Abstract
Protein serine/threonine phosphatase 4 (PP4c) is an essential polypeptide involved in critical cellular processes such as microtubule growth and organization, DNA damage checkpoint recovery, apoptosis, and tumor necrosis factor α signaling. Like other phosphatases of the PP2A family, PP4c interacts with regulatory proteins, which specify substrate targeting and intracellular localization. The identification of these regulatory proteins is, therefore, key to fully understanding the function of this enzyme class. Here, using a sensitive affinity purification/mass spectrometry approach, we identify a novel, stable cytosolic PP4c interacting partner, KIAA1622, which we have renamed PP4R4. PP4R4 displays weak sequence homology with the A (scaffolding) subunit of the PP2A holoenzyme and specifically associates with PP4c (and not with the related PP2Ac or PP6c phosphatases). The PP4c·PP4R4 interaction is disrupted by mutations analogous to those abrogating the association of PP2Ac with PP2A A subunit. However, unlike the PP2A A subunit, which plays a scaffolding role, PP4R4 does not bridge PP4c with previously characterized PP4 regulatory subunits. PP4c·PP4R4 complexes exhibit phosphatase activity toward a fluorogenic substrate and γH2AX, but this activity is lower than that associated with the PP4c·PP4R2·PP4R3 complex, which itself is less active than the free PP4c catalytic subunit. Our data demonstrate that PP4R4 forms a novel cytosolic complex with PP4c, independent from the complexes containing PP4R1, PP4R2·PP4R3, and α4, and that the regulatory subunits of PP4c have evolved different modes of interaction with the catalytic subunit.
Phosphorylation/dephosphorylation cycles regulate the activity of most key cellular processes, and misregulation of phosphorylation can have dramatic consequences, such as overproliferation or apoptosis. A major issue in phosphorylation-dependent signaling is the specificity of the process; that is, a given substrate must be phosphorylated (and dephosphorylated) at a specific time and location to properly regulate a given function. Several mechanisms are in place to ensure this specificity; for example, members of the phosphoprotein phosphatase (PPP)4 family associate with regulatory proteins that confer substrate targeting, subcellular localization, or activation cues (1, 2).
PP4c and PP6c (gene names PPP4C and PPP6C) form, along with PP2Ac (gene names PPP2CA and PPP2CB), a subgroup referred to as the PP2A-type phosphatases. The PP2A-type phosphatases exhibit a high degree of homology and similar sensitivity to all tested chemical inhibitors. PP4c is located throughout the cell (nucleus, cytoplasm, centrosome (3–5)) and regulates a variety of cellular functions independently of PP2Ac; e.g. PP2Ac is unable to compensate for the embryonic lethality of PP4c null mice (6, 7). PP4c has been implicated in a variety of critical biological processes in higher eukaryotes, including regulation of histone acetylation, DNA damage checkpoint signaling, NFκB activation, and microtubule organization at centrosomes (5, 8–11).
PP4c shares >65% identity with PP2Ac but does not associate in significant quantities with the PP2A regulatory subunits. This has prompted several groups to identify specific interacting partners for PP4c. Many PP4c-interacting proteins have now been reported (5); of these, at least three appear to bind to PP4c directly (PP4R1, PP4R2, and α4/IGBP1). PP4R1 (also known as PP4Rmeg; gene name PPP4R1), a ∼125-kDa protein co-purifying with PP4c from bovine testis extracts, was the first of the stable interactors to be characterized (12, 13). PP4R1 weakly resembles the PP2A A scaffolding subunit but specifically associates with PP4c. The PP4c·PP4R1 complex was reported to regulate histone deacetylase 3 activity (9) and was recently linked to microtubule growth at the centrosome via dephosphorylation of NDEL1 (6). A second regulatory protein, PP4R2 (which shares no homology with PP4R1 or to any other human proteins), was subsequently isolated from pig testis, based on co-purification with PP4c, and has been detected in the nucleus and at centrosomes (14, 15). Finally, α4 appears to be a general PP2A family interacting partner, in that it binds to PP4c, PP2Ac, and PP6c (16, 17).
We previously employed tandem-affinity purification (TAP) coupled to liquid chromatography and mass spectrometry to identify stable protein assemblies containing PP4c. This approach readily recapitulated the identification of PP4R1, PP4R2, and α4 as PP4c binding partners. In addition, two highly related proteins, which we termed PP4R3A and PP4R3B (gene names SMEK1 and SMEK2), were found as stoichiometric interactors for PP4c·PP4R2. We also found that the PP4c·PP4R2·PP4R3 complex binds the hTIP41 (human TIP41, gene name TIPRL) protein in substoichiometric amounts (18). The PP4c·PP4R2·PP4R3 complex is linked to sensitivity to the anticancer drug cisplatin (18–20) and is a phosphatase for the histone variant H2AX, a critical protein for the orchestration of DNA damage repair signaling (10, 11, 21). This work highlighted the utility of affinity purification/mass spectrometry to uncover novel complex assemblies for Ser/Thr phosphatases.
The relatively large size of the TAP tag, the lengthy dual purification process, and the sample loss generated as a result of the two purification procedures can, however, affect the recovery of weaker or more transient interactors (22, 23). Here, we sought to determine whether additional PP4c interactors could be identified using a single affinity FLAG tag purification strategy. In addition to recapitulating all binding partners previously detected by TAP purification, we identified several novel PP4c interactors, including a previously uncharacterized protein, KIAA1622 (here renamed PP4R4), which resembles the A scaffolding subunit of PP2A. PP4R4 specifically interacts with PP4c in a manner similar to the PP2Ac·PP2A A interaction. However, PP4R4 does not interact with PP4c in concert with other known binding partners but forms a distinct cytoplasmic complex to target PP4c to as yet unknown substrates.
EXPERIMENTAL PROCEDURES
Plasmids—pcDNA3-FLAG2 was prepared from pcDNA3-NTAP (18) by inserting the FLAG sequence (MDYKDDDDKAA) into the KpnI and PmeI sites of pcDNA3-NTAP (removing the TAP cassette). pcDNA3-FLAG1-PP1c, pcDNA3-FLAG1-PP2Acα, pcDNA3-FLAG1-PP2Acβ, pcDNA3-FLAG1-PP4c, pcDNA3-FLAG1-PP6c, and pcDNA3-NTAP-PP4R1 were described previously (18).
The coding sequence of human PP4R4 was amplified by PCR from the mammalian gene collection (MGC87177; GenBank™ accession number BC068491) and inserted into the BamHI and NotI sites of pcDNA3-FLAG2 and pcDNA3-NTAP. The coding sequences of PP2A Aα (PPP2R1A, MGC786, BC001537), PP2A Aβ (PPP2R1B, MGC26454, BC027596), and murine eIF4E (from starting clone pACTAG-2 (24)) were amplified by PCR and cloned in-frame into the PmeI and PacI sites of pcDNA3-NTAP. The coding sequence of IκBα (NFKBIA, MGC3942, BC004983) was cloned in-frame in the EcoRI and NotI sites of pcDNA3-3HA. Point mutants of PP4R4 and PP4c were generated by two-step PCR and cloned into BamHI/NotI or EcoRI/NotI sites of pcDNA3-FLAG2, respectively. A portion corresponding to amino acids 751–873 of PP4R4 was cloned in-frame into the BamHI and NotI sites of pGEX-6p2 (GE Healthcare) for expression in bacteria and antibody production.
To create pFastBac-FLAG, a 5′ primer containing an RsrII site, an NdeI site, and the sequence for a single FLAG epitope was used to amplify the multiple cloning site (MCS) region of pFastBac HTa (Invitrogen); the 3′ primer was designed to hybridize to the end of the MCS. The resulting PCR product was cloned into the RsrII and XhoI sites of pFastBac HTa, thus replacing the His tag with a FLAG tag. The pFastBac-StreptagII (Strep) was generated using the same strategy, except that the sequence for the Strep-Tactin-binding peptide was used instead of the FLAG sequence. The constructs were sequenced entirely. The coding sequences of PP4R4 and PP4R2 were inserted into the BamHI and XhoI sites of pFastBac-FLAG. The coding sequence of PP4c was cloned in the EcoRI and HindIII sites of pFastBac HTa or pFastBac-StreptagII. The coding sequence of PP4R3A was cloned into the EcoRI and XhoI sites of pFastBac HTa or pFastBac-StreptagII.
Antibodies—Polyclonal antiserum to GST-PP4R4 (rabbit 2) was produced using standard procedures at the animal facility of the University of Toronto. The characterization of the anti-PP4R4 is described below. Rabbit anti-glutathione S-transferase α4 (rabbit 2972, a kind gift from Dr. N. Sonenberg) recognizes a single major band in HEK293 cell extract (not shown). The antiserum to PP4c (rabbit 18335) was raised against a keyhole limpet hemocyanin-conjugated C-terminal peptide (PRF&L, Canadensis, PA); this antiserum cross-reacts with PP2Ac by immunoblotting (not shown). Commercial antibodies were as follows: PP4c (Bethyl A300–835A), PP4R2 (Bethyl, A300–838A), PP4R1 (Bethyl, A300–837A), γH2AX P-Ser-139 (Upstate, clone JBW301), FLAG (Sigma, F3165), and HA (CRP Inc., MMS-101R). Secondary antibodies for immunoblotting were donkey anti-mouse IgG and donkey anti-rabbit IgG, both conjugated to horseradish-peroxidase (GE Healthcare); for immunofluorescence, a goat anti mouse IgG conjugated to Alexa-488 (Invitrogen, A11001) was utilized.
