Abstract
Many G protein-coupled receptors (GPCRs) possess allosteric binding sites distinct from the orthosteric site utilized by their cognate ligands, but most GPCR allosteric modulators reported to date lack signaling efficacy in their own right. McN-A-343 (4-(N-(3-chlorophenyl)carbamoyloxy)-2-butynyltrimethylammonium chloride) is a functionally selective muscarinic acetylcholine receptor (mAChR) partial agonist that can also interact allosterically at the M2 mAChR. We hypothesized that this molecule simultaneously utilizes both an allosteric and the orthosteric site on the M2 mAChR to mediate these effects. By synthesizing progressively truncated McN-A-343 derivatives, we identified two, which minimally contain 3-chlorophenylcarbamate, as pure allosteric modulators. These compounds were positive modulators of the orthosteric antagonist N-[3H]methylscopolamine, but in functional assays of M2 mAChR-mediated ERK1/2 phosphorylation and guanosine 5′-3-O-([35S]thio)triphosphate binding, they were negative modulators of agonist efficacy. This negative allosteric effect was diminished upon mutation of Y177A in the second extracellular (E2) loop of the M2 mAChR that is known to reduce prototypical allosteric modulator potency. Our results are consistent with McN-A-343 being a bitopic orthosteric/allosteric ligand with the allosteric moiety engendering partial agonism and functional selectivity. This finding suggests a novel and largely unappreciated mechanism of “directed efficacy” whereby functional selectivity may be engendered in a GPCR by utilizing an allosteric ligand to direct the signaling of an orthosteric ligand encoded within the same molecule.
GPCRs4 constitute the largest family of cell surface receptors in the human genome (1). Structurally they possess a characteristic seven transmembrane-spanning domain architecture and can be further subdivided into three broad classes, commonly designated Families A, B, and C with the Family A GPCRs representing the largest of the three groups and containing prototypical GPCRs such as rhodopsin, the adrenergic receptors, and the mAChRs (2). Despite their common architecture, GPCRs display significant conformational flexibility as evidenced by their ability to recognize a diverse array of endogenous ligands. Moreover GPCRs are highly promiscuous in their coupling to intracellular pathways, consistent with the hypothesis that the receptors can adopt multiple biologically relevant active states. The ability of a ligand to engender unique conformational states of a given GPCR and consequently activate discrete subsets of intracellular effectors is variously referred to as “stimulus trafficking,” “biased agonism,” or “functional selectivity” (3).
Additional biological complexity but also a novel opportunity for drug discovery arises from the fact that many GPCRs possess allosteric binding sites (4). Allosteric modulators are ligands that bind to these sites to alter the biological properties of the endogenous orthosteric ligand either via changing its affinity, its efficacy, or both (5, 6). Although most allosteric GPCR modulators are pharmacologically quiescent in the absence of orthosteric ligand, it has recently been noted that some allosteric ligands can act as agonists in their own right (5, 7).
The recently recognized phenomenon of allosteric agonism (as opposed to modulation) raises a number of fundamental issues with respect to our understanding of mechanisms of activation of GPCRs and for drug discovery. For example, do allosteric agonists act purely via a topographically distinct allosteric site, or do their effects actually reflect mixed interaction with both orthosteric and allosteric sites? In this regard, the mAChR agonist McN-A-343 (Fig. 1) is an interesting molecule for multiple reasons. First, it is one of the earliest known examples of an mAChR agonist that possesses markedly different degrees of efficacy depending on the mAChR subtype with which it interacts (8, 9). Second, McN-A-343 has recently been found to exhibit true functional selectivity at the M2 mAChR, displaying preferentially greater activity at Gα15 protein-mediated versus Gαi protein-mediated signaling pathways when compared with classic mAChR agonists (10). Third, McN-A-343 can interact allosterically with the orthosteric antagonist [3H]NMS at the M2 mAChR (11–13), although its interaction with other orthosteric ligands, in particular agonists, appears consistent with a competitive mechanism (13, 14).
FIGURE 1.
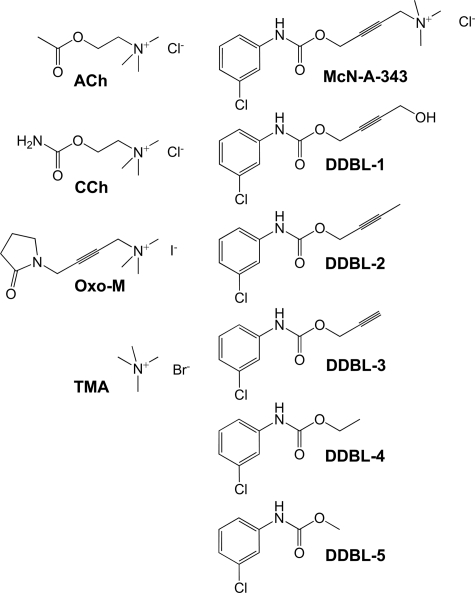
McN-A-343 shares similarities with classic orthosteric ligands of mAChRs. CCh, carbachol; Oxo-M, oxotremorine-M. Also shown are the McN-A-343 derivatives (DDBL-1 to -5) synthesized for this study.
To reconcile these observations and because McN-A-343 shares structural similarities with the well known mAChR orthosteric agonists carbachol and oxotremorine-M (Fig. 1), we hypothesized that it may actually represent a hitherto unappreciated hybrid molecule composed of both an orthosteric and an allosteric moiety, the latter presumably mediated by regions of the molecule diverging from the classic muscarinic pharmacophore, i.e. regions containing the 3-chlorophenylcarbamate moiety; McN-A-343 analogs lacking this structure but retaining the trimethylammonium moiety have been characterized as classic mAChR agonists (15). To address this hypothesis, we synthesized different derivatives of McN-A-343 that retained the 3-chlorophenylcarbamate but exhibited different degrees of truncation from the trimethylammonium end of the molecule (Fig. 1) and characterized the interaction of these ligands alone and in combination with orthosteric ligands in both binding and functional assays of the M2 mAChR. We reveal that small 3-chlorophenylcarbamate-containing moieties are novel allosteric modulators of orthosteric ligand affinity and agonist efficacy at the M2 mAChR and suggest that bitopic allosteric/orthosteric ligand-receptor interactions may be more prevalent than currently appreciated.
EXPERIMENTAL PROCEDURES
Materials—Dulbecco's modified Eagle's medium, penicillin-streptomycin, and hygromycin-B were purchased from Invitrogen. Fetal bovine serum (FBS) was purchased from ThermoTrace (Melbourne, Victoria, Australia). N-[3H]Methylscopolamine (82.0 Ci/mmol) was purchased from PerkinElmer Life Sciences. [35S]GTPγS (>1000 Ci/mmol) was purchased from both PerkinElmer Life Sciences and Amersham Biosciences. The Sure-Fire™ cellular ERK1/2 assay kits were a generous gift from TGR BioSciences (Adelaide, Australia). AlphaScreen™ reagents were from PerkinElmer Life Sciences. McN-A-343 and all other reagents were purchased from Sigma-Aldrich.
