Abstract
FBI-1 (Pokemon/ZBTB7A) is a proto-oncogenic transcription factor of the BTB/POZ (bric-à-brac, tramtrack, and broad complex and pox virus zinc finger) domain family. Recent evidence suggested that FBI-1 might be involved in adipogenic gene expression. Coincidentally, expression of FBI-1 and fatty-acid synthase (FASN) genes are often increased in cancer and immortalized cells. Both FBI-1 and FASN are important in cancer cell proliferation. SREBP-1 is a major regulator of many adipogenic genes, and FBI-1 and SREBP-1 (sterol-responsive element (SRE)-binding protein 1) interact with each other directly via their DNA binding domains. FBI-1 enhanced the transcriptional activation of SREBP-1 on responsive promoters, pGL2-6x(SRE)-Luc and FASN gene. FBI-1 and SREBP-1 synergistically activate transcription of the FASN gene by acting on the proximal GC-box and SRE/E-box. FBI-1, Sp1, and SREBP-1 can bind to all three SRE, GC-box, and SRE/E-box. Binding competition among the three transcription factors on the GC-box and SRE/E-box appears important in the transcription regulation. FBI-1 is apparently changing the binding pattern of Sp1 and SREBP-1 on the two elements in the presence of induced SREBP-1 and drives more Sp1 binding to the proximal promoter with less of an effect on SREBP-1 binding. The changes induced by FBI-1 appear critical in the synergistic transcription activation. The molecular mechanism revealed provides insight into how proto-oncogene FBI-1 may attack the cellular regulatory mechanism of FASN gene expression to provide more phospholipid membrane components needed for rapid cancer cell proliferation.
FBI-1 (factor that binds to the inducer of short transcripts of human immunodeficiency virus-1, also called Pokemon, ZBTB7A) is a member of the BTB/POZ domain class of transcription regulatory proteins (1-2). It was originally isolated as a cellular factor that binds to the inducer of short transcripts of human immunodeficiency virus-1 and the proximal GC-rich sequence of ADH5/FDH (ADH5/FDH is alcohol dehydrogenase 5/formaldehyde dehydrogenase) (3, 4). Molecular cloning revealed that FBI-1 is a transcription factor with a POZ domain at the N terminus and Krüppel-type four C2H2 zinc fingers at the C terminus (4, 5), and Western blot analysis showed that FBI-1 is a 75-kDa protein. FBI-1 has been shown to have variety of biological functions such as human immunodeficiency virus type 1 Tat trans-activation, stimulation of NF-κB activity, potential role in adipogenesis, osteoclastogenesis, B-cell lymphoma, and repression of ADH5/FDH gene transcription (4, 6-10). Serial analysis of gene expression analysis revealed that expression of FBI-1 is increased in multiple cancers (available at www.ncbi.nlm.nih.gov). Recently, FBI-1 was shown to cause cancer in the thymus, liver, and spleen by repressing the tumor suppressor gene ARF that in turn lowers the expression of another tumor suppressor p53 gene (11). Also, mouse FBI-1 (called LRF) was shown to regulate B versus T lymphoid lineage fate decisions (12).
SREBPs3 are bHLH leucine zipper family transcription factors and are the major regulators of transcription of choresterogenic and lipogenic genes (13, 14). There are three isoforms of SREBPs as follows: SREBP-1a, SREBP-1c, and SREBP-2. SREBP-1a and SREBP-1c are transcribed from the same SREBP-1 gene, each by a distinct promoter. The two SREBP-1 proteins are identical except the extreme N termini. SREBP-1a has 28 unique amino acids from its first exon and SREBP-1c has only four (in addition to the initiator Methionine). SREBP-2 is encoded by the separate gene SREBP-2 that encodes a single mRNA (15, 16 and references therein).
The relative levels of SREBP-1a and -1c mRNA were shown to vary over the 50-100-fold range in different tissues. The predominant isoform in adult liver and adipocytes is SREBP-1c, so it may be the key protein involved in SREBP-1-dependent processes in these tissues. SREBP-1a is also the predominant form in spleen and in all cultured cells, including cancer cells (17). It is possible that the more active SREBP-1a isoform is preferentially expressed to meet the cellular need for more cholesterol and fatty acids during periods of rapid cell proliferation.
SREBP-1a has a potent activation domain in the N-terminal region, which interacts efficiently with co-activators such as p300/CREB-binding protein (18), Sp1 (19, 20), and the mammalian mediator complex (21). The shorter N terminus of SREBP-1c is a weak activation domain and interacts weakly with the same transcriptional co-activators. SREBP-1a is a potent transcription activator of all SREBP-responsive genes, including those that mediate the synthesis of cholesterol, fatty acids, and triglycerides. SREBP-1c, with its weak and short N-terminal transcription activation domain, preferentially enhances transcription of genes required for fatty acid synthesis but not cholesterol synthesis. SREBP-2 preferentially activates cholesterol synthesis (15, 16 and references therein).
One of the key metabolic enzymes of which expression is controlled by SREBP-1 is fatty-acid synthase (FASN) (16). FASN catalyzes the condensation of malonyl-CoA and acetyl-CoA to produce the 16-carbon fatty acid palmitate (22). The expression of FASN is very low in almost all nonmalignant adult tissues but is highly expressed in many cancer types (17). Cancer cells show increased de novo fatty acid synthesis, and in fact, FASN is the only enzyme capable of fatty acid synthesis in most carcinomas (23-25 and references therein). Although FASN is active in cancer cells, as in liver and other lipogenic tissues, the fate of the palmitate differs between cancer cells and lipogenic tissues. In liver and adipose tissue, fatty acid synthesis occurs to store excess calories from carbohydrate as triglyceride. But most human cancers do not store significant amounts of triglyceride. Endogenously synthesized fatty acids in cancer cells are esterified to phospholipids to supply membrane components of rapidly proliferating cancer cells (26, 27).
It is important to investigate how the FASN gene is regulated not only in the normal cellular context but also in the context of cancer cells. The transcriptional regulatory network of FASN gene expression under various physiological conditions is complex and involves multiple transcription factors. Transcription factor-wise, specificity proteins 1 and 3 (Sp1 and Sp3), nuclear factor Y (NF-Y), upstream stimulatory factor, retinoic X receptor, liver X receptor, and sterol regulatory element-binding protein-1 (SREBP-1) have binding sites on the promoter of the FASN gene (28 and references therein). Among them, the SREBP-1 and Sp1 are major regulators acting on the proximal promoter and activate transcription synergistically by binding to the SRE (or SRE/E-box) and GC box, respectively (29-34). They interact with each other, and the interaction was suggested to be important in synergistic transcription activation of the FASN gene. Accordingly, other proteins interacting either with SREBP-1 or Sp1 are probably important in the transcriptional regulation of the genes involved in lipid biosynthesis (29-33). We recently found that FBI-1 interacts with both SREBP-1 and Sp1 and that FBI-1 modulates the transcriptional activation by Sp1 (4).
FBI-1 appears to be involved in cell proliferation, preadipocyte differentiation, and adipogenesis (8, 11). The expression of the FBI-1 is gradually increased up to 2 days after the induction of adipocyte differentiation in 3T3L1 cells, and its expression then decreases markedly thereafter. Also, FBI-1 overexpression in 3T3-L1 cells results in more lipid accumulation during differentiation as well. Based on these observations, it was suggested that FBI-1 may be important for adipocyte differentiation where it may activate expression of adipogenic genes, including FASN (8).
In addition, others recently found that FBI-1 stimulates cell proliferation and is a potent proto-oncogene (11). We suspected that the proto-oncogene FBI-1 may increase FASN gene expression in cancer cells and thereby provide phospholipid cell membrane components to meet the lipid needs of rapidly proliferating cancer cells. In this study, we show that the molecular interactions involving Sp1, SREBP, proto-oncogene FBI-1, and proximal promoter DNA elements are important in the transcription activation of the FASN gene that occurs in many cancer cell types. The data may have significant implications for understanding how proto-oncogene FBI-1 plays a role in lipid biosynthesis and cell proliferation during adipogenesis as well as oncogenesis.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents—Fatty-acid synthase promoter-luciferase reporter genes, pGL2-FASN1-Luc (bp -150/+65), pGL2-FASN2-Luc (bp -135/+65), and pGL2-FASN3-Luc (bp -150/-73), are reported elsewhere (34). pGL2-6x (SRE)-tk*-Luc reporter plasmid was prepared by cloning six copies of the SREBP-responsive element (SRE, only top strand shown, 5′-AAAATCACCCCACTGCAA-3′) of the LDLR gene in front of tk* minimal promoter (35). The two Sp1-binding element of the tk promoter were mutated to prepare tk* minimal promoter using site-directed mutagenesis kit (Stratagene, La Jolla, CA).
