Abstract
Previous reports showed that chromatin-associated PCNA couples DNA replication with Cul4-DDB1Cdt2-dependent proteolysis of the licensing factor Cdt1. The CDK inhibitor p21, another PCNA-binding protein, is also degraded both in S phase and after UV irradiation. Here we show that p21 is degraded by the same ubiquitin-proteasome pathway as Cdt1 in HeLa cells. When PCNA or components of Cul4-DDB1Cdt2 were silenced or when the PCNA binding site on p21 was mutated, degradation of p21 was prevented both in S phase and after UV irradiation. p21 was co-immunoprecipitated with Cul4A and DDB1 proteins when expressed in cells. The purified Cul4A-DDB1Cdt2 complex ubiquitinated p21 in vitro. Consistently, p21 protein levels are low during S phase and increase around G2 phase. Mutational analysis suggested that in addition to the PCNA binding domain, its flanking regions are also important for recognition by Cul4-DDB1Cdt2. Our findings provide a new aspect of proteolytic control of p21 during the cell cycle.
Cell cycle progression is driven by the periodic activation of cyclin-dependent kinases (CDKs)3 (1). CDKs associated with G1-cyclins are required for commitment to the cell cycle, whereas CDKs associated with S- and M-cyclins initiate DNA replication and mitotic events, respectively. Checkpoint controls couple cell cycle events with changes in activity of each kinase in order to avoid illegitimate cell cycle progression. Checkpoint control over CDKs is elicited by regulating protein synthesis, degradation, phosphorylation, subcellular localization, and association with a number of CDK inhibitors (2, 3). For example, CDK inhibitors, such as p21 and p27, regulate G1-cyclin-CDKs for progression through G1 phase. Two ubiquitin ligases (E3s), SCF (Skp1/Cullin1/F-box protein) and APC/C (anaphase-promoting complex or cyclosome), are required for proteolysis of key proteins during a cell cycle (4–6). APC/C-dependent ubiquitination operates from M phase to G1 phase. Its targets include mitotic cyclins, securin, and geminin. In contrast, SCF appears to be active from late G1 until G2/M phases. There are a number of F-box proteins, each recognizing specific substrates that are normally marked by phosphorylation. The Skp2 F-box protein, destroyed during late M phase and early G1 phase by APC/C, accumulates around late G1 phase. The CDK inhibitor p27 is a well known substrate for SCFSkp2. p27 is phosphorylated by cyclin E/A-CDK2 at Thr-187 and then recognized by SCFSkp2 for ubiquitination and degradation (7). c-Myc, p21, p57, Cdt1 (Cdc10-dependent transcript 1), and Orc1 (origin recognition complex 1) are also reported to be degraded through SCFSkp2.
DNA replication is initiated by the activation of cyclin E/A-CDK2. Importantly, replication of chromosomal DNA must be limited to only once in a cell cycle (8–10). Before initiation starts, replication origins must be licensed for replication by forming prereplication complexes. This process is performed by loading of the MCM2–7 (minichromosome maintenance 2–7) complex with the aid of Cdc6 and Cdt1 onto chromosomal sites bound by the origin recognition complex. Prereplication complex formation is allowed only during late M phase and G1 phase but is prevented from S phase to the end of mitosis; thus, rereplication of any DNA segment is prevented in the same cell cycle. For this regulation, control of the licensing factor Cdt1 is essential in higher eukaryotes. Cdt1 accumulates during late M phase and G1 phase, but from S phase until the end of M phase it is inactivated by two redundant mechanisms: proteolysis and binding to its inhibitor geminin. When Cdt1 is highly expressed, Cdt1 proteolysis is defective or geminin is knocked down, rereplication is induced (11–14). Although mammalian Cdt1 was shown to associate with Skp2 and be ubiquitinated, subsequent reports indicated that Cdt1 was degraded in the absence of Skp2 (15–17). Recent reports demonstrated that although SCFSkp2 is involved in Cdt1 degradation, at least in mammalian cells, a new ubiquitination system is central to Cdt1 proteolysis from yeast to mammalian cells (18–24). This system uses Cul4-DDB1Cdt2 as an E3 and is dependent on DNA replication. Interestingly, chromatin association of PCNA links DNA replication to Cdt1 degradation, PCNA possibly forming a bridge between Cdt1 and Cul4-DDB1Cdt2 on chromatin. Cdt1 contains a PCNA binding motif at its extreme N terminus. Cdt1 mutated in this site becomes resistant to Cul4-DDB1Cdt2-mediated proteolysis. Cdt1 is also degraded when DNA damage is induced (e.g. by UV irradiation) through a similar pathway.
PCNA, originally characterized as a DNA sliding clamp that helps replicative DNA polymerases, is involved in many aspects of DNA metabolism: replication, repair, chromatin assembly, and cohesion. PCNA interacts with many proteins required for each process, such as DNA polymerase δ and ε, DNA ligase, CAF-1, Ctf7, etc. (25). All of these PCNA-interacting proteins (PIPs) contain a so-called PIP-box, characterized by a consensus sequence, QXX(L/I/M)XX(F/Y)(F/Y). All of these PIPs are essentially not subjected to degradation after association with PCNA for their roles. On the contrary, p21, which also contains a PIP-box and interacts with PCNA, accumulates in G1 phase, but its levels decrease after initiation of S phase (26). In addition, p21 is degraded after UV irradiation (27). Based on these observations, we examined the possibility that p21 was degraded by a pathway similar to Cdt1. We demonstrate that p21 is degraded by a PCNA-dependent Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation in HeLa cells.
