Abstract
Osterix/Sp7, a member of the Sp1 transcription factor family, plays an essential role in bone formation and osteoblastogenesis. Although Osterix has been shown to be induced by BMP2 in a mesenchymal cell line, the molecular basis of the regulation, expression and function of Osterix during osteoblast differentiation, is not fully understood. Thus we examined the role of BMP2 signaling in the regulation of Osterix using the mesenchymal cell lines C3H10T1/2 and C2C12. Osterix overexpression induced alkaline phosphatase activity and osteocalcin expression in C2C12 cells and stimulated calcification of murine primary osteoblasts. Considering that Runx2 overexpression induces Osterix, these results suggest that Osterix functions as downstream of Runx2. Surprisingly, BMP2 treatment induced Osterix expression and alkaline phosphatase activity in mesenchymal cells derived from Runx2-deficient mice. Furthermore, overexpression of Smad1 and Smad4 up-regulated Osterix expression, and an inhibitory Smad, Smad6, markedly suppressed BMP2-induced Osterix expression in the Runx2-deficient cells. Moreover, overexpression of a homeobox gene, Msx2, which is up-regulated by BMP2 and promotes osteoblastic differentiation, induced Osterix expression in the Runx2-deficient cells. Knockdown of Msx2 clearly inhibited induction of Osterix by BMP2 in the Runx2-deficient mesenchymal cells. Interestingly, microarray analyses using the Runx2-deficient cells revealed that the role of Osterix was distinct from that of Runx2. These findings suggest that Osterix is regulated via both Runx2-dependent and -independent mechanisms, and that Osterix controls osteoblast differentiation, at least in part, by regulating the expression of genes not controlled by Runx2.
Osteoblasts are differentiated from multipotent mesenchymal cells (1). This differentiation process is regulated by several cytokines, including bone morphogenetic proteins, transforming growth factor β, Wnt, and hedgehog (2–5). Among them, BMP2 (bone morphogenetic protein 2) is one of the most powerful cytokines that promote differentiation of mesenchymal cells into osteoblasts in vitro and induce bone formation in vivo (2). BMP2 exhibits this osteogenic action by activating Smad signaling and by regulating transcription of osteogenic genes such as ALP, type I collagen, osteocalcin, and bone sialoprotein (Bsp)2(6). Runt-related gene 2 (Runx2)/Core-binding factor 1 (Cbfa1), an essential transcription factor for osteoblast differentiation and bone formation (7) and responsible gene for cleidocranial dysplasia (8), directly regulates the expression of osteocalcin and Bsp (9). BMP2 is known to control the expression and functions of Runx2 through Smad signaling (10–12). These findings have established the importance of the BMP2-Smad-Runx2 axis in osteoblastogenesis.
Osterix, an Sp1 transcription family member, is up-regulated by BMP2 during osteoblastic differentiation (13). Osterix has also been reported to inhibit chondrogenesis (14). Mice deficient in the Osterix gene show no bone formation and complete absence of osteoblasts (13), indicating the indispensable role of Osterix in osteoblastogenesis. Interestingly, Runx2 is expressed in mesenchymal cells of Osterix null mice (13). In contrast, expression of Osterix is not observed in Runx2-deficient mice (13). These findings suggest that Osterix may function as downstream of Runx2 during osteoblast differentiation. On the other hand, another study found that Runx2 is not involved in induction of Osterix (15). To date, the target genes of Osterix, which are responsible for the osteogenic function of Osterix, have not been identified. Therefore, the regulation and functions of Osterix during osteoblast differentiation remain unclear.
In this study, we investigated the molecular mechanisms that regulate the expression and functions of Osterix during osteoblast differentiation. We found that Smad signaling is required for induction of Osterix, and that Osterix expression is regulated via both Runx2-dependent and -independent mechanisms by BMP2 signaling. Furthermore, Osterix promotes osteoblast differentiation of Runx2-deficient mesenchymal cells in association with up-regulation of several genes, which are not up-regulated by Runx2. Thus, we believe that our findings provide novel insights into osteoblastogenesis.
EXPERIMENTAL PROCEDURES
Cells—C3H10T1/2 and C2C12 were purchased from RIKEN Cell Bank and cultured in α-modified minimum Eagle's medium containing 10% fetal bovine serum. IGF-1 was purchased from R&D Systems.
Isolation of Primary Osteoblasts—The calvariae were isolated from 2- or 3-day-old neonatal mice and digested with 0.1% collagenase (Wako, Osaka, Japan) and 0.2% dispase (Dojindo, Tokyo, Japan) for 7 min at 37 °C. The cells were collected by centrifugation as mesenchymal cells. The digested calvariae were sequentially digested four times with 0.1% collagenase and 0.2% dispase for 7 min at 37 °C (16). The last three cell fractions were collected and used as primary osteoblasts (16). The cells showed ALP activity. Runx2-deficient mesenchymal cells were isolated from the calvariae of Runx2-deficient embryos as described previously (17). In brief, the anterior region of calvariae from an embryo at embryonic day 18.5 was minced and cultured for 10–14 days in three-dimensional collagen gel (Cellmatrix, Nitta Gelatin Co.) with α-modified minimum Eagle's medium containing 10% fetal bovine serum. The cells outgrowing from the explants were retrieved by incubation for 30 min with 0.2% collagenase in phosphate-buffered saline (PBS) at 37 °C and then cultured with α-modified minimum Eagle's medium containing 10% fetal bovine serum (17).