The pcDNA3.1-SREBP-1a, pcDNA3.1-SREBP-1a-acidic∼bHLH (aa 1-394), pcDNA3.1-SREBP-1a-acidic∼Pro/Ser (aa 1-178), and pcDNA3.1-SREBP-1a-bHLH (aa 337-374) plasmids were prepared by cloning amplified cDNA fragments from pCMV-SREBP-1a into the pcDNA3.1 plasmid (Invitrogen). The following oligonucleotide PCR primers were used: SREBP-1a acidic∼bHLH (aa 1-394), forward, 5′-GATCGGATCCGACGAGCCACCCTTCAGCGA-3′, and reverse 5′-GATCCTCGAGGCTTTTGTGGACAGCAGTGC-3′; SREBP-1a-acidic∼ Pro/Ser (aa 1-178), forward, 5′-GATCGGATCCGACGAGCCACCCTTCAGCGA-3′, and reverse, 5′-GATCCTCGAGGGGAGGGCTTCCTGTAGAGA-3′; SREBP-1a-bHLH (aa 337-374), forward, 5′-GARCGGATCCGAGAAGCGCACAGCCCACAA-3′, and reverse, 5′-GATCCTCGAGGCTTTTGTGGACAGCAGTGC-3′.
To prepare the recombinant GST-bHLHREBP-1 fusion protein expression vector, the pCMV-SREBP-1a-bHLH (aa 337-374) was cloned into pGEX4T3 vector/BamHI-XhoI (Amersham Biosciences). The oligonucleotide PCR primers used for bHLHSREBP-1 are forward, 5′-GATCGGATCCGCCCCGGCCTCTGCCCAGAGC-3′, and reverse, 5′-GATCCTCGAGTCACAGAGATTTGCTTTTGTGGACAG-3′.
The recombinant GST fusion protein expression vector for FBI-1ZFDBD (zinc finger DNA binding domain) (aa 366-495) was prepared by cloning PCR-amplified cDNA fragments into pGEX4T1/BamHI-SalI. The oligonucleotide PCR primers used for FBI-1ZFDBD are forward, 5′-GATCGGATCCGCCTGGTCGCAGAAGGTGGAG-3′, and reverse, 5′-GATCGTCGACCGAGGGGACGCCGTTGCAGCCGTC-3′.
To prepare various truncated FBI-1 mammalian expression vectors, pcDNA3.0-FLAG-FBI-1, pcDNA3.1-POZNLSFBI-1 (aa 1-131), pcDNA3.0-ZFDBDFBI-1 (aa 371-512), pcDNA3.0-ΔPOZ FBI-1 (aa 132-584), and pcDNA3.0-ZFDBD-C-term (aa 376-584), PCR-amplified cDNA fragments were cloned into pcDNA3 series mammalian expression vectors, and some were reported elsewhere (Invitrogen) (4, 7). The PCR oligonucleotide primers used for the C-terminal (aa 376-584) and zinc finger DBD of FBI-1 (aa 376-505) are as follows: C-terminal forward, 5′-GGATCGAATTCACCATGGACTACAAGGACGACGATGACAAGATCCGAGCCAAGGCCTTC-3′, and reverse, 5′-GGATCTCTAGATCATTAGGCGAGTCCGGCTGT-3′; ZFDBD of FBI-1, forward the same as above and reverse 5′-GGATCTCTAGATCAGCCCCGGACGCGGGGCTT-3′.
Antibodies against SREBP-1, FLAG tag, or control IgG were purchased from Santa Cruz Biotechnology, Upstate (Charlottesville, VA), Cell Signaling, Calbiochem, and Sigma. Antibody against FBI-1 was prepared by us. Most of the chemical reagents were purchased from Sigma.
RT-PCR Analysis of mRNA of FBI-1, SREBP, FASN, Sp1, GAPDH in C57Bl/6J Mice, ob/ob C57Bl/6J Mice, DIO C57Bl/6J Mice, 293Trex, HCT116, and Prostate LNCaP Cells—Male C57BL/6J mice were fed either with high fat chow D12492 or control lean chow (Research Diets Inc., New Brunswick, NJ) for 20 weeks to generate diet-induced obese mice (DIO). Total RNA was isolated from human HEK293T cells, human prostate LNCaP cells, and the abdominal adipose tissues of control and obese ob/ob C57Bl/6J mice or DIO mice using TRIzol reagent (Invitrogen). cDNAs were synthesized using 1 μg of total RNA, random hexamer (10 pmol), and Superscript reverse transcriptase II (200 units) in 20 μl using reverse transcription kit (Invitrogen). PCR were performed by using the following amplification conditions: 94 °C denaturation for 5 min, 35 cycles of amplification reaction, 94 °C for 1 min, 60 °C for 45 s, 72 °C for 1.5 min, and final extension reaction at 72 °C for 10 min. Oligonucleotide primers used for PCR are as follows: FBI-1 (LRF-1), forward, 5′-GCCTGGCTGTGCGACGTGGT-3′, and reverse, 5′-CAGCAGGCGGGCGGCGCTGA-3′; SREBP-1, forward, 5′-GATCGGATCCGAGAAGCGCACAGCCCACAA-3′, and reverse 5′-GATCCTCGAGGCTTTTGTGGACAGCAGTGC-3′; FASN, forward, 5′-GATCGAATTCAACCATGGAGGAGGTGGTGATAGCCGGT-3′, and reverse, 5′-GATCCTCGAGTGCCAGCAAGCTGGAGGAGCAGGC-3′; Sp1, forward, 5′-CGTGGATCCACAGGTGAGCTTGACCTCACAGC-3′, and reverse, 5′-CGTGCGGCCGCCTATATGATCTGTATTTGACCAG-3′; and GAPDH, forward, 5′-ATGGGGAAGGTGAAGGTCGGAGTC-3′, and reverse, 5′-TTACTCCTTGGAGGCCATGTGGGC-3′.
Western Blot Analysis of FBI-1, SREBP, FASN, GAPDH in 293Trex, HCT116, and Prostate LNCaP Cells—Cells were harvested and lysed in RIPA buffer (50 mm Tris-HCl, pH 8.0, 1% Nonidet P-40, 0.25% sodium deoxycholic acid, 150 mm NaCl, 1 mm EGTA, Complete Mini-Protease mixture). Cell extracts (25-50 μg) were separated using 8-12% gradient SDS-PAGE, transferred onto Immun-Blot™ polyvinylidene difluoride Membrane (Bio-Rad), and blocked with 5% skim milk (BD Biosciences). Blotted membranes were incubated with antibodies against FLAG tag (Sigma), FBI-1 (Abcam, Cambridge, UK), GAPDH (Chemicon, CA), SREBP-1 (Santa Cruz Biotechnology), FASN (prepared by us), His tag, and Myc tag (Santa Cruz Biotechnology) and then incubated with horseradish peroxidase-conjugated mouse, rabbit, or goat IgG (Vector Laboratories). Protein bands were visualized with ECL solution (PerkinElmer Life Sciences).
Cell Culture and Transcriptional Analysis of the SREBP-responsive pGL2-6x(SRE)-Luc Promoter by SREBP-1 and FBI-1 by Transient Transfection—Human liver Alexander, human kidney HEK293A, human colon HCT116, mouse embryonic NIH/3T3, and African green monkey kidney CV-1 cells were cultured in the medium recommended by the ATCC (Manassas, VA) and supplemented with 10-15% fetal bovine serum (Invitrogen). The cells were seeded onto 6-well plates and grown for 16 h before transfection.
To test the effect of FBI-1 on the transcriptional activation of the SREBP-1-responsive promoter, the mammalian cells were transfected with the mixture of 1.1 μg of the reporter pGL2-6x(SRE)-tk*-Luc plasmid (0.2 μg), pcDNA3-FBI-1 (0.6 μg), and pcDNA3 SREBP-1a or -1c (0.025 μg), using Lipofectamine Plus reagent (Invitrogen). The cells were harvested 36 h after transfection and lysed, and luciferase activities were analyzed. Luciferase activity was normalized with co-transfected β-galactosidase activity or the protein concentration of the cell lysates.
To investigate the functional consequences of molecular interaction between the SREBP-1 and the ZFDBDFBI-1 on the transcription of pGL2-6x(SRE)-tk*-Luc, the cells (HEK293A or Alexander) were co-transfected with the mixture of 1.1 μg of pGL2-6x(SRE)-tk*-Luc (0.3 μg), pcDNA3-FBI-1, or pcDNA3-ZFDBDFBI-1 (0.6 μg) and pcDNA3 SREBP-1a or -1c (0.025 μg), using Lipofectamine Plus reagent (Invitrogen) and analyzed as described above.
GST Fusion Protein Purification, in Vitro Transcription and Translation, and GST Fusion Protein Pulldown Assays—The Escherichia coli BL21 (DE3) transformed with either GST or GST-bHLHSREBP-1 proteins expression vector (pGEX4T3-bHLHSREBP-1) was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 37 °C. After E. coli pellets were lysed and sonicated in lysis buffer (1× PBS, 1 mm phenylmethylsulfonyl fluoride, 2 mm EDTA, and 0.2 mg/ml lysozyme), recombinant proteins were purified by affinity chromatography using glutathione-agarose 4 beads (Peptron, Daejeon, Korea). Purified proteins were resolved on a 15% SDS-polyacrylamide gel to quantitate and assess purity. The same volume of protein-agarose bead complex was used for all GST fusion protein pulldown assays.