EXPERIMENTAL PROCEDURES
Cell Culture—HeLa, 293T and U2OS cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. For synchronization in early S phase, HeLa cells were treated with thymidine (2 mg/ml), released, and treated with aphidicolin (5 μg/ml). Proteasome inhibitor MG132 was used at 20 μm. UV irradiation was carried out at 20–50 J/m2 using a Stratalinker. For transient transfection into 293T cells, Trans-IT293 (Mirus) was used. To isolate stably expressing HeLa cell lines, cells were transfected with plasmids containing neo genes by Trans-ITL1 (Mirus) and selected in a medium containing 400 μg/ml G418. For analysis of DNA content, flow cytometry was carried out as described (28).
Plasmids—The p21 cDNA containing plasmid pEHP21 was a gift from Dr. Waga and was used to construct the p21 PIP-box mutant pEHP21(PIP), by a QuikChange site-directed mutagenesis method (Stratagene) using the following primers: primer-1 (CGAAAACGGCAGACCAGCGCGACAGCTGCCTACCACTCCAAACGCCGGCTG) and primer-2 (GCGTTTGGAGTGGTAGGCAGCTGTCGCGCTGGTCTGCCGCCGTTTTCG). To make p21-3A, primer-3 (GATTTCTACCACTCCGCGGCCGCACTGATCTTCTCCAAGAGG) and primer-4 (CTTGGAGAAGATCAGTGCGGCCGCGGAGTGGTAGAAATCTGTC) were used. To make p21-4A, primer-5 (GGAGACTCTCAGGGTGCAGCGGCCGCACAGACCAGCATGACAGAT) and primer-6 (TGTCATGCTGGTCTGTGCGGCCGCTGCACCCTGAGAGTCTCCAGGTCC) were used. To express 3FLAG-tagged wild type and mutant forms of p21, these plasmids were used to amplify each form of the p21 fragment by PCR using the primers primer-3 (GGGGAATTCAGAACCGGCTGGGGATGTC) and primer-4 (ATGCTAGTTATTGCTCAGCGG), cut with EcoRI-BamHI, and cloned into the p3FLAG-Myc-CMV-26 plasmid (Sigma), resulting in p3FLAG-p21(WT), -p21(PIP), -p21(3A), and -p21(4A). To insert nuclear localization signals (NLS) between the FLAG tag and the wild type or mutant p21 cDNA, plasmids were cut with HindIII and EcoRI and ligated with a 3×NLS-Myc coding fragment that was amplified using plasmid pCMV/Myc/nuc (Invitrogen) as a template with primer-7 (GGGAAGCTTACGGCCGCAGATCCAAAAAAG) and primer-8 (GGGGAATTCTCGAGTGCGGCCCCATTCAGATCCTC) and was cut with HindIII and EcoRI, resulting in p3FLAG-3NLS-Myc-p21(WT), -p21(PIP), -p21(3A), and -p21(4A). pCMV-Myc-DDB1 was constructed by cloning the PCR-amplified DDB1 into pCMV-Myc plasmid (laboratory stock). The HA-tagged Cul4 expression plasmid was a gift from Dr Chiba.
Antibodies, Western Blotting, and Immunofluorescence—For Western blotting, total whole cell lysates were prepared by lysing cell pellets directly in SDS-PAGE buffer. For immunofluorescence, HeLa cells were fixed in 3.8% paraformaldehyde in PBS for 10 min, permeabilized in 0.25% Triton X-100 in PBS, and stained with the indicated antibodies, as described (20). For double staining, Alexa-488-conjugated anti-mouse and Alexa-592-conjugated anti-rabbit antibodies were used as secondary antibodies with Hoechst 33258 to visualize DNA. Antibodies used were as follows: p21 (mouse; BD Pharmingen), Cdt1 (rabbit; described in Ref. 29), FLAG (rabbit and mouse M2; Sigma), cyclin A (mouse, Ab-6 (Neomarkers) and rabbit, H-432 (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA))), Myc (rabbit, sc-789; Santa Cruz Biotechnology), DDB1 (Santa Cruz Biotechnology), Skp2 (H-435; Santa Cruz Biotechnology), Cul1 (D-5; Santa Cruz Biotechnology), Cul4A (Rockland), Cul4B (C-19; Santa Cruz Biotechnology), PCNA (PC10; Santa Cruz Biotechnology), p21 (556430 (BD Pharmingen) and sc-397 (Santa Cruz Biotechnology)), p27 (F-8; Santa Cruz Biotechnology) BLM (C-18; Santa Cruz Biotechnology), ligIV (a gift from Dr. S. P. Jackson). Anti-Cdt2 antibodies were raised in two rabbits after injecting His-Cdt2 protein (150-amino acid length of C terminus) that was expressed in Escherichia coli and purified on a nickel column.
Immunoprecipitation—p3FLAG-p21 was co-transfected with HA-Cul4A or Myc-DDB1 plasmid into 293T cells. After 3 days, cells were washed three times in phosphate-buffered saline, treated with 0.1% formaldehyde in phosphate-buffered saline for 10 min, and again washed in phosphate-buffered saline three times. The cells were suspended and lysed in 150 μl of 0.1 m NaCl-containing CSK buffer (10 mm PIPES, pH 7.0, 300 mm sucrose, 0.1% Triton X-100, 1 mm MgCl2, 1 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin) and centrifuged for 5 min at 700 × g. The supernatant was used for immunoprecipitation with anti-FLAG antibody-conjugated resins (Sigma). The precipitates were washed with 0.3 m NaCl-containing CSK buffer four times and used for immunoblotting.