Synthesis of McN-A-343 Derivatives—All chemicals used were of reagent grade. Progress of the reactions was monitored by thin layer chromatography on silica gel plates. Extracts were dried over MgSO4, and solvents were removed under reduced pressure. 1H NMR spectra were recorded on a Bruker DPX 300-MHz spectrometer with tetramethylsilane as internal standard, and the values of the chemical shifts are given in ppm. The following derivatives of McN-A-343 (Fig. 1) were synthesized in a manner analogous to that described by Mellin et al. (16): carbamic acid, N-(3-chlorophenyl)-, 4-hydroxy-2-butyn-1-yl ester (DDBL-1), N-2-butynyl-3-chlorophenylcarbamate (DDBL-2), N-propargyl-3-chlorophenylcarbamate (DDBL-3), N-ethyl-3-chlorophenylcarbamate (DDBL-4), and N-methyl-3-chlorophenylcarbamate (DDBL-5). A solution of 3-chlorophenyl isocyanate (1.0 molar eq) and appropriate alcohol (1.2 molar eq, except for DDBL-1 where 1.85 molar eq was used) were stirred in tetrahydrofuran at 4 °C. A catalytic amount of triethylamine was added, and the reaction was stirred at 4 °C for 15 min and then at room temperature for 1–15 h. All compounds were purified by recrystallization from hexane and ethyl acetate (in ratios ranging from 80:20 to 97:3) and characterized via 1H NMR spectroscopy and mass spectrometry.
Mutagenesis—The Y177A point mutation was introduced into the wild type receptor in pEF5/FRT/V5-DEST by site-directed mutagenesis using the QuikChange Multi kit (Stratagene, La Jolla, CA) and the single primer 5′-GAGGATGGGGAGTGCGCCATTCAGTTTTTTTCC-3′. Bold italicized characters denote the nucleotides at which mutations were introduced. Oligonucleotides for site-directed mutagenesis and DNA sequencing were purchased from GeneWorks (Hindmarsh, Australia). The integrity of receptor clones was confirmed by cycle sequencing with the ABI Prism BigDye Terminator v3.1 ready reaction cycle sequencing kit with reactions analyzed using a 3730 Capillary Sequencer (Micromon DNA Sequencing Facility, Monash University, Victoria, Australia). Stable cell lines were then generated as described previously (13, 17).
Cell Culture and Membrane Preparation—Chinese hamster ovary (CHO)-FlpIn cells, either non-transfected or stably transfected with the human M2 mAChR, were grown and maintained as described previously (13, 17). Cell membranes were prepared as described previously (13) with the exception that the final pellet was resuspended in 5 ml of assay buffer (10 mm HEPES, 100 mm NaCl, 10 mm MgCl2, pH 7.4 at 30 °C). Protein content was determined using the method of Bradford (18).
[3H]NMS Dissociation Kinetic Assays—CHO-FlpIn cell membranes (20 μg) were equilibrated with [3H]NMS (0.5 nm) in a 1-ml total volume of assay buffer for 60 min at 37 °C. Atropine (10 μm) alone or in the presence of McN-A-343 derivative was then added at various time points to prevent the reassociation of [3H]NMS with the receptor. In subsequent experiments designed to investigate the effect of a range of derivative concentrations on [3H]NMS dissociation rate, a “two-point kinetic” experimental paradigm was used where the effect of increasing concentrations of derivatives on [3H]NMS dissociation was determined at 6 and 40 min. This approach is valid to determine [3H]NMS dissociation rate constants if the full time course of radioligand dissociation is monophasic both in the absence and presence of modulator (19, 20); this was the case in our current study. All other details of these assays are as described previously (13). Incubation was terminated by rapid filtration through Whatman GF/B filters using a Brandell cell harvester (Gaithersburg, MD). Filters were washed three times with 3-ml aliquots of ice-cold 0.9% NaCl buffer and dried before the addition of 4 ml of scintillation mixture (Ultima Gold, PerkinElmer Life Sciences). Vials were then left to stand until the filters became uniformly translucent before radioactivity was determined in dpm using scintillation counting.
[3H]NMS Association Kinetic Assays—A two-point kinetic experimental paradigm was used where the effect of increasing concentrations of allosteric modulator on [3H]NMS association was determined at 3 and 30 min. CHO-FlpIn cell membranes (20 μg) were added into a 1-ml total volume of assay buffer at 30 °C containing [3H]NMS (0.5 nm) alone or in the presence of a range of concentrations of McN-A-343 or DDBL-4 at 3 and 30 min. Termination of the reaction and determination of radioactivity were performed as described above.
[3H]NMS Equilibrium Binding Assays—Membrane homogenates (20 μg) were incubated in a 1-ml total volume of assay buffer containing [3H]NMS (0.5 nm) and a range of concentrations of ACh, McN-A-343, tetramethylammonium (TMA), or McN-A-343 derivative at 37 °C for 60 min. These experiments were performed in the absence of guanine nucleotides except for those involving the full agonist, ACh, that were performed in the presence of 100 μm Gpp(NH)p. Nonspecific binding was defined using 10 μm atropine. Termination of the reaction and determination of radioactivity were performed as described above. Radioligand saturation binding parameters in these cells have been reported previously (13).
[35S]GTPγS Binding Assay—Membrane homogenates (15 μg) were equilibrated in a 500-μl total volume of assay buffer containing 10 μm guanosine 5′-diphosphate and a range of concentrations of ACh (1 nm–10 μm) in the absence or presence of DDBL-4 (10 and 100 μm) at 30 °C for 60 min. After this time, 50 μl of [35S]GTPγS (1 nm) was added, and incubation continued for 30 min at 30 °C. Termination of the reaction and determination of radioactivity were performed as described above.
ERK1/2 Phosphorylation Assays—These assays were performed using the AlphaScreen-based SureFire kit as described in detail previously (13, 17). All data were expressed as a percentage of ERK1/2 phosphorylation mediated after a 6-min exposure to Dulbecco's modified Eagle's medium containing 3% FBS.
Molecular Modeling—A homology model of the M2 mAChR was constructed based on the high resolution structure of the β2-adrenergic receptor (Protein Data Bank code 2RH1) (21). An alignment between the M2 mAChR and β2 sequences based on conserved TMD residues of biogenic amine receptors was used in the creation of the model. In addition, the M2 mAChR E2 loop residues, Glu172–Cys176, were also aligned with Glu186–Cys191 of the β2 receptor, whereas the remaining loop residues were unaligned and able to move freely in the modeling protocol. The highly conserved disulfide bond between Cys96 and Cys176 at the top of TMD 3 of the M2 mAChR is maintained in the model, which was itself generated using the Molsoft ICM program (22). The protocol involved tethering the M2 mAChR model backbone to the β2-adrenergic receptor template utilizing residues where the sequences were aligned (as well as side chains where coordinates could be obtained from the template). To generate the lowest energy model, the system then underwent multiple rounds of biased probability Monte Carlo (BPMC) simulations. The resultant model was analyzed using the ICM Protein Health algorithm to ensure there were no stereochemical anomalies.
The docking of McN-A-343 to the M2 mAChR homology model was then performed in ICM using the same BPMC algorithm. Initially McN-A-343 was posed manually in an approximate starting position for docking such that Asp103 and the quaternary nitrogen of McN-A-343 could interact based on the critical role of this aspartate residue in the binding of orthosteric ligands for all biogenic amine receptors (23). This position was used to define a 16-Å simulation sphere around the ligand. The ligand and side chains within 11 Å of the ligand were subsequently allowed to move freely in the BPMC simulation. A distance restraint of 4.5 Å between an oxygen of Asp103 and the quaternary nitrogen of McN-A-343 was imposed to ensure that the salt bridge was maintained. Multiple rounds of extensive BPMC simulations were performed (128 million energy evaluations), and the best energy pose was taken as the preferred docking position/binding mode of McN-A-343.