Cell Culture, Transfection, and Leptomycin B Treatment—HEK293, B35 rat neuroblastoma, 293T, and Vero cells were cultured in high glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units of penicillin, and 100 μg of streptomycin/ml in a 5% CO2-humidified atmosphere at 37 °C. Transfections of HEK293, B35, and 293T cells with Lipofectamine PLUS (Invitrogen) and Vero cells with Lipofectamine LTX (Invitrogen) were performed according to the manufacturer's instructions, and cells were collected 48 h post transfection unless otherwise indicated. Where indicated, 30 nm leptomycin B was added to Vero cells 16 h post-transfection, and the cells were fixed and processed for immunofluorescence after the times indicated. Stable cell pools were generated from HEK293 cells as described (18), and the expression was monitored by immunoblotting.
Silencing Experiments—Individual siRNA duplexes to silence KIAA1622 were obtained from Qiagen, and the SMARTpool of siRNA duplexes was obtained from Dharmacon. The target siRNAs against PP4R4 are: PP4R4_1, AAGGAACAAGTGTGATTGCAA (catalog #S100459872); PP4R4_2 ACCGGAAGAAATAGAAAGATT (catalog #S100459879); PP4R4_3, CGCGATGGATTTCAGTCAGAA (catalog #S100459886); PP4R4_4, CTGGTCAAGATGTCCAAGGAA (catalog #S100459893). The sequences for each siRNA in the SMART-pool are as follows: GAACAAGTGTGATTGCAAATT, TGAAAGGGCTGTTTATCTGTT, GATTGACAGTCGATGAAGATT, GCGATGGATTTCAGTCAGATT.
PP4c siRNA recognizes the sequence GACAATCGACCGAAAGCAATT. HEK293 or U2OS cells were transfected with siRNA (18 nm final concentration) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocol. Cells were collected 40–72 h post-transfection.
Immunoprecipitation from Mouse Brain—One mouse brain (∼0.5 g) was homogenized in lysis buffer (1:10 w/v) containing 25 mm Hepes, pH 8, 120 mm NaCl, 0.5% Triton-X100, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 1× of a protease inhibitor mixture (Sigma, P8340). The homogenate was incubated on ice for 40 min followed by centrifugation at 15,000 rpm for 30 min. Cleared lysate (7.5 mg per immunoprecipitation) was incubated with either the preimmune or anti-PP4R4 serum at a 1:500 dilution for 2 h. Next, 10 μl of protein A-Sepharose was added, and the samples were incubated for an additional 3 h. Three washes in lysis buffer were performed, and the samples were eluted directly in three bead volumes of Laemmli sample buffer.
Immunoprecipitation and Purification from Mammalian Cells—Immunoprecipitation from stably transfected HEK293 cells or transiently transfected 293T cells were performed at 4 °C, as described (23). For affinity purification (AP)/mass spectrometry (MS), the immune complexes on anti-FLAG M2 beads (incubation time of 5 h) were washed in lysis buffer then in 50 mm ammonium bicarbonate containing 75 mm KCl and 2 mm EDTA and eluted by 3 bead volumes with 0.5 m freshly prepared ammonium hydroxide (pH > 11.0). For immunoprecipitation/Western experiments, the immune complexes (incubation time of 1 h) were washed in lysis buffer and eluted directly in 3 bead volumes of Laemmli sample buffer.
Baculovirus Expression and Protein Purification—Baculovirus production was performed using the Bac-to-Bac Baculovirus Expression System according to the manufacturer's (Invitrogen) instructions. pFastBac constructs were transformed into DH10Bac cells (Invitrogen) to create bacmids containing the genes of interest. Isolated bacmids were transfected into Sf9 cells (using Cellfectin, Invitrogen) for the generation of baculovirus, which were then amplified by infection of fresh Sf9 cells. Sf9 cells were infected with the indicated virus combinations. After 40–60 h, cells were harvested, washed once with phosphate-buffered saline, pelleted, and frozen on dry ice. To isolate StreptagII (Strep)-PP4c, Sf9 cells infected with baculovirus expressing Strep-PP4c were lysed by syringe lysis with 1 ml per T175 flask of 100 mm Tris, pH 8, 150 mm NaCl, 2 mm EDTA, 0.1% Nonidet P-40, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1:500 Sigma protease inhibitor mixture. Lysis was confirmed by microscopy, and lysates were centrifuged at 14,000 rpm for 15 min at 4 °C. Strep-Tactin-Sepharose (100 μl; IBA 2-1201-010) was added, and the slurry was incubated end-over-end for 2 h at 4 °C. The affinity-purified phosphatases were washed 3 times in lysis buffer and twice in 50 mm Hepes, pH 8, 75 mm NaCl, 0.4 mm EDTA. The purified phosphatases on beads were then stored at -20 °C in 50 mm Hepes, pH 8, 75 mm NaCl, 0.2 mm EDTA, 0.2% β-mercaptoethanol, 50% glycerol. Each purification was monitored by colloidal blue staining of proteins resolved on SDS-PAGE. To purify FLAG-tagged proteins, the same protein was used, except that the lysis buffer contained 50 mm Hepes-KOH, pH 8 (rather than Tris), and 60 μl of FLAG M2-agarose beads were used per T175 flask. In some cases FLAG-tagged proteins were eluted by incubating the immune complexes on beads twice for 15–30 min at 4 °C with 50 mm, pH 7.2, 100 mm NaCl, 0.4 mm EDTA, 1 mm dithiothreitol, supplemented with 100 μg/ml 3×-FLAG peptide (Sigma, F4799). Glycerol was then added to a final concentration of 50%, and the phosphatases were stored at -20 °C.
Phosphatase Assays—Baculovirus-expressed purified protein phosphatases were pre-equilibrated with phosphatase reaction buffer (50 mm Tris, pH 7.2, 0.1 mm CaCl2, 5 mm MnCl2, and 0.2 mg/ml bovine serum albumin) for 10 min at 37 °C. The diluted fluorescent 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) substrate was then added (EnzChek, Invitrogen; final concentration of 60 μm) followed by incubation at 37 °C for 5–60 min. Fluorescence was monitored by using a wavelength excitation of 358 nm and emission detection of 455 nm, according to the manufacturer's instructions. Phosphatase assays using the PP4c protein substrate γH2AX were performed under the same buffer, temperature, and incubation time conditions (11). The γH2AX substrate was delivered to the phosphatase reaction as acid-extracted histones prepared after γ-irradiation and were a kind gift from Drs. S. Nakada and D. Durocher. Detection of the phosphorylation status of H2AX was performed by immunoblotting with a phosphospecific antibody to γH2AX.
Mass Spectrometric Analysis—Samples eluted with ammonium hydroxide were lyophilized in a SpeedVac, resuspended in 50 mm ammonium bicarbonate (pH 8–8.3), and incubated at 37 °C with trypsin (Sigma trypsin singles; 0.75 μg for 16 h and an additional 0.25 μg for 2 h). The ammonium bicarbonate was evaporated, and the samples were resuspended in high performance liquid chromatography buffer A (2% acetonitrile, 0.1% formic acid) then directly loaded onto capillary columns packed in-house with Magic C18AQ, 5 μm, 100A (18). Tandem mass spectrometry data were acquired in data-dependent mode (over a 65-min to 2-h acetonitrile 2–40% gradient) on a ThermoFinnigan LTQ equipped with a Proxeon NanoSource and an Agilent 1100 capillary pump. Acquired RAW files were converted to mgf format, which were searched with the Mascot search engine (Matrix Sciences, London, UK) against the human RefSeq data base (release 24) with a precursor ion mass tolerance of 3.0 and a fragment ion mass tolerance of 0.8. Methionine oxidation was allowed as a variable modification, and trypsin specificity (with two missed cleavages allowed) was selected. The data were exported into Excel files and manually verified. Biological replicates were analyzed for FLAG-PP4c (n = 6), FLAG-PP4R4 (n = 3), and FLAG alone (n = 3). To generate the lists of high confidence-specific interactors, proteins detected in any of the FLAG-alone samples were subtracted from the final list. For the FLAG-PP4c sample, only those proteins that were detected in all biological replicates and with an averaged Mascot score (sum of Mascot scores across the 6 replicates/6) >250 and >5 averaged unique peptides (sum of unique peptides across the 6 replicates/6) are reported. For FLAG-PP4R4, only those proteins that were detected in all biological replicates and with an averaged Mascot score >250 and averaged unique peptides >2 are reported. The high confidence interactions reported here were submitted to the BioGrid.
Immunofluorescence—Cells were grown and transfected on glass coverslips; 24 h post-transfection, cells were fixed either in 4% paraformaldehyde for 10 min (followed by permeabilization with 0.2% Triton-X100 for 5 min) or in cold 90% methanol for 10 min. Fixed cells were washed extensively and blocked with 2% fetal bovine serum followed by incubation with anti-HA.11 antibody (1:1000). Nuclei were visualized by the addition of Hoechst 33342 during the wash before mounting onto a slide. HA-tagged proteins were visualized using Alexa-488 conjugated secondary antibody.
