark incubation time for each 16-flash series is color-coded as follows: 20 s, black squares; 60s, red circles; 120 s, green triangles; 240 s, blue triangles; 480 s, aqua diamonds; and 960 s, magenta triangles. Please note that the flash experiments shown in B are not offset. The addition of 20 mm CaCl2 + 50 μm ionomycin to wild type thylakoids had no apparent effect on the flash oxygen yield pattern and is not shown.
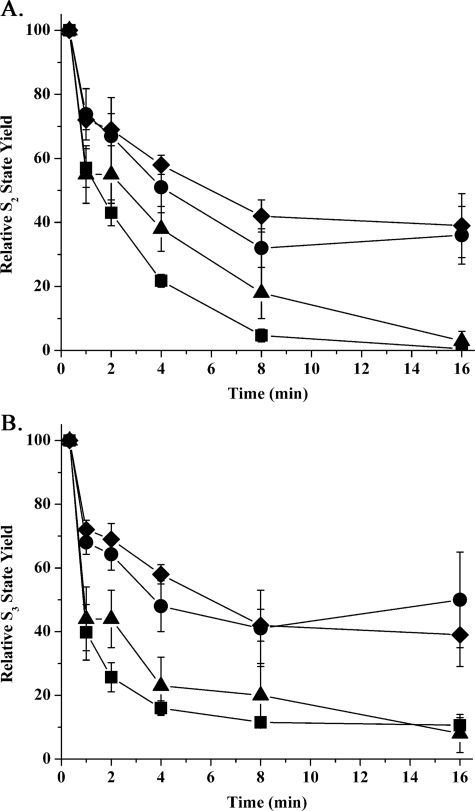
Fig. 2 explicitly illustrates the decay of the S2 and S3 states of the wild type and psbo1 mutant under a variety of ionic conditions. Qualitatively, under the control conditions of 10 mm chloride and no added calcium, the S2 and S3 states decayed rapidly in the wild type but very slowly in the psbo1 mutant. The addition of 150 mm NaCl did not accelerate the rate of decay of these S states in the mutant. The addition of 20 mm CaCl2, however, nearly fully restored the S state decay characteristics in the mutant (a typical flash oxygen yield experiment for the mutant in the presence of 20 mm CaCl2 is shown in Fig. 1C). The addition of 150 mm NaCl or 20 mm CaCl2 had little effect on the decay rates of the S2 and S3 states of the wild type (data not shown).
FIGURE 2.
S2 (A) and S3 (B) state lifetime decay curves for wild type Arabidopsis and psbo1 mutant thylakoids under different ionic conditions. The time axis indicates time of dark incubation at 24 °C. ▪, wild type decay curve under control ionic conditions (10 mm chloride with no added calcium); •, psbo1 control; ♦, psbo1 + 150 mm NaCl; ▴, psbo1 + 20 mm CaCl2 + 50 μm ionomycin (n = 4–13; error bars are ±1 S.E.). In some instances, the symbols are larger than the error bars. Treatment of wild type thylakoids with either 150 mm NaCl or 20 mm CaCl2 + 50 μm ionomycin had little effect on the S state lifetimes or other parameters examined in this study and are not shown.
In spinach, which possesses only a single PsbO protein isoform, chemical removal of this component from PS II membranes leads to increased lifetimes for the S2 and S3 states (14) and a marked slowing of the S3 → [S4] → S0 transition (14, 15). These preparations also exhibit a large loss (75%) of steady state oxygen evolution capability, with high concentrations of calcium and chloride being required to observe even this low activity (2, 16). Additionally, in these experiments, the PsbO protein appears to lower the calcium requirement for oxygen evolution by at least a factor of 2 (2). Similar results were obtained for the cyanobacterium Synechocystis 6803 in mutants from which the single PsbO protein present had been genetically deleted (3, 17). Whereas these mutants can grow photoautotrophically in standard BG-11 medium (albeit more slowly and with lower oxygen evolution rates), they cannot grow in either calcium-depleted (18) or chloride-depleted (19) medium. These findings indicate that, in vivo, the PsbO protein modulates the utilization of calcium in support of normal oxygen-evolving activity. Flash oxygen yield measurements on the Synechocystis ΔpsbO mutant also indicated that the S2 and S3 states are stabilized and that the S3 → [S4] → S0 transition is strongly retarded (3). Mutants containing alterations in other PS II components that cannot bind the PsbO protein normally exhibit similar characteristics (Ref. 21; for an in-depth review, see Ref. 22). Our results indicate that the PsbO-2 protein that is present in the psbo1 mutant is defective in supporting normal operation of the oxygen-evolving complex. Our in vitro characterization of mutant thylakoids indicates that loss of the PsbO-1 protein results in a modified oxygen-evolving complex, which exhibits S state characteristics similar to those obtained by the chemical removal of the PsbO component from higher plant oxygen-evolving membranes and to cyanobacterial cells from which the PsbO protein had been removed genetically. It should be emphasized here, however, that the PsbO-2 protein is, at least, partially functional. The psbo1 mutant grows photoautotrophically (albeit slowly) and can set seed. If the PsbO-2 protein were fully nonfunctional, we would expect the psbo1 mutant to be unable to grow photoautotrophically, as we had observed previously in Arabidopsis that contained RNA interference designed to suppress the expression of both PsbO-1 and PsbO-2 simultaneously (5).