Data Analysis—All computerized nonlinear regression was performed using the program Prism 5.01 (GraphPad Software, San Diego, CA). Radioligand inhibition binding data were empirically fitted to either a one-site or two-site/state inhibition mass action curve to determine inhibitor potency (IC50) estimates, which were then converted to KI values (24) as appropriate. Radioligand potentiation binding curves were fitted to a simple allosteric ternary complex model to derive estimates of allosteric modulator affinity (KB) and cooperativity (α), the latter parameter being a measure of the strength and direction of the allosteric interaction between the orthosteric and allosteric sites (4, 5). Dissociation and association kinetic data were fitted to monoexponential functions to derive observed dissociation and association rate constants. Concentration-response curves were fitted to a three-parameter logistic equation to derive ligand potency (EC50) estimates. Finally for functional ligand combination studies, the interaction between agonists ACh or TMA and the partial allosteric agonist McN-A-343 were fitted to an operational model for the competitive interaction between a full agonist and orthosteric partial agonist (13, 25) to derive estimates of partial agonist affinity (KB) and relative efficacy (τ). All affinities, potencies, efficacies, and cooperativity parameters were estimated as logarithms (26). Results are expressed as means ± S.E. unless otherwise stated. Statistical analyses were performed by unpaired t test, one-way analysis of variance, or by F-test, as appropriate, using Prism 5.01, and statistical significance was taken as p < 0.05.
RESULTS
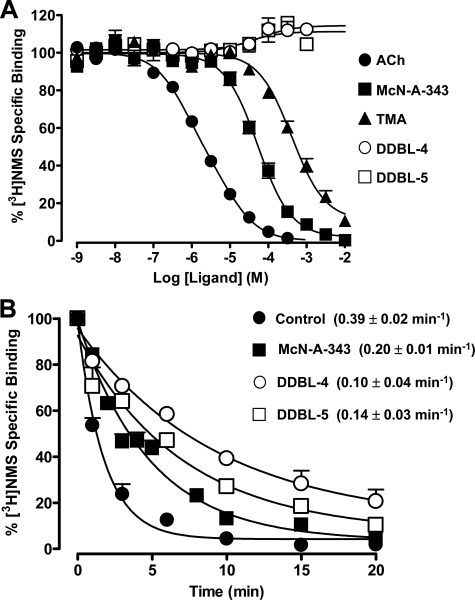
Truncation of McN-A-343 Unmasks Positive Allosteric Modulators of Antagonist Binding—All compounds were initially studied in equilibrium binding assays against the orthosteric antagonist [3H]NMS at the M2 mAChR. In addition to the endogenous agonist ACh, we also tested TMA as a surrogate for the effects of the trimethylammonium moiety common to both ACh and McN-A-343. As shown in Fig. 2A, ACh, McN-A-343, and TMA each caused complete inhibition of [3H]NMS (0.5 nm) specific binding at the M2 mAChR. In contrast, the truncated McN-A-343 derivatives DDBL-1, DDBL-2, and DDBL-3 weakly inhibited the binding of [3H]NMS with lower potencies than the parent compound such that complete inhibition binding curves for these derivatives could not be established (not shown). However, the two shortest derivatives, DDBL-4 and DDBL-5, caused a modest but consistent enhancement in the equilibrium binding of [3H]NMS indicative of a weak allosteric potentiator effect (Fig. 2A).
FIGURE 2.
McN-A-33 and some of its derivatives interact allosterically with the antagonist [3H]NMS at the human M2 mAChR. A, effect of ACh, McN-A-343, TMA, and McN-A-343 derivatives DDBL-4 and DDBL-5 on [3H]NMS equilibrium binding (0.5 nm; 37°C; 1 h) at the M2 mAChR stably expressed in membranes from CHO-FlpIn cells. Data points represent the mean ± S.E. obtained from three experiments conducted in duplicate. B, dissociation of [3H]NMS (0.5 nm; 37 °C) in the absence or presence of 300 μm McN-A-343, DDBL-4, or DDBL-5. Data represent the mean ± S.E. obtained from three experiments conducted in triplicate.
Although the experiments for ACh were performed in the presence of 100 μm Gpp(NH)p to minimize receptor-G protein association, nonlinear regression of the ACh inhibition curve yielded a Hill slope that was significantly less than 1 (0.71 ± 0.03) and preferentially fitted to an empirical two-site binding model (pKHI = 6.61 ± 0.13; pKLO = 5.45 ± 0.17; n = 3). This finding is a general property often associated with inhibition binding curves involving high efficacy agonists (27, 28). In contrast, McN-A-343 was capable of completely inhibiting specific [3H]NMS binding in a manner consistent with interaction with a single class of sites (pKI = 4.67 ± 0.04; n = 3) as observed for many low efficacy agonists (27, 28). Collectively our results suggest that McN-A-343 interacts with [3H]NMS either allosterically with high negative cooperativity and/or recognizes the orthosteric site on the M2 mAChR to interact competitively. The inhibition of [3H]NMS binding by TMA was also consistent with a one-site competitive interaction (pKI = 3.78 ± 0.06; n = 3).
Because DDBL-4 and DDBL-5 both potentiated the binding of [3H]NMS, we applied a simple allosteric ternary complex model to the data, which yielded pKB = 4.19 ± 0.02 for DDBL-4, pKB = 4.36 ± 0.02 for DDBL-5, and log α = 0.19 ± 0.01 (i.e. α = 1.5-fold allosteric enhancement) for both compounds. These findings thus indicate that DDBL-4 and DDBL-5, which both contain a 3-chlorophenylcarbamate moiety, bind simultaneously with the orthosteric radioligand [3H]NMS to the M2 mAChR and potentiate its binding via an allosteric interaction.
Validation of an Allosteric Interaction between DDBL-4 or DDBL-5 and [3H]NMS in Radioligand Kinetic Binding Assays at the M2 mAChR—To more directly probe the ability of McN-A-343 and its derivatives to allosterically modulate orthosteric ligand binding, the effect of these compounds on the rate of orthosteric radioligand dissociation was investigated. In the first instance, a single (high) concentration of each ligand was tested for effects on the control [3H]NMS dissociation rate at the M2 mAChR in a full time course assay (Fig. 2B). The presence of McN-A-343 (300 μm) significantly retarded the dissociation rate of [3H]NMS from the M2 mAChR (Fig. 2B). In contrast, 1 mm TMA had no effect (0.39 ± 0.02 min-1; n = 3) as expected for an orthosteric ligand. Similar experiments were performed with each of the McN-A-343 derivatives at a 300 μm concentration. DDBL-1, DDBL-2, and DDBL-3 did not affect the rate of orthosteric radioligand dissociation, whereas DDBL-4 and DDBL-5 substantially and significantly (p < 0.05) reduced the [3H]NMS dissociation rate (Fig. 2B). These findings unequivocally show that McN-A-343, DDBL-4, and DDBL-5, but not the other McN-A-343 derivatives, are each able to form a ternary complex with the radioligand and the receptor prior to [3H]NMS dissociating from the complex, yielding an altered radioligand dissociation rate constant as a consequence of the resultant conformational change.