RESULTS
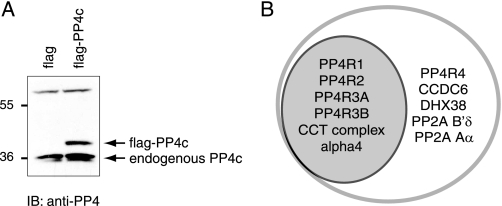
Identification of PP4c-specific Interactors—AP coupled with MS is a robust approach to detect protein-protein interactions surrounding the catalytic subunits of protein serine/threonine phosphatases (18, 22). To capture novel PP4c interactors that have previously escaped detection, a single-step affinity purification was utilized (23). FLAG-tagged PP4c was stably expressed in HEK293 cells. Expression levels of FLAG-PP4c were comparable with that of the endogenous protein (Fig. 1A). AP of the tagged protein was performed as described under “Experimental Procedures.” Purified FLAG-PP4c protein along with its binding partners was eluted from the anti-FLAG resin by incubation with a volatile base (ammonium hydroxide) in the absence of detergents. Eluted proteins were trypsinized and analyzed by liquid chromatography coupled to tandem mass spectrometry. As previously reported, identification of specific interacting partners (as opposed to background contaminants) can be extremely difficult, especially when utilizing single step affinity purification (23); a pool of cells expressing the FLAG epitope alone was, therefore, used as a negative control. Each sample (FLAG alone or FLAG-PP4c) was independently analyzed from at least three biological replicates. Importantly, after subtraction of contaminants, FLAG AP/MS recapitulated all interactions previously detected by tandem affinity purification (Table 1, complete MS data in Table S1). Strikingly, however, several additional interactors were also reproducibly detected with this more sensitive method, including KIAA1622 (an uncharacterized protein here renamed PP4R4, see below), PP2A Aα, PP2A B′δ, DHX38, and CCDC6 (Fig. 1B). PP2A Aα and PP2A B′δ are bona fide PP2Ac interactors (25). CCDC6 is a coiled-coil domain-containing protein that is a frequent fusion partner for the RET oncogene in thyroid cancer and which has also been linked to apoptosis (26); the interaction of PP4c and CCDC6 was previously reported, but the significance of this interaction has not been investigated (27). DHX38 is an ATP-dependent mRNA helicase which is orthologous to the yeast PRP16 splicing component (28). Although all of these proteins are potentially interesting, the sequence analysis of PP4R4 (see below) led us to proceed with the characterization of the PP4c·PP4R4 interaction in further detail.
FIGURE 1.
Identification of interacting partners for FLAG-PP4c. A, HEK293 cells were stably transfected with pcDNA3-FLAG-PP4c or pcDNA3-FLAG alone. After G418 selection, equal amounts of lysate were separated by SDS-PAGE, and immunoblots (IB) were performed using an anti-PP4c antibody. Arrows indicate the positions of endogenous and epitope-tagged PP4c. B, Venn diagram showing the overlap between the interactors identified from TAP-PP4c purifications (gray; as per Gingras et al. (18)) and the FLAG-PP4c purifications (see Table 1 for details).
TABLE 1.
Interactors for FLAG-PP4c FLAG-PP4c was stably expressed in HEK293 cells. AP/MS was performed as described under “Experimental Procedures” (n = 6).
| Name | Gene | Accession | Mra | MSb | UPc | TPd | %Ce | Reference |
|---|---|---|---|---|---|---|---|---|
| DHX38 | DHX38 | NP_054722 | 140.42 | 1558 | 24 | 67 | 25 | This study |
| PP4R3A | SMEK1 | NP_115949 | 93.84 | 1534 | 24 | 94 | 39 | 18 |
| CCT2 | CCT2 | NP_006422 | 57.45 | 1482 | 20 | 75 | 47 | 18 |
| CCT8 | CCT8 | NP_006576 | 59.58 | 1272 | 19 | 73 | 38 | 18 |
| PP4R2 | PPP4R2 | NP_777567 | 46.87 | 1217 | 17 | 67 | 58 | 14 |
| TCP1 | TCP1 | NP_110379 | 60.31 | 1197 | 18 | 59 | 44 | 18 |
| CCT5 | CCT5 | NP_036205 | 59.63 | 1087 | 22 | 58 | 43 | 18 |
| CCT3 | CCT3 | NP_005989 | 60.42 | 993 | 17 | 50 | 35 | 18 |
| CCT6A | CCT6A | NP_001753 | 57.99 | 998 | 16 | 51 | 39 | 18 |
| PP4R3B | SMEK2 | NP_065196 | 87.32 | 969 | 11 | 57 | 29 | 18 |
| CCT4 | CCT4 | NP_006421 | 57.89 | 918 | 13 | 51 | 39 | 18 |
| α4 | IGBP1 | NP_001542 | 39.2 | 866 | 17 | 60 | 50 | 18 |
| PP4R1 | PPP4R1 | NP_001035847 | 105.13 | 863 | 14 | 31 | 22 | 12 |
| CCT7 | CCT7 | NP_006420 | 59.33 | 907 | 14 | 54 | 33 | 18 |
| CCDC6 | CCDC6 | NP_005427 | 53.26 | 562 | 8 | 27 | 22 | 27 |
| KIAA1622 | KIAA1622 | NP_478144 | 99.39 | 334 | 6 | 10 | 10 | This study |
| PP2A Aα | PPP2R1A | NP_055040 | 65.27 | 357 | 6 | 12 | 15 | 27 |
| PP2A B′δ | PPP2R5D | NP_006236 | 69.95 | 257 | 5 | 9 | 14 | This study |
Mr, calculated molecular mass of the protein
MS, averaged Mascot score per analysis
UP, averaged number of unique peptides observed per analysis
TP, averaged total number of peptides (spectral counts) identified per analysis
%C, averaged percentage amino acid coverage per analysis
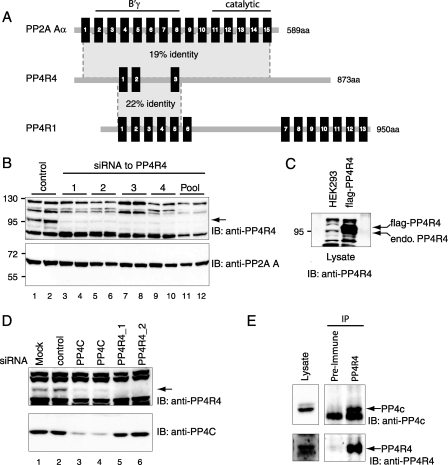
PP4R4 Resembles the PP2A Scaffolding Subunit A—PP4R4 (formerly KIAA1622) is an uncharacterized protein of 873 amino acids in length (29) and is conserved in Metazoa. The gene is located at 14q32.13 and covers 133 kilobases; alternative splice variants have been reported (30). Sequence analysis of PP4R4 revealed the presence of three canonical HEAT repeats (supplemental Fig. S1), and additional lower confidence HEAT repeats were detected by the REP search algorithm (31). HEAT repeats are structural elements thought to mediate protein-protein interactions and are named after the founding members of the family (huntingtin, elongation factor 3, PP2A A subunit, and TOR) (32). Of all HEAT repeat-containing proteins, KIAA1622 exhibits the highest level of homology to the protein phosphatase 2A scaffolding A subunit (which contains 15 such repeats), with ∼19% sequence identity and 40% sequence similarity over 492 amino acids (Fig. 2A). Consistent with this, threading modeling predicted a high degree of homology between the three-dimensional structures of PP2A A and PP4R4 (not shown). PP4R4 also exhibits significant sequence homology to the previously characterized PP4c binding partner, PP4R1, which contains 13 HEAT repeats (22% identity and 42% homology over 265 amino acids).
FIGURE 2.
PP4R4, a novel PP4c interactor, is a ∼100-kDa protein resembling the PP2A A subunit. A, domain architecture similarities between human PP4R4, PP2A Aα, and PP4R1. HEAT repeats are indicated by black boxes. The percentage identity between PP4R4 and the other HEAT repeats over the shaded portion of the sequence is indicated. The PP2A Aα HEAT repeats responsible for interaction with the catalytic subunit and the B′γ subunit are indicated. aa, amino acids. B, chemically synthesized siRNAs directed against PP4R4 were transfected into HEK293 cells. 72 h after transfection, cells were collected. Equal amounts of lysate from each sample were separated by SDS-PAGE, and the proteins were transferred to nitrocellulose. Crude antiserum from a rabbit injected with glutathione S-transferase-PP4R4 C terminus was used for immunoblotting (IB). The arrow indicates the position of the ∼100-kDa band, which disappears after transfection of PP4R4 siRNAs. C, anti-PP4R4 was used in immunoblotting of lysate from untransfected cells (left) or cells stably expressing FLAG-PP4R4 (SDS-7.5%-PAGE). D, effect of silencing PP4c on the expression levels of PP4R4. U2OS cells transfected with PP4c (or PP4R4, two independent siRNAs) siRNAs were collected after 40 h and processed for immunoblotting analysis as in B using antibodies to PP4c or PP4R4. The position of the proteins is indicated. E, anti-PP4R4 or preimmune serum was used to immunoprecipitate (IP) endogenous PP4R4 from mouse brain lysate; the immune complexes were separated via SDS-PAGE, transferred to nitrocellulose, and subjected to immunoblotting with the PP4R4 antiserum or with an antiserum reactive to PP4c (Bethyl A300–835A). The position of PP4c is indicated.