The four full-length and truncated [35S]methionine-labeled FBI-1 polypeptides were prepared by in vitro transcription and translation by incubating 1 μg of the pcDNA3.0 FBI-1 or its deletion mutants [pcDNA3.0 POZ FBI-1 (aa 1-131), pcDNA3.0 FBI-1ΔPOZ (aa 132-584), pcDNA3.0 FBI-1 C-term (aa 376-584)] expression vectors, 2 μl of [35S]methionine (PerkinElmer Life Sciences), and 40 μl of TnT Quick Master Mix (Promega, WI) at 30 °C for 90 min.
Purified GST fusion proteins were incubated with GSH-agarose at 4 °C for 1 h in HEMG buffer (40 mm HEPES, pH 7.9, 100 mm KCl, 0.2 mm EDTA, 5 mm MgCl2, 0.1% Nonidet P-40, 10% glycerol, 1.5 mm 1,4-dithiothreitol, protease inhibitor mixture (1 tablet/50 ml)). After agarose-GST protein complexes were washed, 10 μl of in vitro translated FBI-1 proteins were added and incubated in HEMG buffer at 4 °C for 4 h. The reactions mixtures were centrifuged, and the pellets were washed, denatured, and separated on a 15% SDS-polyacrylamide gel. The gels were exposed to x-ray film using an image-intensifying screen (Eastman Kodak Co.).
Co-immunoprecipitation and Western Blot Analysis—HEK293A cells were harvested and lysed in lysis buffer (50 mm Tris-HCl, pH 7.5, 0.3 m NaCl, 1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (1 tablet/50 ml)). Lysates were clarified by centrifuging at 13,000 rpm for 15 min. 500 μg of the lysates were incubated with anti-FBI-1 (or anti-SREBP-1) antibody (2 μg) and subsequently with protein A-agarose (20 μl) overnight at 4 °C with gentle rotation. The immunoprecipitates were washed, denatured, resolved by 15% SDS-PAGE, and transferred onto Immun-Blot™ polyvinylidene difluoride membrane (Bio-Rad). Blotted membranes were incubated with following antibodies, Ab-SREBP-1 (or Ab-FBI-1) and Ab-GAPDH. Membranes were further incubated with horseradish peroxidase-conjugated anti-mouse IgG (Vector Laboratories) or anti-rabbit IgG (Vector Laboratories) and developed with the ECL reagents (PerkinElmer Life Sciences).
Immunostaining and Cellular Localization of FBI-1 and SREBP-1—Human liver Alexander cells were grown on the coverslips placed in a 6-well culture dish (Sunshine Works, Seoul, Korea), and the cells were transfected with pcDNA3.0-FLAG-FBI-1 expression plasmid (0.6 μg) and pcDNA3.1-SREBP-1a-Myc. After 24 h, the cells were washed with cold PBS and fixed in 97:3 cold methanol/formaldehyde for 20 min at -20 °C. Cells were permeabilized in 0.2% Triton X-100 and washed with PBS. Next, cells were incubated in 5% normal horse serum to block nonspecific binding and further incubated with mouse anti-FLAG primary antibody (0.5 μg/100 μl) for 2 h at room temperature. The cells were washed and incubated with fluorescein isothiocyanate-conjugated anti-mouse IgG secondary antibody (0.5 μg/100 μl) (Jackson Laboratories). For double staining, the cells were fixed with 3.7% formaldehyde for 10 min at room temperature, washed, and blocked with 5% normal horse serum. Washed cells were incubated with rabbit anti-SREBP-1a antibody and subsequently with rhodamine-conjugated anti-rabbit IgG secondary antibody (0.5 μg/100 μl). After washing and mounting, immunostained cells were examined with a Carl Zeiss LSM 510 confocal laser scanning microscope (Carl Zeiss, Germany).
Transcriptional Analysis of FASN WT or Mt Gene Promoter—Because FBI-1 activated the transcription of the SREBP-responsive pGL2-6x(SRE)-tk*-Luc, we investigated whether FBI-1 can also activate the transcription of the SREBP-1-responsive FASN gene promoter. Three different FASN gene promoters, which have short proximal promoter sequences and are differing only in the context of key proximal regulatory elements (pGL2-FASN1-Luc, pGL2-FASN2-Luc, and pGL2-FASN3-Luc), were used in the transcription assays (34). The mammalian cells (Alexander, HEK293A, HCT116, and NIH3T3) were transiently transfected with the mixture of 1.1 μg of pGL2-FASN-Luc reporter plasmid (e.g. pGL2-FASN1-Luc, pGL2-FASN2-Luc, and pGL2-FASN3-Luc (0.3 μg), and pcDNA3-FBI-1 (0.025-0.5 μg), and pcDNA3 SREBP-1a (0.025 μg)) using Lipofectamine Plus reagent (Invitrogen) and analyzed as described above in the transcription analysis by transient transfection.
Also transcriptional regulation of FASN gene by SREBP-1 and FBI-1 was analyzed using much longer -2.7 kb upstream regulatory element with either intact WT or mutated SRE/E-box (bp -65 to -47; WT, 5′-TCAGCCCATGTGGCGTGGC-3′; mutant, 5′-TCAGCCTTTGAAGCGTGGC-3′). The mutations were introduced by site-directed mutagenesis (Stratagene). The cells were transiently transfected with the plasmid mixture of pGL2-FASN(-2.7 kb)-Luc WT (or Mt) (0.3 μg), pcDNA3.1-SREBP1a expression plasmid (0.025 μg), and pcDNA3.1-FBI-1 expression plasmid (0.025-0.5 μg) using Lipofectamine Plus reagent (Invitrogen). After 24 h of incubation, cells were harvested and analyzed as described above.
Electrophoretic Mobility Shift Assay (EMSA)—The oligonucleotide probes (500 pmol each in 83.3 mm Tris-HCl, pH 8.0, 16.7 mm MgCl2, 166.7 mm NaCl) were annealed by heating at 93 °C for 5 min and cooling down slowly to room temperature. After diluting the solution containing the annealed oligonucleotides with water to 50 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 100 mm NaCl, 100 pmol of annealed oligonucleotides for EMSAs were labeled with [α-32P]ATP and Klenow enzyme (Roche Applied Science) by incubating for 30 min at 37 °C. α-32P-Labeled double-stranded oligonucleotides were purified with Sephadex™ G-50 (Amersham Biosciences). Sequences of various oligonucleotides used in EMSA are as follows (only the top strand is shown): FASN SRE 5′-GATCCGGGCATCACCCCACCGACG-3′; FASN SRE/E-Box, 5′-GATCGTCAGCCCATGTGGCGTGGC-3′; and FASN GC-box, 5′-GATCTGGCCGGGCGGCGCAGCCA-3′.
Each binding reaction was carried out in 20 μl of binding buffer containing 10 mm HEPES, pH 7.9, 60 mm KCl, 5 μm ZnCl2, 1 mm dithiothreitol, 1% bovine serum albumin, and 7% glycerol, and purified recombinant His-tagged SREBP (10 ng), Sp1 (50 ng for GC-box, 150 ng for SRE and SRE/E-box), and GST-ZFDBDFBI-1 (300 ng). 1.2 μg of anti SREBP-1 and Sp1 antibodies (Santa Cruz Biotechnology) and 5 μg of anti-GST antibody (Upstate, Charlottesville, VA) were added to the binding mixture in some EMSA binding reactions for supershift assays. The protein-DNA complexes were resolved from free probe by 6% nondenaturing PAGE at room temperature in 0.5× TBE buffer (89 mm Tris borate, 2 mm EDTA, pH 8.3). The dried gels were exposed to x-ray film at -70 °C with a Kodak intensifying screen (Kodak).