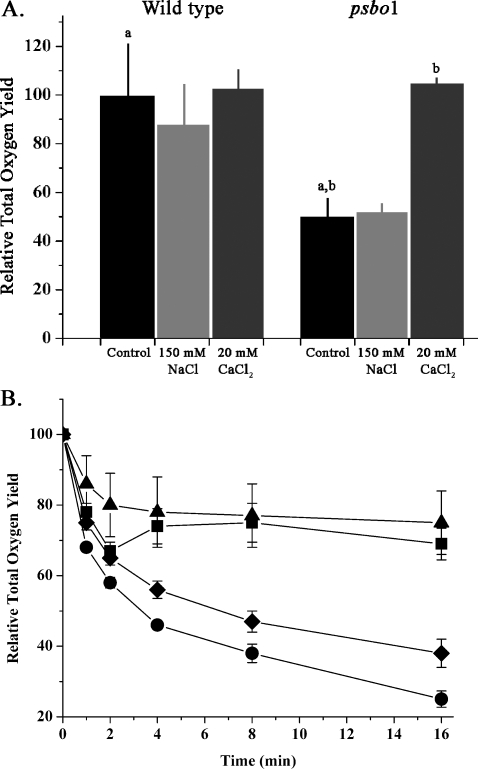
Fig. 3A illustrates the total flash oxygen yield of wild type and psbo1 thylakoids. This was estimated by summing the oxygen yields obtained on the first four flashes after a 20-s dark incubation period for the wild type and mutant under various treatment regimes. The oxygen yield for the wild type was essentially constant for untreated, NaCl-treated, and CaCl2-treated thylakoids. However, the psbo1 thylakoids exhibited a significantly different pattern. Under control and elevated NaCl conditions, the mutant exhibited oxygen yields ∼50% of that observed in the wild type. However, treatment with CaCl2 increased the total oxygen yield to near wild type values. This result was quite surprising. Previously, Murakami et al. (7) reported that the psbo1 mutant accumulated fewer PS II reaction centers (∼75% of the wild type for mature leaves) as measured by immunoquantification of the D1 protein. Consequently, it would be expected that the maximal observable total oxygen yield of the mutant would be 75% of wild type values. However, such an analysis does not take into account the large variability that Murakami et al. (7) observed in their measurements of D1. Our observation of near wild type total oxygen yield in the mutant is fully consistent with Murakami et al. (7), given the level of variability that they report.
FIGURE 3.
Analysis of total flash oxygen yield. A, histogram illustrating the total flash oxygen yield for wild type Arabidopsis and psbo1 mutant thylakoids under different ionic conditions. The CaCl2 treatments also contained 50 μm ionomycin. The total oxygen yield is the sum of the oxygen yields observed on the first four saturating flashes. For the wild type, this value was 10.8 ± 2.6 V, and for psbo1, this was 5.3 ± 1.1 V under control ionic conditions (n = 4–13; error bars indicate ±1 S.E.). a,b, p < 0.05 using Student's t test (one-tailed). B, stability of the total flash oxygen yield during dark incubation at 24 °C under different ionic conditions. The time axis indicates the length of dark incubation. ▪, wild type decay curve under control ionic conditions; •, psbo1 control; ♦, psbo1 + 150 mm NaCl; ▴, psbo1 + 20 mm CaCl2 + 50 μm ionomycin (n = 4–13, error bars ±1 S.E.). In some instances, the symbols are larger than the error bars. Treatment of wild type thylakoids with either 150 mm NaCl or 20 mm CaCl2 had little effect on the stability of the total oxygen yield or other parameters examined in this study and are not shown.