To aid in the characterization of PP4R4, a polyclonal antibody directed against the C-terminal portion of the protein (with no significant homology to any other human protein, including PP2A A) was generated. Although the antibody reacts with multiple bands on an immunoblot, it recognizes a protein of the expected size for PP4R4 (∼100 kDa) in HEK293 whole cell lysates (Fig. 2B, lanes 1 and 2). The 100-kDa protein was confirmed to be PP4R4 by the specific loss of the 100-kDa band after transfection with four independent small interfering RNA duplexes as well as in a separate siRNA mixture directed against PP4R4 (Fig. 2B, lanes 3–12). The antiserum also readily reacts with a FLAG-PP4R4 protein stably expressed in HEK293 cells (Fig. 2C). An immunoprecipitation using the PP4R4 (or preimmune serum) was performed and subjected to immunoblotting with the anti-PP4R4 antiserum; the ∼100-kDa band was readily detected in the anti-PP4R4 sample but not in the preimmune serum (not shown). The PP4R4 antibody is, therefore, able to detect this protein both in immunoprecipitation and immunoblotting experiments.
Consequences of Silencing PP4c and PP4R4—We tested whether silencing PP4c or PP4R4 affected each other's expression. As shown in Fig. 2D, protein levels of PP4R4 were drastically reduced after PP4c silencing, suggesting that interaction with PP4c may maintain stability of PP4R4; alternatively, PP4c may control expression of PP4R4. Conversely, however, silencing PP4R4 has no detectable effect on the levels of endogenous PP4c (supplemental Fig. S2A). Silencing of PP4R4 using two independent siRNAs did not lead to elevated levels of H2AX phosphorylation or accumulation of cells in S-G2/M, as opposed to the silencing of PP4c in the same experiment (supplemental Fig. S2, C and D). The cellular functions of PP4R4, thus, remain to be uncovered.
Endogenous PP4c and PP4R4 Interact in Brain Lysate—The quantity of endogenous PP4R4 protein in HEK293 cells is apparently very low, as the antibody detects severalfold more FLAG-PP4R4 than endogenous PP4R4 in these cells despite the relatively modest expression of the epitope tagged-PP4R4 compared with other FLAG-tagged proteins (not shown). Although the steady-state level of PP4R4 protein in HEK293 cells is likely to be relatively low, it was sufficient for detection in our AP/MS experiments. Two other cell lines tested (293T, U2OS) also exhibited low levels of PP4R4, and the protein was undetectable in HeLa cells (not shown). Consistent with this, in comparison to the other PP4c interactors and to the PP2A A subunits, far fewer expressed sequence tag (EST) sequences have been deposited for PP4R4 in Unigene (PP4R4, 42; PP4R1, 607; PP4R2, 350; α4, 475; PP2A Aα, 3948; PP2A Aβ, 508). Half of the PP4R4 ESTs were derived from hippocampus and other brain libraries (33). This is consistent with the fact that PP4R4 mRNA levels were reported to be highest in several areas of the brain. A reverse transcriptase-enzyme-linked immunosorbent assay detected high levels in the caudate nucleus (part of the striatum), amygdala, and hippocampus, with moderate levels in the thalamus (Kazusa consortium (34)). Similarly, in situ hybridization studies conducted in the Allen Brain Atlas project (35) indicated high levels in the striatum and thalamus as well as hippocampus.
We, therefore, assessed whether endogenous PP4R4 and PP4c could co-immunoprecipitate from mouse brain lysate. Anti-PP4R4 or a preimmune serum was used to immunoprecipitate PP4R4 from brain lysate. Immunoblotting for PP4R4 revealed specific co-precipitation of a doublet at the expected size. Importantly, endogenous PP4c (but not PP2A) was specifically co-precipitated with endogenous PP4R4 from brain lysate (Fig. 2E and data not shown). Although a complete functional characterization of the in vivo function of PP4R4 should include analysis of neuronal cells, here we describe the biochemical characterization of this protein and its interaction with PP4c.
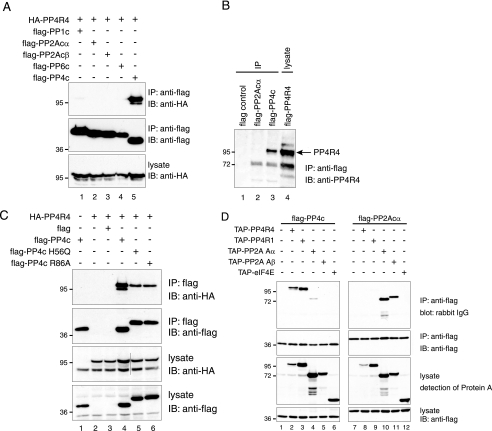
PP4R4 Specifically Interacts with PP4c and Not with Other Phosphatases—Because of the high homology between PP4R4 and PP2A A, we next determined whether PP4R4 also interacts with PP2Ac or the related phosphatase PP6c. Cells (293T) were co-transfected with HA-PP4R4 together with various FLAG-tagged phosphatase catalytic subunits (PP1c, PP2Acα, PP2Acβ, PP4c, and PP6c). Phosphatases were purified via the FLAG epitope, and co-precipitation of HA-PP4R4 was detected by immunoblotting. As shown in Fig. 3A, HA-PP4R4 co-purifies only with FLAG-PP4c and not with the other PP2A family phosphatases. This indicates that PP4R4 interacts specifically with PP4c, which is consistent with the results from endogenous PP4R4 immunoprecipitation from brain. Similar results were observed for the endogenous PP4R4 protein, when the PP4R4 antiserum was used for immunoblotting after immunoprecipitation of FLAG-PP4c and FLAG-PP2Acα from stably transfected cells (Fig. 3B). The PP4R4·PP4c interaction does not depend upon PP4c phosphatase activity; phosphatase-dead PP4c mutants (H56Q and R86A) expressed in HEK293 cells interacted with PP4R4 as efficiently as the wild type protein (Fig. 3C).
FIGURE 3.
PP4R4 specifically interacts with PP4c. A, 293T cells were co-transfected with HA-PP4R4 and FLAG-tagged catalytic subunits as indicated. Cells were harvested 48 h post-transfection, and the levels of HA-PP4R4 in the lysates were monitored by immunoblotting (IB) with the anti-HA antibody. FLAG-tagged phosphatases were next isolated on anti-FLAG M2-Sepharose beads. HA-PP4R4 was detected by immunoblotting with anti-HA, whereas the FLAG-tagged phosphatases were analyzed with anti-FLAG antibody. IP, immunoprecipitate. B, anti-FLAG pulldown was performed from HEK293 cells stably expressing FLAG alone, FLAG-PP2Acα, and FLAG-PP4c, and an immunoblot for endogenous PP4R4 was performed. FLAG-PP4R4 lysate was utilized to confirm the location of the protein. C, the catalytic activity of PP4c is not required for interaction with PP4R4. PP4c wt, H56Q, or R86A were co-expressed in 293T cells with HA-PP4R4 and recovered on FLAG-agarose. Immunoblotting with anti-HA was performed. The wild type protein used for this figure, PP4c, migrates faster than the mutants (and the wild type protein used in Fig. 6), as it was cloned in our older version of the pcDNA3-FLAG vector, which has a shorter linker. This linker does not affect the binding properties of the protein. D, 293T cells were co-transfected with TAP-tagged regulatory subunits and FLAG-tagged catalytic subunits. TAP-eIF4E was used as negative control. Cells were collected 48 h post-transfection. FLAG-PP4c and FLAG-PP2Acα were isolated on anti-FLAG beads, and the protein A moiety of the TAP-tagged proteins was detected by incubation with normal rabbit serum (the rabbit IgGs bind to protein A) followed by detection using donkey anti-rabbit IgG conjugated to horseradish peroxidase.
Because many of the PP4c and PP2Ac regulatory subunits contain HEAT repeats and because PP2A Aα was detected in association with PP4c in AP/MS (Table 1 and (27)), the binding specificity of the HEAT repeat proteins for PP2Ac and PP4c was examined. TAP-tagged versions of PP4R4, PP4R1, PP2A Aα, and PP2A Aβ were co-transfected with FLAG-PP4c or FLAG-PP2Acα. FLAG-tagged phosphatase catalytic subunits were isolated, and the association of the HEAT repeat-containing regulatory subunits was detected by blotting for the protein A component of the TAP tag. As expected, both PP4R1 and PP4R4 associated specifically with PP4c, whereas PP2A Aα and PP2A Aβ preferentially bound PP2Ac (Fig. 3D). Interestingly, however, low amounts of PP2A Aα also co-purify with FLAG-PP4c, consistent with our AP/MS data (similar amounts of FLAG-PP4c and FLAG-PP2Ac were expressed and immunoprecipitated). The biological significance of the interaction of PP4c with PP2A Aα and PP2A B′δ remains to be addressed.