Chromatin Immunoprecipitation (ChIP) Assays—The molecular binding interactions among FBI-1, SREBP-1, Sp1, and FASN proximal promoter in vivo were analyzed using ChIP assay kit (Upstate Biotechnology, Inc., Lake Placid, NY) as reported elsewhere (4). The HEK293A cells grown on a 10-cm culture dish were transfected with pGL2-FASN2-Luc (or pGL2-FASN3-Luc, pGL2-FASN-Luc WT or Mt, -2.7 kb) (2 μg), pcDNA3.1-SREBP-1a (1 μg), and pcDNA3-FLAG-FBI-1 (3 μg) using Lipofectamine Plus. Cells were fixed with formaldehyde (final 1%) to cross-link SREBP1 and FBI-1 onto the FASN promoter. Cells were washed with cold phosphate-buffered saline and lysed with SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1). The lysate was sonicated to shear DNA to a length between 500 and 700 bp. The sonicated supernatant was diluted 10-fold with ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 2 mm Tris-HCl, pH 8.1, 150 mm NaCl) and incubated with antibodies against FLAG, SREBP1, and Sp1 overnight at 4 °C with rotation. To collect DNA-protein-antibody complex, salmon sperm DNA/protein A-agarose slurry was added to the mixture. The mixture was incubated for 2 h at room temperature with rotation and collected DNA-protein A-agarose complex by brief centrifugation (3,000 rpm) at 4 °C. After extensive washing of the pellet with various washing buffers recommended by the manufacturer, the pellet was dissolved with 250 μl of elution buffer and spun to remove agarose. Supernatant was treated with 20 μl of 5 m NaCl and heated to 65°C for 2 h to reverse protein-DNA cross-link. After treatment with EDTA and proteinase K, the supernatant was extracted with phenol/chloroform and precipitated with ethanol to recover DNA. PCRs of immunoprecipitated DNA were carried out using the following oligonucleotide primer sets designed to amplify the proximal promoter region of the FASN gene. FASN WT or Mt primers (forward, 5′-CAGGCGCGTTCCCGCGCAGG-3′, and reverse, 5′-GAGAGCGAGGCTGGAGCGCG-3′). For ChIP assays of the shorter FASN2 or -3 promoter constructs, oligonucleotide PCR amplification primers were designed to bind to the plasmid vector sequences, not to the endogenous FASN gene promoter: forward, 5′-TCCAAACTCATCAATGTA-3′, and reverse, 5′-AAAGCAATTGTTCCAGGAACCAGGG-3′.
Knockdown of FBI-1 Expression by siRNA—Two siRNAs designed to knock down FBI-1 mRNA and a negative control siRNA were designed and purchased from Bioneer (Daejeon, Korea) as follows: siFBI-1 426, 5′-GCUGGACCUUGUAGAUCAAtt-3′ and 5′-UUGAUCUACAAGGUCCAGCtt-3′; siFBI-1 476, 5′-AGUACCUCGAGUUCUUCCAtt-3 and 5′-UGGAAGAACUCGAGGUACUtt-3; and negative control, 5′-CCUACGCCACCAAUUUCGUtt-3′ and 5′-ACGAAAUUGGUGGCGUAGGtt-3′. siRNA (500 pmol of each) were transfected into HEK293A and HCT116 cells (2 × 106 cells/6 well dishes) by using Lipofectamine 2000 reagent (Invitrogen). After 48 h, the cells were harvested, and cell lysates were analyzed by Western blot as described above.
RESULTS
FBI-1 mRNA Expression Is Increased in White Adipose Tissues of ob/ob C57BL/6J Mice and Diet-induced Obese Mice C57BL/6J—Expression of FBI-1 mRNA increased during the early stages of adipocyte differentiation in 3T3L1 cells and then declined as differentiation continued. Also, when 3T3L1 cells were treated with recombinant adenovirus overexpressing FBI-1 during differentiation, lipid accumulation as assessed by Oil Red O staining was significantly enhanced (8). These observations suggested that FBI-1 might play a role in the adipocyte differentiation and adipogenesis.
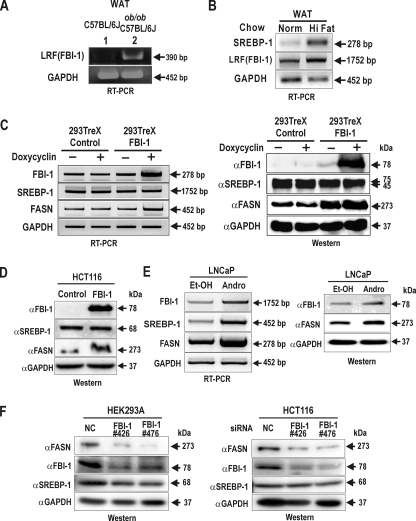
We evaluated the expression level of LRF (mouse FBI-1) mRNA in adipose tissue of lean control versus obese ob/ob C57BL/6J mice to study the possible correlation between fat mass and LRF expression. RT-PCR analysis of total RNA showed that LRF mRNA was higher in obese ob/ob mouse as compared with the lean control mice (Fig. 1A). We also found that LRF mRNA was increased in mice fed with a high fat diet to induce obesity (DIO) compared with the lean control, and SREBP-1, which is a major regulator of fatty acids synthesis, was also significantly increased (Fig. 1B).
FIGURE 1.
The expression of FBI-1 mRNA is increased in genetically obese ob/ob C57BL/6J mice and DIO mice. The stable cells overexpressing FBI-1 or rapid growing LNCaP cells showed increased FASN expression. A, RT-PCR analysis of FBI-1 mRNA using the total RNA was isolated from the abdominal adipose tissues of lean control and obese ob/ob C57BL/6J mice. GAPDH, control. LRF is a murine counterpart of human FBI-1 (also called Pokemon, ZBTB7A). B, RT-PCR analysis of mRNA of LRF (FBI-1) and SREBP-1 using the total RNA isolated from the white adipose tissues (WAT) of control and DIO C57BL/6J mice. Norm, normal; GAPDH, control. C, RT-PCR and Western blot analysis of FBI-1, SREBP-1, and GAPDH in HEK293Trex control and stable cells overexpressing FBI-1. The stable cells were induced to express lacZ or FBI-1 gene by doxycycline treatment. Total RNA or protein was isolated from the cells and analyzed for FBI-1, SREBP-1, and FASN expression at both the mRNA and protein level. GAPDH, control. D, Western blot analysis of human colon cancer stable HCT116 cells established by transfection with lentivirus overexpressing either FBI-1 or lacZ. Cell extracts were analyzed for FBI-1, SREBP-1, and FASN proteins. GAPDH, control. E, RT-PCR and Western blot analysis of LNCaP cells treated with androgen (Andro) or EtOH. Total RNA or protein was isolated from the cells and analyzed for FBI-1, SREBP-1, and FASN expression at both the mRNA and protein level. GAPDH, control. F, Western blot analysis. Knockdown of endogenous FBI-1 decreases FASN gene expression in immortalized HEK293A and HCT116 cancer cells. The cells were transfected with two independent siRNA against FBI-1 or a control siRNA. The cells were harvested at 48 h, and the cellular extracts (35 μg) were analyzed for FASN, FBI-1, SREBP-1a, and GAPDH expression. NC, negative control.
Increased Expression of FBI-1 and FASN in HEK293Trex-FBI-1 Stable Cells and in LNCaP Cells Treated with Androgen—We investigated the expression patterns of SREBP-1 and FASN in cells where FBI-1 expression is elevated. In HEK293-Trex cells overexpressing FBI-1 in an inducible fashion, FASN was significantly increased both at the mRNA and protein level when FBI-1 was induced, whereas expression of SREBP-1 and control GAPDH remained unchanged (Fig. 1C). We observed similar changes in protein expression in the colon cancer cells HCT116 stably overexpressing FBI-1 (Fig. 1D). Prostate cancer LNCaP cells grew much faster in the presence of the male hormone testosterone or androgen. The mRNAs for FBI-1, SREBP-1, and FASN were significantly increased in the LNCaP cells treated with androgen. Immunoblotting showed similar results for the corresponding proteins (Fig. 1E).
Knockdown of Endogenous FBI-1 Decreases FASN Gene Expression in Immortalized HEK293A and HCT116 Cancer Cells—The above data and transcriptional activity measurements presented below rely on overexpressed proteins, and it may be necessary to provide some additional evidence for the FBI-1-SREBP1 interaction in the cells. We investigated the relationship among FASN, FBI-1, SREBP-1 in the immortalized human HEK293A, and HCT116 colon cancer cells. The cells were transfected with two independent siRNA against FBI-1 or a control siRNA. Western blot analysis of the cell lysates showed that knockdown of FBI-1 expression resulted in significant decrease in FASN expression (Fig. 1F). The data demonstrate that FBI-1 is a critical regulator of FASN gene expression not only in the immortalized normal cells but also in the cancer cells, and it also provides more evidence that the interaction between FBI-1 and SREBP-1 is probably of physiological relevance.
FBI-1 Increases Transcription Activation of FASN Gene by SREBP-1a—We co-transfected three versions of FASN promoter constructs with the expression vectors of SREBP-1a and FBI-1 into various mammalian cells such as Alexander, HEK293A, HCT116, and NIH/3T3 cells to determine which promoter regulatory elements might be responsive to FBI-1. The three FASN constructs are different with respect to the inclusion of Sp1 binding GC-box, SRE, and SRE/E-box elements. The promoter constructs have been well characterized, and in previous studies Sp1 and SREBP-1 was shown to synergistically activate transcription by acting on the SRE, GC-box, and SRE/E-box (34). As expected, SREBP-1a activated transcription of the promoter constructs with the upstream GC-box and the proximal SRE/E-box. Although FBI-1 alone had little effect on transcription of any of the promoter constructs, FBI-1 significantly increased the SREBP-1a-dependent stimulation of transcription of both FASN1 and -2 constructs (Fig. 2, C-F). The enhancement was particularly effective in HEK293A and HCT116 cells (Fig. 2, D and E). The FBI-1 was unable to activate transcription of the FASN3 promoter construct that lacked the SRE/E-box (Fig. 2, C-F). In NIH/3T3 cells, FBI-1 repressed transcription of FASN3 promoter in a dose-dependent manner in the presence of SREBP-1a (Fig. 2F). The data suggested that FBI-1 probably stimulates transcription of FASN promoter by modulating the molecular events occurring at the GC-box, SRE/E-box, in cooperation with SREBP-1 and Sp1.