RNA Interference Experiment—The following double-stranded RNAs were made by Dharmacon and transfected at 100 μm using Oligofectamine (Invitrogen), and cells were cultured for 3 days: Skp2, GCAUGUACAGGUGGCUGUU; DDB1, GGACCUGCUGUUUAUCUUG; Cul4A, GAACUUCCGAGACAGACCU; Cul4B, AAGCCUAAAUUACCAGAAA; PCNA, CGGUGACACUCAGUAUGUC; Cdt2, CCAGGAGGUGAUAAACUUU. Small interfering RNA for Luc, known as GL2 (Dharmacon), was used as a control siRNA.
Protein Purification—All proteins were of human origin. Protein expression and reconstitution of Cul4A-containing E3 complex (HA-Cul4A, 3FLAG-DDB1, Cdt2, and His-Myc-Rbx1) with the baculovirus system (pBacPAK6, Clontech), and purification with an anti-FLAG column (anti-FLAG M2-agarose; Sigma), followed by glycerol gradient, were done using Sf21 cells, as described previously (30). PCNA was prepared using an E. coli expression system as described (30). 3FLAG-Myc-p21 was expressed in HeLa cells, purified on an anti-FLAG column, and eluted with 3× FLAG peptides.
In Vitro Ubiquitination Assay—100 ng of FLAG-Myc-p21 was assayed in a 10-μl reaction mixture containing 20 mm HEPES-NaOH (pH 7.5), 20 mm MgCl2, 100 mm NaCl, 0.4 mm dithiothreitol, 2 mm ATP, 0.02 m CP, 250 ng of CPK, 500 ng of PCNA, 100 ng of E1, 200 ng of E2, 25 pmol of DNA (76-mer oligonucleotides synthesized and annealed), and 600 ng of Cul4A-containing E3 complex. After incubation at 37 °C for 30 min, the reaction was terminated with SDS-sample buffer and separated on 12.5% polyacrylamide gels and detected by immunoblotting with anti-p21 (sc-397; Santa Cruz Biotechnology) or anti-Myc (sc-789; Santa Cruz Biotechnology) antibodies. Ubiquitin (U-100), E1 (E-302), and E2 (GST-UbcH5; E2-625) were purchased from BostonBiochem.
RESULTS
Degradation of p21 by a PCNA-dependent Cul4-DDB1Cdt2 Pathway—To identify proteins that are degraded by the same pathway as Cdt1, we examined several PCNA-binding proteins following UV irradiation in HeLa cells. As reported (27), the p21 protein is degraded following UV irradiation (Fig. 1A). When asynchronously growing cells were treated with siRNA for Skp2, total p21 protein levels increased. However, in contrast to the report that UV-induced degradation was dependent on Skp2, its degradation occurred in cells treated with siRNA for Skp2 (Fig. 1B). Instead, its degradation was inhibited when PCNA, DDB1, or Cdt2 was silenced. This was not due to a change in cell cycle profile, because the fraction of BrdUrd-positive cells or profile of flow cytometry following siPCNA or siCul4 treatment was almost the same to control siRNA-treated cells (data not shown, but see supplemental Fig. 1), as previously reported for siSkp2 (20). These results suggest that p21 is degraded through the PCNA-dependent Cul4-DDB1Cdt2 pathway following DNA damage, similar to Cdt1. Since p53 function is defective in HeLa cells, we performed the same silencing experiments using U2OS cells that have a wild type p53. Also in this cell line, p21 degradation was inhibited when PCNA or Cul4A was silenced (Fig. 1C).
FIGURE 1.
p21 degradation following UV irradiation is mediated by a PCNA-dependent Cul4-DDB1Cdt2 system. A, degradation of p21 after UV irradiation. HeLa cells treated (UV+) or not (UV–) with UV irradiation at 20J/m2 were collected 1 h later for immunoblotting with the indicated antibodies. B, silencing of PCNA, DDB1, or Cdt2, but not Skp2, inhibits p21 degradation. HeLa cells were transfected with Cdt2 or Skp2 siRNA. 3 days later, half of the cultures were irradiated, and cells were collected 1 h later for immunoblotting. Another culture of HeLa cells was transfected with PCNA or DDB1 siRNA. 3 days later, cultures were irradiated, and cells were collected at 0, 1, and 3 h postirradiation for immunoblotting. As a control, siRNA for luciferase (siLuc) was used. Cell extracts were blotted with the indicated antibodies. Relative protein levels were quantified by NIH ImageJ and are listed below each panel. The asterisks in the immunoblots mark cross-reacting bands used as loading control. C, U2OS cells were transfected with the indicated siRNAs and analyzed as described in B.
To follow the p21 protein degradation in individual cells, we performed an indirect immunofluorescence analysis as used previously for investigating Cdt1 degradation (20). Initially, asynchronously growing HeLa cells were treated with UV and analyzed 1 h postirradiation. Double staining with anti-p21 and anti-Cdt1 antibodies showed that both p21 and Cdt1 were degraded following UV irradiation (Fig. 2A). Next, asynchronously growing HeLa cells were transfected with siRNA for PCNA, Skp2, or control and fixed for staining with or without UV irradiation. Consistent with the immunoblot analysis, p21 became resistant to UV-induced degradation when PCNA was silenced, but it was still degraded in Skp2-silenced cells (Fig. 2, C–E).