PP4R4 Does Not Bridge Known PP4c Partners—The observation that the mode of interaction between PP4c·PP4R4 and PP2Ac·PP2A A is highly analogous suggests that PP4R4 may, like PP2A A, serve as a scaffolding subunit for PP4c. However, AP/MS of PP4R1, PP4R2, and PP4R3 A and B did not identify PP4R4 (12, 18, 36),5 making it unlikely that PP4R4 represents an essential bridging protein for these subunits or that these proteins tether PP4R4 to PP4c. We also did not find any peptide corresponding to these regulatory subunits in the PP4R4 AP/MS (not shown) nor could we detect co-precipitation of PP4R2, PP4R3, or α4 with PP4R4 by immunoprecipitation/Western (not shown). These results are consistent with the hypothesis that PP4R4·PP4c represents a novel, independent PP4 complex. Furthermore, no novel high confidence regulatory protein for PP4R4 was co-precipitated in significant amounts in this study, which strongly suggests that PP4R4 does not act as a scaffolding protein (at least under the conditions tested here).
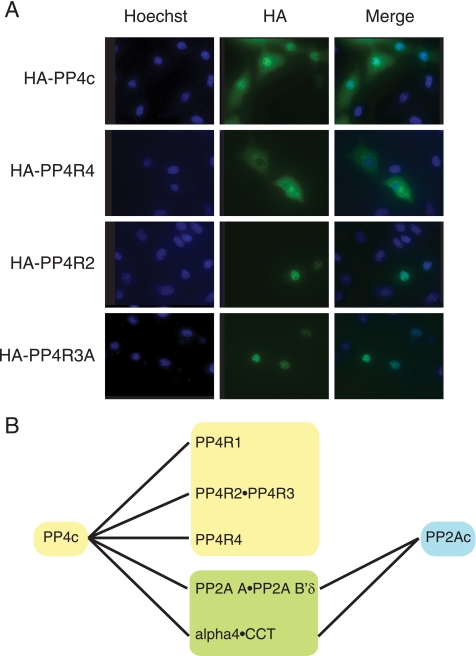
PP4R4 Is a Cytoplasmic PP4c Regulatory Subunit—To examine the intracellular localization of PP4R4, an HA-tagged version of PP4R4 was transiently transfected into Vero cells and visualized by indirect immunofluorescence. As shown in Fig. 4A, HA-PP4R4 displays exclusive cytoplasmic staining. This is in contrast to other PP4c subunits, such as PP4R2 and PP4R3, which are predominately nuclear. PP4c was observed in both the cytoplasm and nucleus, as expected (4, 6, 37). Identical localization of PP4R3A and PP4R4 was also found in HEK293 cells stably expressing each of the proteins (supplemental Fig. S3A). PP4R4 is not likely to shuttle between the cytoplasm and the nucleus, as its localization did not change after treatment with leptomycin B, a potent inhibitor of the CRM1 export pathway (supplemental Fig. S3B). Taken together, these results suggest that PP4R4 is a novel bona fide cytoplasmic regulatory PP4c subunit that interacts with PP4c in a distinct complex separate from PP4R1, PP4R2·PP4R3, and α4 (Fig. 4B).
FIGURE 4.
PP4R4 is a cytoplasmic regulatory subunit for PP4c. A, localization of PP4R4 by indirect immunofluorescence. Vero cells were transfected with HA-tagged constructs as indicated; 24 h post transfection, cells were fixed, permeabilized, and stained with anti-HA antibody, as described under “Experimental Procedures.” B, model depicting multiple PP4c-containing stable complexes. PP4R1, PP4R2-PP4R3, and PP4R4 are all specific interactors for PP4c. α4-CCT interacts with both PP2Ac and PP4c. Although PP2A A-PP2A B′δ exhibits a stronger affinity for PP2Ac, these proteins also weakly interact with PP4c.
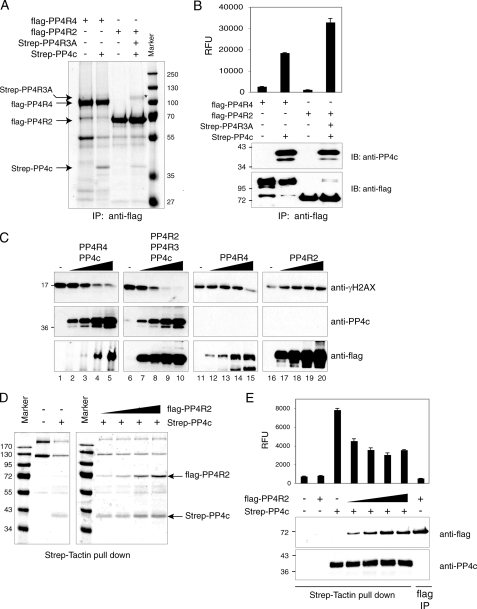
Differential Phosphatase Activity of Various PP4 Subcomplexes—Regulatory subunits of the serine/threonine protein phosphatases are likely to play critical roles in directing substrate specificity and subcellular localization (1). However, whether binding of different regulatory subunits also alters the phosphatase activity of the catalytic subunit has not been fully elucidated. To directly examine phosphatase activity in the PP4c·PP4R4 and PP4c·PP4R3A·PP4R2 complexes, phosphatase assays were performed using baculovirus-expressed proteins. Sf9 cells were co-infected with baculovirus expressing FLAG-PP4R4 (or FLAG-PP4R2 in conjunction with Strep-PP4R3A) and Strep-PP4c. Complexes were purified through the FLAG epitope on the regulatory subunits to prevent isolation of free catalytic subunits. To ensure that the regulatory subunit did not co-precipitate other phosphatases, Sf9 cells were also singly infected with FLAG-PP4R4 or FLAG-PP4R2 alone. As shown in Fig. 5A, Strep-PP4c readily co-purified with either FLAG-PP4R4 or FLAG-PP4R2, whereas no Strep-PP4c was detected in singly infected Sf9 cells. The amount of co-precipitating phosphatase was monitored by immunoblotting before phosphatase assay (not shown). The phosphatase activity of each complex was then monitored using DiFMUP, a generic fluorogenic phosphatase substrate. Surprisingly, the PP4c·PP4R4 complex consistently exhibited approximately half the activity of the PP4c·PP4R2·PP4R3 complex (Fig. 5B). This suggests that binding of specific regulatory subunits to PP4c may directly affect its phosphatase activity. To test whether this differential activity was also mirrored on a protein substrate, the relative activity of the complexes was monitored by immunoblotting using an antibody specific for the phosphorylated form of the histone variant H2AX (γH2AX), an in vivo substrate for PP4c (10, 11). Increasing amounts of the complex were incubated with acid-extracted histones from γ-irradiated cells, and the resulting reactions were separated onto SDS-PAGE gels, transferred onto membranes, and immunoblotted for γH2AX (Fig. 5C, top panel), co-precipitated PP4c (middle panel), and FLAG-tagged protein (bottom panel). As was seen with the fluorogenic substrate, the PP4c·PP4R2·PP4R3A complex was more active (∼2-fold, at equivalent PP4c amounts) than the PP4c·PP4R4 complex (no significant activity was detected in the absence of Strep-PP4c, except with the highest amount of PP4R4).
FIGURE 5.
Differential activity of PP4 subcomplexes. A, Sf9 cells were infected with the indicated combinations of baculoviruses. Protein complexes were purified via FLAG immunoprecipitation (IP), resolved on SDS-PAGE, and visualized by colloidal blue. B, phosphatase activity of purified complexes in A was monitored using a DiFMUP fluorogenic substrate. RFU indicates relative fluorescence units. Levels of Strep-PP4c, FLAG-PP4R4, and FLAG-PP4R2 were determined using anti-PP4c and anti-FLAG antibodies. Note that comparable amounts of Strep-PP4c were used for the phosphatase assay. IB, immunoblot. C, increasing amounts of the purified complexes were incubated with acid-washed histones isolated after γ-irradiation. The relative levels of phosphorylation on H2AX were monitored by immunoblotting with a γH2AX antibody, whereas the levels of Strep-PP4c, FLAG-PP4R4, or FLAG-PP4R2 in the reactions were detected by immunoblotting for PP4c or the FLAG epitope. D, Sf9 cells were infected with baculovirus expressing Strep-PP4c with increasing amount of FLAG-PP4R2. The protein phosphatases were isolated via the StreptagII moiety and analyzed by SDS-PAGE and colloidal blue. E, phosphatase activity in the samples from D was determined using DiFMUP as a substrate, as described in B. Less phosphatase was used for this experiment.