In Fig. 3B, the stability of the total flash oxygen yield of wild type and psbo1 thylakoids at room temperature was examined. The total oxygen yield of wild type thylakoids was quite stable, with ∼70% of the total oxygen yield being retained after 16 min of incubation. The psbo1 thylakoids were, however, very unstable, retaining only 20% of their oxygen yield over the same time period. Treatment of the psbo1 thylakoids with 150 mm NaCl marginally increased the stability of the oxygen yield. However, treatment of the mutant thylakoids with 20 mm CaCl2 dramatically increased the stability of the total flash oxygen yield such that it was indistinguishable from wild type thylakoids.
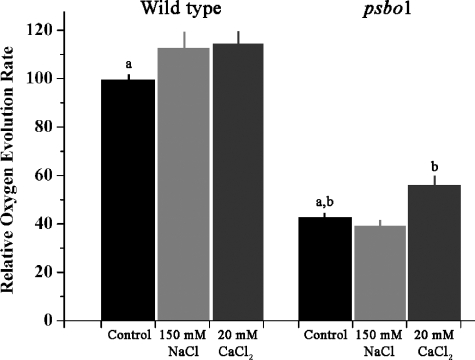
The increase in the total flash oxygen yield observed after CaCl2 treatment of the mutant thylakoids is, in large measure, also reflected in steady state oxygen evolution measurements (Fig. 4). No significant differences were observed in the oxygen evolution rate of wild type thylakoids upon the addition of either NaCl or CaCl2. However, addition of CaCl2 to mutant thylakoids significantly increased the oxygen evolution rate by ∼30%. These results indicate that the defect in the psbo1 mutant affects the ability of the mutant to effectively utilize calcium in support of oxygen evolution, both with respect to total flash oxygen yield and the oxygen evolution rates measured under steady state conditions. Interestingly, the addition of high concentrations of chloride did not appear to restore the total flash oxygen yield of psbo1, nor did it improve the steady state oxygen evolution rates of this mutant.
FIGURE 4.
Comparison of the steady state oxygen evolution rates of wild type and psbo1 mutant thylakoids. The CaCl2 treatments also contained 50 μm ionomycin. The wild type control oxygen evolution rate was 162 ± 12 μmol oxygen mg chlorophyll–1 h–1, whereas the psbo1 control oxygen evolution rate was 72 ± 9 μmol oxygen mg chlorophyll–1 h–1 (n = 9–12; error bars ± 1 S.E.). a,b, p < 0.05 using Student's t test (one-tailed).
Earlier studies on the psbo1 mutant indicated that the absence of PsbO-1 led to a decrease in PS II variable fluorescence yield (FV/FM) to between 70 and 90% of wild type levels (8). Additionally, as noted above, an apparent decrease in assembled PS II reaction centers was observed (monitored by a decrease in the amount of the D1 protein). Our observation that the addition of CaCl2 to mutant thylakoids restored the total oxygen yield to near wild type levels may indicate that the lower quantum yield observed in vivo (8) was principally due to PS II reaction centers that were defective in the sequestration of calcium. It should be noted that whereas in our steady state oxygen evolution measurements the psbo1 rates were significantly increased, they did not achieve wild type levels. This highlights a major difference between flash oxygen yield measurements and steady state experiments. During a steady state experiment, the samples are exposed to saturating light intensities for at least 1 min, whereas during a flash oxygen yield experiment, the sample is exposed to saturating light for <0.5 ms. We hypothesize that the lower level of activity restoration observed in the steady state experiment possibly indicates an increased rate of photoinactivation in the mutant that is not fully complemented by increased calcium concentrations.
Our observations demonstrate that the Arabidopsis psbo1 mutant thylakoids exhibit long-lived S2 and S3 states, lower total oxygen yields, lower stability of the oxygen-evolving complex, and depressed steady state rates of oxygen evolution. These results are very similar to those obtained by the chemical removal of the PsbO protein from higher plant PS II membranes and to those obtained from genetic deletion of the single psbO gene present in Synechocystis. Our results indicate that the PsbO-2 protein, the only PsbO protein present in the psbo1 mutant, is defective in supporting normal operation of the oxygen-evolving complex, even though this protein appears to bind normally to PS II membranes (8). Additionally, the majority of the defects we observed in the psbo1 mutant thylakoids can be nearly fully reversed by calcium in the presence of moderate chloride concentrations. These findings indicate that the PsbO-2 protein is defective in the utilization of these cofactors in support of normal oxygen evolution activity. Consequently, comparisons between the PsbO-1 and PsbO-2 proteins should yield important insights into the function of the competent PsbO-1 protein. PsbO-2 is very similar to PsbO-1, containing only 11 amino acid substitutions (7, 23), the majority of which are conservative replacements. Murakami et al. (8) generated Escherichia coli-expressed chimeric PsbO-1/PsbO-2 proteins and evaluated their ability to reconstitute steady state oxygen-evolving activity in NaCl/urea-washed spinach PS II membranes. They concluded that the observed defects in steady state oxygen evolution rates were due to amino acid substitutions in the C-terminal domain of PsbO-2, specifically the Val186 → Ser and/or Leu246 → Ile substitutions. If their observations are correct, then these residues are responsible either directly or indirectly for the loss of the ability of PsbO-2 to utilize calcium. It should be noted, however, that these steady state oxygen evolution experiments were apparently performed at high calcium and chloride concentrations during the oxygen evolution assays (due to the absence of the PsbP and PsbQ components). Although these authors did not specify the ionic cofactor concentrations used during their assays, they did tangentially cite Betts et al. (24), who included 20 mm Ca2+ and 100 mm Cl– in their assay buffer. It is probable that, under conditions of elevated calcium and chloride, the full extent of the defects associated with only PsbO-2-containing reaction centers would not have been observed (8). Consequently, it is possible that other residues that are present in PsbO-1 but replaced in PsbO-2 may be important with respect to the maintenance of calcium at the active site.