Osterix cDNA—As described previously (13), C2C12 cells were incubated in the presence of BMP2 for 4 days. Because BMP2 showed maximum effect on induction of Osterix at 300 ng/ml (data not shown), we used that dose in this study. Osterix cDNA was isolated from the cells by reverse-transcribed PCR. The sequence of the cDNA was confirmed by DNA sequence analysis. The Osterix cDNA was subcloned into the pcDNA4 expression vector tagged with a Myc epitope at the C terminus (Invitrogen).
Generation of Adenovirus—Recombinant adenovirus carrying Osterix was constructed by homologous recombination between the expression cosmid cassette (pAxCAwt) and the parental virus genome in 293 cells (RIKEN) as described previously using an adenovirus construction kit (Takara) (11, 15, 17). Adenoviruses carrying Runx2, Msx2, Smad1, Smad4, Smad6, and FLAG-Dlx5 were used as described previously (11, 16, 18). The viruses showed no proliferative activity because of a lack of E1A-E1B. Titers of the viruses were determined using a modified point assay (19). Infection of recombinant adenoviruses with C3H10T1/2 cells, C2C12 cells, or primary osteoblasts was performed by incubation with adenovirus at a multiplicity of infection (m.o.i.) of 40, except where indicated otherwise.
Western Blotting—The cells were washed four times with ice-cold PBS and solubilized in lysis buffer (20 mm Hepes (pH 7.4), 150 mm NaCl, 1 mm EGTA, 1.5 mm MgCl2, 10% glycerol, 1% Triton X-100, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 0.2 mm sodium orthovanadate). The lysates were centrifuged at 16,000 × g for 20 min at 4 °C. The supernatants were boiled in SDS sample buffer containing 0.5 m β-mercaptoethanol. These samples were separated by SDS-PAGE, transferred to nitrocellulose membrane, immunoblotted with corresponding antibodies, and visualized with horseradish peroxidase coupled to anti-mouse antibody or horseradish peroxidase coupled to anti-rabbit antibody with enhancement by ECL detection kits.
Reverse-transcribed PCR and Real Time PCR—Total RNA was isolated from cells using the SV total RNA isolation kit (Promega). After denaturation of total RNA at 70 °C for 10 min, cDNA was synthesized with oligo(dT) primer and reverse transcriptase (Invitrogen). PCR amplifications were performed using the specific primers for mouse Osterix (sense primer, 5′-GAAAGGAGGCACAAAGAAG-3′, and antisense primer, 5′-CACCAAGGAGTAGGTGTGTT-3′). PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide. The PCR products were then subcloned into the TA cloning vector and the DNA sequences of the PCR products determined.
To determine the expression levels of Osterix, osteocalcin, Msx2, ALP mRNA, Col1a1, Bsp, Col2a1, Col10a1, and Sox9, the cDNA samples were analyzed by real time PCR using an ABI PRISM 7300 unit (Applied Biosystems) and specific Taqman probes as follows: mouse Osterix (sense primer, 5′-AGCGACCACTTGAGCAAACAT-3′; antisense primer, 5′-GCGGCTGATTGGCTTCTTCT-3′; probe, 5′-CCCGACGCTGCGACCCTCC-3′); mouse Msx2 (sense primer, 5′-CCATATACGGCGCATCCTACC-3′; antisense primer, 5′-CAACCGGCGTGGCATAGAG-3′; probe, 5′-AGACCTGTGCTCCCCATCCCGCC-3′); mouse osteocalcin (sense primer, 5′-GCAATAAGGTAGTGAACAGACTCC-3′; antisense primer, 5′-GTTTGTAGGCGGTCTTCAAGC-3′; probe, 5′-TGGAGCCTCAGTCCCCAGCCCA-3); mouse ALP (sense primer, 5′-ATCTTTGGTCTGGCTCCCATG-3′; antisense primer, 5′-TTTCCCGTTCACCGTCCAC-3′; probe, 5′-TGAGCGACACGGACAAGAAGCCCTT-3′); mouse Col1a1 (sense primer, 5′-GCAACAGTCGCTTCACCTACA-3′; antisense primer, 5′-CAATGTCCAAGGGAGCCACAT-3′; probe, 5′-CCTTGTGGACGGCTGCACGAGTCAC-3′); mouse Bsp (sense primer, 5′-AAGCAGCACCGTTGAGTATGG-3′; antisense primer, 5′-CCTTGTAGTAGCTGTATTCGTCCTC-3′; probe, 5′-CGGTAAGTGTCGCCACGAGGCTCCC-3′); mouse Col2a1 (sense primer, 5′-CCTCCGTCTACTGTCCACTGA-3′; antisense primer, 5′-ATTGGAGCCCTGGATGAGCA-3′; probe, 5′-CTTGAGGTTGCCAGCCGCTTCGTCC-3′); mouse Col10a1 (sense primer, 5′-GCCAAGCAGTCATGCCTGAT-3′; antisense primer, 5′-GACACGGGCATACCTGTTACC-3′; probe, 5′-AGCACTGACAAGCGGCATCCCAGA-3′); and mouse Sox9 (sense primer, 5′-CCTTCAACCTTCCTCACTACAGC-3′; antisense primer, 5′-GGTGGAGTAGAGCCCTGAGC-3′; probe, 5′-CCGCCCATCACCCGCTCGCAATAC-3′). The expression levels of each sample were normalized against β-actin mRNA expression.