FIGURE 2.
FBI-1 and SREBP-1a synergistically activate transcription of the pGL2-FASN-Luc via GC-box and SRE/E-box elements in immortalized or cancer cells. A, nucleotide sequence comparison of three mammalian FASN gene promoters. SRE, upstream SREBP-binding element; GC-box, Sp1-binding GC box; SRE/E-box, downstream SREBP-binding element; TRE, thyroid hormone-response element; TATA, TBP-binding element; Luc, luciferase. Numbers indicate the position of nucleotide upstream of transcription initiation (+1). B, structures of three FASN-Luc reporter promoter constructs tested to investigate the functional elements mediating transcription activation by FBI-1. Tsp, transcription start site (+1). C-F, FBI-1 and SREBP-1a synergistically activate transcription of the pGL2-FASN-Luc via GC-box and SRE/E-box elements in the mammalian cells co-transfected with three different pGL2-FASN-Luc plasmids and expression vectors of SREBP-1a (25 ng) and FBI-1 (25 ng, 125 ng, 500 ng). FBI-1 could not activate transcription of the pGL2-FASN3-luc lacking downstream SRE/E-box.
FBI-1 Synergistically Activates Transcription of the SREBP-1-responsive Promoter, pGL2-6x(SRE)-Luc, in the Presence of SREBP-1—Because FBI-1 is expressed in adipocytes and cancer cells and is correlated with increased lipid accumulation, we reasoned that it might directly stimulate expression of lipogenic genes such as FASN in both cancer derived and immortalized cells. SREBP-1 is an important regulator of many lipogenic genes, including FASN. To specifically evaluate whether FBI-1 might target SREBP-1, we prepared a reporter plasmid construct that contains six copies of the well characterized SRE of the LDLR gene (20) upstream of a minimal tk promoter. The SRE is essentially identical to that of the upstream SRE (bp -145 to -135) of FASN gene promoter, which is a functional SRE in adipocytes (28). The SREs were placed just upstream of the modified tk minimal promoter and luciferase gene (Fig. 3B). The modified tk minimal promoter contains two mutated Sp1-binding GC boxes that do not bind Sp1.
FIGURE 3.
FBI-1 and SREBP-1a or -1c synergistically activate transcription of pGL2-6x(SRE)-tk*-Luc in immortalized or cancer cells. A, domain structures of SREBPs and FBI-1. SREBP-1c and SREBP-1a are identical except N-terminal acidic domain. The bHLH domain are similar among SREBP-1a, -1c, and -2. Acidic, acidic domain; Pro/Ser, Pro/Ser-rich domain; Ser/Gly/Pro, Gly/Pro/Ser-rich-domain; Gln, Gln-rich domain; bHLH, basic helix-loop-helix domain; POZ, POZ domain; ZF, Krüppel-like zinc fingers; NLS, nuclear localization sequence. B, structure of pGL2-6x [SRE]-tk*-Luc plasmid. tk* indicates the thymidine kinase minimal promoter with the mutations at the two Sp1-binding sites. C-F, FBI-1 enhances transcription activation of the pGL2-6x [SRE]-tk*-Luc by SREBP-1 in the mammalian cells co-transfected with pGL2-6x [SRE]-tk*-Luc plasmid and expression vectors of SREBP-1 (25 ng) and FBI-1 (25, 125, and 500 ng). Luciferase activities were normalized with the protein concentration, and data presented are the average of three independent assays. Bars represent standard deviations. ▪, SREBP-1a; □, SREBP-1c.
We co-transfected the human liver Alexander, human kidney HEK293A, human colon HCT116, and mouse embryo NIH/3T3 cells with the plasmid mixture of pGL2-6x (SRE)-tk*-Luc and the expression vectors for SREBP-1a, -1c, and FBI-1 in various combinations. SREBP-1 alone activated transcription potently as expected. FBI-1 further activated transcription of the promoter by SREBP-1 an additional 2-8-fold depending on the cell lines tested. FBI-1 alone did not have a significant effect on the transcription of this promoter (Fig. 3, C-F). Interestingly, in NIH/3T3 cells, FBI-1 did not affect the transcription of this test promoter, although SREBP-1 potently activated transcription as in the other cell lines tested here.
FBI-1 Interacts with SREBP-1 in Vivo—Because FBI-1 and SREBP-1 synergistically activated transcription of the SREBP-responsive promoter, we tested if SREBP-1 and FBI-1 might interact with each other by co-immunoprecipitation. Immunoprecipitation of the cell lysates prepared from the human immortalized HEK293A cells and HCT116 colon cancer cells, using anti-FBI-1 antibody, showed that SREBP-1a forms a complex with FBI-1. Also the immunoprecipitate of the same extract with anti-SREBP-1a antibody showed that FBI-1 is present in the precipitated complex (Fig. 4A). We also find the same protein-protein interaction in HCT116 cells (Fig. 4B).
FIGURE 4.
The FBI-1 interacts with SREBP-1a in immortalized human HEK293A and HCT116 colon cancer cells. A, Western blot (WB) analysis of co-immunoprecipitates of endogenous FBI-1 and SREBP-1. The HEK293A cell lysates (500 μg) were immunoprecipitated (IP) with anti-FBI-1 antibody and analyzed by Western blot using an antibody against SREBP-1a. The same extracts were also immunoprecipitated with anti-SREBP-1a antibody and analyzed by Western blot using an anti-FBI-1 antibody. B, Western blot analysis of the co-immunoprecipitates of endogenous FBI-1 and SREBP-1a in HCT116 cells. The procedures are the same as above. C, immunocytochemistry and cellular co-localization of SREBP-1a and FBI-1. The Alexander cells transfected with the expression vectors of FLAG-FBI-1 and His-tagged SREBP-1a were analyzed by immunocytochemical staining using both mouse anti-FLAG antibody and rabbit anti-His antibody. Fluorescein isothiocyanate-conjugated anti-mouse IgG or rhodamine-conjugated anti-rabbit IgG antibody were used as secondary antibodies.
We investigated whether FBI-1 and SREBP-1a were co-localized in the nucleus of Alexander cells. The cells were transfected with expression vectors of FLAG-FBI-1, and the mature form of SREBP-1a was fused with a His tag and analyzed by immunocytochemistry using anti-FLAG and anti-His antibodies. Transfection of either FBI-1 or SREBP-1a showed both proteins mainly localized to the nucleus. When cells were co-transfected with the two protein expression vectors and analyzed by confocal microscopy, both FBI-1 and SREBP-1 were primarily detected in the nucleus, and the prominent yellow color in the confocal image suggests there is a molecular interaction between the two transcription factors in the cell (Fig. 4C).
Zinc Finger DNA Binding Domain of FBI-1 Interacts with the bHLH Domain of SREBP-1 Directly in Vitro and the ZFDBD of FBI-1 Increases Transcription Activation by SREBP-1a—HEK293A cells were transfected with the expression vectors for FLAG-FBI-1 and SREBP-1a-bHLH-Myc (aa 323-394), and the cell lysates were immunoprecipitated with anti-Myc antibody. The precipitates were analyzed by Western blotting using an antibody against FLAG tag. FBI-1 was co-precipitated with the SREBP-1a-bHLH-Myc fusion protein in vivo (supplement Fig. 2). Because full-length FBI-1 and the bHLH DNA binding domain of SREBP-1a likely interact in vivo, we investigated whether the molecular interaction was direct and also which domain of FBI-1 was interacting with the bHLH of SREBP-1 by GST fusion protein pulldown assays. The GST-bHLH domain fusion protein of SREBP-1a was incubated with in vitro synthesized [35S]methionine-labeled FBI-1 and analyzed by a standard glutathione-agarose pulled down experiment. Autoradiography of the SDS-polyacrylamide gel showed that full-length FBI-1 and the ZFDBD-C-term domain interacted with the bHLH domain of SREBP-1. The data suggest that the molecular interaction is direct and occurs through the DNA binding domains of each partner protein (Fig. 5B).
FIGURE 5.