FIGURE 2.
Analysis of p21 degradation by immunofluorescence. A, asynchronously growing HeLa cells were fixed without irradiation or 1 h after UV irradiation (UV) and stained with anti-p21 and anti-Cdt1 antibodies. B, cell cycle expression of p21. A synchronized culture was made by a thymidine-aphidicolin block. After release from early S phase arrest, cells were collected at the indicated times and used for immunoblotting and flow cytometry analysis. The asterisk marks cross-reacting bands used as a loading control. C–E, HeLa cells were transfected with the indicated siRNAs, and half of the dishes were UV-irradiated and fixed 1 hour postirradiation for immunostaining with anti-p21 and anti-cyclin A. DNA was stained with Hoechst.
Double immunofluorescence analysis with Cdt1 or cyclin A can reveal the cell cycle expression profile of p21. Both Cdt1 and cyclin A are very good cell cycle markers; Cdt1 is present exclusively in G1 phase, whereas cyclin A is present from S phase to early M phase (20). Co-staining of p21 with Cdt1 is consistent with p21 being present in G1 phase (Fig. 2A), whereas p21 was not co-stained with cyclin A (Fig. 2C), indicating that p21 was degraded in S phase. When PCNA was silenced, p21 protein remained stable in cyclin A-positive S phase cells (Fig. 2D). In addition, we performed a same BrdUrd incorporation and double immunofluorescence assay as used for Cdt1 (20). We used a stable HeLa cell line, expressing FLAG-Myc-p21 (see Fig. 5), since the rabbit anti-Myc antibodies used worked well for double staining with mouse anti-BrdUrd antibodies. In asynchronously growing cells, p21 was degraded in BrdUrd-positive cells as expected. When treated with siRNA for PCNA, Cul4, or DDB1, p21 was accumulated in BrdUrd-positive cells but not when treated with control siRNA (supplemental Fig. 1). These results demonstrate that p21 is degraded by the PCNA-dependent Cul4-DDB1Cdt2 pathway both in S phase and following UV irradiation.
FIGURE 5.
PIP-box-surrounding region affects p21 degradation. A, 3FLAG-NLS-Myc-tagged form of p21(WT) and 3A, 4A, and 7A mutants having mutations around the PIP-box. B, analysis of interaction of WT and mutant forms of p21 (PIP, 3A, 4A, and 7A) with PCNA. 293T cells were transfected with each plasmid or mock-transfected (–). Two days later, cell extracts were prepared, immunoprecipitated (IP) with anti-FLAG, and blotted (IB) with anti-PCNA antibody. C, cells stably expressing each plasmid (WT, 3A, or 4A) were treated with UV or not treated as a control. 1 h later, cells were fixed and stained with anti-FLAG and anti-cyclin A antibodies.
SCFSkp2 is active both in S phase and in G2 phase. On the other hand, the Cul4-DDB1Cdt2 pathway requires chromatin-associated PCNA for ubiquitination and is active during S phase. Thus, in contrast to Cdt1, which is degraded both in S and G2 phases, p21 would be expected to be degraded only during S phase. Therefore, we examined p21 protein levels during the HeLa cell cycle. Cells were arrested at early S phase by thymidine-aphidicolin block and released into a synchronous culture. Cdt1 protein was hardly detected in S phase and G2 phase and appeared around the end of M phase, as reported (29) (Fig. 2B). The p27 protein, also a substrate of SCFSkp2, was detected in neither S nor G2 cells. In contrast, p21, which was similarly absent during S phase, appeared around 4 h earlier than Cdt1 and p27, suggesting that degradation of p21 does not take place in G2 phase, when chromatin-associated PCNA is decreased.
PCNA Interaction Is Required for Degradation of p21—The above results suggested that p21 degradation is dependent on PCNA interaction. To address this point, we constructed a p21 bearing mutations in its PIP-box (PIP) (Fig. 3A). To enable efficient detection of the p21 protein, the FLAG tag was fused to the N terminus of p21 (FLAG-p21(PIP) plasmid). A wild type tagged form of p21, FLAG-p21(WT), was similarly constructed. Co-immunoprecipitation assays confirmed that PCNA interacts with FLAG-p21(WT) but not with FLAG-p21(PIP) (Fig. 3A). Using these expression constructs, HeLa cell lines stably expressing each FLAG-tagged p21 at physiological levels were isolated. When each cell line was UV-irradiated, FLAG-p21(WT) was degraded similarly to the endogenous p21 protein, but FLAG-p21(PIP) remained stable (Fig. 3B). On immunofluorescence analysis, FLAG-p21(WT) was detected in cyclin A-negative cells (Fig. 3C, top), and when irradiated with UV, the FLAG-p21(WT) signal was lost. Thus, FLAG-p21(WT) protein is degraded similarly to endogenous p21 in HeLa cells both during the cell cycle and after UV irradiation. In contrast, FLAG-p21(PIP) was stable in S phase cells and was not degraded after UV irradiation (Fig. 3C, bottom). These results indicate that p21 degradation depends on PCNA interaction.
FIGURE 3.