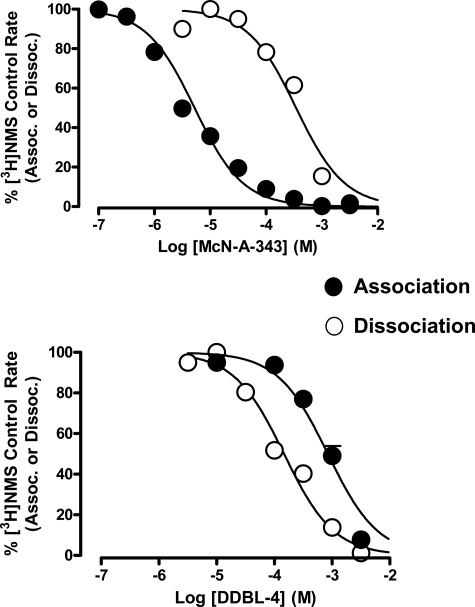
Because DDBL-4 was slightly more effective than DDBL-5 in the initial dissociation kinetic assays, additional experiments were performed with this compound and McN-A-343. By constructing complete concentration-response curves for the inhibition of [3H]NMS dissociation by each of these two modulators, it was found that both caused virtually complete inhibition of [3H]NMS dissociation at the M2 mAChR with DDBL-4 having a slightly higher potency for this effect (pEC50 = 3.84 ± 0.08 (n = 3) compared with 3.46 ± 0.06 (n = 3) for McN-A-343; Fig. 3). We next investigated the effects of the two modulators on the association kinetics of the radioligand. As shown in Fig. 3, both modulators completely prevented the association of radioligand with its binding site on the receptor, but the potency for this effect varied markedly between the two compounds (pEC50 (McN-A-343) = 5.28 ± 0.04; pEC50 (DDBL-4) = 3.07 ± 0.05; n = 3; Fig. 3). Because McN-A-343 was much more potent at preventing [3H]NMS association than dissociation, whereas DDBL-4 was slightly more potent at preventing [3H]NMS dissociation than association, these findings provide a mechanistic explanation for the effects of the ligands on [3H]NMS equilibrium binding affinity because the net effect of McN-A-343 would be a strong inhibition of radioligand binding, whereas the net effect of DDBL-4 would be a modest potentiation.
FIGURE 3.
McN-A-343 and DDBL-4 display reversals in potency for their effects on orthosteric radioligand association and dissociation kinetics. Concentration-effect relationships for McN-A-343 (upper) and DDBL-4 (lower) on the apparent association (Assoc.) rate or dissociation (Dissoc.) rate of [3H]NMS (0.5 nm; 37 °C) at the M2 mAChR stably expressed in membranes from CHO-FlpIn cells are shown. Data are normalized to the percentage of the respective control rate constant (either association or dissociation) determined in the absence of modulator and represent the mean ± S.E. obtained from three experiments conducted in triplicate.
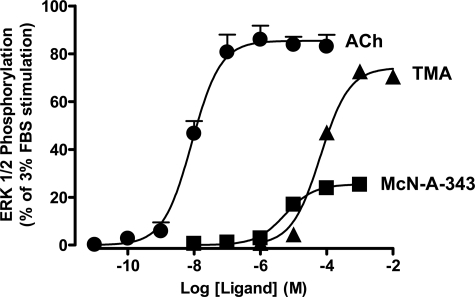
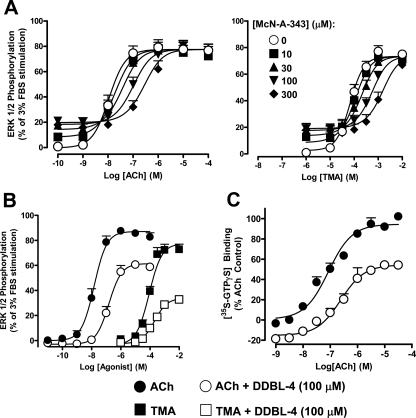
McN-A-343 Derivatives Are Negative Allosteric Modulators of M2 mAChR Agonist Efficacy—ACh and TMA mediated a robust stimulation of ERK1/2 phosphorylation that peaked at 5 min in CHO-FlpIn cells stably expressing the human M2 mAChR, whereas McN-A-343 mediated a much weaker response. No effects were observed in non-transfected cells (not shown). In contrast, none of the McN-A-343 derivatives stimulated ERK1/2 phosphorylation at any time points measured, confirming that they all lack intrinsic efficacy. The time to peak ERK1/2 phosphorylation was then chosen as the stimulation period for subsequent concentration-response experiments (Fig. 4). Although TMA was the least potent of the three agonists tested, it displayed a maximum agonist effect approaching that of ACh (Table 1), whereas McN-A-343 was a much weaker partial agonist (Emax = 13.82 ± 0.65% of the FBS response; n = 4). This latter finding suggested that some aspect of the McN-A-343 structure, away from the trimethylammonium headgroup, has a negative effect on the expression of the efficacy of the agonist but a positive effect on the affinity of the agonist given its higher potency (pEC50 = 5.23 ± 0.06; n = 4) than TMA.
FIGURE 4.
McN-A-343 possesses low efficacy at the wild type human M2 mAChR. Concentration-response curves to ACh, TMA, and McN-A-343 for the stimulation of ERK1/2 phosphorylation are shown. Data points were determined by taking the time to peak response (5 min) for each agonist concentration at 37 °C in CHO-FlpIn cells stably expressing human M2 mAChR. Data represent the mean ± S.E. obtained from four experiments conducted in duplicate and are normalized to the maximum phospho-ERK1/2 response mediated by 3% FBS.
TABLE 1.
Estimates of potency (pEC50) and maximal effect (Emax) of agonists mediating ERK1/2 phosphorylation at the indicated M2 mAChR in the absence or presence of DDBL-4 Values represent the mean ± S.E. obtained from three to six experiments conducted in duplicate.
|
M2 mAChR (wild type)
|
M2 mAChR (Y177A)
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
Control
|
100 μm DDBL-4
|
Control
|
100 μm DDBL-4
|
|||||
| pEC50 | Emaxa | pEC50 | Emax | pEC50 | Emaxa | pEC50 | Emax | |
| Ach | 7.83 ± 0.09 | 87.09 ± 2.31 | 6.82 ± 0.09b | 61.46 ± 2.21b | 8.18 ± 0.09 | 86.50 ± 1.65 | 80.71 ± 2.87 | |
| TMA | 4.01 ± 0.09 | 75.83 ± 3.03 | 3.71 ± 0.07b | 44.22 ± 1.43b | 4.51 ± 0.13 | 82.91 ± 3.46 | 4.11 ± 0.24 | 61.26 ± 5.39b |
Parameter normalized to the maximum response elicited by 3% FBS
Significantly different (p < 0.05) from corresponding control value (Student's t test)
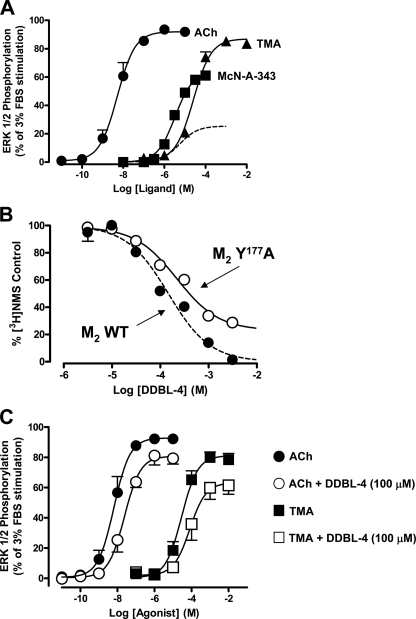
To further probe this phenomenon, interaction studies were performed between various agonists and McN-A-343 derivatives. McN-A-343 caused a parallel rightward shift in ACh-mediated ERK1/2 phosphorylation with no depression in the maximal response at the M2 mAChR (Fig. 5A). An identical effect was also observed on TMA-mediated ERK1/2 phosphorylation (Fig. 5A). As with the equilibrium binding assays, which were performed using a radiolabeled antagonist, these findings suggest that the interaction between McN-A-343 and orthosteric agonists is either highly negatively cooperative or involves direct competition for the orthosteric site. Application of an operational model of interaction between a partial agonist against a full agonist yielded the following operational affinity (pKB) and efficacy (log τ) estimates, respectively, for McN-A-343 against ACh: pKB = 4.88 ± 0.07 and log τ = 0.46 ± 0.08; against TMA, the estimates for McN-A-343 were pKB = 4.87 ± 0.05 and log τ = 0.47 ± 0.06. The pKB values were not significantly different from one another nor from the pKB estimate for McN-A-343 determined in the equilibrium binding assays (p > 0.05, one-way analysis of variance).