The ZFDBD of FBI-1 interacts with the bHLH domain of SREBP-1a directly in vitro, and the ZFDBD of FBI-1 is sufficient to activate transcription of SREBP-1-responsive promoter, pGL2-6x(SRE)-tk*-Luc, by SREBP-1a. A, structures of in vitro translated [35S]methionine-labeled FBI-1 fragments and recombinant GST fusion bHLHSREBP-1a used for pulldown assay. bHLH, basic helix-loop-helix domain (aa 323-394); POZ, POZ domain; ZF, Krüppel-like zinc fingers; NLS, nuclear localization sequence. B, GST fusion protein pulldown assays. Recombinant GST and GST-bHLHSREBP-1 fusion proteins were incubated with the in vitro synthesized [35S]methionine-labeled FBI-1 polypeptides and then were pulled down. The precipitated samples were resolved by 12% SDS-PAGE and exposed to x-ray film. Input, 10% of the labeled polypeptides added in the binding reactions. C, FBI-1 activates transcription of pGL2-6x(SRE)-tk*-Luc by SREBP-1a. The ZFDBD of FBI-1 is sufficient to activate transcription. Alexander or HEK 293A cells were co-transfected with mixtures of pGL2-6x(SRE)-tk*-Luc and the expression vectors of SREBP-1a and FBI-1 full-length or the ZFDBD of FBI-1 in various combinations. Luciferase activities were normalized with the protein concentration, and data presented are the average of three independent assays. Bars represent standard deviations. ▪, Alexander cells; □, 293A cells.
To determine whether the ZFDBD domain was sufficient to stimulate transcription through interaction with SREBP-1, we investigated whether the ZFDBD of FBI could increase the transcription of pGL2-6x(SRE)-tk*-Luc promoter by SREBP-1a in two different cell types, Alexander and HEK293A. The ZFDBD of FBI-1 increased transcription of the reporter gene by SREBP-1a, similar to the full-length FBI-1 protein (Fig. 5C). This result indicates that the ZFDBD domain is sufficient for both interaction with and stimulation of transcription by SREBP-1.
FBI-1 Increases Transcription Activation of Much Longer Promoter of FASN Gene by SREBP-1a—In Fig. 2, we showed that FBI-1 could activate transcription of the short FASN promoter (Fig. 2). We also analyzed transcription of a much longer promoter element (-2.7 kb) to test whether FBI-1 might also stimulate transcription of the FASN gene through other more distal elements in the promoter. In this longer construct, mutations were introduced at the SRE/E-box located immediately downstream of the GC-box to investigate the function of this key element in the longer promoter context (Fig. 6A). SREBP-1a was able to activate transcription of this longer construct strongly as in the shorter constructs (Fig. 6B). As in the shorter promoter constructs, FBI-1 significantly increased transcription of longer FASN promoter in HEK293A, HCT116, and NIH/3T3 cells (Fig. 6, B-D). However, synergistic transcription activation by SREBP and FBI-1 was not observed with the construct containing mutations in the proximal SRE/E-box, suggesting that the proximal SRE/E-box element is essential for the transcriptional regulation by SREBP-1 and FBI-1 even in the presence of more distal regulatory elements. However, FBI-1 was not able to increase the transcription of the longer FASN gene in Alexander and African green monkey kidney CV-1 cells. Rather, inhibited transcription instead, probably representing cell type-specific transcriptional regulation of the gene acting either on the further upstream regulatory sequence or on the proximal promoter (Fig. 6, E and F).
FIGURE 6.
FBI-1 and SREBP-1a activate transcription of pGL2-FASN-Luc (-2.7 kb) via the SRE/E-box elements in immortalized or cancer cells. A, structures of pGL3-FASN-Luc WT or Mt constructs tested to investigate the functional role of SRE/E-box in transcription activation by FBI-1. SRE, SREBP-binding element; GC, Sp1-binding GC box; SRE/E-box, downstream SREBP-binding element; Tsp, transcription start site (+1). In the mutant FASN constructs, two SREBP-binding nucleotide sequences of SRE/E-box were mutated by site-directed mutagenesis. B-F, five mammalian cells were co-transfected with pGL3-FASN-Luc WT or Mt (-2.7 kb) and expression vectors of SREBP-1 (25 ng) and FBI-1 (25, 125, and 375 ng) in various combination indicated. Luciferase activities were normalized with the protein concentration, and data presented are the average of three independent assays. Bars represent standard deviations.
EMSA Analysis of DNA Binding Activities of FBI-1, Sp1, and SREBP-1a on GC-box, and SRE/E-box—FBI-1 appeared to increase transcription of the FASN promoter by modulating the molecular events occurring at the proximal promoter region containing the GC-box, and SRE/E-box elements through the SREBP-1 and Sp1 proteins. Therefore, we investigated the molecular interactions among the DNA elements and the transcription factors, FBI-1, SREBP-1, and Sp1 using EMSA.
Sp1 binds to the proximal GC-box of the FASN promoter and acts as a major activator. Two previous reports showed that the SREBP-1-binding element and Sp1-binding elements of FASN and LDLR genes are close to each other and can be bound by SREBP and Sp1 simultaneously (28, 34). Also, recently we and others showed that FBI-1 is also a GC-box binding transcription factor and can bind some Sp1-binding GC-box elements (11, 36). The results presented here show that FBI-1 and SREBP-1a are interacting with each other via their DNA binding domains (Figs. 4 and 5). Taken together, the data suggested that the three transcription factors may interact with all or parts of the regulatory elements together on the promoter. Using recombinant versions of SREBP-1, Sp1, and the ZFDBD of FBI-1, we carried out EMSA with different FASN promoter fragments to evaluate the binding of all three proteins to DNA. The GC-box and SRE/E-box regulatory elements can be bound by all three transcription factors (Fig. 7, A and B). The protein-probe interactions were abolished by the excess cold probes and also the retarded bands were either supershifted or abolished by the antibodies specific to the proteins (Fig. 7, C and D).
FIGURE 7.
EMSA of the proximal promoter elements, SRE, GC-box, and SRE/E-box of FASN gene. A, EMSA of the GC-box probe. The probes were incubated with recombinant SREBP-1 (200 ng), Sp1 (70 ng), and GST-FBI-1 ZFDBD (150 ng). B, EMSA of the SRE/E-box probe. The probes were incubated with recombinant SREBP-1 (20 ng), Sp1 (200 ng), and GST-FBI-1 ZFDBD (150 ng). C and D, EMSA of the GC-box and SRE/E-box probe. Cold probe competition. E, EMSA. Effect of excess SREBP-1a and FBI-1 proteins on Sp1 binding to the GC-box probe. The probes were incubated with recombinant Sp1 (50 ng), in the presence of increasing amount of SREBP-1 (200, 600, and 1800 ng) or GST-FBI-1 ZFDBD (150, 450, and 1350 ng). F, EMSA. Effect of excess Sp1 and FBI-1 proteins on SREBP-1a binding to the SRE/E-box probe. The probes were incubated with recombinant SREBP-1 (20 ng), in the presence of increasing amount of Sp1 (200, 600, and 1800 ng), and GST-FBI-1 ZFDBD (150, 450, and 1350 ng). Arrowheads (▸), retarded protein-probe complexes; arrowheads with star, supershifted probe-protein-antibody complex. All three recombinant proteins bind to the probes but with different affinity.
Binding competition between SREBP-1a, Sp1, and FBI-1 on the GC-box and SRE/E-box revealed novel features of binding interactions on the two key regulatory elements by the three transcription factors. Although SREBP-1a decreased Sp1 binding to the GC-box, FBI-1 increased Sp1 binding (Fig. 7E). On the contrary, on the SRE/E-box probe, FBI-1 decreased SREBP-1 binding (Fig. 7F). Because of the similar size of the proteins, it is difficult to analyze the effect of excess Sp1 on the SREBP-1a binding. Taken together, the data suggested that interactions among the three transcription factors involves not only the binding competition but also DNA binding enhancement, which might be important in the transcriptional regulation of FASN gene.
ChIP Assays Showed That FBI-1 Changes the Binding Pattern of Sp1 and SREBP-1 in the Presence of Induced SREBP-1 and Drives More Sp1 Binding to the Proximal Promoter Compared with SREBP-1 Binding—The above data suggested that FBI-1 is probably increasing transcription of the FASN promoter by modulating the molecular events occurring at the proximal GC-box, SRE/E-box through both SREBP-1 and Sp1. To extend these observations, we analyzed the protein binding events on the FASN2, and FASN3 constructs in HEK293A cells by a ChIP analysis. The two constructs are different with respect to the GC-box, SRE, and SRE/E-box. FBI-1 activates transcription on the FASN2 but not on the FASN3 promoter. The promoter constructs were well characterized previously, and Sp1 and SREBP-1 were shown to synergistically activate transcription by acting on the GC-box and the SRE/E-box respectively (34).