Microarray Analysis—As we observed that expression of Osterix or Runx2 reached maximum after infection of the corresponding adenovirus and would like to examine the direct effects of Osterix or Runx2 on the target genes, we used total RNA isolated from C3H10T1/2 cells that were infected with or without Runx2 or Osterix adenovirus and incubated for 4 days. Total RNA was isolated from the cells using the SV total RNA isolation kit (Promega). An oligonucleotide microarray (Gene Chip Murine Genome U74Av2, Affymetrix) was used to monitor the relative abundance of transcripts as described previously (20). Double strand cDNA was synthesized from 10 μg of total RNA with oligo(dT) primer, amplified with T7 RNA polymerase up to ∼100 μg of cRNA, and hybridized to the oligonucleotide microarray according to the manufacturer's instructions. For normalization, the average intensity for genes was made equal to 100 to reliably compare variable multiple arrays (20).
Determination of ALP Staining—The cultured C3H10T1/2 cells, C2C12 cells, or Runx2 null mesenchymal cells were washed twice with PBS and stained with nitro blue tetrazolium (Sigma) and 5-bromo-4-chroro-3-indolyl phosphate (Sigma) (18).
Alizarin Red Staining—The cultured calvaria cells were washed twice with PBS and fixed in 10% formalin for 10 min. The cells were then stained with 1% alizarin red solution for 5 min (18).
Sirius Red Staining for Collagen—C3H10T1/2 cells were washed with PBS and fixed in 10% formalin for 10 min. After washing the cells with distilled H2O, the cells were stained with 0.1% Direct Red 80 (Sigma) for 90 min (21).
Immunocytochemical Analysis—C3H10T1/2 cells were washed with PBS and fixed in 10% formalin for 10 min. After blocking with blocking reagent (Vector Laboratories), the cells were stained with anti-collagen antibody (Chemicon), followed by visualization with a VECTASTAIN® ABC kit (Vector Laboratories).
Knockdown Experiment—Cells were transfected with Stealth siRNA (Invitrogen) and Mission siRNA (Sigma) using Lipofectamine 2000 according to the manufacturer's protocol. Two sets of Msx2 siRNA (5′-CCGCCAGAAACAGUACCUGUCCAUA-3′ and 5′-GAUAUGGCAUGUACCAUCUAUCCUA-3′) or Osterix siRNA (5′-CUUCGCAUCUGAAAGCCCA-3′ and 5′-CCUACUUACCCAUCUGACU-3′) were used for knockdown of Msx2 or Osterix, respectively. Control siRNA was purchased from Invitrogen.
RESULTS
Osteoblastogenic Activity of Osterix—To understand the functional role of Osterix in osteoblast differentiation, we examined the effect of Osterix overexpression in C2C12 cells. Osterix overexpression induced ALP activity and osteocalcin expression in C2C12 cells (Fig. 1, A–C). Similar to BMP2 treatment or Runx2 overexpression, Osterix overexpression also induced ALP activity in C3H10T1/2 cells (Fig. 1D). Consistently, knockdown of Osterix inhibited expression of osteocalcin, Bsp, and ALP stimulated by BMP2 in C3H10T1/2 cells (Fig. 1E). In contrast to BMP2 treatment, Osterix had very little effect on the expression of Col1a1 or collagen synthesis (Fig. 1, D and F). In addition, knockdown of Osterix did not suppress Col1a1 expression (Fig. 1E). Moreover, Runx2 overexpression showed marginal effects on Col1a1 expression and collagen synthesis (Fig. 1, A, D, and F). These results suggest that other transcription factors, which would be controlled by BMP2, might be involved in the regulation of Col1a1 expression and maturation. Furthermore, Osterix overexpression in mouse primary osteoblasts clearly stimulated calcification of the cells (Fig. 1G). These findings indicate that Osterix has osteoblastogenic activity. Interestingly, we observed that the osteoblastogenic activity of Osterix differed from that of Runx2 (Fig. 1, B, C, D, F, and G), suggesting that Osterix and Runx2 have distinct and separate roles during osteoblast differentiation. To verify this possibility, we compared the target genes of Runx2 and Osterix by performing microarray analyses. As shown in Fig. 2B, several genes, including Wnt4, Bglap1, and BMP7, were similarly induced by Runx2 or Osterix. However, we found that there were two groups of genes that were induced by either Runx2 or Osterix but not both (Fig. 2, A and C). Together, these findings suggest that Runx2 and Osterix have common roles in osteoblast differentiation but also possess distinct roles in gene regulation during osteoblastogenesis.