FIGURE 5.
The functional properties of McN-A-343 reflect a bitopic mode of orthosteric/allosteric interaction with the human M2 mAChR. A, interaction between ACh (left) or TMA (right) with McN-A-343 on M2 mAChR-mediated phosphorylation of ERK1/2 over 5 min at 37 °C in CHO-FlpIn cells stably expressing the M2 mAChR. Data represent the mean ± S.E. obtained from five experiments conducted in duplicate and are normalized to the maximum phospho-ERK1/2 response mediated by 3% FBS. Curves superimposed on the data points represent the best fit of an operational model of competitive (orthosteric) antagonism. B, ACh- or TMA-mediated ERK1/2 phosphorylation in the absence or presence of DDBL-4 for 5 min at 37 °C in CHO-FlpIn cells stably expressing the M2 mAChR. Data represent the mean ± S.E. obtained from six experiments conducted in duplicate. C, ACh-mediated [35S]GTPγS binding to activated G proteins in the absence or presence of DDBL-4 in CHO-FlpIn cell membranes stably expressing the wild type M2 mAChR. Data represent the mean ± S.E. obtained from three experiments conducted in duplicate.
When similar experiments were performed with the McN-A-343 derivatives, no effects were noted for DDBL-1, DDBL-2, and DDBL-3 (not shown). However, as shown in Fig. 5B, DDBL-4, at a concentration of 100 μm, caused a significant reduction (p < 0.05) in ACh and TMA maximal responses and potencies (Table 1); this was not due to any effects of the dimethyl sulfoxide vehicle as determined by experiments performed with the latter in parallel. To ensure that the functional effect observed with DDBL-4 was not an artifact related to the non-equilibrium nature of the ERK1/2 assay, a second assay measuring ligand-mediated [35S]GTPγS binding to activated G proteins was performed with DDBL-4 against ACh with both agents pre-equilibrated prior to addition of the radiolabel (Fig. 5C). In agreement with the effects on ACh-mediated ERK1/2 phosphorylation, DDBL-4 was again able to reduce the maximum effect (94.6 ± 2.7 versus 54.4 ± 3.2%; n = 3) and the potency (7.07 ± 0.09 versus 6.61 ± 0.11) of the agonist for stimulating [35S]GTPγS binding; these latter experiments also revealed a modest effect of the modulator on basal [35S]GTPγS binding, indicating a possible inverse agonist effect at the level of G protein activation. Collectively the results of the functional experiments reveal that DDBL-4 is a negative allosteric modulator of orthosteric agonist efficacy at the M2 mAChR despite being a weak positive allosteric modulator of antagonist binding. Similar results were obtained when these experiments were repeated using DDBL-5 (see supplemental Fig. 1).
Mutation within an Allosteric Site on the M2 mAChR Reduces the Effects of the McN-A-343 Derivatives on Receptor Function—We have recently shown that the double mutation of Y177A in the E2 loop and T423A near the top of TMD 7 of the M2 mAChR, residues previously implicated in comprising part of an allosteric binding pocket on the M2 mAChR (17, 29), reduces the potency of prototypical mAChR allosteric modulators but, paradoxically, increases the efficacy of McN-A-343 as an agonist (13). We have now found that the single point mutation Y177A is sufficient to recapitulate these findings while having no significant effect on the efficacy or potency of classic orthosteric agonists. Specifically mutation of Y177A significantly reduces the affinity of the prototypical allosteric mAChR modulator gallamine for the M2 mAChR (pKB = 4.90 ± 0.22 compared with 6.17 ± 0.11 for the wild type; n = 3; p < 0.05; see also supplemental Fig. 2) but increases the efficacy of McN-A-343 (Fig. 6A). Moreover [3H]NMS dissociation kinetic studies provided direct evidence for conformational changes in the M2 mAChR that, as a result of this mutation, affect the allosteric binding pocket. As shown in Fig. 6B, DDBL-4 had a reduced ability to maximally retard radioligand dissociation, clearly indicating a change in the binding pocket of the modulator on the dually liganded receptor. However, because the potency of DDBL-4 in the experiments on the mutated receptor (pEC50 = 3.66 ± 0.09; n = 3) was not significantly different (p > 0.05) from its potency at slowing orthosteric dissociation at the wild type receptor (pEC50 = 3.84 ± 0.08; n = 3), it was unlikely that the mutation directly affected the affinity of the modulator, but rather it caused a global conformational perturbation that altered the transmission of cooperativity between the allosteric and orthosteric sites.
FIGURE 6.
Mutation of Y177A in the allosteric pocket affects the efficacy of McN-A-343 and the potency of DDBL-4 and changes the allosteric binding mode of DDBL-4 at the human M2 mAChR. A, concentration-response curves to ACh, TMA, and McN-A-343 for the stimulation of ERK1/2 phosphorylation. Data points were determined by taking the time to peak response (5 min) for each agonist concentration at 37 °C in CHO-FlpIn cells stably expressing human M2 Y177A mutant mAChR. Data represent the mean ± S.E. obtained from four experiments conducted in duplicate. Also shown in the dashed curve is the response of McN-A-343 at the wild type (WT) receptor (taken from Fig. 4). B, concentration-effect relationships for DDBL-4 on the dissociation rate of [3H]NMS (0.5 nm; 37 °C) at the wild type or Y177A M2 mAChR stably expressed in membranes from CHO-FlpIn cells. Data represent the mean ± S.E. obtained from three experiments conducted in triplicate; data points for the wild type receptor are taken from Fig. 3 (lower). C, ACh- or TMA-mediated ERK1/2 phosphorylation in the absence (solid symbols) or presence (open symbols) of 100 μm DDBL-4 for 5 min at 37 °C in CHO-FlpIn cells stably expressing the Y177A mutant M2 mAChR. Data represent the mean ± S.E. obtained from six experiments conducted in duplicate.