PIP-box-dependent proteolysis of p21. A, PIP-box mutant of p21 does not interact with PCNA. PIP-box p21 mutant (PIP) was constructed as indicated. Plasmids FLAG-p21(WT) and -p21(PIP) were transfected into 293T cells. Cell extracts were prepared after 48 h, immunoprecipitated with anti-FLAG, and blotted with anti-PCNA and anti-FLAG antibodies. B, FLAG-p21(PIP) is resistant to UV-induced degradation. HeLa cell lines expressing FLAG-p21(WT) or-p21(PIP) were isolated. Half of each culture was UV-irradiated, and 1 h later, cell extracts were prepared and blotted with anti-p21 antibodies. C, immunofluorescence analysis. Half of the HeLa cells expressing FLAG-p21(WT) or -p21(PIP) were irradiated, fixed 1 h later, and stained with anti-FLAG and anti-cyclin A. D, proteasome inhibitor MG132 stabilizes p21. Cell extracts were prepared from HeLa cells expressing FLAG-p21(WT) or -p21(PIP) treated with DMSO(D) or MG132(M) for 3 h (asy), treated for 2 h, UV-irradiated, and incubated for an additional 1 h (UV) or treated for 3 h following arrest at early S phase with thymidine (thy). Extracts were blotted with anti-p21 antibodies to see FLAG-tagged p21 and endogenous p21 (anti-p21) or anti-FLAG and exposed for a longer time to see the ladder of p21 (long exp). E and F, HeLa cells expressing FLAG-p21(WT) were transfected with siRNAs. Half of each culture was UV-irradiated, and 1 h later, cells were collected for immunoblotting or fixed for immunostaining.
The p21 degradation observed in HeLa cells is ubiquitin-proteasome-dependent. UV-induced degradation of p21, both endogenous and FLAG-tagged, was blocked in the presence of the proteasome inhibitor MG132 (Fig. 3D, lanes 3 and 4). The level of p21 in S phase-arrested cells was low, but it increased following the addition of MG132 (Fig. 3D, lanes 5 and 6). Upon long exposure, a ladder of p21 was observed in MG132-treated cells, especially in UV-irradiated cells, which may correspond to the ubiquitinated form of FLAG-p21. In contrast, FLAG-p21(PIP) levels remained constant with or without treatment of MG132 and both after UV irradiation and S phase arrest, and a high molecular weight ladder was not detected.
To confirm that the FLAG-p21(WT) protein is degraded through the Cul4-DDB1Cdt2 pathway, FLAG-p21(WT)-expressing cells were transfected with siRNAs. Following UV irradiation, FLAG-p21(WT) remained stable when Cul4 or DDB1 was silenced but was still degraded when Skp2 or Cul1 was silenced (Fig. 3, E and F). Double staining with cyclin A also indicated that FLAG-p21(WT) was stable in S phase in siCul4-transfected cells but not in siCul1 transfected cells. p21 Interacts with Cul4A and DDB1 and Is Ubiqitinated by Cul4A-DDB1Cdt2 Complex in Vitro—To confirm that p21 associates with Cul4-DDB1, FLAG-tagged p21 was co-expressed with HA-Cul4A or Myc-DDB1 in 293T cells, and FLAG-p21 was immunoprecipitated with anti-FLAG beads. On immunoblot analysis, HA-Cul4A or Myc-DDB1 was detected in the precipitates but not in the control precipitates (Fig. 4A), indicating that p21 interacts with Cul4A and DDB1.
FIGURE 4.
Interaction and in vitro ubiquitination of p21 with Cul4-DDB1Cdt2. A, interaction of p21 with Cul4A and DDB1. HA-Cul4A expression plasmid (left) or Myc-DDB1 expression plasmid (right) was transfected into 293T cells with FLAG-p21 or transfected alone. After washing with PBS, cells were treated with 0.1% formaldehyde and lysed for immunoprecipitation (IP) with anti-FLAG antibody-conjugated beads. Immunoprecipitates were subjected to immunoblotting (IB) with HA, Myc, or p21 antibodies. The asterisks mark the position of light chain. B, Cul4A-containing E3 complex purified from Sf21 cells co-infected with baculoviruses expressing HA-Cul4A, 3FLAG-DDB1, Cdt2, and His-Myc-Rbx1. The lower panel shows the immunoblotting with antibodies to each component. C, in vitro ubiquitination assay. The 3FLAG-Myc-p21 was incubated alone or with baculopurified Cul4A-DDB1Cdt2 and subjected to immunoblotting with anti-p21 (left) and anti-Myc (right). The asterisks mark nonspecific bands.
Next, we performed an in vitro ubiquitination assay for p21 with Cul4-DDB1Cdt2. For this assay, Cul4A-DDB1Cdt2 complex was purified from Sf21 insect cells that were co-transfected with human Cul4A-, DDB1-, Cdt2-, and Rbx1-expressing baculoviruses (Fig. 4B). The FLAG-Myc-tagged form of p21 (see Fig. 5), purified from HeLa cells, was incubated with or without the purified complex, and the reaction products were analyzed by immunoblotting with anti-p21 or anti-Myc antibodies. In the presence of Cul4A-DDB1Cdt2 E3 ligase, slowly migrating forms of p21 were detected on both immunoblots at similar molecular weight positions (Fig. 4C). In the absence of E3 ligase, such bands were not produced. These data indicate that Cul4A-DDB1Cdt2 ubiquitinates p21.