The higher activity detected in the PP4c·PP4R2·PP4R3A complexes relative to the PP4c·PP4R4 complex may be due to an inhibitory effect of PP4R4 on PP4c or to a stimulatory effect of PP4R2 (or PP4R2·PP4R3A). To discriminate between these possibilities, Sf9 cells were infected with a constant amount of Strep-PP4c and an increasing amount of FLAG-PP4R2; the complexes were recovered by streptactin affinity and analyzed by SDS-PAGE/Coomassie. As seen in Fig. 5D, similar amounts of Strep-PP4c (and contaminating proteins) were recovered in all purifications, but an increasing amount of FLAG-PP4R2 was detected (Fig. 5D). These purified complexes were used in phosphatase assays using DiFMUP, as described above (the presence of FLAG-PP4R2 and Strep-PP4c in these reactions was detected by immunoblotting). Unexpectedly, FLAG-PP4R2 decreased the phosphatase activity of PP4c toward DiFMUP (a maximal decrease of 2-fold was observed), indicating that this protein does not stimulate PP4c activity (Fig. 5E). Rather, binding of PP4R2 appears to decrease PP4c activity (the same phenomenon was observed when transfecting increasing amounts of PP4R2·PP4R3A; not shown). This phenomenon was previously reported with other phosphatase binding partners and may be related to a shift from a high activity/low specificity phosphatase to a less active (in this assay) and more specific phosphatase (38, 39).
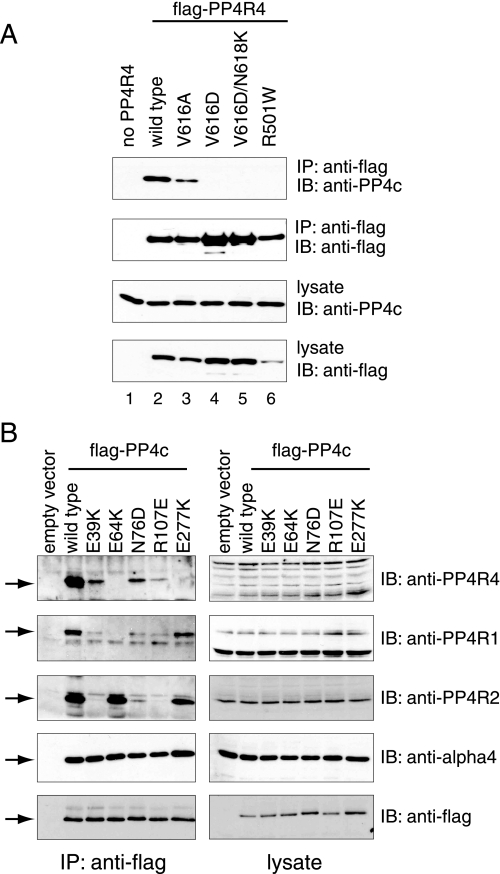
A Similar Mode of Binding to Catalytic Subunits for PP4R4 and PP2A A—Crystal structures of PP2A trimers consisting of PP2Ac, PP2A A, and one of the B′ family regulatory subunits (B56γ, gene name PPP2R5C) has been recently published by independent groups (40, 41). HEAT repeats 2–8 of PP2A A are involved in the binding to the B′ subunit, whereas HEAT repeats 11–15 contact the catalytic subunit (Fig. 2A). These structures clearly highlight the residues at the various binding interfaces. A number of residues in the A subunit of PP2A that are involved in establishing contacts with PP2Ac are conserved in PP4R4 (supplemental Fig. S4A). Specifically, Arg-418 and Val-533 (numbering for PP2A Aα), which are mutated at a high frequency in Aα or Aβ in lung and colon cancer (42–44), correspond to Arg-501 and Val-616 in PP4R4. To determine whether these residues on PP4R4 play a role in the binding to PP4c, point mutants of PP4R4 were generated. Interestingly, mutation of Arg-501 to Trp or of Val-616 to Asp in PP4R4 completely abolished the interaction with PP4c, whereas mutation of Val-616 to Ala diminished but did not abrogate PP4c binding (Fig. 6A). This is consistent with the finding by Xu et al. (40) for the PP2Ac·PP2A A interaction, which strongly suggests that the binding interfaces between these two protein pairs are similar.
FIGURE 6.
Effect of mutation of the PP4R4 or PP4c residues corresponding to the PP2Ac·PP2A A interface on complex formation. A, 293T cells were transiently transfected with FLAG-PP4R4 wild type or point mutants. The levels of PP4c and the PP4R4 mutants in the lysates were monitored by immunoblotting (IB) with PP4c and FLAG antibodies, respectively. FLAG-tagged proteins were next precipitated (IP), and co-precipitation of endogenous PP4c was monitored using an anti-PP4c antibody. B, FLAG-tagged PP4c mutants were transiently transfected in 293T cells, and lysates were prepared 48 h post-transfection. Immunoprecipitation on anti-FLAG-Sepharose beads was performed, and the immune complexes were resolved by SDS-PAGE followed by transfer onto nitrocellulose. Co-precipitation of endogenous PP4R1, PP4R2, and PP4R4 were detected using antibodies to the endogenous proteins.
PP2Ac and PP4c are highly similar (65% identity), and most of the residues in PP2Ac subunit that establish contacts with PP2A A are conserved in PP4c (supplemental Fig. S4B). PP4c mutants in some of the conserved amino acids (E64K, N76D, R107E, and E277K) were generated and tested for their ability to associate with PP4R4 (as well as to the other PP4 regulatory subunits PP4R1, PP4R2, and α4; see below). Substitution (in PP4c) of Glu-64 or Glu-277 to lysine completely abolished the interaction with PP4R4, whereas substitution of Asn-76 by Asp or Arg-107 by Glu decreased, but did not abolish, binding (Fig. 6B). Unexpectedly, mutation of Glu-39, which was predicted to be outside of the PP4c·PP4R4 binding interface, also weakened binding to PP4R4. This suggests that there are additional determinants for association of PP4c with PP4R4. The association of PP4c with α4, a general PP2A-type phosphatase binding partner, was not affected by any of these mutations (Fig. 6B),6 suggesting that these mutants are not grossly misfolded.
Taken together, our mutational analysis suggests that PP4R4 and PP2A A are not only highly similar in primary amino acid sequence but that they interact with their respective catalytic subunits (PP4c and PP2Ac) in a similar fashion. However, several residues in the binding interface of PP2Ac·PP2A A differ from PP4c·PP4R4, likely conferring specificity for a given partner. Only one of the PP2Ac amino acids establishing contact with PP2A A has a non-conservative substitution in PP4c (Lys-71 in PP4c, Asp-74 in PP2Ac). Permutation of this residue (Lys-71 in PP4c to Asp or Gln; Asp-74 in PP2Acα to Lys or Gln) in the catalytic subunits failed to alter binding specificity (not shown). Similarly, our attempts at identifying the binding specificity determinants on PP4R4 and PP2A A have been unsuccessful; substitution of PP4R4 Lys-578 by Pro and double substitution of PP4R4 Leu-540/Pro-541 to Tyr and Ala to mimic PP2A A also did not alter specificity (not shown). Taken together, this suggests that the specificity determinants may be more complex, perhaps involving less-conserved portions of the molecules. In this regard, a large insertion in PP4R4 separates contact regions important for PP2A A binding to PP2Ac, which may also play a role in the specificity of the interaction.
Different Modes of Binding for PP4R1, PP4R2, PP4R4, and α4—The results from mutational analysis in Fig. 6B also confirmed that the PP4R1 and PP4R2 subunits are not required for the association of PP4R4 with PP4c and vice versa and that the association of the three proteins with PP4c is likely independent. PP4R4 is not required for binding of the other proteins to PP4c; both PP4R1 and PP4R2 readily interacted with the PP4c E277K mutant, whereas interaction with PP4R4 was abrogated. This is intriguing, as PP4R1 and PP4R4 are both HEAT repeat-containing proteins with homology to PP2A A; this result indicates some degree of divergence in the mode of binding to PP4c for each of these proteins. PP4R2 is not required for the interaction of PP4c with PP4R4, as the E39K mutation, which completely abrogates PP4R2 interaction, decreased but did not abolish PP4R4 interaction. Similarly, PP4R2 was able to interact with PP4c in the absence of PP4R1 or PP4R4, as the E64K mutant interacted with PP4R2 as efficiently as the wild type protein in the absence of any detectable PP4R1 and PP4R4 interaction. Taken together, these results suggest that α4, PP4R1, PP4R2, and PP4R4 have evolved different modes of association with the catalytic subunit.
DISCUSSION
Here we have identified and characterized PP4R4 (KIAA1622), a new PP4c interactor. The PP4c·PP4R4 complex is independent from PP4R1, PP4R2·PP4R3 or α4·CCT-containing complexes. PP4R4 resembles the PP2A A subunit but specifically targets PP4c. It binds to PP4c in a manner reminiscent of the PP2Ac·PP2A A interaction. In vitro phosphatase assays using complexes isolated from insect cells indicated that PP4c·PP4R4 complexes were less active than PP4c·PP4R2·PP4R3A complexes against a fluorogenic substrate. Although this could indicate that PP4R4 is an inhibitor and/or PP4R2·PP4R3 an activator of the complex, association of increasing amounts of PP4R2 with PP4c also induced a gradual decrease in phosphatase activity. This observation is consistent with the low activity of the PP4c·PP4R2 complex isolated from testis (14). Whether this decreased activity against a nonspecific (DiFMUP) substrate is accompanied by an increase in specific activity against biological targets, as has been proposed (38, 39), is an intriguing question. The differential activity of PP4c in complexes containing PP4R4 and PP4R2 may be due to differing occlusion of the catalytic site by the two proteins; alternatively, it is possible that association of the regulatory subunits also induces long-range conformational changes in the catalytic subunits. Importantly, the γH2AX substrate used here is likely not a physiological target for PP4R4; the two proteins are in different cellular compartments throughout most of the cell cycle. The work presented here also shows that γH2AX levels are not elevated in PP4R4-silenced cells. This is in contrast to recent data showing that silencing of PP4R2 and PP4R3 leads to elevation of γH2AX levels (10, 11, 21). Substrates targeted by PP4c·PP4R4 remain to be identified.