What is the nature of the defect in the PsbO-2 protein? Our observations indicate that the PsbO-2 protein cannot maintain normal levels of calcium in the vicinity of the oxygen-evolving site. Previously, we suggested that the PsbO protein, in concert with the large extrinsic loops of CP47 and CP43, formed a “sequestered domain” for maintaining high concentrations of chloride in the vicinity of the oxygen-evolving site (25). Experimental support for this suggestion was recently provided (26). Our current results indicate that a fully functional PsbO protein is also required for the normal sequestration of calcium at the oxygen-evolving site (for an alternative viewpoint, however, see Ref. 27). One possible hypothesis consistent with the psbo1 phenotype would be that the PsbO-2 protein cannot support a stable, high affinity structural association of PsbP and PsbQ with PS II. It has been shown that the PsbO protein is required for binding of the PsbP component and that PsbP is required for binding of the PsbQ protein (28, 29). More recently, we have identified carboxylates on the PsbO protein that are required for PsbP binding (30). Decreased binding affinity of these two components would lead to lowered affinity for both calcium and chloride at the oxygen-evolving site (for reviews see Refs. 20 and 22). It is also possible that the PsbP and PsbQ proteins can structurally associate normally with PsbO-2-containing PS II reaction centers but that the functional interaction between either PsbO-2 and PsbP/PsbQ or PsbO-2 and intrinsic membrane protein components of the photosystem is compromised. Finally, it is possible that the PsbO-2 protein alone cannot support normal assembly of PS II and that the defective photosystem that does assemble cannot effectively sequester calcium at the active site. Our earlier studies (5) indicated that the PsbO component is required for normal assembly of PS II. We did not, however, examine the relative efficiencies of PsbO-1 versus PsbO-2 in support of normal photosystem assembly. Experimental differentiation among these and other hypotheses is ongoing.
CONCLUSIONS
The psbo1 mutant exhibits significant defects in its ability to evolve oxygen. Steady state oxygen evolution rates are depressed, total flash oxygen yield is significantly lower, oxygen yeild stability is markedly decreased, and the dark decay of both the S2 and S3 states is significantly retarded. The majority of these defects are nearly fully reversed by moderate concentrations of CaCl2. These results indicate that the PsbO-2 protein, the only PsbO protein present in the psbo1 mutant, leads to a defect in the ability of PS II to utilize calcium in support of oxygen evolution.
This work was supported by grants from the National Science Foundation and the Department of Energy (to T. M. B. and L. K. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PS II, Photosystem II; Sx states, oxidation states of the oxygen-evolving complex; Tricine, N-tris(hydroxymethyl)-methylglycine.