FIGURE 1.
Osteoblastogenic activities of Osterix. A, C2C12 cells treated with BMP2 (300 ng/ml) or infected with or without Mock, Runx2, or Osterix adenovirus (40 m.o.i.) were cultured for 4 days. Total RNA isolated from the cells was subjected to real time PCR for expression of Osterix (top panel) and Runx2 (bottom panel) using the specific Taqman probes. The expression of Osterix and Runx2 was normalized withβ-actin expression. Cont, control. B, C2C12 cells treated with BMP2 (300 ng/ml) or infected with or without Mock, Runx2, or Osterix adenovirus (40 m.o.i.) were cultured for a week, and ALP activity was then determined. C, C2C12 cells treated with BMP2 (300 ng/ml) or infected with Runx2 or Osterix adenovirus (40 m.o.i.) were cultured for a week. Total RNA was isolated from the cells and osteocalcin mRNA expression was determined by real time PCR analysis using the Taqman probe specific to osteocalcin. Osteocalcin expression was normalized againstβ-actin expression. D, C3H10T1/2 cells were treated with BMP2 (300 ng/ml) or infected with Runx2 or Osterix adenovirus (40 m.o.i.) and then cultured for a week. The cells were subjected to ALP staining (top panel), Sirius red staining (2nd panel), or immunostaining using anti-Col1 antibody (bottom panel). E, C3H10T1/2 cells were transfected with control siRNA (Cont) or Osterix siRNA (siOsx-1 and siOsx-2), and then cultured in the presence or absence of BMP2 (300 ng/ml) for 4 days. Total RNA isolated from the cells was subjected to real time PCR analyses using the specific Taqman probes for Osterix, Bsp, ALP, Col1a1, and osteocalcin (OCN). The expression of each gene was normalized against β-actin expression. F, C3H10T1/2 cells were treated with BMP2 (300 ng/ml) or infected with Mock, Runx2, or Osterix adenovirus (40 m.o.i.) and then cultured for a week. Total RNA was isolated from the cells, and Col1a1 mRNA expression was determined by real time PCR analysis using the Taqman probe. Col1a1 expression was normalized against β-actin expression. G, primary osteoblasts isolated from mice calvariae were treated with BMP2 (300 ng/ml) or infected with Runx2 or Osterix adenovirus (40 m.o.i.), cultured for a week, and then stained with Alizarin red.
FIGURE 2.
Genes up-regulated by Runx2 or Osterix. C3H10T1/2 cells were infected with Runx2 or Osterix adenovirus (40 m.o.i.) and then cultured for 4 days. Total RNA was isolated from the cells and subjected to microarray analyses.
Regulation of Osterix Expression Independently of Runx2—In contrast to our results (Fig. 1), a previous study reported apparently Runx2-independent Osterix expression (15). As shown above (Fig. 1, B, C, D, F, and G, and Fig. 2), Osterix has distinct roles during osteoblast differentiation. Therefore, we examined whether Runx2 is necessary for induction of Osterix expression using mesenchymal cells isolated from Runx2-deficient mice. These cells were able to differentiate into ALP-positive osteoblastic cells upon BMP2 treatment or Runx2 overexpression within 3 days (Fig. 3, A and B) (16, 17). Although the cells had some ability to differentiate into chondrocytic cells, real time PCR analyses indicated that the cells preferentially differentiated into osteoblastic cells (Fig. 3C). As expected, Runx2 induced Osterix expression in these cells (Fig. 3, D and E). Surprisingly, BMP2 treatment induced Osterix expression in the Runx2-deficient mesenchymal cells (Fig. 3, D and E). These results suggest that Osterix expression is regulated by BMP2 through both Runx2-dependent and -independent mechanisms.
FIGURE 3.
Induction of Osterix expression by BMP2 in Runx2-deficient mesenchymal cells. A and B, Runx2-deficient mesenchymal cells were treated with BMP2 (300 ng/ml) or infected with or without Mock, Runx2, or Osterix adenovirus (40 m.o.i), cultured for 3 or 5 days, and then stained for ALP. Cont, control. C, Runx2-deficient mesenchymal cells were treated with BMP2 (300 ng/ml) or infected with or without Mock, Runx2, or Osterix adenovirus (40 m.o.i.), and cultured for a week. Total RNA was isolated from the cells, and the mRNA expression of ALP, Col1a1, BSP, Col2a1, Sox9, and Col10a1 was determined by real time PCR analysis using the Taqman probes. The expression of each gene was normalized against β-actin expression. D, Runx2-deficient mesenchymal cells were treated with BMP2 (300 ng/ml) or infected with Runx2 adenovirus (40 m.o.i.) and cultured for 4 days. Total RNA was isolated from the cells and subjected to reverse-transcribed PCR analysis for Osterix (top panel) and β-actin mRNA (bottom panel). E, Runx2-deficient mesenchymal cells were treated with BMP2 (300 ng/ml) or infected with or without Mock, Runx2, or Osterix adenovirus (40 m.o.i.) and cultured for 4 days. Total RNA isolated from the cells was subjected to real time PCR analysis using the Taqman probe specific for Osterix. The expression of Osterix was normalized against β-actin expression.