Regions Surrounding the PIP-box Are Implicated in PCNA-dependent Degradation—PCNA binding brings about degradation of Cdt1 and p21. So far, other PCNA-binding proteins appear to be stable. What brings about this difference? One possibility is that amino acids surrounding the PIP-box are important for degradation. To address this possibility, we made alanine-substituted mutants of p21 (Fig. 5A). Since basic amino acid clusters are detected around the PIP-box, we focused on these clusters on p21. Alanine-substituted mutants of p21 (3A, 4A, and 7A) were made and tagged with FLAG at the N terminus. The basic amino acid cluster appears to have a role as a nuclear localization signal, and thus three copies of NLS and Myc tag were inserted as described in the legend to Fig. 5A. Cells stably expressing each construct were isolated. The addition of 3FLAG-NLS-Myc had no effect on p21 degradation (Fig. 5C, WT). p21 with 154KRR156 → AAA or 140RKRR143 → AAAA became stable both in S phase and after UV irradiation (Fig. 5C, 3A and 4A). This was not due to a defect in interacting with PCNA (Fig. 5B). The 7A mutant was also stable, but this mutation resulted in a failure to interact with PCNA (data not shown) (Fig. 5B). These results suggest that in addition to the PIP-box, basic amino acid clusters surrounding the PIP-box are required for ubiquitination.
DISCUSSION
PCNA mediates the ubiquitination and subsequent degradation of Cdt1 by Cul4-DDB1Cdt2. Here, we show that p21 is degraded in a similar fashion in HeLa cells, both during S phase and after UV irradiation. First, degradation of p21, dependent on ubiquitin-proteasome pathway, was blocked when PCNA or components of the Cul4-DDB1Cdt2 complex were silenced. Second, p21 associates with Cul4A and DDB1 and is ubiquitinated in vitro by purified Cul4A-DDB1Cdt2 complex. Third, mutations at the PCNA binding domain of p21 also blocked its degradation. The Cul4-DDB1Cdt2 ubiquitination system is active in S phase, since PCNA mediates substrate recognition by Cul4-DDB1Cdt2 only when associated with chromatin. Consistently, p21 protein levels were very low during S phase, but they increased after completion of DNA replication in G2/M phase and remained high during G1 phase.
There have been several reports addressing the mechanisms regulating p21 proteolysis, both ubiquitin-proteasome-mediated ones and ubiquitin-independent proteasome-mediated ones (27, 31–39). Our finding adds a novel insight into the cell cycle control of the CDK inhibitor p21. We have examined p21 degradation in HeLa cells that have defective p53 function, and thus p53-mediated DNA damage-induced up-regulation of p21 is defective. The FLAG-tagged p21 under the control of the cytomegalovirus promoter, stably integrated into the genome, was regulated similarly to endogenous p21, suggesting that post-translational rather than transcriptional regulation controls the levels of p21 during a cell cycle and following DNA damage. The Xenopus CDK inhibitor Xic1, which exhibits homology to p21 and p27, was also shown to be degraded dependent on PCNA in Xenopus egg extract (40). Therefore, it is possible that the observation found in the HeLa cell line represents a fundamental control of p21 proteolysis in a cell cycle.
Although certain lines of evidence suggested that UV-induced p21 degradation was mediated by Skp2, others indicated that Skp2 may not be involved (27, 31). Our data demonstrate that SCFSkp2 is not essential for p21 degradation following UV irradiation. In contrast, Cul4-DDB1Cdt2 mediates this control. The proteolysis of p21 during S phase is also carried out by Cul4-DDB1Cdt2. The cell cycle expression profile of p21 suggests that Skp2 is not a major mediator of p21 degradation, since the protein appeared to accumulate in G2 cells, in contrast to Cdt1 and p27, which are degraded by SCFSkp2 and are therefore absent in G2 phase (Fig. 2B). However, we cannot rule out the possibility that SCFSkp2- or ubiquitin-independent proteasome-mediated degradation is involved in addition to Cul4-DDB1Cdt2 in p21 degradation during the cell cycle. In addition, dependence of p21 proteolysis on Skp2 may be different between cell lines. In fact, when Skp2 was silenced, the levels of p21 increased on immunoblot and the levels of immunofluorescent signal of p21 also increased (Figs. 1B, 2C, 2E, and 4E), and we detected a population of cells positive both for cyclin A and p21 in Skp2-silenced cells (Fig. 2D). These cells may correspond to G2 phase cells, since in G2 cells, Cul4-DDB1 is not active, and thus Skp2 silencing may have a more pronounced effect on p21 degradation. Since SCFSkp2 mediates proteolysis of p27 CDK inhibitor, its levels increase when Skp2 is silenced, which may have an effect on p21 proteolysis. Recently, it was shown that Skp2 associates not only with Cul1-Skp1 but also with Cul4A-DDB1 to target p27 for proteolysis (41). The same mechanism may operate for p21 proteolysis.