Importantly when the functional interaction between DDBL-4 and either ACh or TMA was investigated at the mutant receptor, a significant blunting was noted in the ability of the modulator to inhibit the efficacy of each of the agonists in comparison with its effects at the wild type M2 mAChR (Fig. 6C and Table 1); similar results were obtained with DDBL-5 (see supplemental Fig. 1). This finding thus suggests a mechanistic explanation for the partial agonism of McN-A-343 at the M2 mAChR, namely the utilization by the 3-chlorophenylcarbamate moiety of an allosteric site to “direct” the efficacy of the orthosteric part of the molecule; mutation of the allosteric site blunts this effect and “unmasks” the full efficacy of the orthosteric moiety, thus explaining the apparent paradox noted in our earlier study (13).
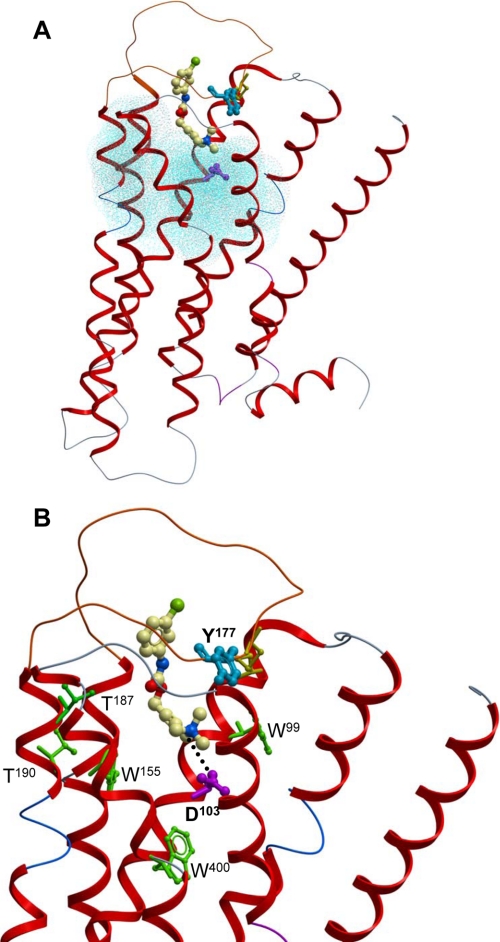
Molecular Modeling Reveals a Pose for McN-A-343 That Is Consistent with a Bitopic Orthosteric/Allosteric Binding Mode—We utilized the recently solved high resolution crystal structure of the β2-adrenergic receptor (21) to generate a homology model of the M2 mAChR. These two GPCRs share closer sequence similarity with each other relative to rhodopsin, which is currently the only other structural template of sufficient resolution for GPCR homology modeling (30). Computerized BPMC-based docking of McN-A-343 using ICM yielded the docking pose depicted in Fig. 7. From this model, a number of salient features were noted. First, as highlighted in Fig. 7A, the bulk of the McN-A-343 molecule sits higher up in the receptor, at the interface of the TMDs and the extracellular loops, relative to the orthosteric binding pocket (blue) that comprises TMD residues known to be critical to the binding of ACh (23); Asp103 (purple) in this pocket, however, interacts with the ammonium headgroup of McN-A-343 (Fig. 7B). Second, the 3-chlorophenylcarbamate moiety of McN-A-343 projects up toward the E2 loop region as expected if this part of the molecule were to engage the allosteric pocket of the M2 mAChR (Fig. 7). It should be noted, however, that a direct interaction between Tyr177 and the 3-chlorophenyl group of McN-A-343 is not predicted from our model. As indicated above, the mutation of Y177A is probably causing a global disruption of the overall conformation of the allosteric pocket, thus blunting the transmission of an allosteric effect between the orthosteric and allosteric sites. Third, although few studies have directly probed the impact of mutations in the orthosteric pocket on the binding of McN-A-343, at least two groups have reported mutations that are known to affect prototypic orthosteric ligands but not the binding of McN-A-343 when examined in parallel (31, 32). The residues implicated in these studies are highlighted in green in Fig. 7B where it can be seen that our model provides an explanation for the results; a bitopic mode of binding by McN-A-343 would result in the ligand being too far away to interact appreciably with the mutated orthosteric site residues.
FIGURE 7.
Molecular modeling and docking at the M2 mAChR suggests a possible bitopic mode of binding for McN-A-343. A, complete view of the minimized M2 mAChR homology model indicating the location of McN-A-343 relative to the TMD-bound orthosteric pocket (blue dots, based on residues known to be critical for ACh binding; see Ref. 23: Asp103 (purple), the highly conserved disulfide bond between the E2 loop and the top of TMD3 (yellow) and Tyr177 in the E2 loop (blue), which was mutated in our study). B, close-up of docked McN-A-343 illustrating the salt bridge between the ammonium headgroup of the molecule and Asp103 (dotted line) as well as amino acids (green) that have been experimentally shown not to affect the binding of McN-A-343 despite affecting other orthosteric ligands. Other details are as for A.
DISCUSSION
The main finding of this study is that McN-A-343, one of the earliest known examples of a subtype-selective GPCR partial agonist (8), is actually a bitopic molecule composed of an orthosteric agonist coupled to an allosteric modulator. Moreover the allosteric moiety within McN-A-343, exemplified by the 3-chlorophenylcarbamate present in both DDBL-4 and DDBL-5, is a positive allosteric modulator of orthosteric antagonist binding but a negative allosteric modulator of orthosteric agonist efficacy. Because allosteric sites are expected to be more divergent between receptor subtypes than orthosteric sites, our findings can thus explain why McN-A-343 has a very low efficacy and displays functional selectivity at the M2 mAChR; both phenomena may be a consequence of the interaction between an M2 mAChR allosteric site and the orthosteric site when simultaneously occupied by one molecule of McN-A-343. This interaction is presumably lacking, or appreciably different, at the other mAChRs because of the different nature of their allosteric sites. Our results also suggest a novel and largely unappreciated mechanism of “directed efficacy” by which GPCR functional selectivity may be engendered, namely through utilizing an allosteric ligand to direct the signaling of an orthosteric ligand within the same molecule.
The classic mAChR pharmacophore for orthosteric agonists such as ACh, carbachol, and muscarine consists of two “active” centers, namely a positively charged amino grouping and an electron donor center. McN-A-343 shares these pharmacophoric elements, and by and large, its pharmacological properties as a partial agonist are consistent with a simple orthosteric mode of action. For example, this compound completely inhibited the specific binding of [3H]NMS at equilibrium (Fig. 2B) and interacted with both ACh and TMA in a manner consistent with competition (Fig. 5A). However, the fact that an allosteric interaction can readily be demonstrated for this ligand at the M2 mAChR when the orthosteric site is already occupied by [3H]NMS (Figs. 2B and 3) suggests that McN-A-343 can also adopt a binding mode on the receptor that involves attachment of at least some part of the ligand to an allosteric site. Our identification of the 3-chlorophenylcarbamate-related structures DDBL-4 and DDBL-5 as pure allosteric modulators at the M2 mAChR now provides conclusive evidence that McN-A-343 contains an allosteric moiety.