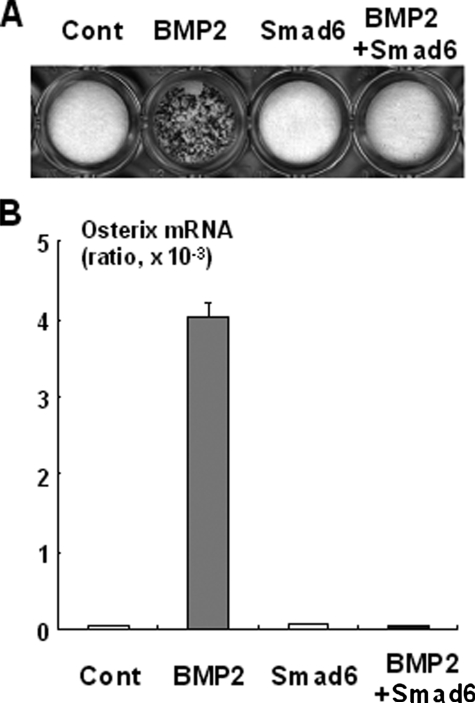
Regulation of Osterix Expression by Smad and Msx2 Signaling—To understand the molecular mechanism by which BMP2 induces Osterix expression in Runx2-deficient cells, we first examined whether Smad signaling is implicated in the regulation of Osterix expression, because Smad signaling plays a central role in BMP2-regulated osteoblast differentiation (22). As shown in Fig. 4, A and B, overexpression of Smad1 and Smad4 stimulated BMP2-induced ALP activity in Runx2-deficient mesenchymal cells. Moreover, overexpression of Smad1 and Smad4 enhanced Osterix expression by BMP2 in these cells (Fig. 4C). To confirm the involvement of Smad signaling in Runx2-independent Osterix expression, we next examined the effect of Smad6 on Runx2-deficient cells. We found that overexpression of Smad6 abolished induction of Osterix and ALP activity by BMP2 in Runx2-deficient cells (Fig. 5, A and B). Collectively, these results indicate that Smad signaling is necessary for Osterix expression in Runx2-deficient mesenchymal cells and subsequent osteoblastic differentiation.
FIGURE 4.
Up-regulation of Osterix expression by Smad signaling in Runx2-deficient mesenchymal cells. A, Runx2-deficient mesenchymal cells were infected with both Smad1 and Smad4 adenoviruses (40 m.o.i.) and then cultured for a week in the presence of BMP2 (50 ng/ml) as indicated. The cells were then stained for ALP. B and C, Runx2-deficient mesenchymal cells were infected with both Smad1 and Smad4 adenoviruses (40 m.o.i.) and then cultured for 4 days in the presence of BMP2 (50 ng/ml) as indicated. Total RNA was isolated from the cells and determined by real time PCR for ALP mRNA (B) and Osterix mRNA (C). ALP and Osterix expression was normalized against β-actin expression.
FIGURE 5.
Inhibition of BMP2-induced Osterix expression by Smad6. A, Runx2-deficient mesenchymal cells were infected with Smad6 adenovirus (40 m.o.i.) and cultured for a week in the presence of BMP2 (300 ng/ml) as indicated. The cells were determined by ALP staining. B, Runx2-deficient mesenchymal cells were infected with Smad6 adenovirus (40 m.o.i.) and cultured for 4 days in the presence of BMP2 (300 ng/ml) as indicated. Total RNA was isolated from the cells and determined by real time PCR analysis for Osterix mRNA expression. Osterix expression was normalized by β-actin expression. Cont, control.
Because we previously reported that Msx2 regulates osteoblast differentiation via a Runx2-independent mechanism (18), we hypothesized that Msx2 may function as upstream of Osterix. First, we determined whether Msx2 is induced by BMP2 treatment in Runx2-deficient mesenchymal cells. As shown in Fig. 6A, BMP2 clearly up-regulated Msx2 expression even in the absence of the Runx2 gene. Importantly, Msx2 overexpression induced Osterix expression in the Runx2-deficient mesenchymal cells (Fig. 6B). Moreover, knockdown of Msx2 clearly inhibited induction of Osterix by BMP2 in the Runx2-deficient mesenchymal cells (Fig. 6, C and D). Although IGF-1 and Dlx5 have been implicated in regulation of Osterix expression (15, 23, 24), we did not observe any effect of IGF-1 or Dlx5 on Osterix expression in Runx2-deficient mesenchymal cells (Fig. 6, E and F). Our findings indicate that Msx2, which is regulated by BMP2, controls Osterix expression via a Runx2-independent mechanism.