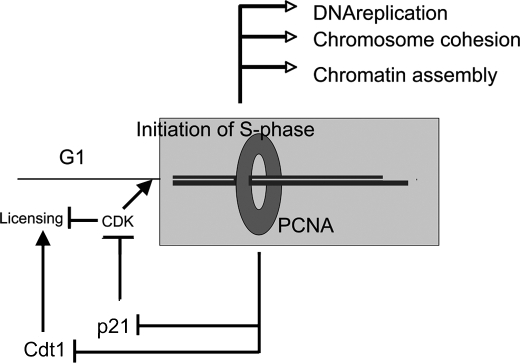
Replication-coupled inactivation of Cdt1 is an ideal mechanism to coordinate initiation of replication and prevention of reinitiation. Cdt1 is present during G1 phase and is required to license DNA for replication, but it must be inactivated after the onset of S phase. E. coli uses a similar system, called RIDA, to inactivate initiator protein DnaA via a sliding clamp, a mammalian homologue of PCNA (42, 43). Our analysis shows that p21 must be added as a mammalian protein inactivated by replication-dependent proteolysis. p21 is present during G1 to inhibit cyclin E/A-CDK activation. High amounts of p21 prevent cells from entering S phase. Once cells are committed to S phase, it is important to keep p21 protein at a low level to maintain CDK activity for replication and preventing rereplication. Chromatin loading of PCNA may signal to the cell that it has entered S phase (Fig. 6). Both Cdt1 and p21 play a role in late M to G1 phase, but their presence in S phase is harmful. When PCNA is loaded on chromatin, it contributes to processes such as replication, chromatin assembly, and chromosome cohesion. In addition, it concomitantly plays a role in Cdt1 and p21 inactivation by proteolysis, signaling entry into S phase.
FIGURE 6.
A model showing that chromatin loading of PCNA may integrate the transition from G1 phase to S phase. Chromatin-loaded PCNA facilitates DNA metabolism, such as DNA replication, chromatin assembly, and cohesion, and at the same time prevents the accumulation of Cdt1 and p21, which are required in G1 but would be harmful in S phase.
Why can PCNA mediate the degradation of Cdt1 and p21 but not of those proteins directly involved in DNA metabolism? The chromatin-associated PCNA provides a platform on which it bridges the PIP and Cul4-DDB1Cdt2 for ubiquitination. If PIP takes part in a multicomponent complex, Cul4-DDB1Cdt2 may not be able to access its substrate. It is also possible that Cul4-DDB1Cdt2 requires a specific motif on the substrate for recognition outside the PIP-box. We show here that mutating sites around the PIP-box blocked the proteolysis of p21 without affecting PCNA interaction. The mutated sites are composed of basic amino acids and are positively charged. These clusters may be required for recognition by Cul4-DDB1Cdt2. However, some PIPs, such as DNA ligase I, also have basic amino acids proximal to their PIP-box. Further analyses are required to address what dictates which of the multiple PCNA binding partners are targeted for proteolysis.
Supplementary Material
Acknowledgments
We are grateful to Dr. Z. Lygerou for corrections and critical reading of the manuscript, Dr. S. Waga for p21 plasmid, Dr. T. Kamura for Rbx1 plasmid, Dr. M. Saijo for FLAG-DDB1 plasmid, Dr. T. Chiba for Cul4A plasmid, and Dr. S. P. Jackson for anti-ligIV antibody.
This work was supported by Grants-in-aid for Scientific Research (C) and Scientific Research on Priority Areas from the Ministry of Education, Culture, Sport, Science, and Technology of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: CDK, cyclin-dependent kinase; PIP, PCNA interacting protein; E3, ubiquitin-protein isopeptide ligase; PCNA, proliferating cell nuclear antigen; PIP, PCNA-interacting protein; PIPES, 1,4-piperazinediethanesulfonic acid; siRNA, small interfering RNA; BrdUrd, bromode-oxyuridine; WT, wild type.
References
- 1.Nurse, P. (1994) Cell 79 547–550 [DOI] [PubMed] [Google Scholar]
- 2.Sherr, C. J., and Roberts, J. M. (1999) Genes Dev. 13 1501–1512 [DOI] [PubMed] [Google Scholar]
- 3.Hartwell, L. H., and Kastan, M. B. (1994) Science 266 1821–1828 [DOI] [PubMed] [Google Scholar]
- 4.Vodermaier, H. C. (2004) Curr. Biol. 14 R787–R796 [DOI] [PubMed] [Google Scholar]
- 5.Cardozo, T., and Pagano, M. (2004) Nat. Rev. 5 739–751 [DOI] [PubMed] [Google Scholar]
- 6.Nakayama, K. I., and Nakayama, K. (2006) Nat. Rev. Cancer 6 369–381 [DOI] [PubMed] [Google Scholar]
- 7.Carrano, A. C., Eytan, E., Hershko, A., and Pagano, M. (1999) Nat. Cell biol. 1 193–199 [DOI] [PubMed] [Google Scholar]
- 8.Nishitani, H., and Lygerou, Z. (2002) Genes Cells 7 523–534 [DOI] [PubMed] [Google Scholar]
- 9.Blow, J. J., and Dutta, A. (2005) Nat. Rev. 6 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arias, E. E., and Walter, J. C. (2007) Genes Dev. 21 497–518 [DOI] [PubMed] [Google Scholar]
- 11.Vaziri, C., Saxena, S., Jeon, Y., Lee, C., Murata, K., Machida, Y., Wagle, N., Hwang, D. S., and Dutta, A. (2003) Mol. Cell 11 997–1008 [DOI] [PubMed] [Google Scholar]