Together with the results of our molecular modeling studies, our experimental observations suggest that McN-A-343 can bind both to a region within the orthosteric site as well extending into regions of the receptor that contribute to an allosteric site in essence displaying a bitopic mode of action. As shown in Fig. 7, this mode of binding can also explain prior data in the literature that found minimal effects of mutation of some key orthosteric site residues on the binding of McN-A-343 with the notable exception of D103A; the aspartic acid is required for all biogenic amines that utilize the orthosteric site (23). If a bitopic mode of action were indeed operative, it is reasonable to hypothesize that agonism can arise from engagement of the orthosteric site, but if that site is already occupied by another orthosteric ligand, then McN-A-343 adopts a different orientation utilizing the allosteric site to modulate the ligand at the orthosteric site. It also follows from this scheme that a single molecule of McN-A-343 should directly compete with orthosteric ligands while simultaneously binding to the allosteric site as any partial overlap for a common (orthosteric) binding domain can still lead to mutually exclusive binding.
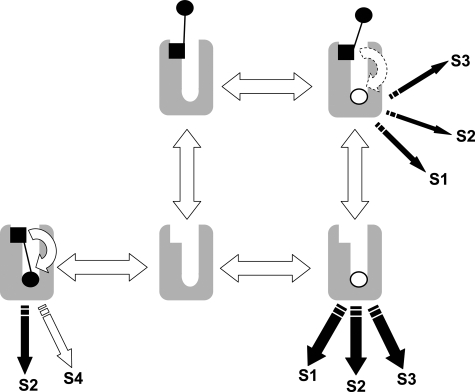
Thus, the high negative cooperativity that is often ascribed to the interaction between McN-A-343 and other orthosteric ligands, especially agonists (13, 14), may in fact be a composite effect reflecting direct orthosteric competition as well as allosteric modulation. Fig. 8 summarizes this mode of interaction schematically where it can be seen that the bitopic ligand is distributed across a variety of receptor states, allowing it to modulate the binding and/or function of an orthosteric molecule while simultaneously directing the efficacy of its own orthosteric moiety. We also show in the supplemental information that it is analytically possible to obtain a reasonable estimate of the binding affinity of a bitopic ligand to the receptor when this mechanism is operative.
FIGURE 8.
A scheme for bitopic orthosteric/allosteric ligand-receptor interaction. The open circle denotes the orthosteric ligand, whereas the solid circle and square denote the orthosteric and allosteric moieties, respectively, of the bitopic ligand. The scheme illustrates the ability for the bitopic ligand to interact allosterically with the orthosteric ligand to modulate its signaling as well as the ability of the bitopic ligand to occupy both sites, thus excluding orthosteric ligand occupancy via simple competition. Also shown is the potential for differential signaling (signal pathways denoted by the letter S) that can arise between an orthosteric and a bitopic ligand because of the ability of the allosteric moiety in the latter to direct the efficacy of the orthosteric moiety encoded in the same molecule.
Perhaps most importantly, our findings shed new light on a possible mechanistic basis for GPCR partial agonism and functional selectivity. McN-A-343 displays highest efficacy at the M1 and M4 mAChRs with lower efficacy at the other mAChR subtypes (9). Given that this agonist binds with similar affinities across mAChR subtypes (33), its display of different efficacies in a variety of functional assays means that it has different degrees of engagement of the intramolecular activation machinery at each mAChR subtype. More recently, Griffin et al. (10) also provided evidence for McN-A-343 being better able to traffic receptor signals to Gα15 rather than Gαi proteins in contrast to other orthosteric mAChR agonists at the M2 mAChR. We propose that the basis for these phenomena may be the engagement by one part of the molecule of an allosteric site on the M2 mAChR to direct the signaling of the other part of the molecule; differences in McN-A-343 efficacy across subtypes can thus reflect differences in the nature and/or location of allosteric sites on other members of the mAChR subfamily.
Our results also have significant implications for drug discovery at GPCRs. First, given the high likelihood of sequence variability between allosteric sites across different GPCR subtypes (4), the directed design of bitopic ligands can increase the probability of discovering both subtype- and pathway-selective agonists via exploiting the unique nature and properties of GPCR allosteric sites to direct the efficacy mediated by the orthosteric site; the phenomenon of functional selectivity posits that it is possible to engender ligand-specific active states that fine tune biological responses by selectively recruiting only a subset of possible signaling pathways available to a given receptor (3). Second, there is a small (but growing) class of novel compounds identified recently from drug discovery programs, such as AC-42, N-desmethylclozapine, and 77-LH-28-1, that have been shown to be functionally selective and to interact allosterically with various mAChRs (7, 13, 34). Although we and others have classed these novel compounds as “allosteric agonists” (7), some of their interactive properties with other orthosteric ligands are consistent with competition. Based on our findings with McN-A-343, we speculate that such ligands may also be bitopic rather than purely allosteric, although it is uncertain whether they adopt a pose similar to that of McN-A-343 or whether they interact with another allosteric site. In addition, we suggest that a key criterion for identifying pure allosteric agonists should be the clear demonstration of interactions with orthosteric ligands that readily deviate from the expectations of orthosteric competition, i.e. interactions that display positive, neutral, or low negative cooperativity as opposed to interactions that appear highly negatively cooperative and thus difficult to distinguish from simple competition. It is possible that pure allosteric agonists, which activate the receptor solely from a novel allosteric site, may themselves display a different spectrum of behaviors to bitopic ligands and/or orthosteric agonists (6); the ability to experimentally discriminate such ligands from classic orthosteric ligands or hybrid bitopic ligands thus assumes major importance.
Finally it is possible that other functionally selective ligands for different classes of GPCRs actually represent hitherto unappreciated bitopic orthosteric/allosteric ligands. Although it has long been established that structural manipulations around a common orthosteric “core” molecule, typically the endogenous agonist, can result in receptor subtype-selective changes in both ligand affinity and efficacy (35), few studies have actually focused on the nature of those elements that constitute the structural manipulations in isolation of the core starting molecule; some of these elements may recognize allosteric binding sites. Interestingly a recent study by Kobilka and co-workers (36) revealed that β2-adrenergic receptor full and partial agonists not only induce different active conformational states of the receptor but adopt very different binding modes within the binding pocket. Of particular relevance to our findings, these authors also reported in that same study that catechol, which has long been considered a canonical moiety within all catecholamines, was itself able to simultaneously occupy a β2 receptor that bound the orthosteric inverse agonist ICI118551, suggesting that a component of a classic orthosteric compound has the potential to bind allosterically relative to another orthosteric compound.
In conclusion, we have identified a novel mechanism of action of a functionally selective GPCR partial agonist that involves the simultaneous bridging of both an orthosteric and an allosteric site on the receptor to direct agonist efficacy. The design and/or detection of similar bitopic ligands is likely to result in greater selectivity of GPCR action as well as providing new insights into mechanisms of receptor activation.
Supplementary Material
Acknowledgments
We thank TGR BioSciences for generously providing the SureFire cellular ERK1/2 assay kits.
This work was supported in part by National Health and Medical Research Council (NHMRC) of Australia Grant 400134 and Australian Research Council Discovery Grant DP0877497. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 as well as other information.