FIGURE 6.
Induction of Osterix by Msx2 in Runx2-deficient mesenchymal cells. A, Runx2-deficient mesenchymal cells were incubated in the presence or absence of BMP2 (300 ng/ml). Total RNA was isolated from the cells and determined by real time PCR analysis for Msx2 mRNA expression. Msx2 expression was normalized against β-actin expression. B, Runx2-deficient mesenchymal cells were infected with or without Msx2 adenovirus (40 m.o.i.), and cultured for 4 days. Total RNA was isolated from the cells, and Osterix mRNA expression was determined by real time PCR analysis. Osterix expression was normalized against β-actin expression. C and D, Runx2-deficient mesenchymal cells were transfected with control (Cont) or Msx2 (site1 or site2) siRNA and then cultured in the presence of absence of BMP2 (300 ng/ml). Total RNA was isolated from the cells and analyzed for Msx2 (C) and Osterix (D) mRNA expression by real time PCR. Msx2 and Osterix expression was normalized against β-actin expression. E, Runx2-deficient mesenchymal cells were incubated in the presence of BMP2 (300 ng/ml) or IGF-1 (200 or 400 ng/ml) and cultured for 4 days. Total RNA was isolated from the cells and Osterix mRNA expression determined by real time PCR analysis. Osterix expression was normalized against β-actin expression. F, Runx2-deficient mesenchymal cells were treated with BMP2 (300 ng/ml) or infected with control (Mock) or Dlx5 adenovirus (40 m.o.i.) and cultured for 4 days. Total RNA was isolated from the cells and Osterix mRNA expression determined by real time PCR analysis. Osterix expression was normalized against β-actin expression.
Osteoblastogenic Activity of Osterix in Runx2-deficient Mesenchymal Cells—We next examined whether Osterix was able to promote osteoblast differentiation in Runx2-deficient mesenchymal cells. We exogenously introduced Osterix into Runx2-deficient mesenchymal cells and found that Osterix overexpression induced ALP activity in these cells (Fig. 3, A, B and E). In addition, Osterix overexpression significantly stimulated osteocalcin and Bsp expression (Fig. 3C). These results suggest that Osterix itself has osteoblastogenic activity.
DISCUSSION
Although a previous genetic study raised the possibility that Osterix functions as downstream of Runx2 (13), the mechanism by which BMP2 regulates Osterix expression during osteoblast differentiation has been unclear. In this study, we demonstrated that Osterix expression is regulated by both Runx2-dependent and -independent mechanisms. Other researchers and we have previously demonstrated that BMP2 up-regulates Runx2 expression during osteoblast differentiation (10, 11, 25), and thus it is likely that BMP2 controls Osterix expression through Runx2. We observed that overexpression of Runx2 induces Osterix expression in mesenchymal cell lines. Notably, we demonstrated that BMP2 and Msx2 induced Osterix expression in Runx2-deficient mesenchymal cells. Furthermore, knockdown of Msx2 blocked the induction of Osterix in the Runx2-deficient mesenchymal cells. Thus, these results indicate the novel paradigm that BMP2 controls Osterix expression independently of Runx2 through Msx2.
The functional role of Msx2 in osteoblasts appears to be complex, as several studies have reported that Msx2 positively and negatively controls osteoblast differentiation (18, 26–28). However, genetic studies have indicated that Msx2 plays critical roles in bone formation and osteoblast differentiation (29, 30). The functional role of Msx2 in bone would be temporally and spatially regulated. Therefore, our present results suggest that Msx2 exhibits osteogenic function by up-regulating Osterix, at least in part, in a Runx2-independent manner.