- 12.Zhu, W., Chen, Y., and Dutta, A. (2004) Mol. Cell. Biol. 24 7140–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melixetian, M., Ballabeni, A., Masiero, L., Gasparini, P., Zamponi, R., Bartek, J., Lukas, J., and Helin, K. (2004) J. Cell Biol. 165 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong, W., Feng, H., Santiago, F. E., and Kipreos, E. T. (2003) Nature 423 885–889 [DOI] [PubMed] [Google Scholar]
- 15.Sugimoto, N., Tatsumi, Y., Tsurumi, T., Matsukage, A., Kiyono, T., Nishitani, H., and Fujita, M. (2004) J. Biol. Chem. 279 19691–19697 [DOI] [PubMed] [Google Scholar]
- 16.Takeda, D. Y., Parvin, J. D., and Dutta, A. (2005) J. Biol. Chem. 280 23416–23423 [DOI] [PubMed] [Google Scholar]
- 17.Li, X., Zhao, Q., Liao, R., Sun, P., and Wu, X. (2003) J. Biol. Chem. 278 30854–30858 [DOI] [PubMed] [Google Scholar]
- 18.Arias, E. E., and Walter, J. C. (2006) Nat. Cell Biol. 8 84–90 [DOI] [PubMed] [Google Scholar]
- 19.Senga, T., Sivaprasad, U., Zhu, W., Park, J. H., Arias, E. E., Walter, J. C., and Dutta, A. (2006) J. Biol. Chem. 281 6246–6252 [DOI] [PubMed] [Google Scholar]
- 20.Nishitani, H., Sugimoto, N., Roukos, V., Nakanishi, Y., Saijo, M., Obuse, C., Tsurimoto, T., Nakayama, K. I., Nakayama, K., Fujita, M., Lygerou, Z., and Nishimoto, T. (2006) EMBO J. 25 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higa, L. A., Banks, D., Wu, M., Kobayashi, R., Sun, H., and Zhang, H. (2006) Cell Cycle 5 1675–1680 [DOI] [PubMed] [Google Scholar]
- 22.Hu, J., and Xiong, Y. (2006) J. Biol. Chem. 281 3753–3756 [DOI] [PubMed] [Google Scholar]
- 23.Jin, J., Arias, E. E., Chen, J., Harper, J. W., and Walter, J. C. (2006) Mol. Cell 23 709–721 [DOI] [PubMed] [Google Scholar]
- 24.Ralph, E., Boye, E., and Kearsey, S. E. (2006) EMBO Rep. 7 1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maga, G., and Hubscher, U. (2003) J. Cell Sci. 116 3051–3060 [DOI] [PubMed] [Google Scholar]
- 26.Nakanishi, M., Adami, G. R., Robetorye, R. S., Noda, A., Venable, S. F., Dimitrov, D., Pereira-Smith, O. M., and Smith, J. R. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 4352–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bendjennat, M., Boulaire, J., Jascur, T., Brickner, H., Barbier, V., Sarasin, A., Fotedar, A., and Fotedar, R. (2003) Cell 114 599–610 [DOI] [PubMed] [Google Scholar]
- 28.Nishitani, H., Lygerou, Z., and Nishimoto, T. (2004) J. Biol. Chem. 279 30807–30816 [DOI] [PubMed] [Google Scholar]
- 29.Nishitani, H., Taraviras, S., Lygerou, Z., and Nishimoto, T. (2001) J. Biol. Chem. 276 44905–44911 [DOI] [PubMed] [Google Scholar]
- 30.Shiomi, Y., Shinozaki, A., Sugimoto, K., Usukura, J., Obuse, C., and Tsurimoto, T. (2004) Genes Cells 9 279–290 [DOI] [PubMed] [Google Scholar]
- 31.Lee, H., Zeng, S. X., and Lu, H. (2006) J. Biol. Chem. 281 26876–26883 [DOI] [PubMed] [Google Scholar]
- 32.Li, X., Amazit, L., Long, W., Lonard, D. M., Monaco, J. J., and O'Malley, B. W. (2007) Mol. Cell 26 831–842 [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. Y., Yu, S. J., Park, Y. G., Kim, J., and Sohn, J. (2007) Mol. Cell. Biol. 27 3187–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, X., Barton, L. F., Chi, Y., Clurman, B. E., and Roberts, J. M. (2007) Mol. Cell 26 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, W., Nacusi, L., Sheaff, R. J., and Liu, X. (2005) Biochemistry 44 14553–14564 [DOI] [PubMed] [Google Scholar]
- 36.Bornstein, G., Bloom, J., Sitry-Shevah, D., Nakayama, K., Pagano, M., and Hershko, A. (2003) J. Biol. Chem. 278 25752–25757 [DOI] [PubMed] [Google Scholar]
- 37.Bloom, J., Amador, V., Bartolini, F., DeMartino, G., and Pagano, M. (2003) Cell 115 71–82 [DOI] [PubMed] [Google Scholar]
- 38.Touitou, R., Richardson, J., Bose, S., Nakanishi, M., Rivett, J., and Allday, M. J. (2001) EMBO J. 20 2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheaff, R. J., Singer, J. D., Swanger, J., Smitherman, M., Roberts, J. M., and Clurman, B. E. (2000) Mol. Cell 5 403–410 [DOI] [PubMed] [Google Scholar]
- 40.Chuang, L. C., and Yew, P. R. (2005) J. Biol. Chem. 280 35299–35309 [DOI] [PubMed] [Google Scholar]
- 41.Bondar, T., Kalinina, A., Khair, L., Kopanja, D., Nag, A., Bagchi, S., and Raychaudhuri, P. (2006) Mol. Cell. Biol. 26 2531–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su'etsugu, M., Takata, M., Kubota, T., Matsuda, Y., and Katayama, T. (2004) Genes Cells 9 509–522 [DOI] [PubMed] [Google Scholar]
- 43.Katayama, T., Kubota, T., Kurokawa, K., Crooke, E., and Sekimizu, K. (1998) Cell 94 61–71 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.