The function of PP4R4 is unknown at present, and we have not found any of the cell cycle defects associated with PP4c silencing in PP4R4-silenced cells. PP4R4 appears to be expressed at relatively low levels in HEK293 cells. However, PP4R4 may be enriched in specific brain structures, since its mRNA levels are highest in striatum, amygdala, hippocampus, and thalamus (by reverse transcriptase-PCR-enzyme-linked immunosorbent assay (34) as well as by in situ hybridization (35)). As shown here, endogenous PP4R4 and PP4c co-precipitate from brain lysate. It is, therefore, plausible that PP4R4 performs cell- or tissue-specific functions and is involved in cognitive processes.
In addition to PP4R4, the AP/MS identified additional PP4c interactors. Other than previously validated interactors, we found that DHX38, CCDC6 (also seen in Ref. 27), PP2A Aα, and PP2A B′δ specifically co-precipitated with FLAG-PP4c in all four biological replicates. The interaction with DHX38 (a splicing factor) points to a possible role for PP4c in controlling splicing events, consistent with previous observations (15, 45). CCDC6 is a protein of unknown function whose N terminus is frequently fused to the RET oncogene in translocation events leading to thyroid cancer (46) and to other proteins (PDGFR-beta, PTEN) in other malignancies (47, 48). The normal physiological role of CCDC6 is poorly characterized but has been linked to the control of apoptosis and ATM (ataxia telangiectasia mutated) signaling (26, 49). Whether CCDC6 and DHX38 bind to PP4c directly or via one of the regulatory subunits (other than PP4R4) remains to be investigated.
In addition to the PP4-specific assemblies, we also showed that although PP2A Aα preferentially associates with PP2Ac, it can also bind to PP4c in lower amounts, both in transient transfection and in FLAG AP/MS. Furthermore, PP2A B′δ (PPP2R5D) was also specifically co-precipitated with PP4c; this is not likely to be an artifact, as the only baits we have observed to pull down PP2A B′δ were PP2Ac and PP4c (n > 40).7 PP2A B′δ was recently shown to be regulated by Cdk1-mediated phosphorylation and to target Cdc25C for dephosphorylation by PP2A (50, 51). Whether a similar mechanism exists to control the activity of a PP4c·PP2A Aα·PP2A B′δ toward an as yet unidentified substrate remains to be explored.
The mode of association between PP4c and PP4R4 is reminiscent of the PP2Ac·PP2A A interaction, but the determinants of the specificity remain unknown. It is interesting that α4, PP4R1, PP4R2, and PP4R4 appear to have evolved different mechanisms for their interaction with PP4c. Although this was expected for α4 and PP4R2 given their sequence differences, the structural homologies between PP4R1 and PP4R4 would have suggested a similar mode of association. Nonetheless, we have found at least one PP4c mutant (E277K) that is deficient in its interaction with PP4R4 yet associates with PP4R1 at wild type levels. Further studies will be required to fully elucidate the structural determinants for these associations.
Supplementary Material
Acknowledgments
We thank Dr. Shinishiro Nakada and Dr. Daniel Durocher for acid-extracted histones, Dr. Nahum Sonenberg for the anti-glutathione S-transferase α4 antibody, Swetha Narala for help with immunoblotting, Yang-Chieh Chou for help with sequence alignments and PyMol, Brett G. Larsen for expert mass spectrometry advice, and Drs. Karen Colwill and Tony Pawson for access to the mammalian gene collection clones. We are grateful to Drs. Brian Raught and Stefan Strack and members of the Gingras laboratory for critical reading of the manuscript.
This work was supported in part by a grant from the Terry Fox Foundation and the National Cancer Institute of Canada (to A. C. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
Footnotes
The abbreviations used are: PPP, phosphoprotein phosphatase; PP4c, catalytic subunit of phosphoprotein phosphatase 4; PP2Ac, catalytic subunit of phosphoprotein phosphatase 2A; PP2A A, A scaffolding subunit of phosphoprotein phosphatase 2A; γH2AX, phosphorylated form of the H2A histone variant X; AP, affinity purification; MS, mass spectrometry; EST, expressed sequence tag; HEAT, huntingtin, elongation factor 3, A subunit of PP2A, TOR; TAP, tandem affinity purification; DiFMUP, 6,8-difluoro-4-methylumbelliferyl phosphate; HA, hemagglutinin; siRNA, small interfering RNA.
G. I. Chen, S. Tisayakorn, and A.-C. Gingras, unpublished results.
M. Mullin, L. M. D'Ambrosio, and A.-C. Gingras, manuscript in preparation.
M. Mullin, G. I. Chen, M. J. Kean, L. M. D'Ambrosio, and A.-C. Gingras, unpublished results.
References
- 1.Janssens, V., Goris, J., and Van Hoof, C. (2005) Curr. Opin. Genet. Dev. 15 34-41 [DOI] [PubMed] [Google Scholar]
- 2.Mumby, M. (2007) Cell 130 21-24 [DOI] [PubMed] [Google Scholar]
- 3.Brewis, N. D., and Cohen, P. T. (1992) Biochim. Biophys. Acta 1171 231-233 [DOI] [PubMed] [Google Scholar]
- 4.Brewis, N. D., Street, A. J., Prescott, A. R., and Cohen, P. T. (1993) EMBO J. 12 987-996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, P. T., Philp, A., and Vazquez-Martin, C. (2005) FEBS Lett. 579 3278-3286 [DOI] [PubMed] [Google Scholar]
- 6.Toyo-oka, K., Mori, D., Yano, Y., Shiota, M., Iwao, H., Goto, H., Inagaki, M., Hiraiwa, N., Muramatsu, M., Wynshaw-Boris, A., Yoshiki, A., and Hirotsune, S. (2008) J. Cell Biol. 180 1133-1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shui, J. W., Hu, M. C., and Tan, T. H. (2007) Mol. Cell. Biol. 27 79-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou, G., Mihindukulasuriya, K. A., MacCorkle-Chosnek, R. A., Van Hooser, A., Hu, M. C., Brinkley, B. R., and Tan, T. H. (2002) J. Biol. Chem. 277 6391-6398 [DOI] [PubMed] [Google Scholar]
- 9.Zhang, X., Ozawa, Y., Lee, H., Wen, Y. D., Tan, T. H., Wadzinski, B. E., and Seto, E. (2005) Genes Dev. 19 827-839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury, D., Xu, X., Zhong, X., Ahmed, F., Zhong, J., Liao, J., Dykxhoorn, D. M., Weinstock, D. M., Pfeifer, G. P., and Lieberman, J. (2008) Mol. Cell 31 33-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakada, S., Chen, G. I., Gingras, A.-C., and Durocher, D. (2008) EMBO Rep., in press [DOI] [PMC free article] [PubMed]
- 12.Kloeker, S., and Wadzinski, B. E. (1999) J. Biol. Chem. 274 5339-5347 [DOI] [PubMed] [Google Scholar]
- 13.Wada, T., Miyata, T., Inagi, R., Nangaku, M., Wagatsuma, M., Suzuki, D., Wadzinski, B. E., Okubo, K., and Kurokawa, K. (2001) J. Am. Soc. Nephrol. 12 2601-2608 [DOI] [PubMed] [Google Scholar]
- 14.Hastie, C. J., Carnegie, G. K., Morrice, N., and Cohen, P. T. (2000) Biochem. J. 347 845-855 [PMC free article] [PubMed] [Google Scholar]
- 15.Carnegie, G. K., Sleeman, J. E., Morrice, N., Hastie, C. J., Peggie, M. W., Philp, A., Lamond, A. I., and Cohen, P. T. (2003) J. Cell Sci. 116 1905-1913 [DOI] [PubMed] [Google Scholar]
- 16.Inui, S., Sanjo, H., Maeda, K., Yamamoto, H., Miyamoto, E., and Sakaguchi, N. (1998) Blood 92 539-546 [PubMed] [Google Scholar]