Because Osterix null mice show complete absence of bone formation and osteoblasts at the embryo stage, Osterix is considered to play an indispensable role in bone development and osteoblastogenesis (13). However, whether Osterix itself is sufficient to stimulate osteoblast differentiation has been unclear. We found that Osterix has osteogenic activity in vitro and stimulates osteoblast differentiation of mesenchymal cells. Interestingly, Osterix exhibited osteoblastogenic activity in Runx2-deficient mesenchymal cells. This result suggests two possibilities. One is that Osterix functions as downstream of Runx2. This idea is well supported by our finding that Runx2 overexpression induced Osterix expression in Runx2-deficient mesenchymal cells as well as in C2C12 cells, in which the Runx2 gene is intact. The other possibility is that Osterix may have functions distinct from those of Runx2. Consistent with this idea, we observed that Osterix showed lesser osteoblastogenic activity than Runx2. To verify these possibilities, we compared the transcriptional target genes of Osterix and Runx2 by performing microarray analyses. Overexpression of either Runx2 or Osterix up-regulated common genes such as Wnt4, Bglap1, and BMP7 in Runx2-deficient cells, suggesting that Osterix and Runx2 possess common roles during osteoblastogenesis. Although deficiency of either the Osterix or Runx2 gene leads to complete absence of bone at the embryo stage, the phenotype of Osterix null mice in bone seems different from that of Runx2 null mice at birth (7, 13). This suggests the possibility that Osterix and Runx2 have distinct functions during bone formation. Indeed, we observed that Osterix and Runx2 also stimulate expression of distinct genes. Collectively, our data suggest that Osterix not only functions as downstream of Runx2 but also regulates osteoblastogenesis independently of Runx2 by controlling Osterix-specific target genes. Because the Runx2 null mice show no bone formation, the Runx2-independent pathway, which is partly regulated by Osterix, could be necessary but not sufficient for osteogenesis in vivo. In other words, both Runx2-dependent and -independent pathways could be required for bone formation. The target genes responsible for the bone phenotype seen in Osterix null mice have not yet been identified. Therefore, identification of the genes that are regulated by Osterix and that play critical roles in osteoblastogenesis may advance our understanding of the molecular mechanism by which BMP2 regulates bone formation and osteoblastogenesis and may contribute to developing therapeutic agents for bone diseases.
It has been demonstrated that Osterix has two isoforms in humans (31). We isolated the longer isoform, which potentially encodes 18 additional amino acids, from C2C12 cells but did not detect the shorter isoform in murine mesenchymal cell lines (data not shown). We generated the two types of Osterix expression vectors that encode Osterix proteins corresponding to the shorter and longer mRNA. However, the longer form of Osterix expression vector failed to show osteoblastogenic activity (data not shown). It is therefore necessary to identify the translational starting site(s) of the Osterix gene.
In conclusion, we have demonstrated that BMP2 regulates Osterix expression independently through two distinct transcription factors, Runx2 and Msx2. We also found that Osterix possesses osteogenic function during osteoblastogenesis. Thus, we believe that our findings contribute to the understanding of the molecular basis by which BMP2 conducts osteoblast differentiation of mesenchymal cells.
This work was supported in part by the Ministry of Education, Science, and Sports of Japan, Culture Grants-in-aid for Scientific Research A 17209059, B15390560, the Naito Foundation, and the Novartis Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Bsp, bone sialoprotein; PBS, phosphate-buffered saline; m.o.i., multiplicity of infection; ALP, alkaline phosphatase; siRNA, small interfering RNA.
References
- 1.Yin, T., and Li, L. (2006) J. Clin. Investig. 116 1195-1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen, M. M. (2002) Am. J. Med. Genet. 109 87-92 [DOI] [PubMed] [Google Scholar]
- 3.Bodine, P. V., Zhao, W., Kharode, Y. P., Bex, F. J., Lambert, A. J., Goad, M. B., Gaur, T., Stein, G. S., Lian, J. B., and Komm, B. S. (2004) Mol. Endocrinol. 18 1222-1237 [DOI] [PubMed] [Google Scholar]
- 4.Baron, R., Rawadi, G., and Roman-Roman, S. (2006) Curr. Top. Dev. Biol. 76 103-127 [DOI] [PubMed] [Google Scholar]
- 5.Hu, H., Hilton, M. J., Tu, X., Yu, K., Ornitz, D. M., and Long, F. (2005) Development (Camb.) 132 49-60 [DOI] [PubMed] [Google Scholar]