In the presence of endogenous low levels of SREBP-1 and Sp1, Sp1 binding was clearly detectable but SREBP-1a binding was very weak. In the presence of overexpressed SREBP-1a, SREBP-1 binding is greatly increased, and Sp1 binding is somewhat decreased (Fig. 8B). Ectopic FBI-1 bound to the promoter strongly, and inhibited binding by SREBP-1 and possibly Sp1 as well. However, Sp1 binding is increased compared with where SREBP-1a only was overexpressed, and the binding pattern is similar to that of endogenous Sp1 and SREBP-1, although somewhat weaker. This may explain why FBI-1 alone did not affect transcription of the FASN2 gene (Figs. 2, 3, and 5). When SREBP-1 and FBI-1 are co-expressed, binding of both FBI-1 and SREBP-1 is decreased compared with the situations where the two proteins are expressed independently, whereas Sp1 binding is increased to the level in lane 1 (Fig. 8B, lanes 1 and 4). The changes in binding pattern either by ectopic FBI-1 or SREBP-1 was not observed in the FASN3 construct, particularly with respect to Sp1 binding, although FBI-1 might still decrease the SREBP-1 binding on the upstream SRE by binding competition. This binding pattern may be important in the synergistic transcription activation of FASN gene by SREBP-1a and FBI-1.
FIGURE 8.
ChIP assays and the molecular binding interaction among FBI-1, SREBP-1, Sp1, and proximal FASN promoter. A, structures of the pGL2-FASN-Luc reporter promoter constructs tested to study in vivo transcription factor binding by SREBP-1, FBI-1, and Sp1. B, ChIP assays of the HEK293A cells transfected with either pGL2-FASN2-Luc or pGL2-FASN3-Luc. C, ChIP assays of the HEK293A cells transfected with pGL3-FASN-Luc WT or Mt (-2.7 kb) plasmid. The HEK293A cells were co-transfected with FASN promoter reporter fusion plasmid, pcDNA3.1-SREBP-1a and/or pcDNA3-FLAG-FBI-1. Sheared chromatin was immunoprecipitated (IP) using antibodies indicated. PCR amplification primers were designed to bind to the plasmid vector sequences, not to the endogenous FASN gene promoter in case of FASN2 and -3. SRE, SREBP-binding element; GC, Sp1-binding GC box; SRE/E-box, downstream SREBP-binding element; Tsp, transcription start site (+1). D, histogram showing the average of independent ChIP assays shown in C. ChIP binding band intensities by SREBP-1, FBI-1 and Sp1, were divided by input band intensities.
We also carried out the same set of ChIP assays using the longer FASN promoter (-2.7 kb) constructs. The results were comparable with the shorter promoter constructs and consistent with the EMSA results. The changes in the transcription factor binding pattern, which appears to be important for the increase in transcription, was clearly evident on the wild type construct but not on the FASN promoter with the mutation at the SRE/E-box (Fig. 8, C and D). These ChIP and EMSA observations suggest the activation of FASN by SREBP-1, Sp1, and FBI-1 occurs through a mechanism that is dependent on the combined interactions between all three proteins.
DISCUSSION
We initially suspected that FBI-1 might be important in lipid metabolism because FBI-1 mRNA expression was increased in ob/ob C57BL/6J and DIO mice compared with the control lean C57BL/6J mice. Others recently found that FBI-1 is a potent proto-oncogene acting at the ARF gene of the p53 pathway (11). FBI-1 caused oncogenic cellular transformation, and the transformed cells proliferated much faster. FBI-1 expression is often increased in cancer tissues, and FASN is critical in the cancer cell proliferation because it is a main supplier of phospholipid, a critical component of cell membrane. FASN gene expression is high in cancer cells and is mainly controlled by SREBP-1 and Sp1 (29, 34).
This led us to suspect that there might be some molecular link between proto-oncogene FBI-1 and FASN gene expression possibly through effects on SREBP-1. Indeed, in stable cancer cells (HCT116) or immortalized stable cells (293T), overexpression of FBI-1 resulted in increased FASN expression. Also, prostate LNCaP cancer cells treated with androgen, and thus proliferating fast, showed increased FBI-1 and FASN expression.
To investigate whether FBI-1 participated in lipid synthesis in immortalized normal and cancer cells, we tested transcriptional regulation of SREBP responsive genes by transient transfection assays. FBI-1 activated (2-3-fold) the transcription of both artificial and natural SREBP-1-responsive promoters, such as pGL2-6x-[SRE]-Luc and FASN gene, in the presence of SREBP-1. Immunoprecipitation and GST fusion protein pulldown showed that SREBP-1a and FBI-1 interact directly through their DNA binding domains, and the interaction is critical in the synergistic activation of FASN gene transcription.
To study the regulatory mechanism of FBI-1 on transcriptional regulation by Sp1 and SREBP-1a on the FASN promoter, we carried out a series of transcription analyses, EMSA and ChIP assays. EMSA showed that the two key regulatory elements (GC-box, SRE/E-box) have unique DNA binding activities recognizing SREBP-1, Sp1, and FBI-1. The two elements critical in transcription activation by FBI-1 and SREBP-1 were bound by all three transcription factors. The SRE/E-box is an SREBP-1-binding element, but it can also be bound by Sp1 and FBI-1. EMSA showed a competition between SREBP-1 and FBI-1 on the SRE/E-box. The EMSA did not provide a clear result for competition on either the GC-box or SRE/E-box because the protein-DNA complexes migrate to close together; however, the ChIP experiments suggest a possible binding competition between Sp1 and SREBP-1 on the SRE/E-box element.
The previously characterized Sp1 binding GC-box can also be bound by SREBP-1 and FBI-1. We recently found that FBI-1 also binds the SRE and some GC-boxes of the p21Waf/Cip1 gene (36, 37). It was shown previously that DNA binding activities of SREBP-1 and Sp1 are overlapping on the two proximal SRE elements of FASN gene (34) and on the closely spaced SRE and Sp1 elements of the LDLR promoter (20). Interestingly, although SREBP-1 decreased the Sp1 binding activity, FBI-1 increased Sp1 binding on the GC-box in our studies of the FASN promoter.
ChIP assay results are basically consistent with EMSA data in terms of binding competition and increases in Sp1 binding by FBI-1. They also revealed information on the dynamics of promoter occupancy by FBI-1, SREBP-1a, and Sp1 on the GC-box, and SRE/E-box under various conditions. Overexpression of FBI-1 decreased the binding activity of SREBP-1 and vice versa, but there were only minor effects on the binding of Sp1. When expression of both FBI-1 and SREBP-1 was high and thus transcription of FASN gene was synergistically activated, FBI-1 and SREBP-1 binding were significantly reduced compared with the situation where they were expressed independently. However, promoter occupancy by both SREBP-1 and Sp1 is increased, and this DNA binding pattern favors transcriptional activation. The changes in the transcription factor binding were probably caused by the binding competition between SREBP-1 and FBI-1 to the GC-box and SRE/E-box and also by the protein-protein interaction between SREBP-1 and FBI, via their DNA binding domains, which could interfere with their DNA binding. The change in Sp1 and SREBP-1 binding appeared to be primarily controlled by the SRE/E-box because the binding pattern for FASN3 and FASN Mt (-2.7 kb), where the proximal SRE/E-box was mutated, was much weaker (Fig. 8, B, lanes 2 and 6, and C, lanes 2 and 6). The increased transcription factor binding resulted in 2-3-fold higher FASN gene transcription compared with conditions where only SREBP-1 was transfected. This situation might reflect robust FASN gene transcription in cancer cells where expression of both FBI-1 and SREBP-1 is increased.
In summary, we found that the molecular interactions between the proximal GC-box, SRE/E-box, SREBP-1a, Sp1, and FBI-1 are important in transcriptional activation of FASN gene by FBI-1 (Fig. 9). The molecular mechanism of transcription activation revealed here provides insight into how the proto-oncogene FBI-1 alters the sensitive cellular regulatory mechanisms of FASN gene expression to provide cellular membrane components needed for rapid cancer cell proliferation.
FIGURE 9.
Hypothetical model on the transcription and changes in binding dynamic of three transcription factors (FBI-1, Sp1, SREBP-1) on the SRE, GC-box, and SRE/E-box on the proximal promoter of FASN gene. A, in the control HEK293A cells where Sp1 is abundant and SREBP-1 is uninduced and low, the promoter elements are mainly bound by Sp1, and transcription is at basal level. No FBI-1 binding is detected. B, upon induction of SREBP-1, Sp1 binding is significantly reduced, and SREBP-1 binding is high, which resulted in potent transcription activation. C, upon introduction of FBI-1, Sp1 and SREBP-1 binding are reduced, and FBI-1 binding is high. The binding pattern with respect to Sp1 and SREBP-1 is similar to that of the control, although at low level. Transcription is at basal level. D, synergistic activation of FASN gene transcription by FBI-1 and SREBP-1. In the presence of high FBI-1 and SREBP-1, moderate level of SREBP-1 binding and high Sp1 binding close to the control is achieved. This is probably because of protein interaction between SREBP-1 and FBI-1, which significantly affects the DNA binding activity of the other protein. Binding by both Sp1 and SREBP-1 of the promoter in a way that favors more Sp1 over SREBP-1 around the GC-box and the SRE/E-box is important in synergistic activation of transcription. Transcription is mainly controlled by the molecular interactions among SRE/E-box, GC-box, FBI-1, Sp1, and SREBP-1. Molecular interaction on the upstream SRE is also expected but has no significant effect on transcription activation by FBI-1. Arrows, DNA-protein interactions. Thickness of the arrow indicated relative binding activity. Tsp (+1), transcription start site.