- 17.Chen, J., Peterson, R. T., and Schreiber, S. L. (1998) Biochem. Biophys. Res. Commun. 247 827-832 [DOI] [PubMed] [Google Scholar]
- 18.Gingras, A. C., Caballero, M., Zarske, M., Sanchez, A., Hazbun, T. R., Fields, S., Sonenberg, N., Hafen, E., Raught, B., and Aebersold, R. (2005) Mol. Cell. Proteomics 4 1725-1740 [DOI] [PubMed] [Google Scholar]
- 19.Hastie, C. J., Vazquez-Martin, C., Philp, A., Stark, M. J., and Cohen, P. T. (2006) FEBS J. 273 3322-3334 [DOI] [PubMed] [Google Scholar]
- 20.Huang, R. Y., Eddy, M., Vujcic, M., and Kowalski, D. (2005) Cancer Res. 65 5890-5897 [DOI] [PubMed] [Google Scholar]
- 21.Keogh, M. C., Kim, J. A., Downey, M., Fillingham, J., Chowdhury, D., Harrison, J. C., Onishi, M., Datta, N., Galicia, S., Emili, A., Lieberman, J., Shen, X., Buratowski, S., Haber, J. E., Durocher, D., Greenblatt, J. F., and Krogan, N. J. (2006) Nature 439 497-501 [DOI] [PubMed] [Google Scholar]
- 22.Gingras, A. C., Gstaiger, M., Raught, B., and Aebersold, R. (2007) Nat. Rev. Mol. Cell Biol. 8 645-654 [DOI] [PubMed] [Google Scholar]
- 23.Chen, G. I., and Gingras, A. C. (2007) Methods 42 298-305 [DOI] [PubMed] [Google Scholar]
- 24.Whalen, S. G., Gingras, A. C., Amankwa, L., Mader, S., Branton, P. E., Aebersold, R., and Sonenberg, N. (1996) J. Biol. Chem. 271 11831-11837 [DOI] [PubMed] [Google Scholar]
- 25.Janssens, V., and Goris, J. (2001) Biochem. J. 353 417-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Celetti, A., Cerrato, A., Merolla, F., Vitagliano, D., Vecchio, G., and Grieco, M. (2004) Oncogene 23 109-121 [DOI] [PubMed] [Google Scholar]
- 27.Ewing, R. M., Chu, P., Elisma, F., Li, H., Taylor, P., Climie, S., McBroom-Cerajewski, L., Robinson, M. D., O'Connor, L., Li, M., Taylor, R., Dharsee, M., Ho, Y., Heilbut, A., Moore, L., Zhang, S., Ornatsky, O., Bukhman, Y. V., Ethier, M., Sheng, Y., Vasilescu, J., Abu-Farha, M., Lambert, J. P., Duewel, H. S., Stewart, II, Kuehl, B., Hogue, K., Colwill, K., Gladwish, K., Muskat, B., Kinach, R., Adams, S. L., Moran, M. F., Morin, G. B., Topaloglou, T., and Figeys, D. (2007) Mol. Syst. Biol. 3 89-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou, Z., and Reed, R. (1998) EMBO J. 17 2095-2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagase, T., Kikuno, R., Nakayama, M., Hirosawa, M., and Ohara, O. (2000) DNA Res. 7 273-281 [DOI] [PubMed] [Google Scholar]
- 30.Thierry-Mieg, D., and Thierry-Mieg, J. (2006) Genome Biology 7 Suppl. 1, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrade, M. A., Ponting, C. P., Gibson, T. J., and Bork, P. (2000) J. Mol. Biol. 298 521-537 [DOI] [PubMed] [Google Scholar]
- 32.Andrade, M. A., and Bork, P. (1995) Nat. Genet. 11 115-116 [DOI] [PubMed] [Google Scholar]
- 33.Wheeler, D. L., Church, D. M., Federhen, S., Lash, A. E., Madden, T. L., Pontius, J. U., Schuler, G. D., Schriml, L. M., Sequeira, E., Tatusova, T. A., and Wagner, L. (2003) Nucleic Acids Res. 31 28-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kikuno, R., Nagase, T., Nakayama, M., Koga, H., Okazaki, N., Nakajima, D., and Ohara, O. (2004) Nucleic Acids Res. 32 (database issue) 502-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., Boe, A. F., Boguski, M. S., Brockway, K. S., Byrnes, E. J., Chen, L., Chen, L., Chen, T. M., Chin, M. C., Chong, J., Crook, B. E., Czaplinska, A., Dang, C. N., Datta, S., Dee, N. R., Desaki, A. L., Desta, T., Diep, E., Dolbeare, T. A., Donelan, M. J., Dong, H. W., Dougherty, J. G., Duncan, B. J., Ebbert, A. J., Eichele, G., Estin, L. K., Faber, C., Facer, B. A., Fields, R., Fischer, S. R., Fliss, T. P., Frensley, C., Gates, S. N., Glattfelder, K. J., Halverson, K. R., Hart, M. R., Hohmann, J. G., Howell, M. P., Jeung, D. P., Johnson, R. A., Karr, P. T., Kawal, R., Kidney, J. M., Knapik, R. H., Kuan, C. L., Lake, J. H., Laramee, A. R., Larsen, K. D., Lau, C., Lemon, T. A., Liang, A. J., Liu, Y., Luong, L. T., Michaels, J., Morgan, J. J., Morgan, R. J., Mortrud, M. T., Mosqueda, N. F., Ng, L. L., Ng, R., Orta, G. J., Overly, C. C., Pak, T. H., Parry, S. E., Pathak, S. D., Pearson, O. C., Puchalski, R. B., Riley, Z. L., Rockett, H. R., Rowland, S. A., Royall, J. J., Ruiz, M. J., Sarno, N. R., Schaffnit, K., Shapovalova, N. V., Sivisay, T., Slaughterbeck, C. R., Smith, S. C., Smith, K. A., Smith, B. I., Sodt, A. J., Stewart, N. N., Stumpf, K. R., Sunkin, S. M., Sutram, M., Tam, A., Teemer, C. D., Thaller, C., Thompson, C. L., Varnam, L. R., Visel, A., Whitlock, R. M., Wohnoutka, P. E., Wolkey, C. K., Wong, V. Y., Wood, M., Yaylaoglu, M. B., Young, R. C., Youngstrom, B. L., Yuan, X. F., Zhang, B., Zwingman, T. A., and Jones, A. R. (2007) Nature 445 168-17617151600 [Google Scholar]
- 36.Hastie, C. J., and Cohen, P. T. (1998) FEBS Lett. 431 357-361 [DOI] [PubMed] [Google Scholar]
- 37.Helps, N. R., Brewis, N. D., Lineruth, K., Davis, T., Kaiser, K., and Cohen, P. T. (1998) J. Cell Sci. 111 1331-1340 [DOI] [PubMed] [Google Scholar]
- 38.Hombauer, H., Weismann, D., Mudrak, I., Stanzel, C., Fellner, T., Lackner, D. H., and Ogris, E. (2007) PLoS Biol. 5 e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Usui, H., Imazu, M., Maeta, K., Tsukamoto, H., Azuma, K., and Takeda, M. (1988) J. Biol. Chem. 263 3752-3761 [PubMed] [Google Scholar]
- 40.Xu, Y., Xing, Y., Chen, Y., Chao, Y., Lin, Z., Fan, E., Yu, J. W., Strack, S., Jeffrey, P. D., and Shi, Y. (2006) Cell 127 1239-1251 [DOI] [PubMed] [Google Scholar]
- 41.Cho, U. S., and Xu, W. (2007) Nature 445 53-57 [DOI] [PubMed] [Google Scholar]
- 42.Wang, S. S., Esplin, E. D., Li, J. L., Huang, L., Gazdar, A., Minna, J., and Evans, G. A. (1998) Science 282 284-287 [DOI] [PubMed] [Google Scholar]
- 43.Ruediger, R., Pham, H. T., and Walter, G. (2001) Oncogene 20 1892-1899 [DOI] [PubMed] [Google Scholar]
- 44.Takagi, Y., Futamura, M., Yamaguchi, K., Aoki, S., Takahashi, T., and Saji, S. (2000) Gut 47 268-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi, Y., Reddy, B., and Manley, J. L. (2006) Mol. Cell 23 819-829 [DOI] [PubMed] [Google Scholar]
- 46.Grieco, M., Cerrato, A., Santoro, M., Fusco, A., Melillo, R. M., and Vecchio, G. (1994) Oncogene 9 2531-2535 [PubMed] [Google Scholar]
- 47.Drechsler, M., Hildebrandt, B., Kundgen, A., Germing, U., and Royer-Pokora, B. (2007) Ann. Hematol. 86 353-354 [DOI] [PubMed] [Google Scholar]
- 48.Puxeddu, E., Zhao, G., Stringer, J. R., Medvedovic, M., Moretti, S., and Fagin, J. A. (2005) Mutat. Res. 570 17-32 [DOI] [PubMed] [Google Scholar]
- 49.Merolla, F., Pentimalli, F., Pacelli, R., Vecchio, G., Fusco, A., Grieco, M., and Celetti, A. (2007) Oncogene 26 6167-6175 [DOI] [PubMed] [Google Scholar]
- 50.Forester, C. M., Maddox, J., Louis, J. V., Goris, J., and Virshup, D. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19867-19872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Margolis, S. S., Perry, J. A., Forester, C. M., Nutt, L. K., Guo, Y., Jardim, M. J., Thomenius, M. J., Freel, C. D., Darbandi, R., Ahn, J. H., Arroyo, J. D., Wang, X. F., Shenolikar, S., Nairn, A. C., Dunphy, W. G., Hahn, W. C., Virshup, D. M., and Kornbluth, S. (2006) Cell 127 759-773 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.