- 6.Chen, D., Harris, M. A., Rossini, G., Dunstan, C. R., Dallas, S. L., Feng, J. Q., Mundy, G. R., and Harris, S. E. (1997) Calcif. Tissue Int. 60 283-290 [DOI] [PubMed] [Google Scholar]
- 7.Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R. T., Gao, Y. H., Inada, M., Sato, M., Okamoto, R., Kitamura, Y., Yoshiki, S., and Kishimoto, T. (1997) Cell 89 755-764 [DOI] [PubMed] [Google Scholar]
- 8.Otto, F., Thornell, A. P., Crompton, T., Denzel, A., Gilmour, K. C., Rosewell, I. R., Stamp, G. W., Beddington, R. S., Mundlos, S., Olsen, B. R., Selby, P. B., and Owen, M. J. (1997) Cell 89 765-771 [DOI] [PubMed] [Google Scholar]
- 9.Ducy, P., Zhang, R., Geoffroy, V., Ridall, A. L., and Karsenty, G. (1997) Cell 89 747-754 [DOI] [PubMed] [Google Scholar]
- 10.Hanai, J., Chen, L. F., Kanno, T., Ohtani-Fujita, N., Kim, W. Y., Guo, W. H., Imamura, T., Ishidou, Y., Fukuchi, M., Shi, M. J., Stavnezer, J., Kawabata, M., Miyazono, K., and Ito, Y. (1999) J. Biol. Chem. 274 31577-31582 [DOI] [PubMed] [Google Scholar]
- 11.Nishimura, R., Hata, K., Harris, S. E., Ikeda, F., and Yoneda, T. (2002) Bone (Elmsford) 31 303-312 [DOI] [PubMed] [Google Scholar]
- 12.Javed, A., Barnes, G. L., Jasanya, B. O., Stein, J. L., Gerstenfeld, L., Lian, J. B., and Stein, G. S. (2001) Mol. Cell. Biol. 21 2891-2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakashima, K., Zhou, X., Kunkel, G., Zhang, Z., Deng, J. M., Behringer, R. R., and de Crombrugghe, B. (2002) Cell 108 17-29 [DOI] [PubMed] [Google Scholar]
- 14.Kaback, L. A., Soung do, Y., Naik, A., Smith, N., Schwarz, E. M., O'Keefe, R. J., and Drissi, H. (2008) J. Cell. Physiol. 214 173-182 [DOI] [PubMed] [Google Scholar]
- 15.Lee, M. H., Kwon, T. G., Park, H. S., Wozney, J. M., and Ryoo, H. M. (2003) Biochem. Biophys. Res. Commun. 309 689-694 [DOI] [PubMed] [Google Scholar]
- 16.Hata, K., Nishimura, R., Ueda, M., Ikeda, F., Matsubara, T., Ichida, F., Hisada, K., Nokubi, T., Yamaguchi, A., and Yoneda, T. (2005) Mol. Cell. Biol. 25 1971-1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, T., Gao, Y., Sakamoto, K., Minamizato, T., Furukawa, K., Tsukazaki, T., Shibata, Y., Bessho, K., Komori, T., and Yamaguchi, A. (2007) J. Cell. Physiol. 211 728-735 [DOI] [PubMed] [Google Scholar]
- 18.Ichida, F., Nishimura, R., Hata, K., Matsubara, T., Ikeda, F., Hisada, K., Yatani, H., Cao, X., Komori, T., Yamaguchi, A., and Yoneda, T. (2004) J. Biol. Chem. 279 34015-34022 [DOI] [PubMed] [Google Scholar]
- 19.Miyake, S., Makimura, M., Kanegae, Y., Harada, S., Sato, Y., Takamori, K., Tokuda, C., and Saito, I. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 1320-1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishii, M., Hashimoto, S., Tsutsumi, S., Wada, Y., Matsushima, K., Kodama, T., and Aburatani, H. (2000) Genomics 68 136-143 [DOI] [PubMed] [Google Scholar]
- 21.Maniatopoulos, C., Sodek, J., and Melcher, A. H. (1988) Cell Tissue Res. 254 317-330 [DOI] [PubMed] [Google Scholar]
- 22.Nishimura, R., Kato, Y., Chen, D., Harris, S. E., Mundy, G. R., and Yoneda, T. (1998) J. Biol. Chem. 273 1872-1879 [DOI] [PubMed] [Google Scholar]
- 23.Celil, A. B., and Campbell, P. G. (2005) J. Biol. Chem. 280 31353-31359 [DOI] [PubMed] [Google Scholar]
- 24.Celil, A. B., Hollinger, J. O., and Campbell, P. G. (2005) J. Cell. Biochem. 95 518-528 [DOI] [PubMed] [Google Scholar]
- 25.Lee, K. S., Kim, H. J., Li, Q. L., Chi, X. Z., Ueta, C., Komori, T., Wozney, J. M., Kim, E. G., Choi, J. Y., Ryoo, H. M., and Bae, S. C. (2000) Mol. Cell. Biol. 20 8783-8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng, S. L., Shao, J. S., Charlton-Kachigian, N., Loewy, A. P., and Towler, D. A. (2003) J. Biol. Chem. 278 45969-45977 [DOI] [PubMed] [Google Scholar]
- 27.Kim, Y. J., Lee, M. H., Wozney, J. M., Cho, J. Y., and Ryoo, H. M. (2004) J. Biol. Chem. 279 50773-50780 [DOI] [PubMed] [Google Scholar]
- 28.Igarashi, M., Yogiashi, Y., Mihara, M., Takada, I., Kitagawa, H., and Kato, S. (2007) Mol. Cell. Biol. 27 7947-7954 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Satokata, I., Ma, L., Ohshima, H., Bei, M., Woo, I., Nishizawa, K., Maeda, T., Takano, Y., Uchiyama, M., Heaney, S., Peters, H., Tang, Z., Maxson, R., and Maas, R. (2000) Nat. Genet. 24 391-395 [DOI] [PubMed] [Google Scholar]
- 30.Wilkie, A. O., Tang, Z., Elanko, N., Walsh, S., Twigg, S. R., Hurst, J. A., Wall, S. A., Chrzanowska, K. H., and Maxson, R. E., Jr. (2000) Nat. Genet. 24 387-390 [DOI] [PubMed] [Google Scholar]
- 31.Milona, M. A., Gough, J. E., and Edgar, A. J. (2003) BMC Genomics 4 43. [DOI] [PMC free article] [PubMed] [Google Scholar]