References
- 1.Murata, N., Mijao, M., Omata, T., Matsunami, H., and Kuwabara, T. (1984) Biochim. Biophys. Acta 765 363–369 [Google Scholar]
- 2.Bricker, T. M. (1992) Biochemistry 31 4623–4628 [DOI] [PubMed] [Google Scholar]
- 3.Burnap, R., Shen, J. R., Jursinic, P. A., Inoue, Y., and Sherman, L. A. (1992) Biochemistry 31 7404–7410 [DOI] [PubMed] [Google Scholar]
- 4.Bricker, T. M., and Ghanotakis, D. F. (1996) in Oxygenic Photosynthesis: The Light Reactions (Ort, D. R., and Yocum, C. F., eds) pp. 113–136, Kluwer Academic Publishers, Dordrecht, The Netherlands
- 5.Yi, X., McChargue, M., Laborde, S. M., Frankel, L. K., and Bricker, T. M. (2005) J. Biol. Chem. 280 16170–16174 [DOI] [PubMed] [Google Scholar]
- 6.Yi, X., Hargett, S., Liu, H., Frankel, L. K., and Bricker, T. M. (2007) J. Biol. Chem. 282 24833–24841 [DOI] [PubMed] [Google Scholar]
- 7.Murakami, R., Ifuku, K., Takabayashi, A., Shikanai, T., Endo, T., and Sato, F. (2002) FEBS Lett. 523 138–142 [DOI] [PubMed] [Google Scholar]
- 8.Murakami, R., Ifuku, K., Takabayashi, A., Shikanai, T., Endo, T., and Sato, F. (2005) FEBS J. 272 2165–2175 [DOI] [PubMed] [Google Scholar]
- 9.Liu, H., Frankel, L. K., and Bricker, T. M. (2007) Biochemistry 46 7607–7613 [DOI] [PubMed] [Google Scholar]
- 10.Meunier, P. C. (1993) Photosynth. Res. 36 111–118 [Google Scholar]
- 11.Messinger, J., Schroder, W. P., and Renger, G. (1993) Biochemistry 32 7658–7668 [DOI] [PubMed] [Google Scholar]
- 12.Bricker, T. M., Lowrance, J., Sutton, H., and Frankel, L. K. (2001) Biochemistry 40 11483–11489 [DOI] [PubMed] [Google Scholar]
- 13.Arnon, D. I. (1949) Plant Physiol. 24 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyao, M., Murata, M., Lavorel, J., Maison-Petri, B., Boussac, A., and Etienne, A.-L. (1987) Biochim. Biophys. Acta 890 151–159 [Google Scholar]
- 15.Ono, T.-A., and Inoue, Y. (1985) Biochim. Biophys. Acta 806 331–340 [Google Scholar]
- 16.Miyao, M., and Murata, N. (1984) FEBS Lett. 170 350–354 [Google Scholar]
- 17.Vass, I., Cool, K. M., Zsuzsanra, D., Mayes, S. R., and Barber, J. (1992) Biochim. Biophys. Acta 1102 195–201 [Google Scholar]
- 18.Philbrick, J. B., Diner, B. A., and Zilinskas, B. A. (1991) J. Biol. Chem. 266 13370–13376 [PubMed] [Google Scholar]
- 19.Engels, D. H., Lott, A., Schmid, G. H., and Pistorious, E. K. (1994) Photosynth. Res. 42 227–244 [DOI] [PubMed] [Google Scholar]
- 20.Miqyass, M., van Gorkom, H. J., and Yocum, C. F. (2007) Photosynth. Res. 92 275–287 [DOI] [PubMed] [Google Scholar]
- 21.Putnam-Evans, C., Wu, J., Burnap, R., Whitmarsh, J., and Bricker, T. M. (1996) Biochemistry 35 4046–4053 [DOI] [PubMed] [Google Scholar]
- 22.Popelkova, H., and Yocum, C. F. (2007) Photosynth. Res. 93 111–121 [DOI] [PubMed] [Google Scholar]
- 23.Bricker, T. M., and Burnap, R. L. (2005) in Photosystem II: The Water/Plastoquinone Oxido-Reductase of Photosynthesis (Wydrzynski, T., and Satoh, K., eds) pp. 95–120, Springer, Dordrecht, the Netherlands
- 24.Betts, S. D., Ross, J. R., Pichersky, E., and Yocum, C. F. (1997) Biochemistry 36 4047–4053 [DOI] [PubMed] [Google Scholar]
- 25.Bricker, T. M., and Frankel, L. K. (1998) Photosynth. Res. 56 157–173 [Google Scholar]
- 26.Popelkova, H., Betts, S. D., Lydakis Simantiris, N., Im, M. M., Swenson, E., and Yocum, C. F. (2006) Biochemistry 45 3107–3115 [DOI] [PubMed] [Google Scholar]
- 27.Seidler, A. (1996) Biochim. Biophys. Acta 1277 35–60 [DOI] [PubMed] [Google Scholar]
- 28.Andersson, B., Larsson, C., Jansson, C., Ljungberg, U., and Akerlund, H.-E. (1984) Biochim. Biophys. Acta 766 21–26 [Google Scholar]
- 29.Kavelaki, K., and Ghanotakis, D. F. (1991) Photosynth. Res. 29 149–155 [DOI] [PubMed] [Google Scholar]
- 30.Bricker, T. M., and Frankel, L. K. (2003) Biochemistry 42 2056–2061 [DOI] [PubMed] [Google Scholar]