FASN, a key multifunctional enzyme of de novo fatty acid synthesis, is highly expressed in most human carcinomas. FASN is associated with poor prognosis in breast, lung, and prostate cancers, and its inhibition is selectively cytotoxic to human cancer cells. Thus, FASN and fatty acid metabolism in cancer has become a focus for the potential diagnosis and treatment of cancer (38). Because regulating FASN gene expression can be important in the control of cancer cell growth, the molecular mechanism revealed here might provide a novel target for the development of future anti-cancer drugs.
Supplementary Material
This work was supported by a Korea Research Foundation Grant E00046 (2005), Korea Ministry of Education, and National Research Laboratory Grant R0A-2003-000-10318-0 from Korea Science and Engineering Foundation, Korea Ministry of Science and Technology. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: SREBP, sterol-responsive element-binding protein; Ab, antibody; bHLH, basic helix loop helix DNA binding domain; ChIP, chromatin immuno precipitation; DBD, DNA binding domain; EMSA, electrophoretic mobility shift assay; FASN, fatty-acid synthase; SRE, SREBP-responsive element; GC-box, Sp1 binding GC-rich box; GST, glutathione S-transferase; ZFDBD, zinc finger DNA binding domain; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RT, reverse transcription; SRE, SREBP-response element; aa, amino acid; PBS, phosphate-buffered saline; tk, thymidine kinase; LDLR, low density lipoprotein receptor; siRNA, small interfering RNA; Mt, mutant; DIO, diet-induced obese.
References
- 1.Bardwell, V. J., and Treisman, R. (1994) Genes Dev. 8 1664-1677 [DOI] [PubMed] [Google Scholar]
- 2.Kelly, K. F., and Daniel, J. M. (2006) Trends Cell Biol. 16 578-587 [DOI] [PubMed] [Google Scholar]
- 3.Pessler, F., Pendergrast, P. S., and Hernandez, N. (1997) Mol. Cell. Biol. 17 3786-3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee, D. K., Suh, D., Edenberg, H. J., and Hur, M. W. (2002) J. Biol. Chem. 277 26761-26768 [DOI] [PubMed] [Google Scholar]
- 5.Morrison, D. J., Pendergrast, P. S., Stavropoulos, P., Colmenares, S. U., Kobayashi, R., and Hernandez, N. (1999) Nucleic Acids Res. 27 1251-1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pendergrast, P. S., Wang, C., Hernandez, N., and Huang, S. (2002) Mol. Biol. Cell 13 915-929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, D. K., Kang, J. E., Park, H. J., Kim, M. H., Yim, T. H., Kim, J. M., Heo, M. K., Kim, K. Y., Kwon, H. J., and Hur, M. W. (2005) J. Biol. Chem. 280 27783-27791 [DOI] [PubMed] [Google Scholar]
- 8.Laudes, M., Christodoulides, C., Sewter, C., Rochford, J. J., Considine, R. V., Sethi, J. K., Vidal-Puig, A., and O'Rahilly, S. (2004) J. Biol. Chem. 279 11711-11718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kukita, A., Kukita, T., Ouchida, M., Maeda, H., Yatsuki, H., and Kohashi, O. (1999) Blood 94 1987-1997 [PubMed] [Google Scholar]
- 10.Davies, J. M., Hawe, N., Kabarowski, J., Huang, Q. H., Zhu, J., Brand, N. J., Leprince, D., Dhordain, P., Cook, M., Morriss-Kay, G., and Zelent, A. (1999) Oncogene 18 365-375 [DOI] [PubMed] [Google Scholar]
- 11.Maeda. T., Hobbs, R. M., Merghoub, T., Guernah, I., Zelent, A., Cordon-Cardo, C., Teruya-Feldstein, J., and Pandolfi, P. P. (2005) Nature 433 278-285 [DOI] [PubMed] [Google Scholar]
- 12.Maeda, T., Merghoub, T., Hobbs, R. M., Dong, L., Maeda, M., Zakrzewski, J., van den Brink, M. R., Zelent, A., Shigematsu, H., Akashi, K., Teruya-Feldstein, J., Cattoretti, G., and Pandolfi, P. P. (2007) Science 316 860-866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama, C., Wang, X., Briggs, M. R., Admon, A., Wu, J., Hua, X., Goldstein, J. L., and Brown, M. S. (1993) Cell 75 185-197 [PubMed] [Google Scholar]
- 14.Tontonoz, P., Kim, J. B., Graves, R. A., and Spiegelman, B. M. (1993) Mol. Cell. Biol. 13 4753-4759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osborne, T. F. (2000) J. Biol. Chem. 275 32379-32382 [DOI] [PubMed] [Google Scholar]
- 16.Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002) J. Clin. Investig. 109 1125-1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimomura, L., Shimano, H., Horton, J. D., Goldstein, J. L., and Brown, M. S. (1997) J. Clin. Investig. 99 838-845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliner, J. D., Andresen, J. M., Hansen, S. K., Zhou, S., and Tjian, R. (1996) Genes Dev. 10 2903-2911 [DOI] [PubMed] [Google Scholar]
- 19.Yieh, L., Sanchez, H. B., and Osborne, T. F. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 6102-6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez, H. B., Yieh, L., and Osborne, T. F. (1995) J. Biol. Chem. 270 1161-1169 [DOI] [PubMed] [Google Scholar]
- 21.Yang, F., Vought, B. W., Satterlee, J. S., Walker, A. K., Jim, Sun, Z. Y., Watts, J. L., DeBeaumont, R., Saito, R. M., Hyberts, S. G., Yang, S., Macol, C., Iyer, L., Tjian, R., van den Heuvel, S., Hart, A. C., Wagner, G., and Näär, A. M. (2006) Nature 442 700-704 [DOI] [PubMed] [Google Scholar]
- 22.Wakil, S. J. (2004) Lipids 39 1045-1053 [DOI] [PubMed] [Google Scholar]
- 23.Medes, G., Thomas, A., and Weinhouse, S. (1953) Cancer Res. 13 27-29 [PubMed] [Google Scholar]
- 24.Kuhajda, F. P. (2000) Nutrition 16 202-208 [DOI] [PubMed] [Google Scholar]
- 25.Menendez, J. A., and Lupu, R. (2007) Nat. Rev. Cancer 7 763-777 [DOI] [PubMed] [Google Scholar]
- 26.Costello, L. C., and Franklin, R. B. (2005) Mol. Cell. Biochem. 280 1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swinnen, J. V., Van Veldhoven, P. P., Timmermans, L., De Schrijver, E., Brusselmans, K., Vanderhoydonc, F., Van de Sande, T., Heemers, H., Heyns, W., and Verhoeven, G. (2003) Biochem. Biophys. Res. Commun. 302 898-903 [DOI] [PubMed] [Google Scholar]
- 28.Griffin, M. J., and Sul, H. S. (2004) IUBMB Life 56 595-600 [DOI] [PubMed] [Google Scholar]
- 29.Magana, M. M., and Osborne, T. F. (1996) J. Biol. Chem. 271 32689-32694 [DOI] [PubMed] [Google Scholar]
- 30.Kim, J. B., Sarraf, P., Wright, M., Yao, K. M., Mueller, E., Solanes, G., Lowell, B. B., and Spiegelman, B. M. (1998) J. Clin. Investig. 101 1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latasa, M. J., Moon, Y. S., Kim, K. H., and Sul, H. S. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 10619-10624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Athanikar, J. N., Sanchez, H. B., and Osborne, T. F. (1997) Mol. Cell. Biol. 17 5193-5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf, S. S., Roder, K. H., and Schweizer, M. (1998) Biochem. Soc. Trans. 26 S95. [DOI] [PubMed] [Google Scholar]
- 34.Bennett, M. K., Lopez, J. M., Sanchez, H. B., and Osborne, T. F. (1995) J. Biol. Chem. 270 25578-25583 [DOI] [PubMed] [Google Scholar]
- 35.Smith, J. R., Osborne, T. F., Goldstein, J. L., and Brown, M. S. (1990) J. Biol. Chem. 265 2306-2310 [PubMed] [Google Scholar]
- 36.Pessler, F., and Hernandez, N. (2003) J. Biol. Chem. 278 29327-29335 [DOI] [PubMed] [Google Scholar]
- 37.Inoue, N., Shimano, H., Nakakuki, M., Matsuzaka, T., Nakagawa, Y., Yamamoto, T., Sato, R., Takahashi, A., Sone, H., Yahagi, N., Suzuki, H., Toyoshima, H., and Yamada, N. (2005) Mol. Cell. Biol. 25 8938-8947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuhajda, F. P. (2006) Cancer Res. 66 5977-5980 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.