Footnotes
The abbreviations used are: GPCR, G protein-coupled receptor; ACh, acetylcholine; BPMC, biased probability Monte Carlo; CHO, Chinese hamster ovary; DDBL-1, carbamic acid, N-(3-chlorophenyl)-, 4-hydroxy-2-butyn-1-yl ester; DDBL-2, N-2-butynyl-3-chlorophenylcarbamate; DDBL-3, N-propargyl-3-chlorophenylcarbamate; DDBL-4, N-ethyl-3-chlorophenylcarbamate; DDBL-5, N-methyl-3-chlorophenylcarbamate; E2, second extracellular loop; ERK, extracellular signal-regulated kinase; FBS, fetal bovine serum; Gpp(NH)p, guanosine 5′-(βγ-imido)triphosphate; GTPγS, guanosine 5′-3-O-(thio)triphosphate; mAChR, muscarinic acetylcholine receptor; McN-A-343, 4-(N-(3-chlorophenyl)carbamoyloxy)-2-butynyltrimethylammonium chloride; NMS, N-methylscopolamine; TMA, tetramethylammonium; TMD, transmembrane-spanning domain; ICM, internal coordinate mechanics.
References
- 1.Lagerstrom, M. C., and Schioth, H. B. (2008) Nat. Rev. Drug Discov. 7 339-357 [DOI] [PubMed] [Google Scholar]
- 2.Kristiansen, K. (2004) Pharmacol. Ther. 103 21-80 [DOI] [PubMed] [Google Scholar]
- 3.Urban, J. D., Clarke, W. P., von Zastrow, M., Nichols, D. E., Kobilka, B., Weinstein, H., Javitch, J. A., Roth, B. L., Christopoulos, A., Sexton, P. M., Miller, K. J., Spedding, M., and Mailman, R. B. (2007) J. Pharmacol. Exp. Ther. 320 1-13 [DOI] [PubMed] [Google Scholar]
- 4.Christopoulos, A., and Kenakin, T. (2002) Pharmacol. Rev. 54 323-374 [DOI] [PubMed] [Google Scholar]
- 5.May, L. T., Leach, K., Sexton, P. M., and Christopoulos, A. (2007) Annu. Rev. Pharmacol. Toxicol. 47 1-51 [DOI] [PubMed] [Google Scholar]
- 6.Leach, K., Sexton, P. M., and Christopoulos, A. (2007) Trends Pharmacol. Sci. 28 382-389 [DOI] [PubMed] [Google Scholar]
- 7.Langmead, C. J., and Christopoulos, A. (2006) Trends Pharmacol. Sci. 27 475-481 [DOI] [PubMed] [Google Scholar]
- 8.Roszkowski, A. P. (1961) J. Pharmacol. Exp. Ther. 132 156-170 [PubMed] [Google Scholar]
- 9.Caulfield, M. P., and Birdsall, N. J. (1998) Pharmacol. Rev. 50 279-290 [PubMed] [Google Scholar]
- 10.Griffin, M. T., Figueroa, K. W., Liller, S., and Ehlert, F. J. (2007) J. Pharmacol. Exp. Ther. 321 1193-1207 [DOI] [PubMed] [Google Scholar]
- 11.Birdsall, N. J., Burgen, A. S., Hulme, E. C., Stockton, J. M., and Zigmond, M. J. (1983) Br. J. Pharmacol. 78 257-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waelbroeck, M. (1994) Mol. Pharmacol. 46 685-692 [PubMed] [Google Scholar]
- 13.May, L. T., Avlani, V. A., Langmead, C. J., Herdon, H. J., Wood, M. D., Sexton, P. M., and Christopoulos, A. (2007) Mol. Pharmacol. 72 463-476 [DOI] [PubMed] [Google Scholar]
- 14.Christopoulos, A., and Mitchelson, F. (1997) Eur. J. Pharmacol. 339 153-156 [DOI] [PubMed] [Google Scholar]
- 15.Roszkowski, A. P., and Yelnosky, J. (1967) J. Pharmacol. Exp. Ther. 156 238-245 [PubMed] [Google Scholar]
- 16.Mellin, C., Vargas, H. M., and Ringdahl, B. (1989) J. Med. Chem. 32 1590-1593 [DOI] [PubMed] [Google Scholar]
- 17.Avlani, V. A., Gregory, K. J., Morton, C. J., Parker, M. W., Sexton, P. M., and Christopoulos, A. (2007) J. Biol. Chem. 282 25677-25686 [DOI] [PubMed] [Google Scholar]
- 18.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 19.Kostenis, E., Botero Cid, H. M., Holzgrabe, Y., and Mohr, K. (1996) Eur. J. Pharmacol. 314 385-392 [DOI] [PubMed] [Google Scholar]
- 20.Lazareno, S., and Birdsall, N. J. (1995) Mol. Pharmacol. 48 362-378 [PubMed] [Google Scholar]
- 21.Cherezov, V., Rosenbaum, D. M., Hanson, M. A., Rasmussen, S. G., Thian, F. S., Kobilka, T. S., Choi, H. J., Kuhn, P., Weis, W. I., Kobilka, B. K., and Stevens, R. C. (2007) Science 318 1258-1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abagyan, R., and Totrov, M. (1994) J. Mol. Biol. 235 983-1002 [DOI] [PubMed] [Google Scholar]
- 23.Lu, Z. L., Saldanha, J. W., and Hulme, E. C. (2002) Trends Pharmacol. Sci. 23 140-146 [DOI] [PubMed] [Google Scholar]
- 24.Cheng, Y., and Prusoff, W. H. (1973) Biochem. Pharmacol. 22 3099-3108 [DOI] [PubMed] [Google Scholar]
- 25.Leff, P., Dougall, I. G., and Harper, D. (1993) Br. J. Pharmacol. 110 239-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christopoulos, A. (1998) Trends Pharmacol. Sci. 19 351-357 [DOI] [PubMed] [Google Scholar]
- 27.Christopoulos, A., and El-Fakahany, E. E. (1999) Biochem. Pharmacol. 58 735-748 [DOI] [PubMed] [Google Scholar]
- 28.Christopoulos, A., Grant, M. K., and El-Fakahany, E. E. (2000) J. Pharmacol. Toxicol. Methods 43 55-67 [DOI] [PubMed] [Google Scholar]
- 29.Wess, J. (2005) Mol. Pharmacol. 68 1506-1509 [DOI] [PubMed] [Google Scholar]
- 30.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., Yamamoto, M., and Miyano, M. (2000) Science 289 739-745 [DOI] [PubMed] [Google Scholar]
- 31.Schwarz, R. D., Spencer, C. J., Jaen, J. C., Mirzadegan, T., Moreland, D., Tecle, H., and Thomas, A. J. (1995) Life Sci. 56 923-929 [DOI] [PubMed] [Google Scholar]
- 32.Heitz, F., Holzwarth, J. A., Gies, J. P., Pruss, R. M., Trumpp-Kallmeyer, S., Hibert, M. F., and Guenet, C. (1999) Eur. J. Pharmacol. 380 183-195 [DOI] [PubMed] [Google Scholar]
- 33.Jakubik, J., Bacakova, L., El-Fakahany, E. E., and Tucek, S. (1995) J. Pharmacol. Exp. Ther. 274 1077-1083 [PubMed] [Google Scholar]
- 34.Sur, C., Mallorga, P. J., Wittmann, M., Jacobson, M. A., Pascarella, D., Williams, J. B., Brandish, P. E., Pettibone, D. J., Scolnick, E. M., and Conn, P. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13674-13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Black, J. (1989) Science 245 486-493 [DOI] [PubMed] [Google Scholar]
- 36.Swaminath, G., Deupi, X., Lee, T. W., Zhu, W., Thian, F. S., Kobilka, T. S., and Kobilka, B. (2005) J. Biol. Chem. 280 22165-22171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.